Abstract
Poultry has long been cited as a reservoir for Campylobacter spp., and litter has been implicated as a vehicle in their transmission. Chicks were raised on litter removed from a broiler house positive for Campylobacter jejuni. Litter was removed from the house on days 0, 3, and 9 after birds were removed for slaughter. Chicks were raised on these three litters under controlled conditions in flocks of 25. None of these birds yielded C. jejuni in their cecal droppings through 7 weeks. Two successive flocks from the same Campylobacter-positive broiler house were monitored for Campylobacter colonization. Campylobacter jejuni prevalence rates were determined for each flock. Randomly amplified polymorphic DNA (RAPD)-PCR and 23S rRNA-PCR typing methods were used to group isolates. A high prevalence (60%) of C. jejuni in flock 1 coincided with the presence of an RAPD profile not appearing in flock 2, which had a lower rate of prevalence (28%). A 23S rRNA-PCR typing method was used to determine if strains with different RAPD profiles and different prevalence rates contained different 23S sequences. RAPD profiles detected with higher prevalence rates contained a spacer in the 23S rRNA region 100% of the time, while RAPD profiles found with lower prevalence rates contained an intervening sequence less than 2% of the time. Data suggest varying colonizing potentials of different RAPD profiles and a source other than previously used litter as a means of transmission of C. jejuni. These molecular typing methods demonstrate their usefulness, when used together, in this epidemiologic investigation.
The mode of transmission of Campylobacter jejuni into broiler flocks has been an issue for investigation in recent years. Although the literature supports litter as a possible means of transmission of C. jejuni to susceptible chicks under controlled conditions (9), the role of litter as a medium for the maintenance of the organism and its subsequent transmission to later flocks has not been fully evaluated. Hatcheries have been implicated in the introduction of Salmonella spp. to broiler flocks (2). However, because of the suggested viable but nonculturable characteristics of C. jejuni (4, 13), such a connection between breeder flocks and C. jejuni colonization of broiler flocks has proven elusive to date. Furthermore, this type of vertical transmission is doubted by many researchers (1, 16). Recent advances in molecular microbiology have resulted in genetic typing methods that lend themselves to epidemiological studies. The ability to trace organisms from environmental sources to broiler flocks by using genetic information may prove to be a powerful investigative tool in determining the mode of transmission of a given organism. Randomly amplified polymorphic DNA (RAPD)-PCR and 23S rRNA-PCR were used to establish genetic profiles of C. jejuni isolates cultured during this study. Genetic typing of recovered C. jejuni isolates was used to trace strains of the organism through successive flocks to pursue the epidemiologic objectives of this study. One objective of the investigation was to determine if previously used litter in a poultry house acts as a reservoir for C. jejuni under experimental conditions. The study also investigated the role of used litter in the transmission of C. jejuni to successive flocks in the same poultry house.
MATERIALS AND METHODS
Litter study.
A commercial broiler flock found to be positive for C. jejuni (flock 1) and the successive flock placed in the same house (flock 2) were used in this study. On days 0, 3, and 9 after the first flock was taken to slaughter, used litter was removed and transferred to PDRC (Poultry Disease Research Center, University of Georgia, Athens, Ga.), a controlled study site. Newly hatched chicks obtained from the area commercial hatchery used to supply flocks 1 and 2 were placed on the used litter at the site. The controlled house contained no physical barriers between individual pens except for wire. All pens used for the study were separated by at least one empty pen. The pens, approximately 5 by 10 ft, were disinfected prior to the introduction of chicks, and 2 to 3 in. of the collected used litter was placed in each pen.
The study began on the day flock 1 was taken to slaughter, designated day 0 of the experiment. Twenty-five chicks were placed in a pen on day 0 litter. In a separate pen, 25 chicks were placed on fresh wood shavings as a control group. An additional 25 chicks were sacrificed for evidence of C. jejuni in their ceca. On day 3, a second load of litter was transferred from the poultry house to the controlled site and 25 more hatchlings were placed on the used litter. On day 9, the final load of litter was transferred to the controlled site and 25 hatchlings were again obtained and placed on this litter. Each group was raised for 49 days. Biosecurity measures, such as wearing of sterilized plastic boot covers and smocks, were used when pens were entered. No antibiotics were administered to the birds with the exception of feed containing bacitracin, which is widely used throughout the industry as a growth promoter. Fresh, chlorinated tap water was supplied to the pens daily, and all birds in the study were fed in a manner consistent with common industry standards.
Sample collection.
Cecal droppings were sampled on a weekly basis during the rearing of the two successive commercial broiler flocks. Grow-out time for each flock was 49 days. The first samples were taken from each flock during the 3rd week of rearing, and the last samples were taken on the 7th week. This sampling protocol was the same for both farm flocks and for the controlled flock located at PDRC. Freshly deposited cecal droppings were collected with a sterile swab, placed in a 17- by 100-mm snap-cap culture tube containing 9 ml of 0.1% sterile buffered peptone water, and placed on ice for transport to the laboratory. Sample collection was random but covered the entire length and breadth of the broiler house. Forty samples were collected on each sampling day for the farm broiler house flocks (flocks 1 and 2). Samples were pooled, two per pool, for a total of 20 samples for laboratory analysis. Ten samples were collected from the controlled flocks on each sampling day, and samples were not pooled.
Laboratory analysis.
Cecal droppings were placed in 1-quart Ziploc bags containing 100 ml of Hunt’s enrichment broth (5) with antibiotics and supplements, with recovery and culturing of the organism according to the U.S. Department of Agriculture method (12).
Isolates confirmed as C. jejuni were stored at −70°C on Microbank beads (ProLab Diagnostics, Austin, Tex.). Single colonies were picked from C. jejuni growth plates and placed on beads per the manufacturer’s instructions. Isolates were recultured on brucella-FBP agar for genetic typing procedures.
Genetic typing.
DNA template was made by the whole-cell technique for gram-negative organisms proposed by Woods et al. (18). A hot air thermocycler (Rapidcycler; Idaho Technology, Idaho Falls, Idaho) was used to amplify the DNA.
RAPD-PCR.
RAPD-PCR typing was performed with the OPA 11 primer, 5′-CAATCGCCGT-3′, with 4 mM MgCl2 (5). Cycling parameters for RAPD-PCR consisted of three cycles. The first cycle was comprised of two cycles with a denaturing step at 92°C for 30 s, an annealing step at 36°C for 7 s, and an extension step at 27°C for 70 s, with a slope of 1.0. This cycle had two repetitions. The second cycle shared the same temperatures as the first cycle, but the denaturing step had a duration of 1 s, the annealing step had a duration of 7 s, and the extension step had a duration of 60 s. The second cycle used a slope of 6.0 and had 38 repetitions. A holding cycle of 72°C for 4 min followed the first two cycles and ended the amplification process.
23S rRNA-PCR.
23S rRNA-PCR typing was performed with 2 mM MgCl2, forward primer 5′-TCGGCGGAAAATATAACGGGGCTA-3′, and reverse primer 5′-CTCAACTTAATTATCGCTACTCAT-3′, according to the method of Trust et al. (15). Amplification of 23S rRNA consisted of two cycles. The first cycle was a holding step at 96°C for 5 min, with no repetitions. The second cycle was a denaturation step at 94°C for 10 s, an annealing step at 50°C for 10 s, and an elongation step at 72°C for 35 s, with a slope of 2.0 and 30 repetitions.
After amplification, reaction tube contents were placed in wells cut in 1.5% agarose gels and electrophoresis was conducted at 80 V for 60 min. After electrophoresis, gels were stained with ethidium bromide (Sigma, St. Louis, Mo.). A 100-bp ladder standard (Boehringer Mannheim, Indianapolis, Ind.) was used to measure DNA banding patterns. DNA banding profiles were viewed with a UV light source and photographed.
RESULTS
Prevalence.
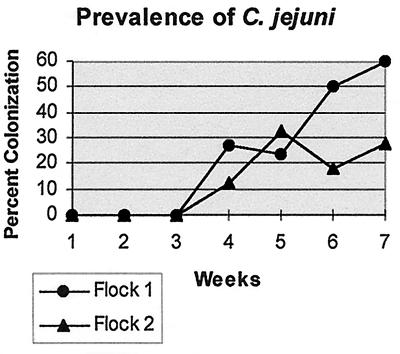
Prevalence rates for C. jejuni colonization were calculated as percentages of positive cecal droppings. None of the groups in the controlled flock reared on previously used litter yielded C. jejuni in their cecal droppings, including the control group and cecal samples taken from day-old hatchlings. Neither flock 1 nor flock 2 yielded Campylobacter isolates prior to the 4th week of sampling. Flock 1 yielded the higher prevalence rate for C. jejuni throughout the study except during the 5th week of sampling, when flock 2 experienced an increase in prevalence. At the time of harvest, flock 1 yielded a 60% prevalence rate for C. jejuni colonization while flock 2 yielded only 28% prevalence at harvest time, as shown in Fig. 1. All quality assurance parameters during isolation and identification were satisfied for data accuracy and consistency, and percentages derived from sampling are believed to be reflective of actual C. jejuni colonization profiles for each flock sampled.
FIG. 1.
Prevalence of C. jejuni recovered from cecal droppings in flocks 1 (n = 67) and 2 (n = 28).
RAPD-PCR typing.
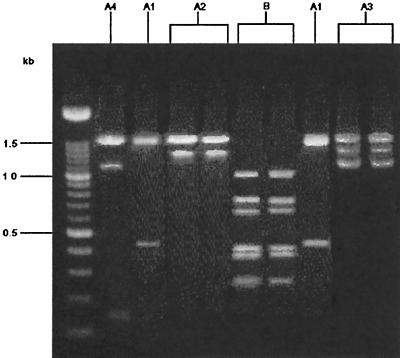
Campylobacter isolates recovered from flocks 1 and 2 were grouped by profiles obtained by RAPD-PCR. Groupings were made by observation of banding patterns throughout RAPD-PCR runs. RAPD types with amplicons of 1.5 kb were denoted group A. Subgroups of group A were identified as types having amplicons that appeared in addition to the 1.5-kb band. Group A1 consisted of isolates that had a 1.5-kb band only. Group A2 organisms had a 1.3-kb band that appeared with the 1.5-kb band, and group A3 organisms were Campylobacter sp., with a 1.1-kb band that appeared in addition to the 1.3- and 1.5-kb bands. Group A4 organisms comprised a group that had only a 1.1- and a 1.5-kb band. Group B organisms, on the other hand, did not express a 1.5-kb band but expressed a banding pattern consisting of 0.6-, 0.7-, and 1.0-kb amplicons. Bands less than 0.6 kb were not used for grouping purposes because their presence was inconsistent and not reproducible. The difficulty with these minor bands has been substantiated by others (10). Banding patterns observed were consistent and repeatable among triplicate RAPD-PCR experiments. Bands other than those specified above were observed, but only those amplicons mentioned were used for grouping. Figure 2 illustrates these RAPD profiles.
FIG. 2.
RAPD-PCR profiles of C. jejuni.
A total of 67 isolates from flock 1 were examined, of which 63 (94%) produced an RAPD product. Fifty-one of these products (81%) were identified as subgroups of group A, which occurred throughout the sampling period from the 4th to the 7th week. Of these 51 isolates, 10 isolates (19%) were placed in group A1, 20 (39%) were placed in group A2, and 21 (41%) were placed in group A3. The remaining 12 isolates (23%), which were characterized as group B, were found only during the 7th week of sampling, immediately preceding the harvest of flock 1. However, group B isolates constituted 54% of the isolates recovered that week.
Only 28 isolates were recovered from flock 2. Of these, 27 (96%) yielded RAPD products. All patterns observed were placed in group A. Twenty (74%) of these isolates yielded products characteristic of group A1, while the remaining seven isolates (26%) were identified as group A4. No group B isolates were identified in flock 2. The occurrences of group A isolates, as determined by the RAPD-PCR method, are presented in Table 1.
TABLE 1.
Recovery of RAPD types from flocks 1 and 2 during weeks 4 through 7
| RAPD Type | % Recovery in indicated flock
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wk 4
|
Wk 5
|
Wk 6
|
Wk 7
|
|||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| A1 | 100 | 100 | 42 | 100 | 65 | |||
| A2 | 100 | 66 | 4 | 36 | ||||
| A3 | 33 | 54 | 9 | |||||
| A4 | 35 | |||||||
| B | 54a | |||||||
Represents the only recovery of RAPD group B isolates.
23S rRNA typing.
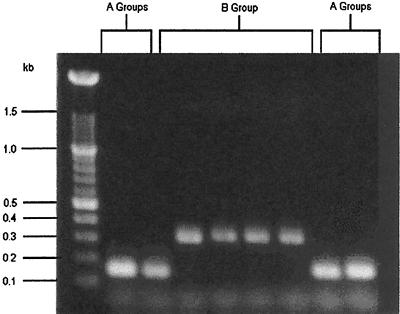
The presence or absence of an intervening sequence (7) in the 23S genome of the isolates was determined to detect differences in parentage between groupings based on RAPD results. Isolates having a 167-bp product do not have a nontranscribed spacer region in the 23S gene. Isolates that yield a 310-bp product have the spacer. Figure 3 illustrates 167- and 310-bp products and their absence or presence within the 23S rRNA region.
FIG. 3.
23S rRNA-PCR of C. jejuni.
Of the 52 isolates recovered from flock 1 that were grouped into RAPD group A (inclusive to all subgroups of group A), all but one yielded 167-bp amplicons. However, of isolates identified as group B, 12 (100%) yielded 310-bp amplicons. Only 1 (1.9%) of the 51 group A isolates contained the 23S spacer. In flock 2, 16 of the 27 group A isolates were examined with 23S rRNA-PCR. None of these isolates contained the spacer.
DISCUSSION
The role of litter.
One objective of this study was to further investigate the role of litter in the transmission of C. jejuni to subsequent flocks reared in the same broiler house environment. Because no chickens raised in a controlled environment on previously used litter were positive for C. jejuni, the evidence in this study does not support an association with previously used litter and the maintenance or transmission of C. jejuni to successive flocks, nor does it support an earlier conclusion by Willis and Murray (17) that broilers raised on used litter become colonized. The limited data presented here suggest that C. jejuni is not transmitted from the hatchery to the controlled flock. This supports evidence reported by Berndtson et al. (3) in an epidemiological study in which parent flocks did not seem to transmit the organism to farm flocks.
Genetic typing.
All isolates obtained from flock 1 and flock 2 were genotyped by the RAPD-PCR and 23S rRNA-PCR techniques for identification of genetic profiles. Band patterns produced by RAPD-PCR were grouped in terms of similar patterns developed on agarose gel electrophoresis. A comparison of genetic information acquired with the use of these procedures was made to determine if isolates belonging to certain RAPD-PCR groups originated from a common source, or a common parental line, by the presence or absence of a spacer within the 23S rRNA genome. Genomic identification methods using the 23S rRNA sequence have potential as epidemiological tools because of the conserved nature of 23S genes. As reported by Ludwig and Schleifer (8), there seems to be greater discrimination potential with 23S rRNA genes than with 16S rRNA genes. Campylobacter has been shown to have three loci of 23S genes in its genome (11). Also, the literature presents no evidence of there being some 23S loci which do not contain the spacer present with loci which do. Our data supports this assumption, since none of the strains produced multiple products in 23S rRNA-PCR.
Campylobacter isolates with RAPD group A profiles appeared throughout flock 1 and flock 2, suggesting a maintenance of RAPD group A organisms through successive flocks. It seems plausible that C. jejuni group A types may be maintained from a common source.
Our study determined that C. jejuni colonization prevalence was much greater at the time of harvest of flock 1 than of flock 2, with a second RAPD profile appearing only in flock 1. Stern et al. (14), during a study of competitive exclusion on cecal colonization of chicks, observed that different isolates of C. jejuni exhibited different potentials for colonization, possibly caused by differences in phenotypic expression between strains. Group B RAPD profiles appeared only in the 7th week of sampling in flock 1 but comprised 54% of all recovered C. jejuni isolates for that sampling period. This suggests that this group of isolates may have a greater ability to colonize than do group A isolates, which were common to both flock 1 and flock 2. The presence of the 23S spacer in the genome of isolates in RAPD group B demonstrates that these isolates are of a different parental line and that they may have been introduced from a source internal or external to the broiler house environment but one not common to the source of group A.
Our study suggests that while the RAPD-PCR method is useful in epidemiologic investigations, it may be too sensitive when used alone to group C. jejuni isolates, because differences found in the RAPD group A data were not confirmed by the 23S rRNA-PCR method. The 23S rRNA method did not differentiate between subgroups A1, A2, A3, and A4. Therefore, it may be that these isolates are of the same parental lineage and that the differences expressed in the RAPD patterns may be evidence of genetic recombination that occurred in the organism through the grow-out periods of flock 1 and flock 2. It seems evident that when RAPD-PCR methods are used, they should be complemented by a PCR method, such as the 23S rRNA-PCR method, that examines a more conserved region of the genome. Our RAPD-PCR (group B) data and its 100% correlation to the presence of spacer regions demonstrate that the 23S rRNA-PCR method is complementary to the RAPD-PCR method.
Our data suggest that the common source of Campylobacter isolates was not previously used litter. C. jejuni is present in a variety of types of samples in a broiler house environment, such as water, litter, and dust, etc. Application of the appropriate PCR techniques would appear to be useful not only in the investigation of Campylobacter in other areas of the broiler house but also in other applications in the poultry industry.
ACKNOWLEDGMENTS
This work was supported in part by the U.S. Poultry and Egg Association and the U.S. Department of Agriculture.
REFERENCES
- 1.Annan-Prah S, Jane M. The mode of spread of Campylobacter jejuni/coli to broiler flocks. J Vet Med. 1988;35:11–18. doi: 10.1111/j.1439-0450.1988.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 2.Barnhart H M, Dreesen D W, Bastien R, Pancorbo O C. Prevalence of Salmonella enteritidis and other serovars in ovaries of layer hens at time of slaughter. J Food Prot. 1991;54:448–491. doi: 10.4315/0362-028X-54.7.488. [DOI] [PubMed] [Google Scholar]
- 3.Berndtson E, Emanuelson U, Engvall A, Danielsson-Tham M L. A 1-year epidemiological study of campylobacters in 18 Swedish farms. Prev Vet Med. 1996;26:167–185. [Google Scholar]
- 4.Beumer R R, de Vries J, Rombouts F M. Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol. 1992;15:153–163. doi: 10.1016/0168-1605(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez J, Fayos A, Ferrus M A, Owen R J. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and C. coli isolated from human faeces, seawater and poultry products. Res Microbiol. 1995;146:685–696. doi: 10.1016/0923-2508(96)81065-5. [DOI] [PubMed] [Google Scholar]
- 6.Hunt J M. FDA bacteriological analytical Manual. 7th ed. Arlington, Va: Association of Official Analytical Chemists; 1992. Campylobacter; pp. 77–94. [Google Scholar]
- 7.Konkel M E, Marconi R T, Mead D J, Cleplak W. Identification and characterization of an intervening sequence within the 23S ribosomal RNA gene of Campylobacter jejuni. Mol Microbiol. 1994;14:235–241. doi: 10.1111/j.1365-2958.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 9.Montrose M S, Shane S M, Harrington K S. Role of litter in the transmission of Campylobacter jejuni. Avian Dis. 1984;29:392–399. [PubMed] [Google Scholar]
- 10.Myers L E, Silva S V P S, Procunier J D, Little P B. Genomic fingerprinting of “Haemophilus somnus” isolates by using a random-amplified polymorphic DNA assay. J Clin Microbiol. 1993;31:512–517. doi: 10.1128/jcm.31.3.512-517.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newnham E, Chang N, Taylor D E. Expanded genomic map of Campylobacter jejuni UA580 and localization of 23S ribosomal rRNA genes by I-CeuI restriction endonuclease digestion. FEMS Microbiol Lett. 1996;142:223–239. doi: 10.1111/j.1574-6968.1996.tb08434.x. [DOI] [PubMed] [Google Scholar]
- 12.Ransom G M, Rose B E. USDA/FSIS microbiology laboratory guidebook. 3rd ed. Washington, D.C: U.S. Department of Agriculture; 1995. Isolation, identification, and enumeration of Campylobacter jejuni/coli from meat and poultry products; pp. 6-1–6-10. [Google Scholar]
- 13.Rollins D M, Colwell R R. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern N J, Bailey J S, Blankenship L C, Cox N A, McHan F. Colonization characteristics of Campylobacter jejuni in chick ceca. Avian Dis. 1988;32:330–334. [PubMed] [Google Scholar]
- 15.Trust T J, Logan S M, Gustafson C E, Romaniuk P J, Kim N W, Chan V L, Ragan M A, Guerry P, Gutell R R. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J Bacteriol. 1994;176:4597–4609. doi: 10.1128/jb.176.15.4597-4609.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Giessen A, Mazurier S, Jacobs-Reitsma W, Jansen W, Berkers P, Ritmeester W, Wernars K. Study on the epidemiology and control of Campylobacter jejuni in poultry broiler flocks. Appl Environ Microbiol. 1992;58:1913–1917. doi: 10.1128/aem.58.6.1913-1917.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis, W. L., and C. L. Murray. 1996. Campylobacter jejuni colonization assessment of broilers on different litter. Poult. Sci. 1(Suppl.):24.
- 18.Woods C R, Versalovic J, Koeuth T, Lupski J R. Whole-cell repetitive element sequence-based polymerase chain reaction allows rapid assessment of clonal relationships of bacterial isolates. J Clin Microbiol. 1993;31:1927–1931. doi: 10.1128/jcm.31.7.1927-1931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]