Abstract
Simple Summary
Teat number is important for the mothering ability of a sow, but this trait has seldom been investigated by high-depth genomic data-based genome-wide association study (GWAS). Here, we performed GWAS for the teat number-related traits in 100 Chinese native Qingping pigs by recording their left and right teat numbers and analyzing their genetic variations through 10-fold whole-genome sequencing. T-Box Transcription Factor 3 (TBX3) on Sus scrofa chromosome (SSC) 14 and Wnt signaling pathway are revealed to be associated with teat number-related traits, with important roles in mammary gland morphogenesis and development.
Abstract
Teat number plays an important role in the reproductive performance of sows and the growth of piglets. However, the quantitative trait loci (QTLs) and candidate genes for the teat number-related traits in Qingping pigs remain unknown. In this study, we performed GWAS based on whole-genome single-nucleotide polymorphisms (SNPs) and insertions/deletions (Indels) for the total number of teats and five other related traits in 100 Qingping pigs. SNPs and Indels of all 100 pigs were genotyped using 10× whole genome resequencing. GWAS using General Linear Models (GLM) detected a total of 28 SNPs and 45 Indels as peak markers for these six traits. We also performed GWAS for the absolute difference between left and right teat number (ADIFF) using Fixed and random model Circulating Probability Unification (FarmCPU). The most strongly associated SNP and Indel with a distance of 562,788 bp were significantly associated with ADIFF in both GLM and FarmCPU models. In the 1-Mb regions of the most strongly associated SNP and Indel, there were five annotated genes, including TRIML1, TRIML2, ZFP42, FAT1 and MTNR1A. We also highlighted TBX3 as an interesting candidate gene for SSC14. Enrichment analysis of candidate genes suggested the Wnt signaling pathway may contribute to teat number-related traits. This study expanded significant marker-trait associations for teat number and provided useful molecular markers and candidate genes for teat number improvement in the breeding of sows.
Keywords: teat number, Qingping pig, whole genome resequencing, TBX3, Wnt signaling pathway
1. Introduction
Porcine teats are located from the anterior to posterior limb bud and symmetrical to the abdominal midline, and teat number is an important trait for the mothering ability of a sow, which can affect the piglets’ weight gain and mortality. It has been shown that teat number is a quantitative trait with a medium level of heritability (0.32) [1]. Using genetic markers can speed up the genetic improvement of teat number. Thus far, 655 teat number quantitative trait loci (QTLs) in pigs have been reported and included in the PigQTL database [2]. Previous genome-wide association studies (GWAS) found microsatellite markers or single nucleotide polymorphisms (SNPs) in popular breeds, including Durocs [1,3], Large Whites [4], and their crosses with Meishan pigs [5,6]. GWAS were also performed for teat number-related traits in several Chinese native pig breeds, including Erhualian [7], Sushan [8] and Beijing black [9]. However, the genetic architecture for teat number in Qingping pig, a Chinese native pig breed, is still not clear.
In 2000, Wada et al. revealed two QTLs for teat number on SSC1 and SSC7 by QTL analysis of 265 F2 offspring of the Meishan and Göttingen miniature pig [10]. Since then, an increasing number of studies have used microsatellite markers to confirm the QTL on SSC7 in other pig populations, including Meishan × Duroc F2 resource population [5], F2 populations of Yorkshire boars and Meishan sows [11], and Meishan × Large White F2 pigs [12]. Additionally, a number of studies detected associated SNPs for teat number near or within this QTL on SSC7 in Duroc pig [1,3,13,14], White Duroc × Erhualian F2 resource population [15], a commercial swine population [16], and Large White pig [17]. Notably, VRTN (located at 103.4 Mb on SSC7 on Sscrofa10.2) was a credible candidate gene in this major QTL for teat number on SSC7. These studies also indicated that QTLs for teat number-related traits are distributed across the genome on every chromosome. Several other genes have also been annotated as teat number-associated genes, such as Lysine Demethylase 6B (KDM6B) [15], TOX High Mobility Group Box Family Member 3 (TOX3) [17], Estrogen Receptor 1 (ESR1) and Nuclear Receptor Subfamily 5 Group A Member 1 (NR5A1) [8]. However, most previous studies have not used the high-density single-nucleotide polymorphisms (SNPs) detected by high-throughput sequencing data to investigate teat number-related traits.
The purpose of the current study was to detect genome-wide associations for teat number-related traits in Qingping pigs using whole-genome SNPs and insertions/deletions (Indels) based on whole-genome resequencing.
2. Materials and Methods
2.1. Sample and Sequencing
In this study, all 100 pigs were raised indoors in the Qingping pig Conservation Farm in Yichang, Hubei, China. Ear tissues were collected and stored in liquid nitrogen until further analysis. Genomic DNA samples were extracted from ear tissues, using a standard phenol–chloroform method. Sequencing libraries were constructed and sequenced by the Novogene Bioinformatics Institute (Novogene, Beijing, China). High-throughput sequencing was performed as paired-end 150 sequencing using a HiSeq 4000 sequencing system (Illumina, San Diego, CA, USA).
2.2. Phenotypic Data
In this study, three independent persons observed all 100 pigs and counted the teat number of the left and right lines. A total of six teat number-related traits were obtained: (i) the number of teats on the left side (LTN); (ii) the number of teats on the right side (RTN); (iii) the total number of teats (TNUM = LTN + RTN); (iv) the maximum number of teats in LTN and RTN (MAXAP); (v) the difference between the two sides (L-R = LTN − RTN) and (vi) the absolute difference between left and right teat number (ADIFF = |LTN − RTN|). The mean, standard deviation, minimum value, maximum value, and coefficient of variance for each trait were calculated using R (version 3.6.0) (R Core Team, Vienna, Austria).
2.3. Genotyping and Quality Control
Clean reads from all 100 pigs were aligned to the Sscrofa11.1 reference genome by the BWA software (version: 0.7.8) (Wellcome Trust Sanger Institute, Hinxton, UK) [18]. To reduce mismatches generated by PCR amplification before sequencing, duplicated reads were removed using SAMtools (Wellcome Trust Sanger Institute, Hinxton, UK) [19]. SNPs and Indels calling were initially performed to generate a gvcf file using UnifiedGenotyper in GATK (version 3.6) (Broad Institute, Cambridge, MA, USA) [20]. SNPs and Indels were divided using SelectVariants in GATK. Hard filtering of SNPs was applied to the raw variant set using “QUAL < 30.0 || QD < 2.0 || FS > 60.0 || MQ < 40.0 || SOR > 3.0 || ReadPosRankSum < −8.0”. Hard filtering of Indels was applied to the raw variant set using “QUAL < 30.0 || QD < 2.0 || FS > 200.0 || SOR > 10.0 || ReadPosRankSum < −20.0 || MQ < 40.0 || MQRankSum < −12.5”. SNPs and Indels in the VCF files were quality-filtered using VCFtools (v0.1.16) (Wellcome Trust Sanger Institute, Hinxton, UK) to remove variants with sequencing depth less than 3 [21]. SNPs and Indels were further filtered using PLINK 2.0 (Complete Genomics, Mountain View, CA, USA) [22]. Missing genotypes were imputed using beagle.03Jul19.b33.jar [23], followed by filtering SNPs and Indels again using PLINK 2.0 to obtain the high-quality common SNPs and Indels of 100 pigs for further analysis (individual call rate > 0.90; minor allele frequency > 0.05; call rate > 0.90, SNPs and Indels in Hardy–Weinberg equilibrium (p > 1 × 10−6) and excluding SNPs and Indels located on the sex chromosomes).
2.4. SNP-Based Heritability
The phenotypic variance explained by genome-wide SNPs (SNP-based heritability) was estimated using GREML in Genome-wide Complex Trait Analysis (GCTA) [24]. Briefly, the genetic relationships between pairwise individuals from all the autosomal SNPs were estimated using the genetic relationship matrix (GRM) based on high-quality common SNPs, followed by GRM and phenotype for restricted maximum likelihood (REML) analysis to estimate the variance explained by the SNPs.
2.5. Principal Component Analysis
Principal component analysis (PCA) was used to explore the population structure of Qingping pigs and determine whether principal components (PCs) should be added to the GWAS. PCA was performed using MVP.Data.PC in rMVP R (version 3.6.0) (Huazhong Agricultural University, Wuhan, China) [25] package based on high quality common SNPs. PC1 and PC2 were visualized using MVP.PCAplot in rMVP R (version 3.6.0) package.
2.6. GWAS Using General Linear Model (GLM)
In the present study, GLM in rMVP R (version 3.6.0) package was used to perform the GWAS for teat number-related traits based on high-quality common SNPs and Indels [26]. Principal component analysis showed no discernible clustering. Therefore, no principal component was adjusted in the subsequent association analysis. Each SNP or Indel for teat number-related traits was tested by GLM as follows:
where is the vector of each teat number-related trait in Qingping pigs; , a matrix of test SNP or Indel; , an incidence vector for ; , a vector of residuals following a normal distribution with a mean of zero and covariance, where is the identity matrix and is the residual variance.
2.7. GWAS Using FarmCPU
The Fixed Effect Model (FEM) and the Random Effect Model (REM) are used iteratively in FarmCPU [27], and FarmCPU in rMVP R (version 3.6.0) package was also used to perform GWAS for ADIFF. In the GLM model, some genetic markers were significantly associated with ADIFF with a whole-genome significant p-value (2.16 × 10−9 for SNPs and 2.44 × 10−8 for Indels). These markers can be used to define kinship in the REM step of FarmCPU to avoid the model over-fitting problem in FEM. The FEM is used to test each genetic marker, one at a time. Pseudo QTNs are included as covariates to control false positives. Specifically, the FEM could be expressed by the following equation:
where is the phenotype of the individual; , , …, , the genotypes of . pseudo QTNs, initiated with no QTN; , , …, , the corresponding effects of the pseudo QTNs; , the genotype of the individual and genetic marker; , the corresponding effect of the genetic marker; , the residual having a distribution with zero mean and variance of .
The REM is used to optimize the selection of pseudo QTNs from markers with whole genome significant p-values (2.16 × 10−9 for SNPs and 2.44 × 10−8 for Indels) and positions by using the SUPER algorithm [28]. The REM could be expressed by the following equation:
where and . are the same as in FEM, and indicates the total additive genetic effect of the individual. The expectations of the individuals’ total genetic effects are zeros. The variance and covariance matrix of the individuals’ total genetic effects can be expressed by , where is an unknown genetic variance and is the kinship matrix calculated by pseudo QTNs. The FEM and REM are iterated until no new pseudo QTNs are added, or the specified maximum number of iterations is reached.
2.8. Comparison with Known QTLs and Haplotype Analysis
Pig QTLs based on Sscrofa11.1 were downloaded from the PigQTL database. The information of QTLs for teat number-related traits was obtained using R (version 3.6.0), followed by comparing the significant SNPs and Indels with these QTLs using R (version 3.6.0). For the strongest significant SNP (rs322863105) for ADIFF on SSC17, SNPs with suggestive significant p-values (1/557,540, 1.79 × 10−6) around rs322863105 were used for haplotype block analysis to evaluate the linkage disequilibrium (LD) patterns of selected SNPs within this region using Haploview version 4.2 [29]. The effects of this SNP on ADIFF were plotted using ggplot2 R (version 3.6.0) package.
2.9. Annotation of Candidate Genes and Functional Enrichment Analysis
Candidate genes, including or close to the significant SNPs and Indels, were annotated using the biomaRt [30] R (version 3.6.0) package based on Sscrofa11.1. Candidate genes located in 1-Mb regions of significant SNPs and Indels were also annotated. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses and visualizations were performed using the clusterProfiler R (version 3.6.0) package [31].
3. Results
3.1. Genotyping and Phenotypic Statistics
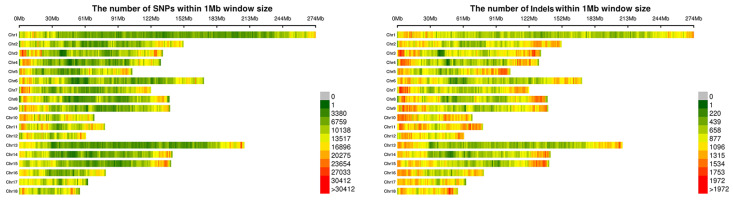
After whole-genome sequencing, 2.76 TB of sequences were generated, with a mean coverage of 98.47% at an average of 9.66-fold depth for 100 Qingping pigs in this study (Table S1). Clean data were mapped to the pig reference genome (Sscrofa11.1), with 36,482,281 SNPs and 4,859,001 Indels being called with GATK. After filtering, 23,193,931 SNPs and 2,053,221 Indels, with a distribution roughly proportional to autosomal chromosomes of pigs, were retained for subsequent analyses (Figure 1).
Figure 1.
The distribution of SNPs and Indels on autosomal chromosomes of pigs.
Table 1 shows the descriptive statistics of teat number-related traits of Qingping pigs. The average teat number [standard deviation (SD)] was seen to be 14.78 (0.97), ranging from 14 to 17, which was higher than the values of Beijing Black Pig (13.6) [9], Japanese Duroc (13.73) [3], American (10.90) and Canadian Duroc (10.92) [14], but lower than the value of Erhualian pigs (19.13) [7]. The meansvalues (SD) for the other teat number-related traits, were 7.36 (0.48), 7.42 (0.61), 7.50 (0.59), −0.06 (0.51) and 0.22 (0.46) for LTN, RTN, MAXAP, L-R and ADIFF, with coefficient of variation (CV) values of 6.55, 8.17, 6.56 and 7.93% for LTN, RTN, TNUM and MAXAP, respectively.
Table 1.
Summary of phenotypic data in terms of mean, standard deviation, minimum, maximum, coefficient of variance, and SNP-based heritability and standard error (SE) for each trait.
| Trait | N | Mean | SD | Min | Max | C.V. | SE | |
|---|---|---|---|---|---|---|---|---|
| LTN | 100 | 7.36 | 0.48 | 7.00 | 8.00 | 6.55 | 0.29 | 0.21 |
| RTN | 100 | 7.42 | 0.61 | 6.00 | 9.00 | 8.17 | 0.19 | 0.21 |
| TNUM | 100 | 14.78 | 0.97 | 14.00 | 17.00 | 6.56 | 0.36 | 0.23 |
| MAXAP | 100 | 7.50 | 0.59 | 7.00 | 9.00 | 7.93 | 0.38 | 0.22 |
| L-R | 100 | −0.06 | 0.51 | −2.00 | 2.00 | NA | 0.00 | 0.20 |
| ADIFF | 100 | 0.22 | 0.46 | 0.00 | 2.00 | NA | 0.24 | 0.20 |
LTN: the number of teats on the left side; RTN: the number of teats on the right side; TNUM: the total number of teats (TNUM = LTN + RTN); MAXAP: the maximum number of teats in LTN and RTN (MAXAP); L-R: the difference between the two sides (L-R = LTN − RTN); ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
In Table 1, it was shown that the values of SNP-based heritability () for teat number-related traits were 0.29, 0.19, 0.36, 0.38, 0.00 and 0.24 for LTN, RTN, TNUM, MAXAP, L-R and ADIFF in Qingping pigs, respectively. Unfortunately, the standard errors (SE) were large, which reflects the low accuracy of the heritability estimates. Therefore, we compared the results of the teat number-related heritability estimates in Qingping pigs with those of other pig breeds. In Qingping pigs, the narrow-sense heritability for TNUM (0.36) (only considering the contribution of additive genetic effects) was consistent with the values of purebred Korean Yorkshire pigs (0.37) [4] and Duroc pigs (0.34 ± 0.05) [3]. In previous studies, the values of LTN, RTN, MAXAP and L-R were 0.16, 0.26, 0.261 and 0.00, which were consistent with our results [17]. Unexpectedly, ADIFF had a medium value (0.24), which was virtually null in previous studies [16,17]. These results suggest that estimates of the heritability of teat number-related traits in Qingping pigs were reliable, but caution is required due to the large standard errors.
3.2. GLM GWAS for Teat Number-Related Traits
PCA results indicated that Qingping pigs could not be clustered into groups (Figure S1), so GWAS was performed using GLM with no principal component. The Bonferroni correction assumes that each of the tests is independent, an is thereby inherently conservative when considering SNPs in LD. We calculated the effectively independent tests based on the estimated number of independent markers [32]. A total of 557,540 SNP and 28,629 Indel independent tests were suggested, with the threshold p-value of 8.97 × 10−8 (0.05/557,540) for SNPs and 1.75 × 10−6 (0.05/28,629) for Indels.
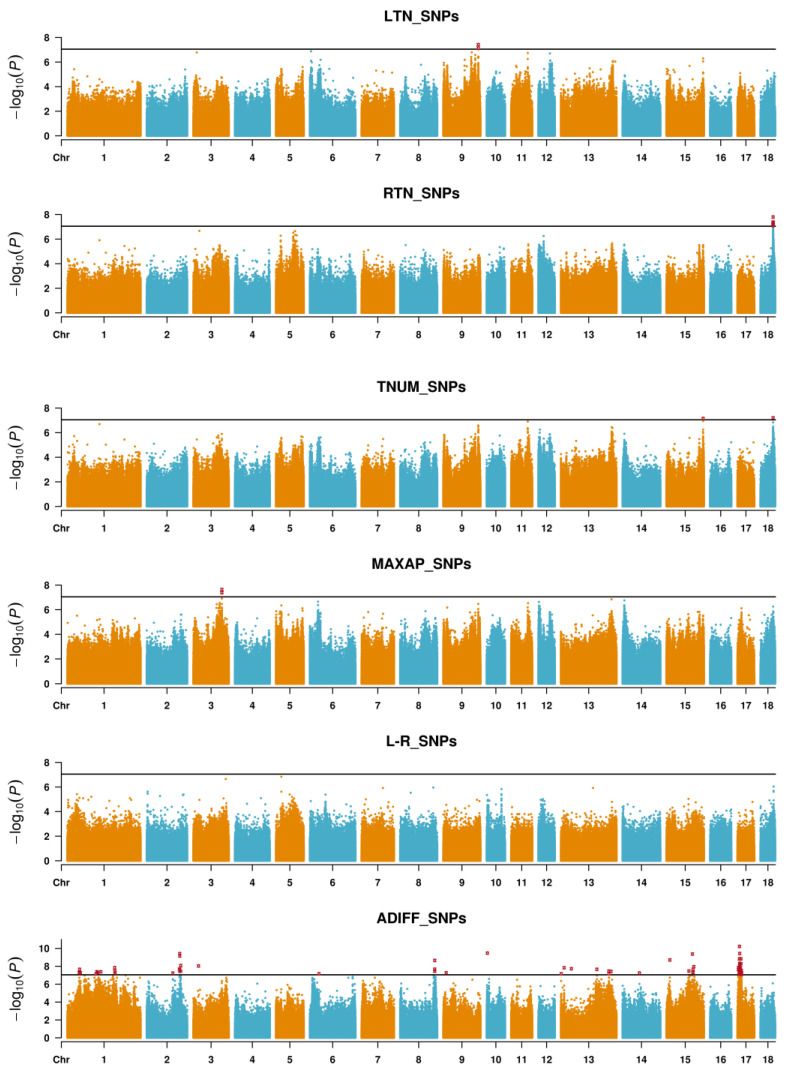
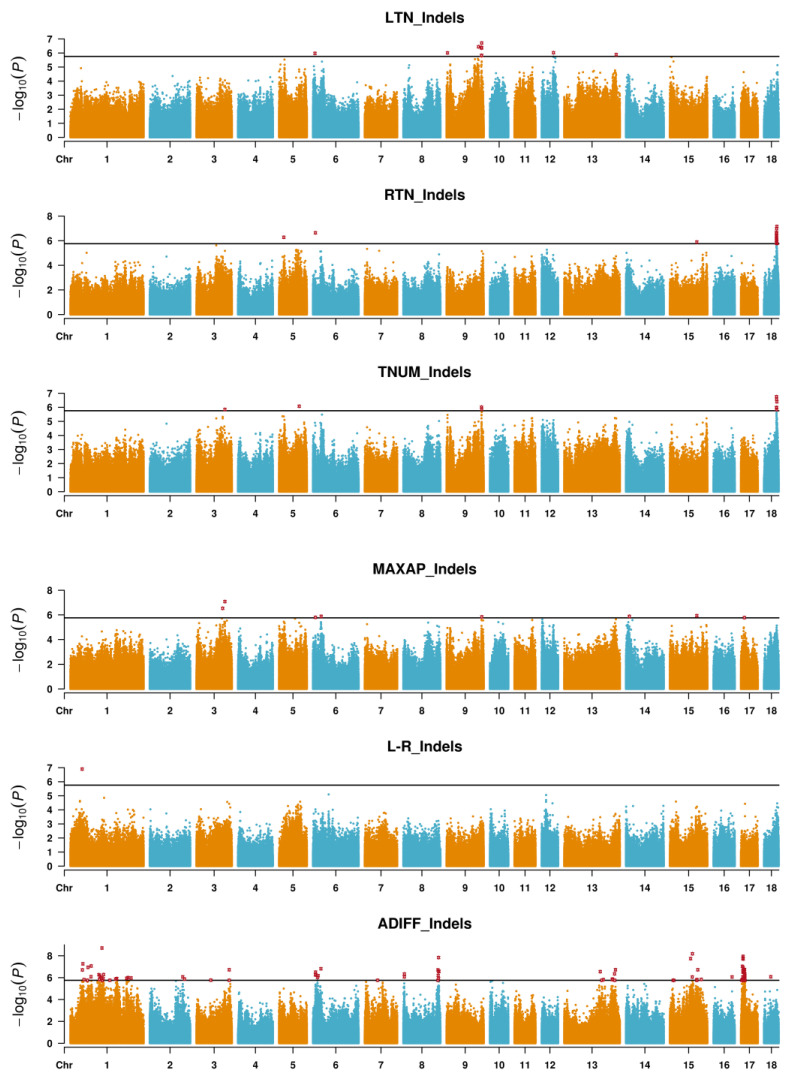
Figure 2 and Figure 3 show the Manhattan plots for LTN, RTN, TNUM, MAXAP, L-R and ADIFF, and Figure S2 presents the Q-Q plots for these traits. A total of 28 significant SNPs and 45 significant Indels were detected as peak associated variants for LTN, RTN, TNUM, MAXAP, L-R and ADIFF (Table 2). Among the 28 SNPs identified, 10 SNPs were located within 11 genes and the others (18 SNPs) were located at 3798 to 46,070,668 bp from the nearest genes (Table 2). Among the 45 Indels identified, 21 Indels were located within 22 genes and the others (24 Indels) were located 2660 to 137,303 bp from the nearest genes (Table 2).
Figure 2.
Manhattan plots of GLM GWAS for teat number-related traits in Qingping pigs, including LTN, RTN, TNUM, MAXAP, L-R and ADIFF based on SNPs. LTN: the number of teats on the left side; RTN: the number of teats on the right side; TNUM: the total number of teats (TNUM = LTN + RTN); MAXAP: the maximum number of teats in LTN and RTN (MAXAP); L-R: the difference between the two sides (L-R = LTN − RTN); ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
Figure 3.
Manhattan plots of GLM GWAS for teat number-related traits in Qingping pigs, including LTN, RTN, TNUM, MAXAP, L-R and ADIFF based on Indels. LTN: the number of teats on the left side; RTN: the number of teats on the right side; TNUM: the total number of teats (TNUM = LTN + RTN); MAXAP: the maximum number of teats in LTN and RTN (MAXAP); L-R: the difference between the two sides (L-R = LTN − RTN); ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
Table 2.
Significant SNPs and Indels of GLM GWAS for teat number-related traits.
| SNP | Trait | SSC | Position | Effect | p-Value | QTLs * | Annotation | Gene (Distance from the Gene in bp) |
|---|---|---|---|---|---|---|---|---|
| chr9:130956708 | LTN | 9 | 130956708 | 0.64 | 3.85 × 10−8 | intronic | PACC1(within), NENF(within) | |
| rs345573243 | RTN | 18 | 47399908 | 0.62 | 1.60 × 10−8 | 24,290, 7470 | intronic | OSBPL3(within) |
| rs703282466 | MAXAP | 3 | 106788730 | 0.45 | 2.29 × 10−8 | 5224, 8797, 8798, 4250, 4256 | intergenic | LTBP1(63796), ENSSSCG00000050704(9889) |
| chr15:137183045 | TNUM | 15 | 137183045 | −0.79 | 7.02 × 10−8 | 223,293 | intronic | MLPH(within) |
| rs345573243 | TNUM | 18 | 47399908 | 0.96 | 6.25 × 10−8 | 24,290, 7470 | intronic | OSBPL3(within) |
| rs1108940033 | ADIFF | 1 | 45796983 | 0.61 | 2.19 × 10−8 | intergenic | PHF3(500583),ENSSSCG00000049526(273685) | |
| rs321204530 | ADIFF | 1 | 177056841 | 0.59 | 1.44 × 10−8 | 5223, 5255, 822, 845, 1250 | intronic | MDGA2(within) |
| rs318957512 | ADIFF | 2 | 97849348 | 0.50 | 5.79 × 10−8 | intronic | ADGRV1(within) | |
| rs326371568 | ADIFF | 2 | 123586226 | 0.62 | 3.87 × 10−10 | 4255 | intergenic | FAM170A(119517),PRR16(613282) |
| rs342451777 | ADIFF | 2 | 127224122 | 0.57 | 8.31 × 10−9 | 4255 | intergenic | ENSSSCG00000042143(13463),ENSSSCG00000040936(55836) |
| rs325963999 | ADIFF | 3 | 19301837 | 0.66 | 9.15 × 10−9 | 5224, 7455, 7472 | intronic | KATNIP(within) |
| rs326276043 | ADIFF | 6 | 34051848 | 0.55 | 6.98 × 10−8 | 24,289 | intergenic | ENSSSCG00000050973(273191),CYLD(8093) |
| rs338649298 | ADIFF | 8 | 129552162 | 0.59 | 2.22 × 10−9 | intergenic | SNCA(163855),ENSSSCG00000043431(95940) | |
| rs321470648 | ADIFF | 9 | 10769337 | 0.45 | 5.38 × 10−8 | intergenic | ENSSSCG00000046278(36887),ENSSSCG00000045225(26362) | |
| rs1109963100 | ADIFF | 10 | 2911179 | 0.80 | 3.37 × 10−10 | intergenic | ENSSSCG00000042899(210778),BRINP3(52348) | |
| rs339887165 | ADIFF | 13 | 12135388 | 0.68 | 1.46 × 10−8 | 7479 | intergenic | ENSSSCG00000044771(16544),ENSSSCG00000051554(31818) |
| chr13:39266305 | ADIFF | 13 | 39266305 | 0.72 | 1.88 × 10−8 | 7479 | intronic | DNAH12(within) |
| rs701874475 | ADIFF | 13 | 134665423 | −0.51 | 2.19 × 10−8 | 7479 | intergenic | LMLN(8675),ENSSSCG00000050583(21280) |
| rs338558804 | ADIFF | 13 | 179683904 | −0.45 | 3.49 × 10−8 | intergenic | ENSSSCG00000038062(120975),NRIP1(140958) | |
| rs343864506 | ADIFF | 13 | 187708682 | −0.54 | 3.72 × 10−8 | intergenic | ENSSSCG00000047308(444803),ENSSSCG00000050420(500018) | |
| rs334271954 | ADIFF | 14 | 62959726 | 0.52 | 5.70 × 10−8 | intergenic | FAM13C(33877),SLC16A9(66937) | |
| rs1109225784 | ADIFF | 15 | 12581324 | 0.64 | 1.96 × 10−9 | intergenic | U6(133156),U6(186459) | |
| rs326978910 | ADIFF | 15 | 84015934 | 0.38 | 3.38 × 10−8 | 7468 | intronic | OSBPL6(within) |
| rs334746473 | ADIFF | 15 | 97200323 | 0.75 | 4.23 × 10−10 | 7468 | intergenic | ENSSSCG00000046205(186079),U2(688281) |
| rs322863105 | ADIFF | 17 | 7610979 | −0.88 | 6.01 × 10−11 | intergenic | ENSSSCG00000045345(132678),ENSSSCG00000047202(438984) | |
| chr17:8221026 | ADIFF | 17 | 8221026 | 0.79 | 1.47 × 10−9 | intergenic | U6(19092),FAT1(227734) | |
| rs330045817 | ADIFF | 17 | 8536301 | −0.71 | 3.76 × 10−10 | ncRNA_intronic | FAT1(within) | |
| rs324534432 | ADIFF | 17 | 13364668 | 0.79 | 1.47 × 10−9 | intergenic | PSD3(100469),ENSSSCG00000046441(3798) | |
| Indel | ||||||||
| chr6:7472906 | LTN | 6 | 7472906 | −0.41 | 1.06 × 10−6 | 24,289 | intronic | CDYL2(within) |
| rs695882779 | LTN | 9 | 3690507 | 0.39 | 9.84 × 10−7 | ncRNA_intronic | ENSSSCG00000049604(within) | |
| chr9:119650540 | LTN | 9 | 119650540 | −0.30 | 3.62 × 10−7 | intergenic | ENSSSCG00000044083(337170),ENSSSCG00000050832(4698) | |
| rs792699200 | LTN | 9 | 130899869 | 0.51 | 4.41 × 10−7 | intronic | PACC1(within) | |
| chr12:44292044 | LTN | 12 | 44292044 | −0.37 | 9.55 × 10−7 | 5227, 5261, 6472, 6479, 595, 2929 | intronic | NOS2(within) |
| chr13:194617904 | LTN | 13 | 194617904 | −0.31 | 1.30 × 10−6 | intergenic | KRTAP11-1(111382),ENSSSCG00000047315(5813) | |
| rs709659410 | RTN | 5 | 17969771 | 0.42 | 5.28 × 10−7 | 2927 | intergenic | KRT73(2682),KRT2(14544) |
| chr6:9186279 | RTN | 6 | 9186279 | 0.89 | 2.28 × 10−7 | 24,289 | intronic | WWOX(within) |
| rs790747253 | RTN | 15 | 100810568 | −0.61 | 1.28 × 10−6 | 7468 | intronic | PGAP1(within) |
| chr18:48316684 | RTN | 18 | 48316684 | 0.41 | 7.00 × 10−8 | 24,290, 7470 | intronic | STK31(within) |
| chr3:98429885 | MAXAP | 3 | 98429885 | 0.50 | 2.93 × 10−7 | 5224, 8797, 8798 | intergenic | ENSSSCG00000045166(59604),ENSSSCG00000046007(449247) |
| rs793312568 | MAXAP | 3 | 106792080 | 0.43 | 8.28 × 10−8 | 5224, 8797, 8798, 4250, 4256 | intergenic | LTBP1(67153),ENSSSCG00000050704(6530) |
| chr6:9186279 | MAXAP | 6 | 9186279 | 0.82 | 1.59 × 10−6 | 24,289 | intronic | WWOX(within) |
| chr6:30642639 | MAXAP | 6 | 30642639 | 0.43 | 1.31 × 10−6 | 24,289 | intergenic | ENSSSCG00000047270(70049),ENSSSCG00000041426(189382) |
| chr9:131996847 | MAXAP | 9 | 131996847 | −0.49 | 1.46 × 10−6 | intronic | ENSSSCG00000040650(within) | |
| chr14:12750600 | MAXAP | 14 | 12750600 | 0.43 | 1.31 × 10−6 | intronic | HMBOX1(within) | |
| rs790747253 | MAXAP | 15 | 100810568 | −0.60 | 1.17 × 10−6 | 7468 | intronic | PGAP1(within) |
| rs711984029 | MAXAP | 17 | 13041965 | −0.47 | 1.68 × 10−6 | intronic | PSD3(within) | |
| rs793312568 | TNUM | 3 | 106792080 | 0.63 | 1.41 × 10−6 | 5224, 8797, 8798, 4250, 4256 | intergenic | LTBP1(67153),ENSSSCG00000050704(6530) |
| chr5:75592729 | TNUM | 5 | 75592729 | 0.98 | 8.38 × 10−7 | intronic | NELL2(within) | |
| rs792699200 | TNUM | 9 | 130899869 | 1.00 | 9.67 × 10−7 | intronic | PACC1(within) | |
| chr18:48316684 | TNUM | 18 | 48316684 | 0.62 | 4.01 × 10−7 | 24,290, 7470 | intronic | STK31(within) |
| chr1:44096236 | LR | 1 | 44096236 | 0.69 | 1.25 × 10−7 | intergenic | ENSSSCG00000042072(159101),ENSSSCG00000045405(51729) | |
| chr1:44973455 | ADIFF | 1 | 44973455 | 0.70 | 1.96 × 10−7 | intronic | ZUP1(within),RSPH4A(within) | |
| chr1:46840905 | ADIFF | 1 | 46840905 | 0.71 | 5.46 × 10−8 | intergenic | ENSSSCG00000050391(123491),U6(392319) | |
| chr1:65957275 | ADIFF | 1 | 65957275 | 0.43 | 1.14 × 10−7 | intergenic | FBXL4(21578),FAXC(274880) | |
| chr1:77875820 | ADIFF | 1 | 77875820 | 0.42 | 8.57 × 10−8 | intergenic | FYN(65243),U6(107381) | |
| chr1:118273285 | ADIFF | 1 | 118273285 | −0.64 | 1.96 × 10−9 | 5223, 6481, 5255 | intergenic | ENSSSCG00000049391(9019),ENSSSCG00000045826(5556) |
| rs1113667849 | ADIFF | 2 | 123584099 | 0.55 | 8.50 × 10−7 | 4255 | intergenic | FAM170A(117391),PRR16(615405) |
| chr3:54080763 | ADIFF | 3 | 54080763 | 0.44 | 1.70 × 10−6 | 5224, 7455, 7472, 6465 | intergenic | LONRF2(99093),REV1(54761) |
| chr3:122749868 | ADIFF | 3 | 122749868 | 0.76 | 1.91 × 10−7 | intergenic | LRATD1(17941),ENSSSCG00000045589(179131) | |
| chr6:9702570 | ADIFF | 6 | 9702570 | 0.61 | 3.15 × 10−7 | 24,289 | intronic | WWOX(within) |
| chr6:29725029 | ADIFF | 6 | 29725029 | −0.41 | 1.54 × 10−7 | 24,289 | intergenic | ENSSSCG00000034192(132519),CES5A(117783) |
| chr7:48226837 | ADIFF | 7 | 48226837 | 0.67 | 1.74 × 10−6 | 5257 | intronic | RASGRF1(within) |
| chr8:3878010 | ADIFF | 8 | 3878010 | −0.48 | 4.61 × 10−7 | 7477 | intronic | ENSSSCG00000027349(within) |
| chr8:131927486 | ADIFF | 8 | 131927486 | 0.68 | 1.46 × 10−8 | intergenic | AFF1(2666),ENSSSCG00000032190(41799) | |
| chr13:134712100 | ADIFF | 13 | 134712100 | −0.50 | 2.83 × 10−7 | 7479 | intergenic | ENSSSCG00000050583(20384),OSBPL11(6327) |
| chr13:188309252 | ADIFF | 13 | 188309252 | 0.35 | 4.51 × 10−7 | intergenic | ENSSSCG00000050420(95234),ENSSSCG00000043493(500979) | |
| chr13:191714830 | ADIFF | 13 | 191714830 | 0.76 | 1.91 × 10−7 | intergenic | ENSSSCG00000051384(248028),ENSSSCG00000048685(53357) | |
| chr15:12585471 | ADIFF | 15 | 12585471 | 0.67 | 1.74 × 10−6 | intergenic | U6(137304),U6(182308) | |
| chr15:84277014 | ADIFF | 15 | 84277014 | 0.53 | 6.57 × 10−9 | 7468 | intergenic | ENSSSCG00000036052(46519),ENSSSCG00000038561(122000) |
| rs792656057 | ADIFF | 16 | 69550278 | −0.60 | 8.69 × 10−7 | 5228 | intronic | GRIA1(within) |
| rs793561441 | ADIFF | 17 | 5622328 | −0.33 | 9.14 × 10−8 | intronic | PCM1(within) | |
| rs700363122 | ADIFF | 17 | 8173767 | 0.60 | 1.13 × 10−8 | intergenic | ENSSSCG00000047202(116838),U6(28063) | |
| rs789477433 | ADIFF | 18 | 25541841 | 0.43 | 8.35 × 10−7 | 24,290 | intergenic | ENSSSCG00000048651(270433),FAM3C(27695) |
* QTL number in PigQTL database. Effect means additive effect. LTN: the number of teats on the left side; RTN: the number of teats on the right side; TNUM: the total number of teats (TNUM = LTN + RTN); MAXAP: the maximum number of teats in LTN and RTN (MAXAP); L-R: the difference between the two sides (L-R = LTN − RTN); ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
Compared with known QTLs for teat number-related traits in the PigQTL database, 14 significant SNPs and 24 significant Indels overlapped with known QTLs (Table 2). Interestingly, Manhattan plots showed similar trends between SNPs and Indels. In Table 2, there were several significant SNPs and Indels were located at the same teat number-related traits QTLs in the PigQTL database. For RTN, significant SNP (rs345573243) and Indel (chr18:48316684) on SSC18 were located at QTL 7470 and QTL 24290, and coincidentally, rs345573243 and chr18:48316684 also showed significant associations with TNUM. For MAXAP, significant SNP (rs703282466) and Indel (rs793312568) on SSC3 were located at QTL 4250 and 4256, and coincidentally, rs793312568 also exhibited a significant association with TNUM. For ADIFF, significant SNPs (rs326371568 and rs342451777) and Indel (rs1113667849) on SSC2 were located at QTL 4255; significant SNP (rs701874475) and Indel (chr13:134712100) on SSC13 at QTL 7479; significant SNP (rs326978910) and Indel (chr15:84277014) on SSC15 at QTL 7468. Additionally, five new significant SNPs were closed to significant Indels, including rs1108940033 was closed to chr1:44973455 and chr1:46840905 on SSC1, rs338649298 was closed to chr8:131927486 on SSC8, rs343864506 was closed to chr13:188309252 on SSC13, rs1109225784 was closed to chr15:12585471 on SSC15, chr17:8221026 was closed to rs700363122 on SSC17.
3.3. FarmCPU GWAS for ADIFF
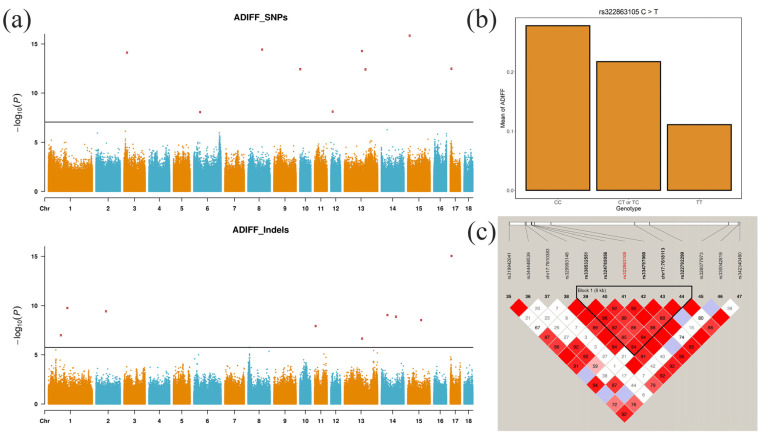
The GLM GWAS results of ADIFF showed significant associations even using whole genome SNPs or Indels to decide the thresholds (SNP: 0.05/2,319,3931; Indels: 0.05/2,053,221). However, Q-Q plots and genomic inflation factors (λSNP = 1.37, λIndel = 1.36) indicated possible false positives (Figure S2). FarmCPU, a powerful and efficient GWAS model, was used to control false positives and retain true positives. Q-Q plots and genomic inflation factors (λSNP = 0.98, λIndel = 0.86) were improved by FarmCPU (Figure S3). In Figure 4a and Table 3, 9 SNPs and 9 Indels were shown to be significantly associated variants for ADIFF on SSC1, 2, 3, 6, 8, 10, 11, 12, 13, 14, 15 and 17, with six SNPs and five Indels included in known QTLs (Table 3).
Figure 4.
FarmCPU GWAS for ADIFF. (a) Manhattan plots of the GWAS based on SNPs and Indels. (b) Difference analysis of the strongest significant SNP (rs322863105) on SSC17 in GLM, which was retested in FarmCPU. (c) Haplotype block analysis of selected suggestive significant SNPs associated with ADIFF on SSC17 in GLM, including rs322863105. ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
Table 3.
Significant SNPs and Indels of FarmCPU GWAS for ADIFF.
| SNP | SSC | Position | Effect | p-Value | QTLs * | Annotation | Gene (Distance from the Gene in bp) |
|---|---|---|---|---|---|---|---|
| rs325963999 # | 3 | 19301837 | 0.27 | 7.82 × 10−15 | 5224, 7455, 7472 | intronic | KATNIP(within) |
| rs693622708 | 6 | 39540583 | 0.17 | 8.58 × 10−9 | 24,289 | intergenic | UQCRFS1(166710), ENSSSCG00000050718(41760) |
| rs326134805 | 8 | 88543901 | 0.13 | 3.81 × 10−15 | 7477, 1100 | intergenic | ENSSSCG00000044017(86061),SLC7A11(63983) |
| rs1109963100 # | 10 | 2911179 | 0.33 | 3.74 × 10−13 | intergenic | ENSSSCG00000042899(210778),BRINP3(52348) | |
| rs1113875395 | 12 | 11463144 | 0.16 | 7.51 × 10−9 | 5227, 1128 | intergenic | ABCA8(15848),ENSSSCG00000045738(20026) |
| rs343773900 | 13 | 110154062 | 0.24 | 5.42 × 10−15 | 7479 | intronic | PLD1(within) |
| rs333970515 | 13 | 132158230 | −0.18 | 3.99 × 10−13 | 7479 | ncRNA_exonic | ENSSSCG00000047632(within) |
| rs1109225784 # | 15 | 12581324 | 0.32 | 1.46 × 10−16 | intergenic | U6(133156),U6(186459) | |
| rs322863105 # | 17 | 7610979 | −0.34 | 3.34 × 10−13 | intergenic | ENSSSCG00000045345(132678),ENSSSCG00000047202(438984) | |
| Indel | |||||||
| chr1:77875820 # | 1 | 77875820 | 0.17 | 1.02 × 10−7 | intergenic | FYN(65243),U6(107381) | |
| chr1:118273285 # | 1 | 118273285 | −0.35 | 1.74 × 10−10 | 5223, 6481, 5255 | intergenic | ENSSSCG00000049391(9019),ENSSSCG00000045826(5556) |
| rs788352632 | 2 | 62598411 | −0.12 | 3.81 × 10−10 | 909 | ncRNA_intronic | ENSSSCG00000048292(within) |
| chr11:6896071 | 11 | 6896071 | −0.13 | 1.22 × 10−8 | 5260 | ncRNA_intronic | ENSSSCG00000036846(within) |
| rs787621311 | 13 | 110898097 | 0.23 | 2.27 × 10−7 | 7479 | intronic | FNDC3B(within) |
| rs701717756 | 14 | 38222283 | 0.26 | 9.42 × 10−10 | intergenic | RBM19(17536),ENSSSCG00000042669(3247) | |
| chr14:91416556 | 14 | 91416556 | −0.30 | 1.34 × 10−9 | intergenic | ENSSSCG00000047278(204580),CXCL12(99865) | |
| chr15:84277014 # | 15 | 84277014 | 0.22 | 2.92 × 10−9 | 7468 | intergenic | ENSSSCG00000036052(46519),ENSSSCG00000038561(122000) |
| rs700363122 # | 17 | 8173767 | 0.40 | 9.10 × 10−16 | intergenic | ENSSSCG00000047202(116838),U6(28063) |
* QTL number in in PigQTL database. # Duplicate signals between GLM and FarmCPU. Effect means additive effect. ADIFF: the absolute difference between left and right teat number (ADIFF = |LTN − RTN|).
Three of the 9 significant SNPs were located within four genes and six SNPs were located 15,848 to 133,156 bp from the nearest genes (Table 3). Three of the 9 significant Indels were located within three genes and six Indels were located 3267 to 99,873 bp from the nearest genes (Table 3). Compared with the GLM GWAS results for ADIFF, 4 SNPs (rs325963999, rs1109963100, rs1109225784, rs322863105) and four Indels (chr1:77875820, chr1:118273285, chr15:84277014, rs700363122) were duplicated in the FarmCPU GWAS results, suggesting the reliability of the results (Table 3).
The strongest significant SNP in GLM for ADIFF on SSC17 (rs322863105, p-value = 6.01 × 10−11) also showed significant association with ADIFF in FarmCPU (p-value = 3.34 × 10−13). The effect of rs322863105 on the ADIFF was estimated by genotyping Qingping pigs for this SNP. Individuals with the TT genotype had a lower ADIFF, suggesting LTN and RTN were more symmetrical (Figure 4b). Linkage analysis of the suggestive significant SNPs around this SNP identified one haplotype block of 8 kb between rs338532551 and rs322792299, including rs322863105 (Figure 4c). Three annotated genes were contained in the 1-Mb region around rs322863105, including tripartite motif family like 1 (TRIML1), tripartite motif family like 2 (TRIML2), and ZFP42 zinc finger protein (ZFP42). Moreover, a peak Indel (rs700363122) close to this SNP showed significant associations with ADIFF in the results of both GLM (1.13 × 10−8) and FarmCPU (9.10 × 10−16), with two annotated genes in the 1-Mb region around this Indel, including FAT Atypical Cadherin 1 (FAT1), and Melatonin Receptor 1A (MTNR1A).
3.4. Functional Enrichment of Candidate Genes
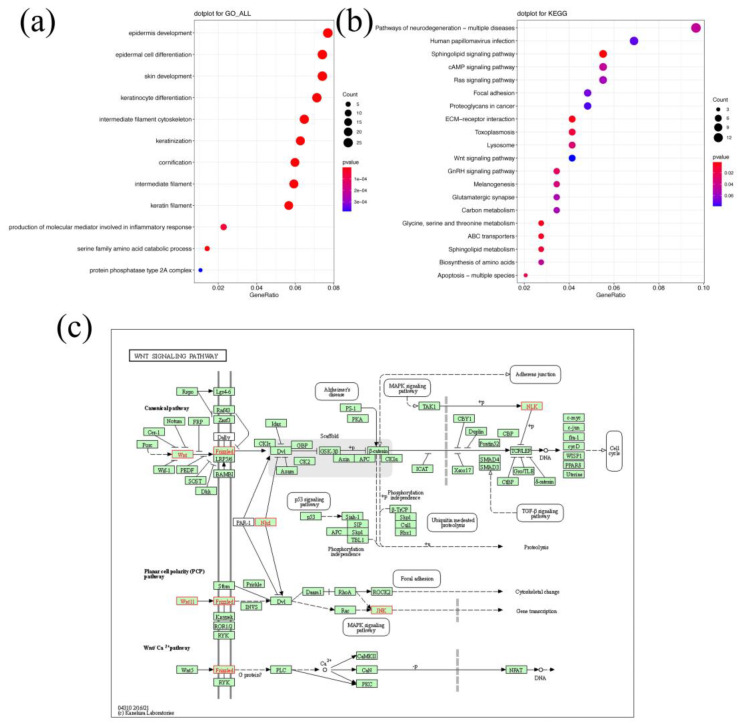
Annotated genes within 1-Mb regions of significant SNPs and Indels were defined as candidate genes. A total of 397 annotated genes were found in these regions (Table S2). In Figure 5a, GO enrichment analysis showed the enrichment of these candidate genes in epidermis development (p = 2.31 × 10−7), epidermal cell differentiation (p = 4.61 × 10−9), and skin development (p = 1.12 × 10−7). In Figure 5b KEGG pathway analysis revealed the enrichment of candidate genes in the pathways, such as the Sphingolipid signaling pathway (p = 1.27 × 10−3), ECM-receptor interaction (p = 4.79 × 10−3), and Glycine, serine and threonine metabolism (p = 5.43 × 10−3). Furthermore, we also paid attention to the Wnt signaling pathway (Figure 5c), due to its important role in initiating mammary morphogenesis and all subsequent stages of mammary formation as previously reported [33].
Figure 5.
Enrichment results of candidate genes within 1-Mb regions of significant SNPs and Indels. (a) Dotplot of GO term enrichment. (b) Dotplot of KEGG pathway enrichment. (c) Wnt signaling pathway.
4. Discussion
During the first month after birth, piglets only have sow milk as a source of nutrients, which contributes to the regulation of their basal metabolism and temperature. Therefore, the sow’s ability to produce milk can influence the health and growth of piglets, probably with a long-term effect post-weaning. The sows’ lactation performance can be improved by enhancing the growth of the mammary gland and sows with a low prolactin/progesterone ratio before farrowing were reported to have a higher colostrum yield [34]. Additionally, milk production could be increased by adding L-arginine to the diets of lactating primiparous sows [35]. Moreover, for primiparous sows, teat suckling only for the first 2 days postpartum ensures the optimal mammary development and milk yield in the next lactation [36]. Furthermore, the lactating ability of sows can also be improved by increasing the teat number. As shown in the present study, teat number is heritable, with a genetic correlation to numerous microsatellite sites and SNPs (especially a QTL on SSC7).
The of most teat number-related traits in Qingping pigs was moderate, except for L-R, suggesting it is feasible to improve teat number in pigs through genetic selection. SNPs and Indels associated with teat number-related traits might play an essential role in teat number improvement. Several candidate genes were reported to be related to mammary gland development and breast cancer. CDYL2, including an Indel significantly associated with LTN on SSC6, positively regulates breast cancer cell migration, invasion and epithelial-to-mesenchymal transition through p65/NF-κB and STAT3 [37]. FAM3C, which is in the 1-Mb region of an Indel on SSC18 and associated with ADIFF, encodes Interleukin-Like Epithelial-Mesenchymal Transition Inducer for the proliferation and migration of breast cancer cells [38]. WWOX, including an Indel (chr6:9186279) significantly associated with RTN, is known to play a role in breast cancer [39]. TRIML2 and MTNR1A were close to SNP (rs330045817) and Indel (rs700363122), respectively, on SSC17. As mentioned above, these two different types of variants were close to each other and significantly associated with ADIFF in both GLM and FarmCPU models. TRIML2 was significantly associated with prognosis, with a higher expression in triple-negative breast cancer cell lines than in normal mammary cell lines [40]. Common variants in MTNR1A may contribute to breast cancer susceptibility [41]. TBX5, encoding T-Box Transcription Factor 5, was close to the significant Indel (rs701717756) for ADIFF on SSC14. In a large German family, TBX3 and TBX5 duplication was reported to be associated with Ulnar-Mammary syndrome [42]. Importantly, TBX3 was the placode marker required for the formation of mammary placodes and the development of fetal mammary glands in all mammals [43]. Although not located in the 1-Mb region of rs701717756, TBX3 was close to this Indel with a distance of 625,516 bp.
Unlike earlier studies, the present study failed to detect significant association signals in genome regions around VRTN on SSC7, which was reported as a credible candidate gene for the teat number and the vertebra number [17,44]. Zhuang et al. suggested that the genetic heterogeneity of variants in VRTN may exist in different populations and VRTN may not be a strong or the only causal gene for teat number based on their finding that VRTN mutation was significantly associated with the teat number in Canadian Duroc pigs, but not in American Duroc pigs [14]. Moreover, VRTN mutation on SSC7 was also not significantly associated with the teat number in Chinese pig breeds, including Beijing Black pig and Sushan pig [8,9], but significantly associated with the vertebra number in Beijing Black pig. Furthermore, VRTN was reported to modulate somite segmentation [44]. These reports suggested that VRTN plays a more important role in vertebra number and varies in its role in teat number among populations with different genetic backgrounds. Therefore, the teat number in Qingping pigs is speculated to involve other genes or pathways.
The GO enrichment results showed significant enrichment in skin development, epidermis development, and epidermal cell differentiation. The mammary gland is an epithelial organ, and epithelial–stromal crosstalk is a key aspect of mammary morphogenesis [45]. In KEGG enrichment analysis, the Wnt signaling pathway did not reach a significance level of 0.05 (0.078). Interestingly, candidate genes, including WNT11, WNT16 and FZD3, were located at key positions in this pathway (Figure 5c). WNT16 and FZD3 were also involved in GO terms, skin development and epidermis development. The Wnt signaling cascade is implicated in almost all stages of mammary development and is pivotal for the specification and morphogenesis of the mammary gland [46]. WNT11 was expressed in stromal cells and basal cells in the adult mammary gland [46]. Chu et al. reported that WNT11 and FZD3 were expressed in mammary buds at E12.5 and E15.5 [47]. Importantly, Wnt signaling interacted with TBX3 in mammary placode development [47]. These reports suggested that candidate genes might affect teat number during mammary gland morphogenesis. Unlike other breeds, the teat number of Qingping pigs showed a medium heritability (0.24) for ADIFF, implying the potential involvement of candidate genes in mammary gland morphogenesis. Therefore, our candidate genes related to mammary gland morphogenesis and development can be assumed to contribute to teat number improvement and even may influence milk production.
5. Conclusions
In this study, GLM and FarmCUP GWAS were carried out to detect associated SNPs and Indels for 6 teat number-related traits. We found a total of 33 SNPs and 50 Indels for teat number. The most significant SNP and Indel were located on SSC17. Six candidate genes were enriched in the Wnt signaling pathway with an important role in mammary gland morphogenesis and development. A novel candidate gene on SSC14, TBX3, was detected as a mammary placode marker. These findings contribute to our understanding of the genetic architecture of teat number and provide genetic markers for genetic improvement of teat number in Qingping pigs.
Acknowledgments
We thank the Qingping pig Conservation Farm for providing samples and information. The computations in this paper were run on the bioinformatics computing platform of the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani12091057/s1. Figure S1: Principal component analysis of Qingping pigs; Figure S2: The Q-Q plots of the GLM GWAS for teat number-related traits in Qingping pigs, including LTN, RTN, TNUM, MAXAP, L-R and ADIFF. (a–f) The Q-Q plots of GLM GWAS based on SNPs. (g–l) The Q-Q plots of GLM GWAS based on Indels; Figure S3: The Q-Q plots of the FarmCPU GWAS for ADIFF in Qingping pigs. (a) The Q-Q plot of FarmCPU GWAS based on SNPs. (b) The Q-Q plot of FarmCPU GWAS based on Indels; Table S1: Sequencing reads, alignment statistics, and mean coverage for each sample in this study; Table S2: Annotated genes within 1-Mb regions of significant SNPs and Indels.
Author Contributions
Conceptualization, Z.L. and S.J.; methodology, Z.L. and H.L.; software, Z.L.; formal analysis, Z.L.; investigation, Z.L.; resources, Z.L. and Z.Z.; data curation, Z.L.; writing—original draft preparation, S.J.; writing—review and editing, Z.L.; visualization, S.J.; supervision, S.J.; project administration, S.J.; funding acquisition, S.J. All authors have read and agreed to the published version of the manuscript.
Funding
S.J. was funded by the National Key R & D Program of China (2021YFD1301200, 2017YFD0502000), China Agriculture Research System (CARS-36), The Fundamental Research Funds for the Central Universities (2662020DKPY012), Research Funds for Hubei Key Laboratory of Animal Embryo Engineering and Molecular Breeding (KLAEMB-2019-01) and Key R & D pro-jects of Hubei Province (2021BBA082, 2020BBB069, 2020ABA016).
Institutional Review Board Statement
The experimental protocol used in this study was reviewed and approved by the Animal Experimental Ethical Inspection of Laboratory Animal Centre, Huazhong Agriculture University. All sample collection was conducted under a permit (No. HZAUSW-2017-004) approved by the Attitude of the Animal Management and Ethics Committee.
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw sequences for the 100 QP pigs have been deposited into the NCBI Sequence Read Archive under PRJNA489520 and will be available on 30 December 2023. Phenotypes and genotypes are deposited in OSF of Center for Open Science (https://osf.io/szu86/) (will be accessible on 30 December 2023).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li Y., Pu L., Shi L., Gao H., Zhang P., Wang L., Zhao F. Revealing New Candidate Genes for Teat Number Relevant Traits in Duroc Pigs Using Genome-Wide Association Studies. Animals. 2021;11:806. doi: 10.3390/ani11030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pig Quantitative Trait Locus (Qtl) Database (Pig Qtldb) [(accessed on 27 December 2021)]. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/SS/index.
- 3.Arakawa A., Okumura N., Taniguchi M., Hayashi T., Hirose K., Fukawa K., Ito T., Matsumoto T., Uenishi H., Mikawa S. Genome-Wide Association Qtl Mapping for Teat Number in a Purebred Population of Duroc Pigs. Anim. Genet. 2015;46:571–575. doi: 10.1111/age.12331. [DOI] [PubMed] [Google Scholar]
- 4.Lee J., Lee S., Park J.E., Moon S.H., Choi S.W., Go G.W., Lim D., Kim J.M. Genome-Wide Association Study and Genomic Predictions for Exterior Traits in Yorkshire Pigs1. J. Anim. Sci. 2019;97:2793–2802. doi: 10.1093/jas/skz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato S., Atsuji K., Saito N., Okitsu M., Sato S., Komatsuda A., Mitsuhashi T., Nirasawa K., Hayashi T., Sugimoto Y., et al. Identification of Quantitative Trait Loci Affecting Corpora Lutea and Number of Teats in a Meishan X Duroc F2 Resource Population. J. Anim. Sci. 2006;84:2895–2901. doi: 10.2527/jas.2006-176. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez S.C., Finlayson H.A., Ashworth C.J., Haley C.S., Archibald A.L. A Genome-Wide Linkage Analysis for Reproductive Traits in F2 Large White X Meishan Cross Gilts. Anim. Genet. 2014;45:191–197. doi: 10.1111/age.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Zhang Y., Zhang T., Zhang L., Yan H., Liu X., Wang L. Genotyping by Sequencing Reveals a New Locus for Pig Teat Number. Anim. Genet. 2017;48:470–472. doi: 10.1111/age.12547. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L., Zhao W., Fu Y., Fang X., Ren S., Ren J. Genome-Wide Detection of Genetic Loci and Candidate Genes for Teat Number and Body Conformation Traits at Birth in Chinese Sushan Pigs. Anim. Genet. 2019;50:753–756. doi: 10.1111/age.12844. [DOI] [PubMed] [Google Scholar]
- 9.Niu N., Wang H., Shi G., Liu X., Liu H., Liu Q., Yang M., Wang L., Zhang L. Genome Scanning Reveals Novel Candidate Genes for Vertebral and Teat Number in the Beijing Black Pig. Anim. Genet. 2021;52:734–738. doi: 10.1111/age.13111. [DOI] [PubMed] [Google Scholar]
- 10.Wada Y., Akita T., Awata T., Furukawa T., Sugai N., Inage Y., Ishii K., Ito Y., Kobayashi E., Kusumoto H., et al. Quantitative Trait Loci (Qtl) Analysis in a Meishan X Göttingen Cross Population. Anim. Genet. 2000;31:376–384. doi: 10.1046/j.1365-2052.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- 11.Jinghu Z., Xiong Y., Zuo B., Lei M., Jiang S., Li F., Zheng R., Li J., Xu D. Detection of Quantitative Trait Loci Associated with Several Internal Organ Traits and Teat Number Trait in a Pig Population. J. Genet. Genom. 2007;34:307–314. doi: 10.1016/S1673-8527(07)60032-0. [DOI] [PubMed] [Google Scholar]
- 12.Bidanel J.P., Rosendo A., Iannuccelli N., Riquet J., Gilbert H., Caritez J.C., Billon Y., Amigues Y., Prunier A., Milan D. Detection of Quantitative Trait Loci for Teat Number and Female Reproductive Traits in Meishan X Large White F2 Pigs. Animal. 2008;2:813–820. doi: 10.1017/S1751731108002097. [DOI] [PubMed] [Google Scholar]
- 13.Tan C., Wu Z., Ren J., Huang Z., Liu D., He X., Prakapenka D., Zhang R., Li N., Da Y., et al. Genome-Wide Association Study and Accuracy of Genomic Prediction for Teat Number in Duroc Pigs Using Genotyping-by-Sequencing. Genet. Sel. Evol. 2017;49:35. doi: 10.1186/s12711-017-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang Z., Ding R., Peng L., Wu J., Ye Y., Zhou S., Wang X., Quan J., Zheng E., Cai G., et al. Genome-Wide Association Analyses Identify Known and Novel Loci for Teat Number in Duroc Pigs Using Single-Locus and Multi-Locus Models. BMC Genom. 2020;21:344. doi: 10.1186/s12864-020-6742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J., Zhang Z., Yang B., Guo Y., Ai H., Long Y., Su Y., Cui L., Zhou L., Wang X., et al. Identification of Loci Affecting Teat Number by Genome-Wide Association Studies on Three Pig Populations. Asian-Australas. J. Anim. Sci. 2017;30:1–7. doi: 10.5713/ajas.15.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohrer G.A., Nonneman D.J. Genetic Analysis of Teat Number in Pigs Reveals Some Developmental Pathways Independent of Vertebra Number and Several Loci Which Only Affect a Specific Side. Genet. Sel. Evol. 2017;49:4. doi: 10.1186/s12711-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moscatelli G., Dall’Olio S., Bovo S., Schiavo G., Kazemi H., Ribani A., Zambonelli P., Tinarelli S., Gallo M., Bertolini F., et al. Genome-Wide Association Studies for the Number of Teats and Teat Asymmetry Patterns in Large White Pigs. Anim. Genet. 2020;51:595–600. doi: 10.1111/age.12947. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Subgroup Genome Project Data Processing The Sequence Alignment/Map Format and Samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The Variant Call Format and Vcftools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-Generation Plink: Rising to the Challenge of Larger and Richer Datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browning B.L., Zhou Y., Browning S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common Snps Explain a Large Proportion of the Heritability for Human Height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Team, R Core . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
- 26.Yin L., Zhang H., Tang Z., Xu J., Yin D., Zhang Z., Yuan X., Zhu M., Zhao S., Li X., et al. Rmvp: A Memory-Efficient, Visualization-Enhanced, and Parallel-Accelerated Tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021;19:619–628. doi: 10.1016/j.gpb.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., Huang M., Fan B., Buckler E.S., Zhang Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016;12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Tian F., Pan Y., Buckler E.S., Zhang Z. A Super Powerful Method for Genome Wide Association Study. PLoS ONE. 2014;9:e107684. doi: 10.1371/journal.pone.0107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and Visualization of Ld and Haplotype Maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Durinck S., Spellman P.T., Birney E., Huber W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package Biomart. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.G., Han Y., He Q.Y. Clusterprofiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicodemus K.K., Liu W., Chase G.A., Tsai Y.Y., Fallin M.D. Comparison of Type I Error for Multiple Test Corrections in Large Single-Nucleotide Polymorphism Studies Using Principal Components Versus Haplotype Blocking Algorithms. BMC Genet. 2005;6((Suppl. 1)):S78. doi: 10.1186/1471-2156-6-S1-S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spina E., Cowin P. Embryonic Mammary Gland Development. Semin. Cell Dev. Biol. 2021;114:83–92. doi: 10.1016/j.semcdb.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Loisel F., Farmer C., van Hees H., Quesnel H. Relative Prolactin-to-Progesterone Concentrations around Farrowing Influence Colostrum Yield in Primiparous Sows. Domest. Anim. Endocrinol. 2015;53:35–41. doi: 10.1016/j.domaniend.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.W., Wu G. Regulatory Role for Amino Acids in Mammary Gland Growth and Milk Synthesis. Amino Acids. 2009;37:89–95. doi: 10.1007/s00726-008-0151-5. [DOI] [PubMed] [Google Scholar]
- 36.Farmer C. Review: Mammary Development in Lactating Sows: The Importance of Suckling. Animal. 2019;13:s20–s25. doi: 10.1017/S1751731118003464. [DOI] [PubMed] [Google Scholar]
- 37.Siouda M., Dujardin A.D., Barbollat-Boutrand L., Mendoza-Parra M.A., Gibert B., Ouzounova M., Bouaoud J., Tonon L., Robert M., Foy J.P., et al. Cdyl2 Epigenetically Regulates Mir124 to Control Nf-Kappab/Stat3-Dependent Breast Cancer Cell Plasticity. iScience. 2020;23:101141. doi: 10.1016/j.isci.2020.101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W., Feng B., Meng Y., Wang J., Geng B., Cui Q., Zhang H., Yang Y., Yang J. Fam3c-Yy1 Axis Is Essential for Tgfbeta-Promoted Proliferation and Migration of Human Breast Cancer Mda-Mb-231 Cells Via the Activation of Hsf1. J. Cell. Mol. Med. 2019;23:3464–3475. doi: 10.1111/jcmm.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pospiech K., Pluciennik E., Bednarek A.K. Wwox Tumor Suppressor Gene in Breast Cancer, a Historical Perspective and Future Directions. Front. Oncol. 2018;8:345. doi: 10.3389/fonc.2018.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X., Zhang C., Liu Z., Liu Q., He K., Yu Z. Characterization of Cerna Network to Reveal Potential Prognostic Biomarkers in Triple-Negative Breast Cancer. PeerJ. 2019;7:e7522. doi: 10.7717/peerj.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deming S.L., Lu W., Beeghly-Fadiel A., Zheng Y., Cai Q., Long J., Shu X.O., Gao Y.T., Zheng W. Melatonin Pathway Genes and Breast Cancer Risk among Chinese Women. Breast Cancer Res. Treat. 2012;132:693–699. doi: 10.1007/s10549-011-1884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cenni C., Andres S., Hempel M., Strom T.M., Thomas E., Davies A., Timoney N., Frigiola A., Logan M., Holder-Espinasse M. Tbx3 and Tbx5 Duplication: A Family with an Atypical Overlapping Holt-Oram/Ulnar-Mammary Syndrome Phenotype. Eur. J. Med. Genet. 2021;64:104213. doi: 10.1016/j.ejmg.2021.104213. [DOI] [PubMed] [Google Scholar]
- 43.Slepicka P.F., Somasundara A.V.H., Santos C.O.D. The Molecular Basis of Mammary Gland Development and Epithelial Differentiation. Semin. Cell Dev. Biol. 2021;114:93–112. doi: 10.1016/j.semcdb.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan Y., Zhang H., Zhang Z., Gao J., Yang J., Wu Z., Fan Y., Xing Y., Li L., Xiao S., et al. Vrtn Is Required for the Development of Thoracic Vertebrae in Mammals. Int. J. Biol. Sci. 2018;14:667–681. doi: 10.7150/ijbs.23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gjorevski N., Nelson C.M. Integrated Morphodynamic Signalling of the Mammary Gland. Nat. Rev. Mol. Cell Biol. 2011;12:581–593. doi: 10.1038/nrm3168. [DOI] [PubMed] [Google Scholar]
- 46.Yu Q.C., Verheyen E.M., Zeng Y.A. Mammary Development and Breast Cancer: A Wnt Perspective. Cancers. 2016;8:65. doi: 10.3390/cancers8070065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu E.Y., Hens J., Andl T., Kairo A., Yamaguchi T.P., Brisken C., Glick A., Wysolmerski J.J., Millar S.E. Canonical Wnt Signaling Promotes Mammary Placode Development and Is Essential for Initiation of Mammary Gland Morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequences for the 100 QP pigs have been deposited into the NCBI Sequence Read Archive under PRJNA489520 and will be available on 30 December 2023. Phenotypes and genotypes are deposited in OSF of Center for Open Science (https://osf.io/szu86/) (will be accessible on 30 December 2023).