Abstract
Before leaving the house, it is a good idea to check for road closures that may affect the morning commute. Otherwise, one may encounter significant delays arriving at the destination. While this is commonly true, motorists may be able to consult a live interactive traffic map and pick an alternate route or detour to avoid being late. However, this is not the case if one needs to catch the train which follows a single track to the terminus; if something blocks the track, there is a delay. Such is the case for the DNA replisome responsible for copying the genetic information that provides the recipe of life. When the replication machinery encounters a DNA roadblock, the outcome can be devastating if the obstacle is not overcome in an efficient manner. Fortunately, the cell’s DNA synthesis apparatus can bypass certain DNA obstructions, but the mechanism(s) are still poorly understood. Very recently, two papers from the O’Donnell lab, one structural [1] and the other biochemical [2], have challenged the conventional thinking of how the replicative CMG helicase is arranged on DNA, unwinds double-stranded DNA, and handles barricades in its path. These new findings raise important questions in the search for mechanistic insights into how DNA is copied, particularly when the replication machinery encounters a roadblock.
Keywords: CMG, MCM, helicase, DNA replication, DNA damage, interstrand cross-link
Graphical Abstract
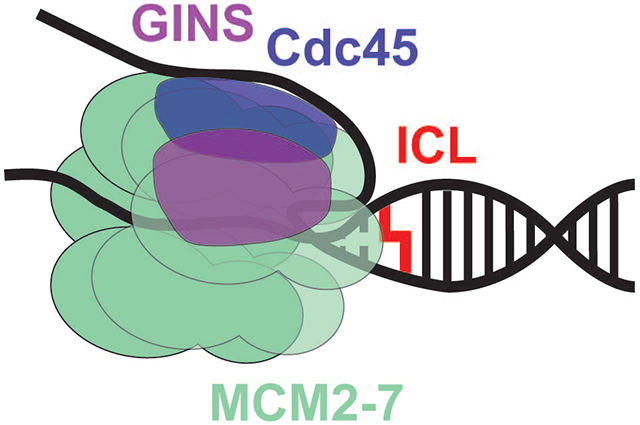
Introduction
Nucleic acid helicases are essential enzymes for the genomic maintenance of all forms of life. DNA specific helicases act on a variety of substrates encountered during DNA replication, repair, and recombination to destabilize and separate duplex DNA by coupling ATP hydrolysis to unwinding. Even though hexameric DNA helicases structurally converged into a common ring-shaped architecture [3], they arose separately into two major families classified by conserved folds and unwinding polarities [4,5]. Superfamily (SF) 4 DNA replication helicases from bacteria and many phages, including E. coli DnaB, bacteriophage T7 gp4, and mitochondrial Twinkle, utilize RecA-folds to unwind DNA in the 5′-3′ direction. Alternatively, eukaryotes and archaea rely on SF 5 helicases, including minichromosomal maintenance (MCM) proteins, derived from ATPases associated with a variety of activities (AAA+) clade that translocate with opposite 3′-5′ polarity. In spite of these evolutionary and functional differences, both families utilize a common P-loop ATPase motif that organizes ATP binding motifs from one subunit to arginine fingers contained in an adjacent subunit catalyzing ATP hydrolysis to power DNA unwinding [6].
Organization of the MCM Helicase on DNA
As a consequence of the measured unwinding polarities of SF 4 and SF 5 helicases, the hexameric helicase will encircle and translocate along the lagging strand in bacteria (DnaB) and along the leading strand in archaea and eukaryotes (MCM). Binding of bacterial DnaB to a replication fork arranges the C-terminal RecA motor domain (CTD) adjacent to the duplex with the lagging strand traversing the central channel of the helicase towards the N-terminal domain (NTD) [7]. Loading of the MCM2-7 helicase in eukaryotes proceeds by sequential binding of two individual hexamers to origin DNA followed by closing of the ring around double-stranded DNA upon ATP hydrolysis and concomitant release of Cdt1 to form an off-set double hexamer ring [8–10]. The active form of the MCM2-7 helicase includes melting of the duplex DNA and transitioning to the encircling of one strand of DNA coincident with sequential binding of Cdc45 and GINS subunits across the MCM2-5 gate to form the CMG complex effecting bidirectional topological separation of the leading and lagging strands [11–14].
Binding of the homo-hexameric MCM from archaea to fork DNA was also shown using various fluorescence experiments to orientate the CTD (AAA+ domain) towards the duplex [15,16]. From these initial experiments, it seemed that, like bacterial DnaB, the motor domain acts to separate the duplex, while the NTD is free to interact with other replisome proteins. Like archaeal MCM, the eukaryotic D. melanogaster CMG complex was shown by electron microscopy (EM) to have an identical binding orientation placing the CTD towards the duplex DNA [17]. However, the recent high resolution EM structure of the S. cerevisiae CMG complex from the O’Donnell laboratory challenges this orientation and instead shows the NTD leading the way along the leading strand [1]. The EM visualization is quite clear and as such has created a controversy over how eukaryotic origins are activated and propagated to form bidirectional replication forks after loading of the MCM2-7 double hexamer.
There are significant differences between experimental systems that need to be explored. First and foremost, the yeast CMG complex includes both GINS and Cdc45 that are absent from the archaeal experiments that may help to properly orientate the MCM complex on DNA. If so, then answers on how Cdc45 and GINS help to direct complex binding are of utmost importance. Of course, there could be actual orientation differences between archaeal and eukaryotic MCM on DNA; however, as most of the other features of DNA replication are conserved, this would be very surprising. More surprising would be species-specific differences in MCM2-7 activation/orientation with the eukaryotic domain as seen between the fly and yeast CMG EM structures. One caveat with the yeast CMG orientation is that the authors utilized five phosphorothioate deoxythymidines on the 3′ end of the leading strand, presumably to prevent degradation when the leading strand DNA polymerase epsilon was included in future structures. However, archaeal MCM has been shown to load and slide onto the 3′ end of a DNA strand [15], and whether eukaryotic MCM2-7 is influenced by these phosphorothioates or, on the other hand, whether archaeal MCM is influenced by fluorescent labels is not known. There are also DNA sequence-specific differences between the three systems. The archaeal MCM orientation (CTD first) was determined using both random sequence [15] and poly-dT [16] fork arms. The Drosophila (CTD first) [17] and yeast (NTD first) [1] CMG orientations also utilized poly-dT for the leading strand, however, the yeast structure also had a short random 16-nucleotide excluded strand. Whether the excluded strand influences CMG loading in a particular orientation onto artificial fork substrates needs to be addressed.
What is confirmed is that a double hexamer of MCM2-7 is loaded at an origin of replication with the NTDs arranged head-to-head. Although the 3′-5′ unwinding polarity of CMG is not under question, how the double hexamer MCM2-7 becomes activated for unwinding would be considering this new orientation. In addition to the foremost unanswered question in the DNA replication field for how a double hexamer goes from encircling double-stranded DNA to individual CMG hexamers encircling opposing single-strands, biophysics of helicase fork activation also become probing. Should the NTD lead the way for unwinding, the two hexamers would have to pass by one another to form the individual forks by having one CMG encircle the displaced strand of the other (Fig. 1A). This is not impossible but would require somewhat complicated protein-DNA reorganization that includes a larger DNA bubble to accommodate this transversion. Alternatively, individual CMGs that translocate with the CTD leading the way would just have to destabilize dodecamer association before single-stranded DNA translocation and unwinding commences (Fig. 1B).
Figure 1. Models of hexameric helicase activation and unwinding.
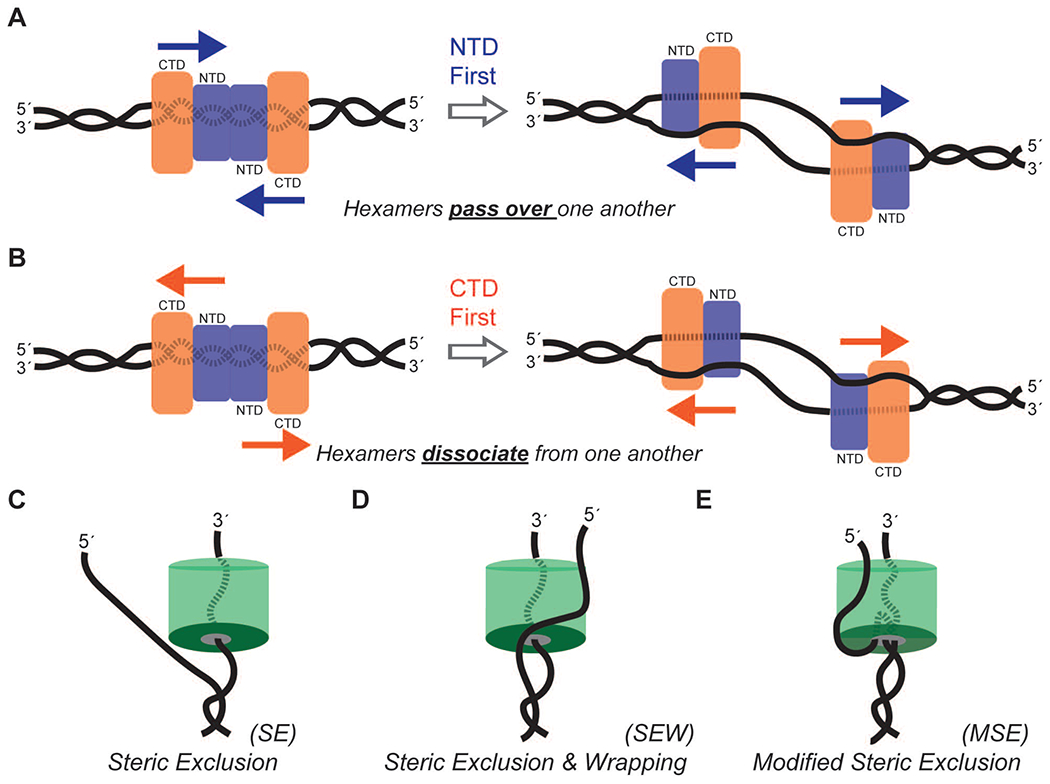
A double hexamer MCM complex forms with the N-terminal domain (NTD) head to head at the origin of replication where single-stranded DNA selection and unwinding can proceed either with the A) NTD or B) C-terminal domain (CTD) leading the way. During active unwinding, the MCM helicase proceeds in the 3′-5′ direction on the translocating strand C) excluding single-stranded DNA from the central channel that can D) interact on the exterior surface to control the unwinding rate and efficiency. E) A modification of these models includes initial double-stranded DNA separation occurring within the central channel.
Hexameric Helicase Structure-Function Unwinding Mechanism
The steric exclusion (SE) model of DNA unwinding by hexameric helicases seems to be conserved across both superfamilies of enzymes (Fig. 1C). Here, single-stranded DNA contacts conserved positively charged hairpins within the central channel to pass the DNA through or the hexameric helicase along effectively translocating on one strand. Recent high resolution EM data of a CMG-Pol epsilon complex from the Costa lab is consistent with the SE model with CMG behaving as a single-stranded DNA translocase [18]. The opposing excluded strand is separated prior to entry into the central channel using a physical toroidal wedge to unwind DNA. Previous models where single-stranded DNA is extruded out a side channel within the interior of the hexamer or double-stranded DNA is pumped and twisted through the central channel have been almost universally discounted. Instead, more information on the precise contacts with DNA have led to slight modifications of the SE model. Electrostatic interactions on the exterior surface with the excluded single-stranded DNA has led to the steric excluding and wrapping (SEW) model (Fig. 1D) [19]. The mechanistic role and binding dynamics of the excluded strand may vary with different classes of helicases; however, this interaction clearly modulates the rate and efficiency of DNA unwinding in all hexameric helicases tested [20,21].
Recently, more high resolution EM data from the O’Donnell laboratory has visualized a small stretch of duplex DNA within the NTD of the S. cerevisiae CMG complex [1] (O’Donnell, unpublished). They propose a somewhat different modified steric exclusion (MSE) model of unwinding, whereby double-stranded DNA enters the NTD to be separated inside the central channel only to be extruded back out the same path and excluded along the exterior of the CMG (Fig. 1E). Whether the excluded strand at this point binds specific MCM subunits or interacts specifically with Cdc45 or GINS or other components of the eukaryotic replisome remains to be determined.
Biochemical experiments using purified CMG proteins and biotin-streptavidin blocks attached to either the translocating or excluded strand seem to support the MSE model [2]. It was observed that adducts on either strand blocked or reduced unwinding by the CMG complex. Most tellingly, the covalent methyltransferase adduct on the lagging strand was an even stronger block to unwinding. They further showed that streptavidin can be displaced from biotin on the lagging strand providing key support for entry of duplex DNA into the CMG complex followed by an internal unwinding mechanism. Thus, their results did not support a strict SE model. This MSE model is consistent with both EM and In vitro biochemical data from the O’Donnell laboratory and will have strong implications for the ability of the replisome to overcome and bypass a multitude of noncovalent and covalent roadblocks during DNA replication.
Helicase Encounters with DNA Adducts or Protein Obstacles
Even though CMG had a hard time progressing past large bulky lesions on either the translocating or nontranslocating strand, it should be stressed that emerging evidence indicates that inhibition of DNA unwinding activity is dependent on the type of adduct and the specific helicase under investigation [22,23]. It is generally thought that relatively small base lesions that only minimally affect DNA structure are well tolerated by ring-like helicases that possess a central channel for DNA translocation. For example, the sequence-related SF 4 helicases E. coli DnaB and human mitochondrial Twinkle, implicated in replication of the bacterial [24,25] and mitochondrial [26] genomes, respectively, were unaffected by a cyclopurine (cPu) lesion (that perturbs helix twist and base stacking) in either the translocating or non-translocating strands of the duplex DNA substrate [21,27]. Similarly, a single alkyl phosphotriester modification that introduces a hydrophobic group into the DNA backbone and neutralizes the negative charge of the phosphodiester moiety displayed no apparent effect on DnaB or M. thermoautotrophicum MCM [28].
Even DNA helicases sharing sequence homology within the helicase core domain may display differences in how they are affected by DNA modifications, suggesting subtle and specialized differences in unwinding mechanisms [29]. It should be kept in mind that the size of the biotin-SA moiety used for interrogation of strand-specific inhibition of DNA unwinding by the yeast CMG helicase [2] really represents an extreme case in the type of DNA block because it is considerably bulkier than DNA lesions such as those described above. Therefore, it may not be surprising that the biotin-SA positioned on either the translocating or non-translocating strands negatively affects yeast CMG-catalyzed DNA unwinding. Nonetheless, protein-DNA covalent adducts can be quite large, so the findings from biotin-SA modified DNA substrates are informative. As more interacting proteins are identified that collaborate with helicases, it seems likely that such factors will regulate their activity and potentially direct pausing or bypass at specific lesions.
The mammalian DNA tumor virus SV40 large T antigen is a hexameric DNA helicase demonstrated to have the capability of unwinding past a DNA-protein (Hpall methyltransferase conjugate) cross-link on the helicase translocating strand, leaving the authors to speculate that the helicase ring can open to bypass the protein barrier [30]; however, ring-opening was not formally shown. It remains to be seen if other replicative DNA helicases can behave in a similar manner to bypass a significantly large bulky covalent adduct. The possibility that CMG helicase complex can bypass a bulky lesion in the translocating strand by a ring-opening mechanism would certainly be relevant to its encounter with an interstrand cross-link (ICL), a hot topic of debate (see next section).
The finding from the O’Donnell lab that the purified yeast CMG helicase is sensitive to biotin-SA in either strand [2] is in contrast to a 2011 report from the Walter lab using Xenopus laevis egg extract protein showing that CMG-supported replisome-catalyzed DNA synthesis on a plasmid-based DNA substrate is sensitive to biotin-SA positioned in the translocating strand but not the non-translocating strand [31]. In the latter case, it was argued that the results strongly supported a classic SE model, which is now in doubt for the purified yeast CMG helicase [2]. Aside from the unlikely (but possible) explanation of species-specific differences, one must keep in mind that replisome/CMG-interacting proteins in the Xenopus extract may mediate bypass of the bulky lesion positioned in the strand opposite to the one that helicase is believed to thread. Although likely by a different mechanism, the single-stranded DNA binding protein Replication Protein A (RPA), which physically interacts with FANCJ [32] or RECQ1 [33], can enable either helicase to efficiently unwind past a TG lesion positioned in the non-translocating strand, but not the translocating strand [29]. It will be important to identify the putative protein factors that enable the Xenopus replisome/CMG to bypass a bulky DNA lesion or protein obstacle in a strand-specific manner, and determine if a similar mechanism operates for other eukaryotic CMG complexes in a highly purified reconstituted system. Such studies are pertinent to many forms of biologically relevant DNA lesions, including ICLs (next section).
A reasonably good class of candidates to mediate CMG bypass of a DNA adduct or protein obstacle is accessory DNA helicases. The McGlynn lab has made significant advances to comprehend the genetic and biochemical roles of accessory helicases as components of the replisome to bypass protein-bound DNA complexes (for review, see [34–36]). In E. coli, the 3′ to 5′ SF 1 DNA helicase Rep actively collaborates with the 5′ to 3′ replicative DnaB helicase to provide an efficient mechanism for genome duplication [37–39]. It is yet unclear if the helicases operate together to bypass any types of bulky DNA adducts; moreover, to our knowledge the identification and characterization of an accessory 5′ to 3′ helicase(s) that operates in conjunction with the 3′ to 5′ CMG helicase is not reported. Among the potential candidates are the SF 1 helicases Pif1 [40,41] or human DNA helicase B (HDHB) [42].
Replication Fork and CMG Dynamics at Bulky Blocks
Replication forks are frequently challenged by a host of impediments including alternate DNA structures and DNA covalent adducts. Lesions, such as ultraviolet photoproducts, that can be included within the central channel of CMG may not impede the passage of the fork, although they inhibit polymerases. The consequence, shown in an early investigation of replication in E. coli exposed to UV, is gaps in the daughter strands. That study supported a model of replication restart past polymerase blocking photoproducts [43]. That view has been extended in more recent studies, and the concept of leading daughter strand synthesis interrupted by DNA adducts on the template strand is well established [44–47].
On the other hand, blocks on the leading strand template, too large to be accommodated in the central channel of the CMG, would be expected to stop forward progress of the CMG. Indeed, as summarized above, placement of bulky adducts, too large for inclusion in the central channel, on leading or lagging strand templates, or on both strands, has been used to interrogate the disposition of the template strand in the architecture of the CMG. Two different conclusions have emerged from these studies. Walter and colleagues concluded that the “CMG encircles single-stranded DNA with no duplex DNA remaining in the central channel” [31], in accord with a SE model. An additional implication was that when the CMG complex encountered an ICL, the DNA immediately adjacent to the lesion would be single-stranded since one strand would be within the central channel, while the other would be excluded [31] (see below).
The recent publications from the O’Donnell lab [1,2], cited above, provide a somewhat different view of duplex DNA and leading and lagging templates strands within and on the surface of the CMG. Their MSE model explained why a block on either strand would challenge forward movement of the helicase. They suggested that the different conclusions of the two groups were due to the use of purified proteins (O’Donnell) versus extracts (Walter).
The experiments from both the Walter and O’Donnell laboratories support the conclusion that that bulky adducts on the leading strand template cannot be bypassed by the replisome. This would also extend to ICLs that link one strand to an enormous bulky adduct, i.e., the other DNA strand. Consequently, ICLS have always been considered absolute blocks to the replication fork. However, this view was challenged with the description of what was termed “replication traverse”, in which DNA synthesis restarted on the side of an ICL distal to that of the fork encounter as measured in cells [48]. Assuming that the CMG that collided with an ICL also drives the restart, this pathway is reminiscent of the discontinuous synthesis pathway reported almost 50 years ago by Dean Rupp [43]. However, restart past the ICL by a CMG topologically locked around the leading strand template is not possible without structural adjustments.
Replication Restart Past an Interstrand Cross-link
A series of steps must occur for replication restart past an ICL. (1) The topological constraints imposed by the CMG lock around the leading strand template would have to be temporarily relieved, i.e., the ring must open; (2) The channel of the “open” replisome must be able to include the crosslinked DNA; (3) The complex would have to translocate past the ICL; (4) The replisome would have to be reactivated and DNA synthesis resumed.
(1). How Is the topological constraint Imposed by CMG relieved?
By analogy, as alluded to above for the bypass of a bulky lesion by Large T antigen [30], an alternative but simple explanation for the bypass of an ICL is that the CMG helicase opens its ring upon encounter with the ICL, and then closes again in a timely manner once the ICL is traversed, suggested by studies with Xenopus extracts [31] (Fig. 2A). Deciphering the putative mechanism of ring opening/closing will be insightful. Along these lines, there may be a specific factor(s) interacting with CMG and/or DNA substrate that mediates bypass of the ICL in a yet unknown manner. For example, as mentioned above and by analogy to the bacterial system, an accessory DNA helicase or factor may promote CMG bypass of a replisome obstacle like the ICL (Fig. 2B). These alternatives should be approachable from an experimental standpoint.
Figure 2. ICL lesion bypass by a CMG helicase in a unidirectional manner.

A) CMG converts from a single-stranded DNA translocase to a double-stranded DNA translocase as it passes the ICL. B) CMG helicase opens its ring upon encounter with the ICL, and then closes again once the ICL is traversed. C) An accessory DNA helicase (triangle) may promote CMG bypass of the ICL.
A possible solution to the first problem, i.e., relief of the topological constraint, was provided by an observation from the Costanzo laboratory in experiments using Xenopus egg extracts. They found that the GINS complex was lost from the CMG when it encountered a pause imposed by a double-stranded DNA break. On the other hand, Cdc45, the other protein involved in ring closure, was retained [49]. It has been suggested that the retention of Cdc45 could prevent the loss of the MCM complex from the DNA, thus facilitating eventual reconstruction of a functional CMG [50]. Loss of the GINS could open either the N-tier or the N- and C-tiers, as depicted by Langston and O’Donnell [2], enabling reannealing of the lagging strand template to the leading strand template within the central channel. Consequently, the reannealing would be anchored by this duplex element, and the “CM” complex could move past the ICL without the complexity of a transition from an exclusively single-strand binding to a double-strand binding mode. On the other hand, if the CMG complex proceeds by strict steric exclusion, it would have to transition from single-strand binding to double-strand binding, presumably in such a way that double-stranded DNA (with an ICL contained within the barrel of the helix) enters the CMG. Eventually, a GINS complex would have to re-associate, reconstructing an active CMG which would reestablish the fork.
(2). Is the channel large enough to include the crosslinked duplex?
There is a precedent for translocation of a replicative DNA helicase past an ICL lesion in the DNA. Bastia et al. determined that E. coli DnaB could translocate past a duplex DNA region containing an ICL in vitro [51]. Similarly, DnaB was shown to displace protein bound to duplex DNA via its double-strand DNA translocase activity [52]. T7 hexameric gp4 was shown to be capable of switching from traveling on single-stranded DNA to translocating on double-stranded DNA while continuously encircling the DNA tract, be it one-stranded or two annealed strands [53]. Therefore, it is a reasonable hypothesis that the same CMG complex acting as the replicative helicase to unwind parental duplex strands might convert to a double-stranded DNA translocase mode upon encountering a DNA ICL (Fig. 2C).
(3). How would an open replisome be translocated past an ICL?
Replication traverse of an ICL was shown to be facilitated by the translocase activity of the Fanconi Anemia protein FANCM, in association with interacting partners PCNA and the BLM helicase [48,54,55]. Thus, it is possible that the “open” replisome might be moved past a block by the action of perhaps transiently associated DNA translocases. Multiple translocases are associated with the cellular response to replication stress [56–58]. These translocases, in addition to FANCM, may contribute to traverse of ICLs or other potent blocks to the replisome.
(4). How would the replisome be reactivated?
Because the mechanism of CMG activation is not yet known, it may even be possible that a single MCM2-7 hexamer with the associated Cdc45, GINS, and CDK/DDK activity could be reactivated to encircle a single-strand after double-strand translocation of an ICL. Although there is very little known about such factors that would mediate CMG bypass of a bulky DNA adduct or an ICL, and subsequent replisome reactivation, some clues may be obtained from studies that examined the association of protein factors with CMG during replication initiation, keeping in mind that the mechanisms of origin activation and replisome reactivation after DNA damage bypass are inherently different. The RECQL4 helicase (defective in Rothmund-Thomson syndrome, Baller-Gerald syndrome, and RAPADILINO [59]) was found to be associated with the CMG complex and MCM10 at replication origins in a cell cycle dependent manner [60,61]. Furthermore, the interaction of RECQL4 with MCM10 was found to play an important role in efficient DNA replication origin firing [62]. Using a Xenopus egg extract with plasmid DNA harboring a site-specific origin, it was found that RECQL4 promotes the conversion of the pre-initiation complex to an active replisome via DNA unwinding [63]. Taken together, these results suggest that RECQL4 may be a candidate to mediate CMG bypass of an ICL or bulky lesion and replisome reactivation. Another candidate is HDHB which upon its depletion was shown to cause inhibition of DNA replication initiation [64]. Consistent with its role in the commencement of DNA synthesis, HDHB was found to interact with Cdc45 and aid its loading onto chromatin, suggesting involvement in the assembly of the pre-replication complex [64].
Contrasting Implications of the Modified Steric Exclusion and Strict Steric Exclusion Models
The two models have differing implications for fork encounters with replication blocks. The inclusion of some stretch of duplex DNA into the replisome, proposed in the MSE, implies that any DNA modification or alternate structure, that is too large to for the entry port of CMG would have the potential to block progress of the complex. This consideration would be independent of the strand on which the structure/adduct was formed. For example, a G quadruplex (G4) structure on the lagging strand template, R-loops in either orientation relative to the fork, protein-DNA adducts, etc., all could be serious impediments to the CMG. Consequently, completion of S phase may be even more challenging than the prior focus on leading strand impediments would suggest. Thus, the transient “opening” of the CMG may occur quite frequently during replication to overcome blocks on either strand. This would occur without loss of CMG, which is quite stable on DNA with a very slow off-rate compared with other replication factors [14].
Alternatively, in the strict steric exclusion model only leading strand structures could block the CMG. Thus, the two models make quite different predictions for the influence of lagging strand blocks on the progress of the replication fork. If the MSE model is correct the transient “opening” of the CMG may occur quite frequently during replication to overcome blocks on either strand.
Following this logic, further characterization of DNA helicases with structural features like that of MCM helicase will be valuable to ascertain if the proposed MSE model applies to them. For example, the aforementioned mitochondrial replicative DNA helicase Twinkle has a ring-like structure [65,66], and recent experimental evidence indicates that it dynamically interacts with both strands [21]. Twinkle is poorly active on a variety of intermolecular and intramolecular G4 DNA substrates [67], suggesting that its architecture is not conducive to overcoming this type of DNA roadblock even though predicted G4-forming sequences are highly abundant in the human mitochondrial genome [67,68]. Surprisingly, the sequence-related E. coli DnaB, which is also a 5′ to 3′ hexameric helicase [69], was shown to unwind a two-stranded anti-parallel G-quadruplex DNA substrate (that Twinkle does not) [67], suggesting specialization of substrate recognition and/or mechanism. To our knowledge, there are no reports on G4 DNA unwinding by CMG (MCM).
Replication Forks Converging on an Interstrand Cross-link
Above, we addressed potential lesion bypass mechanisms for a fork coming from a single direction; however, converging forks stalled at an ICL pose a unique structural dynamic that may operate by a more elaborate or entirely different mechanism (Fig. 3). Indeed, experimental evidence from the Xenopus egg extract system with a plasmid-based ICL DNA substrate in which two replication forks converge on the ICL demonstrated that ubiquitin signaling targets BRCA1 to the ICL-stalled fork, and together with leading strand DNA polymerase extension, these two elements promote CMG helicase complex unloading from the stalled replication fork [70]. CMG unloading allows dual DNA incisions in one DNA strand on opposite sides of the ICL, a prerequisite for recombination-mediated error-free repair of the cross-linked DNA. In subsequent work, it was demonstrated that post BRCA1 recruitment to the ICL, the p97/Cdc48/VCP segregase unloads the stalled CMG complex by a degradation-independent mechanism involving MCM7 polyubiquitylation [71]. This occurs upon replication completion and disassembly [72], suggesting it is not likely for ICL bypass and reactivation. Therefore, it is yet unclear if the described p97/BRCA1 mechanism is applicable to bypass of and ICL or other forms of replication stress.
Figure 3. ICL lesion bypass by CMG helicases at converging forks.
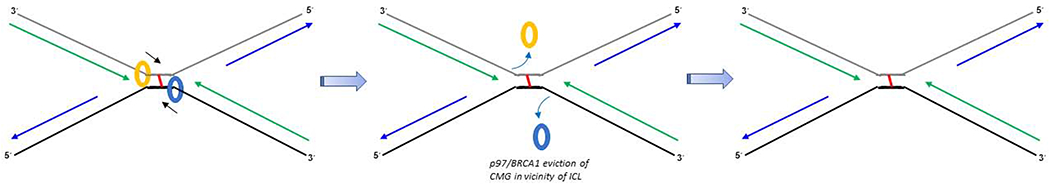
p97 and BRCA1, together with leading strand DNA synthesis, promote CMG helicases to unload at the converging fork. Reloading occurs on the distal side of the ICL.
Summary
Although the current literature and latest developments suggest that as in the case of a train track barricade, replication past DNA roadblocks is complicated, it may very well be that the cell uses more than one mechanism for lesion bypass that is dependent on several factors. Firstly, mechanisms between prokaryote and eukaryote, and even between individual species, may be in part conserved, but also diverge in subtle ways. Secondly, the mechanism for lesion bypass is likely to be dependent on the type of DNA adduct or protein obstacle. Thirdly, purified components of the replisome (e.g., helicase) behave differently when acting alone versus in the context of interacting proteins. Cell type and transformation status that dictate the expression profile of various proteins influencing the replisome would likely come into play. Despite the seemingly myriad of factors influencing replication bypass, progress in the field is evident and points toward a more comprehensive view of biological and potentially clinical relevance, yet more work is needed to clarify specific mechanisms.
Highlights.
Interaction of CMG helicase with DNA substrate dictates fate of replication fork
Modified versus strict steric exclusion model has implications for CMG block bypass
Replisome-associated factors may affect replisome bypass of bulky adducts
CMG structural adjustments are required for fork traverse of interstrand cross-link
Multiple mechanisms are responsible for bypass of replication obstacles in vivo
Acknowledgments
This work was supported by the National Science Foundation MCB-1613534 and Baylor University to M.A.T. and by the National Institutes of Health, National Institute on Aging, Intramural Research Program to R.M.B. and M.M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest.
References
- 1.Georgescu R; Yuan Z; Bai L; de Luna Almeida Santos R; Sun J; Zhang D; Yurieva O; Li H; O’Donnell ME Structure of eukaryotic cmg helicase at a replication fork and implications to replisome architecture and origin initiation. Proceedings of the National Academy of Sciences of the United States of America 2017, 114, E697–e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langston L; O’Donnell M Action of cmg with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trakselis MA Structural mechanisms of hexameric helicase loading, assembly, and unwinding. F1000Research 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer LM; Leipe DD; Koonin EV; Aravind L Evolutionary history and higher order classification of aaa+ atpases. Journal of structural biology 2004, 146, 11–31. [DOI] [PubMed] [Google Scholar]
- 5.Thomsen ND; Berger JM Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell 2009, 139, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leipe DD; Koonin EV; Aravind L Evolution and classification of p-loop kinases and related proteins. Journal of molecular biology 2003, 333, 781–815. [DOI] [PubMed] [Google Scholar]
- 7.Jezewska MJ; Rajendran S; Bujalowski W Complex of escherichia coli primary replicative helicase dnab protein with a replication fork: Recognition and structure. Biochemistry 1998, 37, 3116–3136. [DOI] [PubMed] [Google Scholar]
- 8.Li N; Zhai Y; Zhang Y; Li W; Yang M; Lei J; Tye BK; Gao N Structure of the eukaryotic mcm complex at 3.8 a. Nature 2015, 524, 186–191. [DOI] [PubMed] [Google Scholar]
- 9.Ticau S; Friedman LJ; Champasa K; Correa IR Jr.; Gelles J; Bell SP Mechanism and timing of mcm2-7 ring closure during DNA replication origin licensing. Nature structural & molecular biology 2017, 24, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Y; Cheng E; Wu H; Li N; Yung PY; Gao N; Tye BK Open-ringed structure of the cdt1-mcm2-7 complex as a precursor of the mcm double hexamer. Nature structural & molecular biology 2017, 24, 300–308. [DOI] [PubMed] [Google Scholar]
- 11.Bochman ML; Schwacha A The mcm2-7 complex has in vitro helicase activity. Molecular cell 2008, 31, 287–293. [DOI] [PubMed] [Google Scholar]
- 12.Costa A; Ilves I; Tamberg N; Petojevic T; Nogales E; Botchan MR; Berger JM The structural basis for mcm2-7 helicase activation by gins and cdc45. Nature structural & molecular biology 2011, 18, 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilves I; Petojevic T; Pesavento JJ; Botchan MR Activation of the mcm2-7 helicase by association with cdc45 and gins proteins. Molecular cell 2010, 37, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeeles JT; Deegan TD; Janska A; Early A; Diffley JF Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGeoch AT; Trakselis MA; Laskey RA; Bell SD Organization of the archaeal mcm complex on DNA and implications for the helicase mechanism. Nature structural & molecular biology 2005, 12, 756–762. [DOI] [PubMed] [Google Scholar]
- 16.Rothenberg E; Trakselis MA; Bell SD; Ha T Mcm forked substrate specificity involves dynamic interaction with the 5′-tail. The Journal of biological chemistry 2007, 282, 34229–34234. [DOI] [PubMed] [Google Scholar]
- 17.Costa A; Renault L; Swuec P; Petojevic T; Pesavento JJ; Ilves I; MacLellan-Gibson K; Fleck RA; Botchan MR; Berger JM DNA binding polarity, dimerization, and atpase ring remodeling in the cmg helicase of the eukaryotic replisome. eLife 2014, 3, e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou JC; Janska A; Goswami P; Renault L; Abid Ali F; Kotecha A; Diffley JF; Costa A Cmg-pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proceedings of the National Academy of Sciences of the United States of America 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham BW; Schauer GD; Leuba SH; Trakselis MA Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during mcm helicase unwinding. Nucleic acids research 2011, 39, 6585–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carney SM; Trakselis MA The excluded DNA strand is sew important for hexameric helicase unwinding. Methods (San Diego, Calif.) 2016, 108, 79–91. [DOI] [PubMed] [Google Scholar]
- 21.Khan I; Crouch JD; Bharti SK; Sommers JA; Carney SM; Yakubovskaya E; Garcia-Diaz M; Trakselis MA; Brosh RM Jr. Biochemical characterization of the human mitochondrial replicative twinkle helicase: Substrate specificity, DNA branch migration, and ability to overcome blockades to DNA unwinding. The Journal of biological chemistry 2016, 291, 14324–14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan I; Sommers JA; Brosh RM Jr. Close encounters for the first time: Helicase interactions with DNA damage. DNA repair 2015, 33, 43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suhasini AN; Brosh RM Jr. Mechanistic and biological aspects of helicase action on damaged DNA. Cell cycle (Georgetown, Tex.) 2010, 9, 2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaguni JM; Kornberg A Replication initiated at the origin (oric) of the e. Coli chromosome reconstituted with purified enzymes. Cell 1984, 38, 183–190. [DOI] [PubMed] [Google Scholar]
- 25.LeBowitz JH; McMacken R The escherichia coli dnab replication protein is a DNA helicase. The Journal of biological chemistry 1986, 261, 4738–4748. [PubMed] [Google Scholar]
- 26.Milenkovic D; Matic S; Kuhl I; Ruzzenente B; Freyer C; Jemt E; Park CB; Falkenberg M; Larsson NG Twinkle is an essential mitochondrial helicase required for synthesis of nascent d-loop strands and complete mtdna replication. Human molecular genetics 2013, 22, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan I; Suhasini AN; Banerjee T; Sommers JA; Kaplan DL; Kuper J; Kisker C; Brosh RM Jr. Impact of age-associated cyclopurine lesions on DNA repair helicases. PloS one 2014, 9, e113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhasini AN; Sommers JA; Yu S; Wu Y; Xu T; Kelman Z; Kaplan DL; Brosh RM Jr. DNA repair and replication fork helicases are differentially affected by alkyl phosphotriester lesion. The Journal of biological chemistry 2012, 287, 19188–19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suhasini AN; Sommers JA; Mason AC; Voloshin ON; Camerini-Otero RD; Wold MS; Brosh RM Jr. Fancj helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein a to unwind the damaged DNA substrate in a strand-specific manner. The Journal of biological chemistry 2009, 284, 18458–18470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yardimci H; Wang X; Loveland AB; Tappin I; Rudner DZ; Hurwitz J; van Oijen AM; Walter JC Bypass of a protein barrier by a replicative DNA helicase. Nature 2012, 492, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu YV; Yardimci H; Long DT; Ho TV; Guainazzi A; Bermudez VP; Hurwitz J; van Oijen A; Scharer OD; Walter JC Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R; Sharma S; Sommers JA; Kenny MK; Cantor SB; Brosh RM Jr. Fancj (bach1) helicase forms DNA damage inducible foci with replication protein a and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 2007, 110, 2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui S; Arosio D; Doherty KM; Brosh RM Jr.; Falaschi A; Vindigni A Analysis of the unwinding activity of the dimeric recq1 helicase in the presence of human replication protein a. Nucleic acids research 2004, 32, 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruning JG; Howard JL; McGlynn P Accessory replicative helicases and the replication of protein-bound DNA. Journal of molecular biology 2014, 426, 3917–3928. [DOI] [PubMed] [Google Scholar]
- 35.McGlynn P Helicases that underpin replication of protein-bound DNA in escherichia coli. Biochemical Society transactions 2011, 39, 606–610. [DOI] [PubMed] [Google Scholar]
- 36.Syeda AH; Atkinson J; Lloyd RG; McGlynn P The balance between recombination enzymes and accessory replicative helicases in facilitating genome duplication. Genes 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson J; Gupta MK; McGlynn P Interaction of rep and dnab on DNA. Nucleic acids research 2011, 39, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson J; Gupta MK; Rudolph CJ; Bell H; Lloyd RG; McGlynn P Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic acids research 2011, 39, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy CP; Atkinson J; Gupta MK; Mahdi AA; Gwynn EJ; Rudolph CJ; Moon PB; van Knippenberg IC; Cadman CJ; Dillingham MS, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Molecular cell 2009, 36, 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochman ML Roles of DNA helicases in the maintenance of genome integrity. Molecular & cellular oncology 2014, 1, e963429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochman ML; Sabouri N; Zakian VA Unwinding the functions of the pif1 family helicases. DNA repair 2010, 9, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taneja P; Gu J; Peng R; Carrick R; Uchiumi F; Ott RD; Gustafson E; Podust VN; Fanning E A dominant-negative mutant of human DNA helicase b blocks the onset of chromosomal DNA replication. The Journal of biological chemistry 2002, 277, 40853–40861. [DOI] [PubMed] [Google Scholar]
- 43.Rupp WD; Howard-Flanders P Discontinuities in the DNA synthesized in an excision-defective strain of escherichia coli following ultraviolet irradiation. Journal of molecular biology 1968, 31, 291–304. [DOI] [PubMed] [Google Scholar]
- 44.Heller RC; Marians KJ Replication fork reactivation downstream of a blocked nascent leading strand. Nature 2006, 439, 557–562. [DOI] [PubMed] [Google Scholar]
- 45.Lehmann AR; Fuchs RP Gaps and forks in DNA replication: Rediscovering old models. DNA repair 2006, 5, 1495–1498. [DOI] [PubMed] [Google Scholar]
- 46.Mouron S; Rodriguez-Acebes S; Martinez-Jimenez MI; Garcia-Gomez S; Chocron S; Blanco L; Mendez J Repriming of DNA synthesis at stalled replication forks by human primpol. Nature structural & molecular biology 2013, 20, 1383–1389. [DOI] [PubMed] [Google Scholar]
- 47.Yeeles JT; Marians KJ Dynamics of leading-strand lesion skipping by the replisome. Molecular cell 2013, 52, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J; Liu S; Bellani MA; Thazhathveetil AK; Ling C; de Winter JP; Wang Y; Wang W; Seidman MM The DNA translocase fancm/mhf promotes replication traverse of DNA interstrand crosslinks. Molecular cell 2013, 52, 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimoto Y; Puddu F; Costanzo V Rad51- and mre11-dependent reassembly of uncoupled cmg helicase complex at collapsed replication forks. Nature structural & molecular biology 2011, 19, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petojevic T; Pesavento JJ; Costa A; Liang J; Wang Z; Berger JM; Botchan MR Cdc45 (cell division cycle protein 45) guards the gate of the eukaryote replisome helicase stabilizing leading strand engagement. Proceedings of the National Academy of Sciences of the United States of America 2015, 112, E249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastia D; Zzaman S; Krings G; Saxena M; Peng X; Greenberg MM Replication termination mechanism as revealed by tus-mediated polar arrest of a sliding helicase. Proceedings of the National Academy of Sciences of the United States of America 2008, 105, 12831–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan DL; O’Donnell M Dnab drives DNA branch migration and dislodges proteins while encircling two DNA strands. Molecular cell 2002, 10, 647–657. [DOI] [PubMed] [Google Scholar]
- 53.Jeong YJ; Rajagopal V; Patel SS Switching from single-stranded to double-stranded DNA limits the unwinding processivity of ring-shaped t7 DNA helicase. Nucleic acids research 2013, 41, 4219–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling C; Huang J; Yan Z; Li Y; Ohzeki M; Ishiai M; Xu D; Takata M; Seidman M; Wang W Bloom syndrome complex promotes fancm recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell discovery 2016, 2, 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohleder F; Huang J; Xue Y; Kuper J; Round A; Seidman M; Wang W; Kisker C Fancm interacts with pcna to promote replication traverse of DNA interstrand crosslinks. Nucleic acids research 2016, 44, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciccia A; Nimonkar AV; Hu Y; Hajdu I; Achar YJ; Izhar L; Petit SA; Adamson B; Yoon JC; Kowalczykowski SC, et al. Polyubiquitinated pcna recruits the zranb3 translocase to maintain genomic integrity after replication stress. Molecular cell 2012, 47, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Couch FB; Bansbach CE; Driscoll R; Luzwick JW; Glick GG; Betous R; Carroll CM; Jung SY; Qin J; Cimprich KA, et al. Atr phosphorylates smarcal1 to prevent replication fork collapse. Genes & development 2013, 27, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kile AC; Chavez DA; Bacal J; Eldirany S; Korzhnev DM; Bezsonova I; Eichman BF; Cimprich KA Hltf’s ancient hiran domain binds 3′ DNA ends to drive replication fork reversal. Molecular cell 2015, 58, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu L; Jin W; Wang LL Aging in rothmund-thomson syndrome and related recql4 genetic disorders. Ageing research reviews 2017, 33, 30–35. [DOI] [PubMed] [Google Scholar]
- 60.Im JS; Park SY; Cho WH; Bae SH; Hurwitz J; Lee JK Recql4 is required for the association of mcm10 and ctf4 with replication origins in human cells. Cell cycle (Georgetown, Tex.) 2015, 14, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X; Rochette PJ; Feyissa EA; Su TV; Liu Y Mcm10 mediates recq4 association with mcm2-7 helicase complex during DNA replication. The EMBO journal 2009, 28, 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kliszczak M; Sedlackova H; Pitchai GP; Streicher WW; Krejci L; Hickson ID Interaction of recq4 and mcm10 is important for efficient DNA replication origin firing in human cells. Oncotarget 2015, 6, 40464–40479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanuki Y; Kubota Y; Kanemaki MT; Takahashi TS; Mimura S; Takisawa H Recq4 promotes the conversion of the pre-initiation complex at a site-specific origin for DNA unwinding in xenopus egg extracts. Cell cycle (Georgetown, Tex.) 2015, 14, 1010–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerhardt J; Guler GD; Fanning E Human DNA helicase b interacts with the replication initiation protein cdc45 and facilitates cdc45 binding onto chromatin. Experimental cell research 2015, 334, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez-Millan P; Lazaro M; Cansiz-Arda S; Gerhold JM; Rajala N; Schmitz CA; Silva-Espina C; Gil D; Bernado P; Valle M, et al. The hexameric structure of the human mitochondrial replicative helicase twinkle. Nucleic acids research 2015, 43, 4284–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziebarth TD; Gonzalez-Soltero R; Makowska-Grzyska MM; Nunez-Ramirez R; Carazo JM; Kaguni LS Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. The Journal of biological chemistry 2010, 285, 14639–14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bharti SK; Sommers JA; Zhou J; Kaplan DL; Spelbrink JN; Mergny JL; Brosh RM Jr. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form g-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative twinkle helicase. The Journal of biological chemistry 2014, 289, 29975–29993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong DW; Pereira F; Barrett SP; Kolesar JE; Cao K; Damas J; Yatsunyk LA; Johnson FB; Kaufman BA Association of g-quadruplex forming sequences with human mtdna deletion breakpoints. BMC genomics 2014, 15, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel SS; Picha KM Structure and function of hexameric helicases. Annual review of biochemistry 2000, 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 70.Long DT; Joukov V; Budzowska M; Walter JC Brca1 promotes unloading of the cmg helicase from a stalled DNA replication fork. Molecular cell 2014, 56, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fullbright G; Rycenga HB; Gruber JD; Long DT P97 promotes a conserved mechanism of helicase unloading during DNA cross-link repair. Molecular and cellular biology 2016, 36, 2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonneville R; Moreno SP; Knebel A; Johnson C; Hastie CJ; Gartner A; Gambus A; Labib K Cul-2lrr-1 and ubxn-3 drive replisome disassembly during DNA replication termination and mitosis. Nature cell biology 2017, 19, 468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]


