Abstract
We report DNA-scaffolded synergistic catalysis, a concept that combines the diverse reaction scope of synergistic catalysis with the ability of DNA to precisely pre-organize abiotic groups and undergo stimuli-triggered conformational changes. As an initial demonstration of this concept, we focus on Cu-TEMPO-catalyzed aerobic alcohol oxidation, using DNA as a scaffold to hold a copper co-catalyst and an organic radical co-catalyst (TEMPO) in proximity. The DNA-scaffolded catalyst maintained high turnover number upon dilution and exhibited 190-fold improvement in catalyst turnover number relative to the unscaffolded co-catalysts. By incorporating the co-catalysts into a DNA hairpin-containing scaffold, we demonstrate that the rate of the synergistic catalytic reaction can be controlled through a reversible DNA conformational change that alters the distance between the co-catalysts. This work demonstrates the compatibility of synergistic catalytic reactions with DNA scaffolding, opening future avenues in reaction discovery, sensing, responsive materials, and chemical biology.
Graphical Abstract
INTRODUCTION
Creation of catalysts that mimic enzymes through the pre-organization of multiple reactive groups is a long-standing goal.1 In natural enzymes, protein scaffolds hold multiple functional groups in proximity, engendering massive rate accelerations that are not observed when the same functional groups are unscaffolded.2 While matching the sophistication of enzymes has been elusive in synthetic systems, supramolecular enzyme mimics offer a broader palette of functional groups, since they are not limited to the natural amino acids and cofactors. Impressive catalytic properties have been demonstrated in a variety of enzyme-mimicking supramolecular scaffolds.3–10
An exciting area of recent progress in supramolecular catalysis is the pre-organization of synergistic catalytic reactions. Synergistic catalysis combines two distinct, abiotic co-catalysts that work in concert to carry out a reaction that would not be possible using either co-catalyst alone.11 Examples include a variety of synergistic combinations of transition metal catalysts, photocatalysts, organocatalysts, radical catalysts, and Lewis acid catalysts.12–20 In scaffolded synergistic catalysis, the proximity afforded by precise positioning of the co-catalysts accelerates the reaction compared to unscaffolded reactions, in which the large mean distance between the co-catalysts limits synergistic interactions, often necessitating high loadings.21 Successful demonstrations of scaffolded synergistic catalysis have utilized peptoids,22 peptides,23,24 foldamers,25 soluble proteins,26,27 metal–organic frameworks,28,29 covalent–organic frameworks,30 and polymers.21 However, it remains challenging to fine-tune the spacing of each co-catalyst. Furthermore, the previously demonstrated scaffolds for synergistic catalytic reactions are difficult to engineer for stimuli-triggered conformational changes to alter the rate of the reaction. This important property of enzyme catalysis, allostery, is of fundamental interest for enzyme-mimicking systems and for the development of sensors and responsive materials
We envisioned that DNA-scaffolded synergistic catalysis would offer multiple advantages: 1) precise positioning of the co-catalysts due to the programmability of DNA and the ease of site-specific modification, 2) convergent and facile synthesis of many catalyst variants through one-step bioconjugation followed by self-assembly, and 3) allosteric switching using DNA strand displacements (Figure 1). Indeed, the outstanding advantages of DNA have previously been demonstrated in templating synthetic reactions and influencing the rates of mono-catalytic processes. DNA has long been used as a template to accelerate bond formation in a variety of reactions31–40 and to bring substrates proximal to catalysts.41–46 DNA has also been used as a scaffold to activate mono-catalysts47–49 and to influence mono-catalytic processes for asymmetric synthesis.50–52 Conformational switching of DNA has been used to regulate catalytic activity of mono-catalytic gold bound to DNA,53 catalytic activity of a PNA-peptide hybrid,54 to activate heme-catalyzed peroxidation,55,56 and to alter the distance between two enzymes in a cascade reaction.57,58 However, to the best of our knowledge, DNA-scaffolded synergistic catalysis has not been demonstrated, and more broadly, no scaffolded synergistic catalysis has been performed that incorporates the advantages of DNA (convergent synthesis, precise positioning, and switching in response to stimuli) and it is unclear whether this will work with DNA: for example, there may be problematic DNA-catalyst interactions or DNA-DNA interactions that confound the desired control over co-catalyst-co-catalyst interactions. Here, we report DNA-scaffolded Cu/TEMPO oxidation of alcohols. We show that synergistic catalysis can be optimized through precise orientation of the co-catalysts on DNA scaffolds, and activity can be controlled through conformational switching of DNA.
Figure 1.
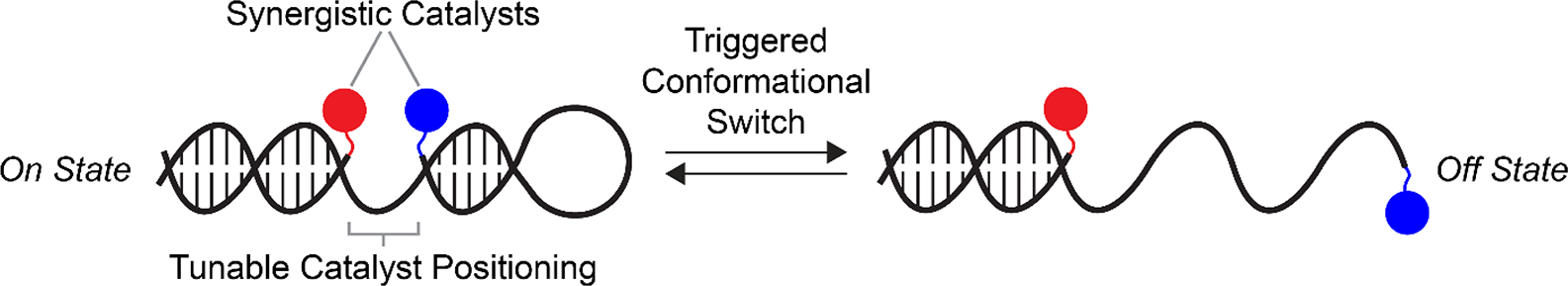
Overview of DNA-scaffolded synergistic catalysis. Abiotic synergistic co-catalysts (red and blue) are tethered site-specifically to DNA structures with tunable co-catalyst spacings. Inter-co-catalyst spacing—and as a result, catalytic reaction rate—can be altered through DNA conformational changes triggered by chemical stimuli.
We chose Cu/TEMPO alcohol oxidation as a proof-of-principle reaction to demonstrate DNA scaffold-dependent rate enhancement because of its remarkably mild reaction conditions59–62 and the synergistic cooperation of the two cocatalysts.63,64 Additionally, Cu/TEMPO oxidations have been accelerated through arranging the two co-catalysts on several different scaffolds, including peptoids,22 silica particles,65,66 and simple synthetic tethers67,68 In contrast to these prior examples, we envisioned that the unique advantages of a DNA scaffold could allow for similar enhancements in reactivity while also allowing highly tunable co-catalyst positioning and stimuli-responsiveness. Our goal in this communication is to report a fundamental study of the effects of DNA-scaffolding on synergistic catalysis, not to report a synthetic method. For further reading on the synthetic applications of this synergistic system see the reviews by Stahl and Swarts.69,70
RESULTS AND DISCUSSION
We prepared a simple DNA duplex with the two co-catalysts (Cu and TEMPO) attached to the end of the helix via sufficiently long tethers to span the 2 nm diameter of B-form DNA (Figure 2A). First, we bioconjugated a carboxylate-bearing stable nitroxyl radical (4-carboxy-TEMPO) and a carboxylate-bearing bipyridine ligand (4,4′-dicarboxy-2,2′-bipyridine) to two separate amine-bearing DNA oligonucleotides with complementary 26-nucleotide sequences. Attachment of the nitroxyl radical using DMTMM as the coupling reagent led to significant multiple addition products, so attachment of 4-carboxy-TEMPO was performed using a milder EDC/HOAt/DIPEA protocol71 that gave higher yields of the desired product with lower yields of over-coupled impurities (Figure S10). The resulting small molecule-DNA conjugates were HPLC purified, and their identities were confirmed using electrospray ionization coupled with high resolution and mass accuracy mass spectrometry along with negative electron transfer dissociation tandem mass spectrometry (Figures S1–12).72 Following intact mass analysis, tandem MS confirmed that the modifications were localized to the expected oligonucleotide termini. Further investigation of over-coupled impurities showed that attachment of co-catalytic moieties to weakly nucleophilic nucleobases was not site-specific. After purification and characterization, the complementary DNA-co-catalyst conjugates were assembled into a duplex with thermal annealing in the presence of NaCl (Figure S15).73 Copper was added immediately prior to catalytic reactions to metalate the bipyridine ligand.
Figure 2.
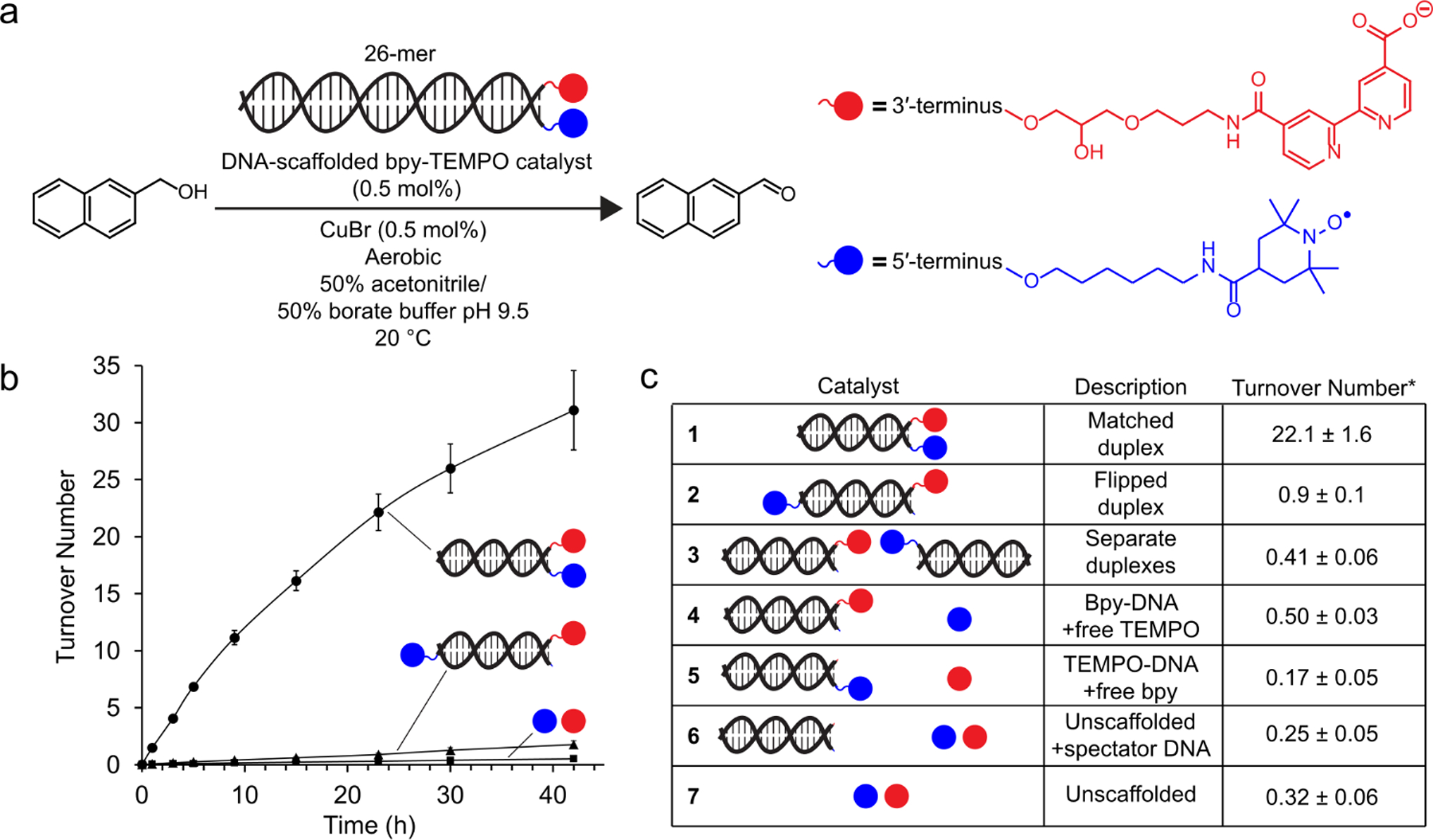
DNA duplex-scaffolded Cu-TEMPO alcohol oxidation. (a) 2-napthalenemethanol (10 mM) was oxidized in the presence of 0.5 mol % of a DNA scaffold functionalized with bpy and TEMPO (50 μM), with 0.5 mol % copper (I) (50 μM). (b) Alcohol oxidation was measured for the DNA-scaffolded Cu-TEMPO catalyst, a DNA scaffold with the two co-catalysts tethered to opposite ends, and the unscaffolded catalytic system. (c) Catalyst turnover number for 2-napthalenemethanol oxidation at 23 h for negative control catalysts. Data are presented as the mean ± standard deviation of 3 independent experiments.
Catalytic activity of the scaffolded DNA duplex (0.5 mol %) was evaluated by monitoring oxidation of 2-napthalenemethanol (Figure 2A–C). The activity of the DNA-scaffolded co-catalysts (condition 1) was dramatically enhanced relative to that of the unscaffolded reaction (condition 7); at 30 h, the scaffolded reaction showed a 70-fold enhancement in catalyst turnover number (TON). Tethering the co-catalysts to opposite ends of the DNA duplex (~9 nm) resulted in a similar TON to the unscaffolded reaction, indicating that proximity of the co-catalysts was essential for enhanced activity (condition 2). The slight difference in activity between conditions 2 and 7 can be attributed to the presence of a minor impurity presenting multiple bipyridine ligands (Figure S7). To confirm that the DNA remained structurally undamaged and annealed under reaction conditions, we recovered DNA from the reaction solution by alcohol precipitation and analyzed it by agarose gel electrophoresis. The mobility of the DNA was unchanged, and no fragmentation was observed (Figure S16). While the reactions depicted in Figure 2 were performed in a semi-aqueous solvent system, optimized from a panel of water/acetonitrile mixtures (Figure S17), we also observed DNA-scaffold-dependent activity enhancement under fully aqueous conditions, although the TON was significantly reduced (Figure S18).
Control reactions confirmed that scaffold-induced proximity of the co-catalysts was responsible for the observed activity enhancement (Figure 2C). When one co-catalyst was tethered to the DNA duplex and the other was not (conditions 4 and 5), or when the co-catalysts were tethered to separate duplexes (condition 3), the TON was similar to that of the unscaffolded reaction. These results suggest that the activity enhancement observed in condition 1 was not due to an intrinsic increase in the activity of either co-catalyst upon conjugation to DNA. They further suggest that substrate recruitment to the DNA scaffold does not contribute significantly to the activity enhancement observed in 1. The DNA-scaffolded rate acceleration was not specific to 2-napthalenemethanol; it was also observed for six benzyl alcohol derivatives (Figure S19). During the scaffolded catalyst time-course reactions, we observed a gradual decrease in reaction rate (Figure 2C), which we determined was not due to product inhibition (Figure S20). Spiking in additional copper after the reaction rate had plateaued increased catalytic activity, suggesting that a co-catalyst deactivation pathway such as copper aggregation may be operative (Figure S21).74,75
To further confirm that the DNA-scaffolded synergistic co-catalysts operate through an intramolecular reaction mechanism (i.e., Cu and TEMPO interact on the same DNA helix), we compared the TONs of the scaffolded and unscaffolded reactions at different catalyst loadings (Figure 3). The TON of the unscaffolded reaction decreased as the catalysts were diluted, presumably due to diminished frequency of synergistic co-catalyst–co-catalyst interactions. In contrast, the TON of the scaffolded reaction increased, apparently insensitive to the change in absolute catalyst concentration. The difference in reactivity was especially pronounced at lower catalyst loadings: at 0.1 mol % loading, the scaffolded co-catalysts exhibited a 190-fold increase in turnover number relative to the unscaffolded co-catalysts.
Figure 3.
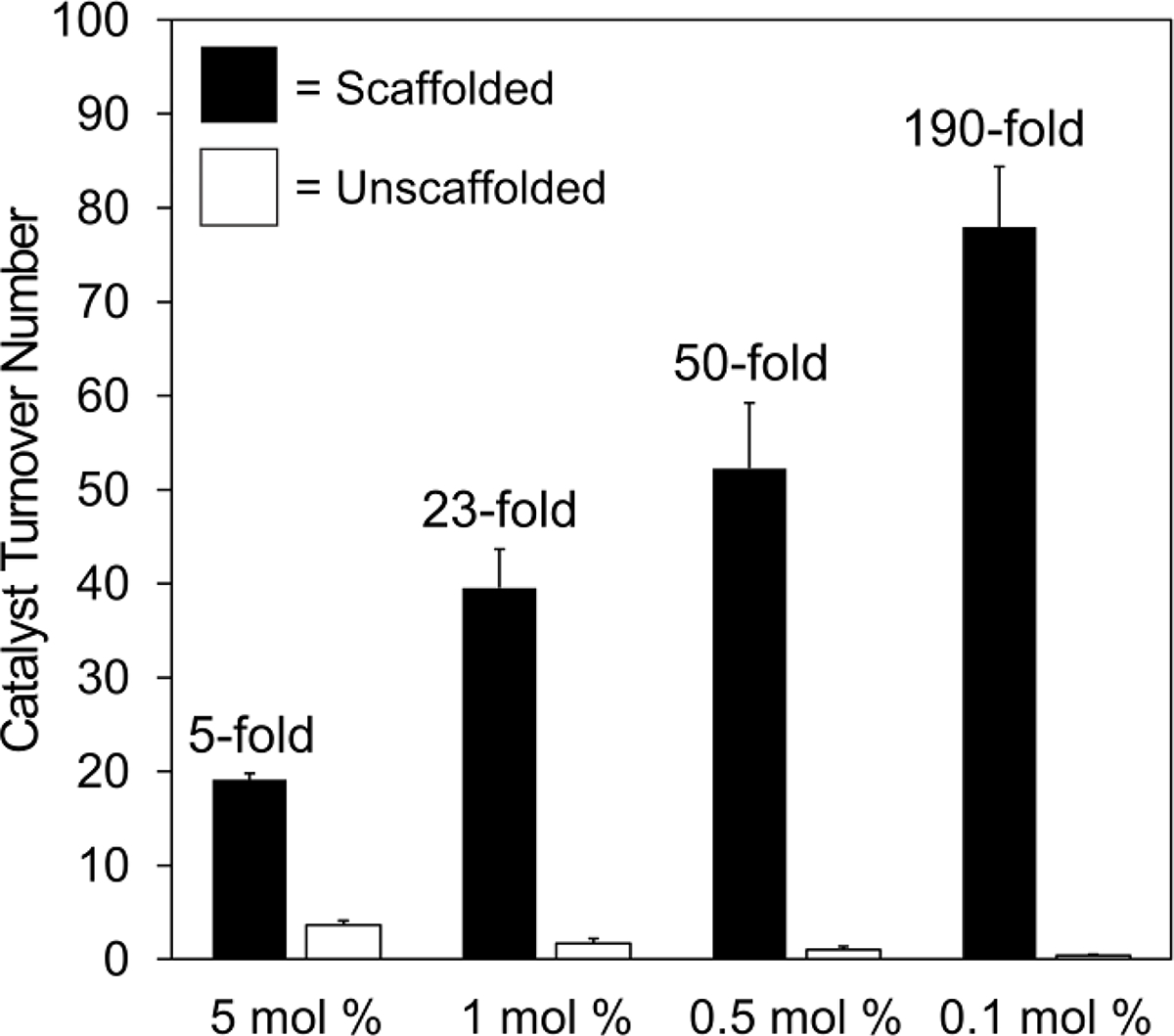
Effect of catalyst loading on the TON of scaffolded and unscaffolded reactions. 2-Napthalenemethanol was oxidized as in Figure 2, except that catalyst loadings were varied. Catalyst loadings indicate the mol % loading for bpy ligand, TEMPO, and copper(I). The fold increase in catalyst TON due to DNA scaffolding is indicated for each catalyst loading. Data are presented as the mean ± standard deviation of 3 independent experiments.
To further investigate the DNA-scaffolded Cu-TEMPO activity enhancement, we estimated the increase in effective concentration experimentally and using a simple theoretical model. We measured the initial rates of alcohol oxidation by a single co-catalyst tethered to DNA in the presence of non-tethered co-catalyst at varying concentrations, reasoning that a sufficiently high concentration of non-tethered co-catalyst could potentially mimic the high local concentration of the DNA-tethered co-catalyst. We determined that 50 μM of the DNA-scaffolded Cu-TEMPO catalyst exhibited a reaction rate equivalent to that expected for 7 mM of the non-tethered dcbpy-Cu complex and 34 mM of the non-tethered TEMPO co-catalyst (Figures S22–23). These results suggest that DNA scaffolding provided an increase in effective concentration of over two orders of magnitude. These experimental results matched well with a simple geometric model, from which we calculated an effective concentration of approximately 13 mM of the Cu-bpy and TEMPO co-catalysts tethered to the DNA helix (Figure S31)).
A key advantage of DNA as a scaffold for synergistic catalysis is the ability to control the relative spacing of the co-catalysts using templating strands (analogous to DNA molecular rulers),76–79 which facilitate modular assembly of 2D and 3D architectures.80–82 Distance-dependent reactivity has been demonstrated in DNA-templated reactions,83,84 and we hypothesized that this distance-dependence could be exploited for control over synergistic catalysis. We explored this advantage for Cu-TEMPO catalysis by annealing DNA-co-catalyst conjugates to a series of unmodified templating strands, with varying single-stranded spacers controlling the inter-co-catalyst distance (Figure 4A). We used polythymidine spacers to prevent the formation of unwanted secondary structures, which have been reported in DNA-templated synthesis.85 The architecture with no gap between the co-catalysts exhibited the fastest rate, and as the length of the intervening spacer region increased, the rate of alcohol oxidation steadily decreased until it matched that of the unscaffolded reaction (Figure 4B). To determine whether unpaired nucleobases in proximity to the co-catalyst attachment sites contribute to the decrease in rate with longer template strands (presumably through Cu-coordination) we performed control experiments in which one co-catalyst was tethered to a DNA duplex near a polyT overhang and the other co-catalyst was added exogenously at high concentration (Figures S24–25). Longer polyT overhangs caused a moderate decrease in reaction rate (Figure S26). The identity of the unpaired nucleobase also mattered, with guanine demonstrating the greatest decrease in reactivity and adenine having a negligible effect (Figure S27). These observations suggest that the DNA architecture used in Figure 4 governs the rate of catalysis through at least two mechanisms: inter-co-catalyst spacing and nucleobase-co-catalyst interactions. We anticipate that more advanced 2D and 3D scaffolding architectures should enable even finer control over the rate of synergistic catalysis.
Figure 4.
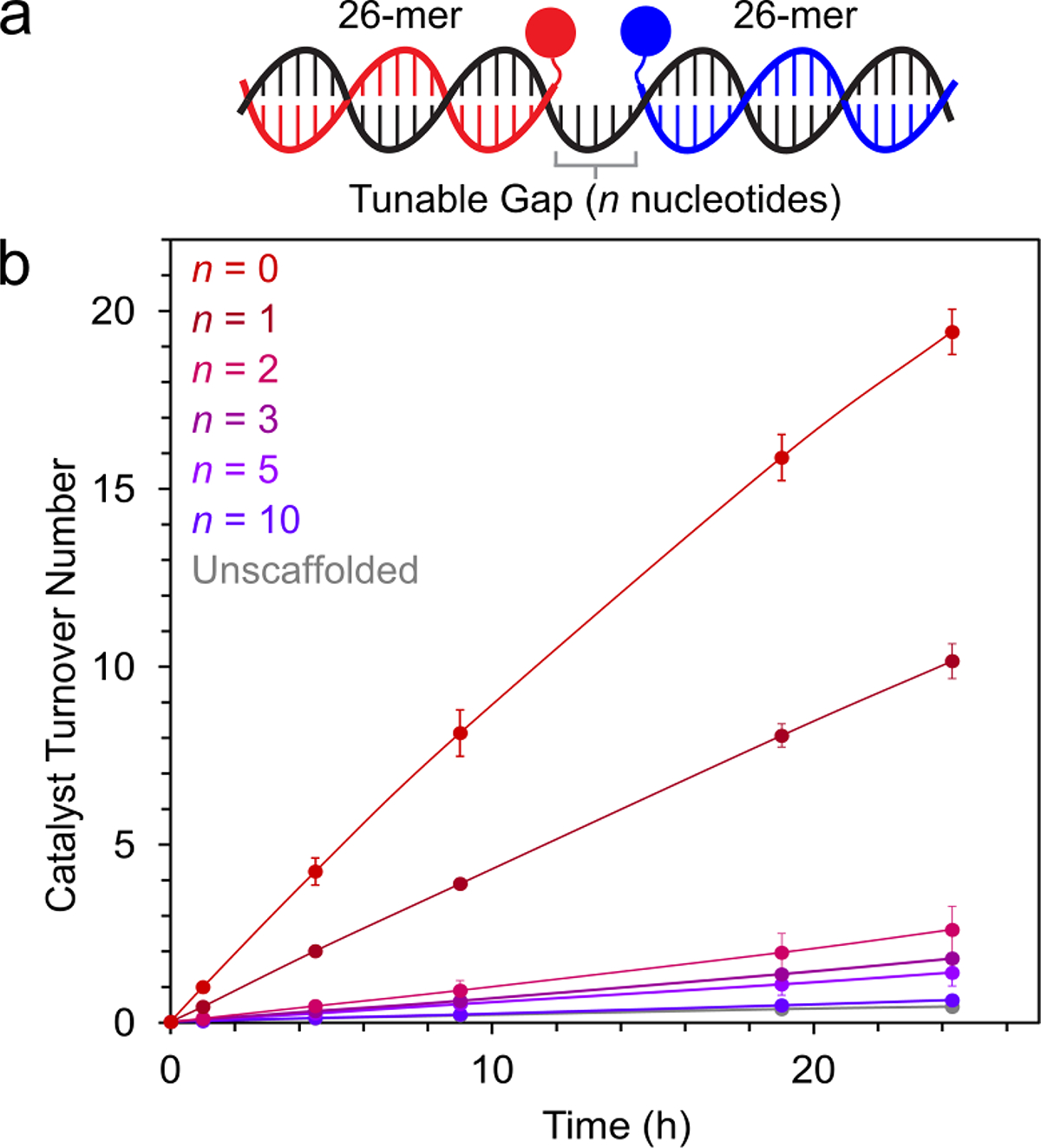
Tuning inter-co-catalyst distance using template strands with single-stranded spacers. (a) Co-catalyst-bearing ssDNA oligomers (red and blue) were annealed to a template ssDNA (black), bringing the co-catalysts into proximity. (b) 2-Napthalenemethanol oxidation by 3-strand DNA-scaffolded Cu-TEMPO catalysts with polythymidine spacers of varying lengths. Data are plotted as the mean ± standard deviation of 3 independent experiments.
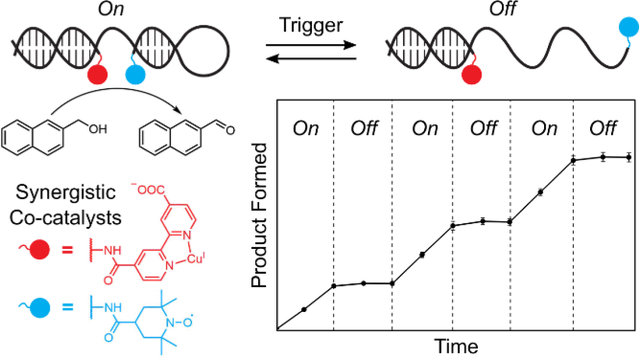
Taking inspiration from dynamic strand interchange reactions,86 the widespread application of FRET-based molecular beacons,87 and the tuning of BRET enzyme-catalyst distance to turn off photocatalysis88 we envisioned that proximity-dependent synergistic catalysis might be incorporated into a stimulus-responsive switch architecture (Figure 5A). By incorporating a stem-loop motif into the sequence of one of the co-catalyst conjugates, we created a dynamic DNA structure to link catalytic activity to sequence recognition. In the presence of a trigger sequence complementary to the stem-loop motif, the structure opens, holding the co-catalysts apart and turning off synergistic catalysis. When a corresponding anti-trigger sequence recognizes an 8-nucleotide toehold region on the trigger strand and displaces the catalytic assembly, the DNA-scaffolded co-catalysts refold, and catalytic activity is restored. Adding the trigger strand to the reaction solution after 1 h completely turned off the catalytic oxidation of 2-napthalenemethanol, and catalytic activity was fully restored upon addition of the anti-trigger (Figure 5B). Catalytic performance was unaffected after 3 cycles of in situ activation/deactivation over 6 h. These results represent the first demonstration of switchable synergistic catalysis through tuning the geometric relationship between co-catalysts. DNA-templated mono-catalytic systems have been used to demonstrate nucleic-acid-dependent generation of fluorescent reporter molecules.42,48,49,53 Similarly the DNA-scaffolded co-catalysts exhibited activity in the oxidation of the fluorogenic probe 6-methoxy-2-napthalenemethanol (Figure S28), demonstrating that switchable catalytic activity can be coupled to a fluorescent output. In the future, we anticipate that incorporating various recognition elements into the stem-loop of this switchable DNA architecture will allow catalysts to be developed that respond to small molecule or protein stimuli.89,90
Figure 5.
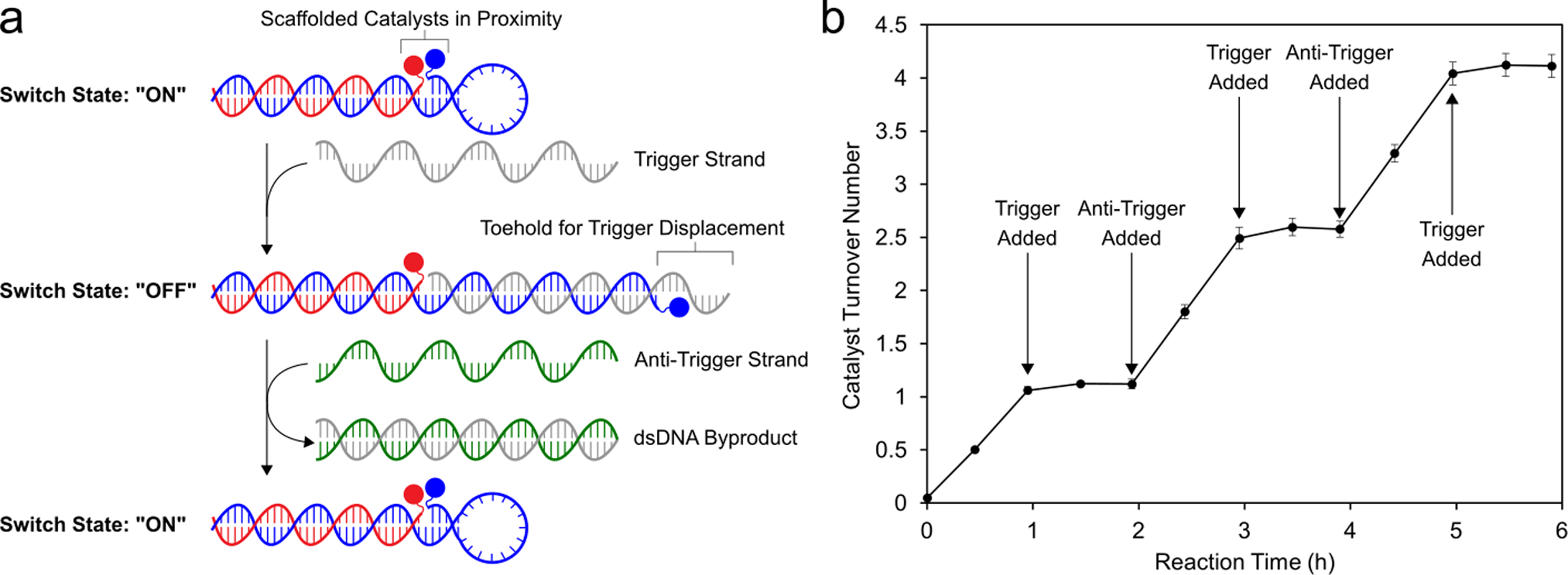
Switchable DNA-scaffolded synergistic catalysis. (a) Design of the switchable DNA architecture. The distance between the two co-catalysts is dramatically increased upon introduction of an ssDNA “trigger” strand. The original catalyst conformation is restored upon addition of an “anti-trigger” strand. (b) Oxidation of 2-napthalenemethanol quantified through multiple cycles of DNA conformational changes, controlled by successive addition of the trigger and anti-trigger strands. Data are plotted as the mean ± standard deviation of 3 independent experiments.
CONCLUSION
We have successfully demonstrated the first DNA-scaffolded acceleration of synergistic catalysis, using Cu/TEMPO oxidation of alcohols as a model reaction. The DNA backbone allowed for systematic exploration of the scaffolded reactivity, and our data suggest that the reactivity enhancement is due to the increased effective concentration afforded by proximity of the scaffolded co-catalysts. The DNA backbone allowed for dynamic activity switching using a strand-displacement approach. While this initial demonstration was not intended to pursue a new synthetic method and the yields and rate are modest at high dilution, with further optimization this scaffolding approach may provide solutions to challenges in synergistic catalysis. DNA-scaffolded synergistic catalysis will be a promising strategy to accelerate other synergistic catalytic reactions (beyond Cu-TEMPO oxidation) in which interactions between the two co-catalysts are rate-limiting.12 This acceleration may allow for low catalyst loadings in reactions that currently require high concentrations of both co-catalysts and may even unlock new reactions that are inaccessible in an unscaffolded format. Furthermore, information-encoding properties of DNA could enable DNA-encoded discovery of synergistic catalysts. Finally, the groundwork laid in this study could be applied to create functional DNA nanomaterials containing synergistic catalytic sites.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Profs. Samuel Gellman, Shannon Stahl, and Tehshik Yoon for helpful discussions. This work was financially supported by the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin–Madison with funding from the Wisconsin Alumni Research Foundation. We thank the Army Research Office for support to J.D.M. (W911NF2110073). We gratefully acknowledge the National Institutes of Health for support to J.J.C. (P41 GM108538 and 5T32HG002760). T.M.P.-C. thanks the National Human Genome Research Institution for a training grant through the Genomic Sciences Training Program (5T32HG002760). E.B.P thanks the UW-Madison Department of Chemistry for support through a Sam C. Slifkin Award. The Bruker AVANCE 500 NMR spectrometer used in the Paul Bender Chemical Instrumentation Center was supported by a generous gift from Paul J. and Margaret M. Bender.
Footnotes
CONFLICT OF INTEREST STATEMENT
J.J.C. is a consultant for Thermo Fisher Scientific. J.D.M. and E.B.P. have submitted a provisional patent application related to this work.
SUPPORTING INFORMATION
Experimental methods, DNA sequences, characterization data for DNA-co-catalyst conjugates, supplemental experiments, and supplemental notes on estimating effective concentration and reaction mechanism.
REFERENCES
- (1).Obexer R; Godina A; Garrabou X; Mittl PRE; Baker D; Griffiths AD; Hilvert D Emergence of a Catalytic Tetrad during Evolution of a Highly Active Artificial Aldolase. Nat Chem 2017, 9 (1), 50–56. [DOI] [PubMed] [Google Scholar]
- (2).Silverman RB The Organic Chemistry of Enzyme-Catalyzed Reactions; Academic press, 2002, 1–36. [Google Scholar]
- (3).Cram DJ The Design of Molecular Hosts, Guests, and Their Complexes (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 1988, 27 (8), 1009–1020. 10.1002/anie.198810093. [DOI] [PubMed] [Google Scholar]
- (4).Breslow R Biomimetic Chemistry and Artificial Enzymes: Catalysis by Design. Acc. Chem. Res. 1995, 28 (3), 146–153. [Google Scholar]
- (5).Matsumoto M; Lee SJ; Waters ML; Gagné MR A Catalyst Selection Protocol That Identifies Biomimetic Motifs from β-Hairpin Libraries. J. Am. Chem. Soc. 2014, 136 (45), 15817–15820. 10.1021/ja503012g. [DOI] [PubMed] [Google Scholar]
- (6).Wiester MJ; Ulmann PA; Mirkin CA Enzyme Mimics Based Upon Supramolecular Coordination Chemistry. Angew. Chem. Int. Ed. 2011, 50 (1), 114–137. 10.1002/anie.201000380. [DOI] [PubMed] [Google Scholar]
- (7).VanEtten RL; Sebastian JF; Clowes GA; Bender ML Acceleration of Phenyl Ester Cleavage by Cycloamyloses. A Model for Enzymic Specificity. J. Am. Chem. Soc. 1967, 89 (13), 3242–3253. [Google Scholar]
- (8).Kang J; Rebek J Acceleration of a Diels-Alder Reaction by a Self-Assembled Molecular Capsule. Nature 1997, 385 (6611), 50–52. 10.1038/385050a0. [DOI] [PubMed] [Google Scholar]
- (9).Hastings CJ; Pluth MD; Bergman RG; Raymond KN Enzymelike Catalysis of the Nazarov Cyclization by Supramolecular Encapsulation. J. Am. Chem. Soc. 2010, 132 (20), 6938–6940. 10.1021/ja102633e. [DOI] [PubMed] [Google Scholar]
- (10).Coric I; List B Asymmetric Spiroacetalization Catalysed by Confined Bronsted Acids. Nature 2012, 483 (7389), 315–319. 10.1038/nature10932. [DOI] [PubMed] [Google Scholar]
- (11).Martínez S; Veth L; Lainer B; Dydio P Challenges and Opportunities in Multicatalysis. ACS Catal. 2021, 11 (7), 3891–3915. 10.1021/acscatal.0c05725. [DOI] [Google Scholar]
- (12).Allen AE; MacMillan DWC Synergistic Catalysis: A Powerful Synthetic Strategy for New Reaction Development. Chem. Sci. 2012, 3 (3), 633–658. 10.1039/C2SC00907B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Nicewicz DA; MacMillan DWC Merging Photoredox Catalysis with Organocatalysis: The Direct Asymmetric Alkylation of Aldehydes. Science 2008, 322 (5898), 77. 10.1126/science.1161976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Du J; Skubi KL; Schultz DM; Yoon TP A Dual-Catalysis Approach to Enantioselective [2 + 2] Photocycloadditions Using Visible Light. Science 2014, 344 (6182), 392. 10.1126/science.1251511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tellis JC; Primer DN; Molander GA Single-Electron Transmetalation in Organoboron Cross-Coupling by Photoredox/Nickel Dual Catalysis. Science 2014, 345 (6195), 433–436. 10.1126/science.1253647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zuo Z; Ahneman DT; Chu L; Terrett JA; Doyle AG; MacMillan DWC Merging Photoredox with Nickel Catalysis: Coupling of α-Carboxyl Sp3-Carbons with Aryl Halides. Science 2014, 345 (6195), 437–440. 10.1126/science.1255525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Pfund B; Steffen DM; Schreier MR; Bertrams M-S; Ye C; Börjesson K; Wenger OS; Kerzig C UV Light Generation and Challenging Photoreactions Enabled by Upconversion in Water. J. Am. Chem. Soc. 2020, 142 (23), 10468–10476. 10.1021/jacs.0c02835. [DOI] [PubMed] [Google Scholar]
- (18).Skubi KL; Blum TR; Yoon TP Dual Catalysis Strategies in Photochemical Synthesis. Chem. Rev. 2016, 116 (17), 10035–10074. 10.1021/acs.chemrev.6b00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Park J; Hong S Cooperative Bimetallic Catalysis in Asymmetric Transformations. Chem. Soc. Rev. 2012, 41 (21), 6931–6943. 10.1039/C2CS35129C. [DOI] [PubMed] [Google Scholar]
- (20).Cui H-L; Peng J; Feng X; Du W; Jiang K; Chen Y-C Dual Organocatalysis: Asymmetric Allylic–Allylic Alkylation of α,α-Dicyanoalkenes and Morita–Baylis–Hillman Carbonates. Chem. – Eur. J. 2009, 15 (7), 1574–1577. 10.1002/chem.200802534. [DOI] [PubMed] [Google Scholar]
- (21).Pan Y; Zhang N; Liu C-H; Fan S; Guo S; Zhang Z-M; Zhu Y-Y Boosting Photocatalytic Activities for Organic Transformations through Merging Photocatalyst and Transition-Metal Catalyst in Flexible Polymers. ACS Catal. 2020, 10 (20), 11758–11767. 10.1021/acscatal.0c03597. [DOI] [Google Scholar]
- (22).Prathap KJ; Maayan G Metallopeptoids as Efficient Biomimetic Catalysts. Chem. Commun. 2015, 51 (55), 11096–11099. 10.1039/C5CC04266F. [DOI] [PubMed] [Google Scholar]
- (23).Kinghorn MJ; Valdivia-Berroeta GA; Chantry DR; Smith MS; Ence CC; Draper SRE; Duval JS; Masino BM; Cahoon SB; Flansburg RR; Conder CJ; Price JL; Michaelis DJ Proximity-Induced Reactivity and Product Selectivity with a Rationally Designed Bifunctional Peptide Catalyst. ACS Catal. 2017, 7 (11), 7704–7708. 10.1021/acscatal.7b02699. [DOI] [Google Scholar]
- (24).Müller CE; Hrdina R; Wende RC; Schreiner PR A Multicatalyst System for the One-Pot Desymmetrization/Oxidation of Meso-1,2-Alkane Diols. Chem. – Eur. J. 2011, 17 (23), 6309–6314. 10.1002/chem.201100498. [DOI] [PubMed] [Google Scholar]
- (25).Girvin ZC; Andrews MK; Liu X; Gellman SH Foldamer-Templated Catalysis of Macrocycle Formation. Science 2019, 366 (6472), 1528–1531. 10.1126/science.aax7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhou Z; Roelfes G Synergistic Catalysis in an Artificial Enzyme by Simultaneous Action of Two Abiological Catalytic Sites. Nat. Catal. 2020, 3 (3), 289–294. 10.1038/s41929-019-0420-6. [DOI] [Google Scholar]
- (27).Christoffel F; Igareta NV; Pellizzoni MM; Tiessler-Sala L; Lozhkin B; Spiess DC; Lledós A; Maréchal J-D; Peterson RL; Ward TR Design and Evolution of Chimeric Streptavidin for Protein-Enabled Dual Gold Catalysis. Nat. Catal. 2021. 10.1038/s41929-021-00651-9. [DOI] [Google Scholar]
- (28).Zhu Y-Y; Lan G; Fan Y; Veroneau SS; Song Y; Micheroni D; Lin W Merging Photoredox and Organometallic Catalysts in a Metal–Organic Framework Significantly Boosts Photocatalytic Activities. Angew. Chem. Int. Ed. 2018, 57 (43), 14090–14094. 10.1002/anie.201809493. [DOI] [PubMed] [Google Scholar]
- (29).Quan Y; Song Y; Shi W; Xu Z; Chen JS; Jiang X; Wang C; Lin W Metal–Organic Framework with Dual Active Sites in Engineered Mesopores for Bioinspired Synergistic Catalysis. J. Am. Chem. Soc. 2020, 142 (19), 8602–8607. 10.1021/jacs.0c02966. [DOI] [PubMed] [Google Scholar]
- (30).Li H; Pan Q; Ma Y; Guan X; Xue M; Fang Q; Yan Y; Valtchev V; Qiu S Three-Dimensional Covalent Organic Frameworks with Dual Linkages for Bifunctional Cascade Catalysis. J. Am. Chem. Soc. 2016, 138 (44), 14783–14788. 10.1021/jacs.6b09563. [DOI] [PubMed] [Google Scholar]
- (31).Gryaznov SM; Letsinger RL Template Controlled Coupling and Recombination of Oligonucleotide Blocks Containing Thiophosphoryl Groups. Nucleic Acids Res. 1993, 21 (6), 1403–1408. 10.1093/nar/21.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Herrlein MK; Letsinger RL Selective Chemical Autoligation on a Double-Stranded DNA Template. Nucleic Acids Res. 1994, 22 (23), 5076–5078. 10.1093/nar/22.23.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Xu Y; Kool ET A Novel 5′-Iodonucleoside Allows Efficient Nonenzymatic Ligation of Single-Stranded and Duplex DNAs. Tetrahedron Lett. 1997, 38 (32), 5595–5598. 10.1016/S0040-4039(97)01266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Fujimoto K; Matsuda S; Takahashi N; Saito I Template-Directed Photoreversible Ligation of Deoxyoligonucleotides via 5-Vinyldeoxyuridine. J. Am. Chem. Soc. 2000, 122 (23), 5646–5647. 10.1021/ja993698t. [DOI] [Google Scholar]
- (35).Gartner ZJ; Liu DR The Generality of DNA-Templated Synthesis as a Basis for Evolving Non-Natural Small Molecules. J. Am. Chem. Soc. 2001, 123 (28), 6961–6963. 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mattes A; Seitz O Mass-Spectrometric Monitoring of a PNA-Based Ligation Reaction for the Multiplex Detection of DNA Single-Nucleotide Polymorphisms. Angew. Chem. Int. Ed. 2001, 40 (17), 3178–3181. . [DOI] [PubMed] [Google Scholar]
- (37).Li X; Liu DR DNA-Templated Organic Synthesis: Nature’s Strategy for Controlling Chemical Reactivity Applied to Synthetic Molecules. Angew. Chem. Int. Ed. 2004, 43 (37), 4848–4870. 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- (38).Gartner ZJ; Tse BN; Grubina R; Doyon JB; Snyder TM; Liu DR DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles. Science 2004, 305 (5690), 1601. 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hansen MH; Blakskjær P; Petersen LK; Hansen TH; Højfeldt JW; Gothelf KV; Hansen NJV A Yoctoliter-Scale DNA Reactor for Small-Molecule Evolution. J. Am. Chem. Soc. 2009, 131 (3), 1322–1327. 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- (40).Trinh T; Saliba D; Liao C; de Rochambeau D; Prinzen AL; Li J; Sleiman HF “Printing” DNA Strand Patterns on Small Molecules with Control of Valency, Directionality, and Sequence. Angew. Chem. Int. Ed. 2019, 58 (10), 3042–3047. 10.1002/anie.201809251. [DOI] [PubMed] [Google Scholar]
- (41).Brunner J; Mokhir A; Kraemer R DNA-Templated Metal Catalysis. J. Am. Chem. Soc. 2003, 125 (41), 12410–12411. 10.1021/ja0365429. [DOI] [PubMed] [Google Scholar]
- (42).Röthlingshöfer M; Gorska K; Winssinger N Nucleic Acid Templated Uncaging of Fluorophores Using Ru-Catalyzed Photoreduction with Visible Light. Org. Lett. 2012, 14 (2), 482–485. 10.1021/ol203029t. [DOI] [PubMed] [Google Scholar]
- (43).Golub E; Albada HB; Liao W-C; Biniuri Y; Willner I Nucleoapzymes: Hemin/G-Quadruplex DNAzyme–Aptamer Binding Site Conjugates with Superior Enzyme-like Catalytic Functions. J. Am. Chem. Soc. 2016, 138 (1), 164–172. 10.1021/jacs.5b09457. [DOI] [PubMed] [Google Scholar]
- (44).Ouyang Y; Biniuri Y; Fadeev M; Zhang P; Carmieli R; Vázquez-González M; Willner I Aptamer-Modified Cu2+-Functionalized C-Dots: Versatile Means to Improve Nanozyme Activities-“Aptananozymes.” J. Am. Chem. Soc. 2021, 143 (30), 11510–11519. 10.1021/jacs.1c03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chang D; Lindberg E; Winssinger N Critical Analysis of Rate Constants and Turnover Frequency in Nucleic Acid-Templated Reactions: Reaching Terminal Velocity. J. Am. Chem. Soc. 2017, 139 (4), 1444–1447. 10.1021/jacs.6b12764. [DOI] [PubMed] [Google Scholar]
- (46).Flanagan ML; Arguello AE; Colman DE; Kim J; Krejci JN; Liu S; Yao Y; Zhang Y; Gorin DJ A DNA-Conjugated Small Molecule Catalyst Enzyme Mimic for Site-Selective Ester Hydrolysis. Chem. Sci. 2018, 9 (8), 2105–2112. 10.1039/C7SC04554A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Czlapinski JL; Sheppard TL Nucleic Acid Template-Directed Assembly of Metallosalen−DNA Conjugates. J. Am. Chem. Soc. 2001, 123 (35), 8618–8619. 10.1021/ja0162212. [DOI] [PubMed] [Google Scholar]
- (48).Prusty DK; Kwak M; Wildeman J; Herrmann A Modular Assembly of a Pd Catalyst within a DNA Scaffold for the Amplified Colorimetric and Fluorimetric Detection of Nucleic Acids. Angew. Chem. Int. Ed. 2012, 51 (47), 11894–11898. 10.1002/anie.201206006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Anzola M; Winssinger N Turn On of a Ruthenium(II) Photocatalyst by DNA-Templated Ligation. Chem. – Eur. J. 2019, 25 (1), 334–342. 10.1002/chem.201804283. [DOI] [PubMed] [Google Scholar]
- (50).Roelfes G; Feringa BL DNA-Based Asymmetric Catalysis. Angew. Chem. Int. Ed. 2005, 44 (21), 3230–3232. 10.1002/anie.200500298. [DOI] [PubMed] [Google Scholar]
- (51).Dey S; Jäschke A Tuning the Stereoselectivity of a DNA-Catalyzed Michael Addition through Covalent Modification. Angew. Chem. Int. Ed. 2015, 54 (38), 11279–11282. 10.1002/anie.201503838. [DOI] [PubMed] [Google Scholar]
- (52).Punt PM; Langenberg MD; Altan O; Clever GH Modular Design of G-Quadruplex MetalloDNAzymes for Catalytic C–C Bond Formations with Switchable Enantioselectivity. J. Am. Chem. Soc. 2021, 143 (9), 3555–3561. 10.1021/jacs.0c13251. [DOI] [PubMed] [Google Scholar]
- (53).Green SA; Montgomery HR; Benton TR; Chan NJ; Nelson HM Regulating Transition-Metal Catalysis through Interference by Short RNAs. Angew. Chem. Int. Ed. 2019, 58 (46), 16400–16404. 10.1002/anie.201905333. [DOI] [PubMed] [Google Scholar]
- (54).Machida T; Dutt S; Winssinger N Allosterically Regulated Phosphatase Activity from Peptide–PNA Conjugates Folded Through Hybridization. Angew. Chem. Int. Ed. 2016, 55 (30), 8595–8598. 10.1002/anie.201602751. [DOI] [PubMed] [Google Scholar]
- (55).Hu L; Lu C-H; Willner I Switchable Catalytic DNA Catenanes. Nano Lett. 2015, 15 (3), 2099–2103. 10.1021/nl504997q. [DOI] [PubMed] [Google Scholar]
- (56).Aleman Garcia MA; Hu Y; Willner I Switchable Supramolecular Catalysis Using DNA-Templated Scaffolds. Chem. Commun. 2016, 52 (10), 2153–2156. 10.1039/C5CC08873A. [DOI] [PubMed] [Google Scholar]
- (57).Xin L; Zhou C; Yang Z; Liu D Regulation of an Enzyme Cascade Reaction by a DNA Machine. Small 2013, 9 (18), 3088–3091. 10.1002/smll.201300019. [DOI] [PubMed] [Google Scholar]
- (58).Fu J; Yang YR; Johnson-Buck A; Liu M; Liu Y; Walter NG; Woodbury NW; Yan H Multi-Enzyme Complexes on DNA Scaffolds Capable of Substrate Channelling with an Artificial Swinging Arm. Nat. Nanotechnol. 2014, 9 (7), 531–536. 10.1038/nnano.2014.100. [DOI] [PubMed] [Google Scholar]
- (59).Hoover JM; Stahl SS Highly Practical Copper(I)/TEMPO Catalyst System for Chemoselective Aerobic Oxidation of Primary Alcohols. J. Am. Chem. Soc. 2011, 133 (42), 16901–16910. 10.1021/ja206230h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Semmelhack MF; Schmid CR; Cortes DA; Chou CS Oxidation of Alcohols to Aldehydes with Oxygen and Cupric Ion, Mediated by Nitrosonium Ion. J. Am. Chem. Soc. 1984, 106 (11), 3374–3376. 10.1021/ja00323a064. [DOI] [Google Scholar]
- (61).Gamez P; Arends IWCE; Reedijk J; Sheldon RA Copper(Ii)-Catalysed Aerobic Oxidation of Primary Alcohols to Aldehydes. Chem. Commun. 2003, No. 19, 2414–2415. 10.1039/B308668B. [DOI] [PubMed] [Google Scholar]
- (62).Kumpulainen ETT; Koskinen AMP Catalytic Activity Dependency on Catalyst Components in Aerobic Copper–TEMPO Oxidation. Chem. – Eur. J. 2009, 15 (41), 10901–10911. 10.1002/chem.200901245. [DOI] [PubMed] [Google Scholar]
- (63).Hoover JM; Ryland BL; Stahl SS Mechanism of Copper(I)/TEMPO-Catalyzed Aerobic Alcohol Oxidation. J. Am. Chem. Soc. 2013, 135 (6), 2357–2367. 10.1021/ja3117203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Rabeah J; Bentrup U; Stößer R; Brückner A Selective Alcohol Oxidation by a Copper TEMPO Catalyst: Mechanistic Insights by Simultaneously Coupled Operando EPR/UV-Vis/ATR-IR Spectroscopy. Angew. Chem. Int. Ed. 2015, 54 (40), 11791–11794. 10.1002/anie.201504813. [DOI] [PubMed] [Google Scholar]
- (65).Fernandes AE; Riant O; Jensen KF; Jonas AM Molecular Engineering of Trifunctional Supported Catalysts for the Aerobic Oxidation of Alcohols. Angew. Chem. Int. Ed. 2016, 55 (37), 11044–11048. 10.1002/anie.201603673. [DOI] [PubMed] [Google Scholar]
- (66).Chandra P; Jonas AM; Fernandes AE Spatial Coordination of Cooperativity in Silica-Supported Cu/TEMPO/Imidazole Catalytic Triad. ACS Catal. 2018, 8 (7), 6006–6011. 10.1021/acscatal.8b01399. [DOI] [Google Scholar]
- (67).Lu Z; Costa JS; Roubeau O; Mutikainen I; Turpeinen U; Teat SJ; Gamez P; Reedijk J A Copper Complex Bearing a TEMPO Moiety as Catalyst for the Aerobic Oxidation of Primary Alcohols. Dalton Trans. 2008, No. 27, 3567–3573. 10.1039/B802109K. [DOI] [PubMed] [Google Scholar]
- (68).Liu X; Xia Q; Zhang Y; Chen C; Chen W Cu-NHC-TEMPO Catalyzed Aerobic Oxidation of Primary Alcohols to Aldehydes. J. Org. Chem. 2013, 78 (17), 8531–8536. 10.1021/jo401252d. [DOI] [PubMed] [Google Scholar]
- (69).Ryland BL; Stahl SS Practical Aerobic Oxidations of Alcohols and Amines with Homogeneous Copper/TEMPO and Related Catalyst Systems. Angew. Chem. Int. Ed. 2014, 53 (34), 8824–8838. 10.1002/anie.201403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Marais L; Swarts AJ Biomimetic Cu/Nitroxyl Catalyst Systems for Selective Alcohol Oxidation. Catalysts 2019, 9 (5), 395. 10.3390/catal9050395. [DOI] [Google Scholar]
- (71).Li Y; Gabriele E; Samain F; Favalli N; Sladojevich F; Scheuermann J; Neri D Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb. Sci. 2016, 18 (8), 438–443. 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Peters-Clarke TM; Quan Q; Brademan DR; Hebert AS; Westphall MS; Coon JJ Ribonucleic Acid Sequence Characterization by Negative Electron Transfer Dissociation Mass Spectrometry. Anal. Chem. 2020, 92 (6), 4436–4444. 10.1021/acs.analchem.9b05388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Rozenman MM; Liu DR DNA-Templated Synthesis in Organic Solvents. ChemBioChem 2006, 7 (2), 253–256. 10.1002/cbic.200500413. [DOI] [PubMed] [Google Scholar]
- (74).Hoover JM; Ryland BL; Stahl SS Copper/TEMPO-Catalyzed Aerobic Alcohol Oxidation: Mechanistic Assessment of Different Catalyst Systems. ACS Catal. 2013, 3 (11), 2599–2605. 10.1021/cs400689a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Gamez P; Simons C; Steensma R; Driessen WL; Challa G; Reedijk J A Spectacular Increase in the Polymerisation Rate of 2,6-Dimethylphenol Induced by Acetonitrile. Eur. Polym. J. 2001, 37 (7), 1293–1296. 10.1016/S0014-3057(00)00259-7. [DOI] [Google Scholar]
- (76).Eberhard H; Diezmann F; Seitz O DNA as a Molecular Ruler: Interrogation of a Tandem SH2 Domain with Self-Assembled, Bivalent DNA–Peptide Complexes. Angew. Chem. 2011, 123 (18), 4232–4236. 10.1002/ange.201007593. [DOI] [PubMed] [Google Scholar]
- (77).Diezmann F; Seitz O DNA-Guided Display of Proteins and Protein Ligands for the Interrogation of Biology. Chem. Soc. Rev. 2011, 40 (12), 5789–5801. 10.1039/C1CS15054E. [DOI] [PubMed] [Google Scholar]
- (78).Rinker S; Ke Y; Liu Y; Chhabra R; Yan H Self-Assembled DNA Nanostructures for Distance-Dependent Multivalent Ligand–Protein Binding. Nat. Nanotechnol. 2008, 3 (7), 418–422. 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Gorska K; Huang K-T; Chaloin O; Winssinger N DNA-Templated Homo- and Heterodimerization of Peptide Nucleic Acid Encoded Oligosaccharides That Mimick the Carbohydrate Epitope of HIV. Angew. Chem. Int. Ed. 2009, 48 (41), 7695–7700. 10.1002/anie.200903328. [DOI] [PubMed] [Google Scholar]
- (80).Goodman RP; Schaap IAT; Tardin CF; Erben CM; Berry RM; Schmidt CF; Turberfield AJ Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science 2005, 310 (5754), 1661. 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- (81).McLaughlin CK; Hamblin GD; Hänni KD; Conway JW; Nayak MK; Carneiro KMM; Bazzi HS; Sleiman HF Three-Dimensional Organization of Block Copolymers on “DNA-Minimal” Scaffolds. J. Am. Chem. Soc. 2012, 134 (9), 4280–4286. 10.1021/ja210313p. [DOI] [PubMed] [Google Scholar]
- (82).Gartner ZJ; Grubina R; Calderone CT; Liu DR Two Enabling Architectures for DNA-Templated Organic Synthesis. Angew. Chem. Int. Ed. 2003, 42 (12), 1370–1375. 10.1002/anie.200390351. [DOI] [PubMed] [Google Scholar]
- (83).Gartner ZJ; Kanan MW; Liu DR Expanding the Reaction Scope of DNA-Templated Synthesis. Angew. Chem. 2002, 114 (10), 1874–1878. . [DOI] [PubMed] [Google Scholar]
- (84).Grossmann TN; Röglin L; Seitz O Target-Catalyzed Transfer Reactions for the Amplified Detection of RNA. Angew. Chem. Int. Ed. 2008, 47 (37), 7119–7122. 10.1002/anie.200801355. [DOI] [PubMed] [Google Scholar]
- (85).Snyder TM; Tse BN; Liu DR Effects of Template Sequence and Secondary Structure on DNA-Templated Reactivity. J. Am. Chem. Soc. 2008, 130 (4), 1392–1401. 10.1021/ja076780u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Zhang DY; Winfree E Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J. Am. Chem. Soc. 2009, 131 (47), 17303–17314. 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- (87).Tyagi S; Kramer FR Molecular Beacons: Probes That Fluoresce upon Hybridization. Nat. Biotechnol. 1996, 14 (3), 303–308. 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- (88).Lindberg E; Angerani S; Anzola M; Winssinger N Luciferase-Induced Photoreductive Uncaging of Small-Molecule Effectors. Nat. Commun. 2018, 9 (1), 3539. 10.1038/s41467-018-05916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Keefe AD; Pai S; Ellington A Aptamers as Therapeutics. Nat. Rev. Drug Discov. 2010, 9 (7), 537–550. 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Pfeiffer F; Mayer G Selection and Biosensor Application of Aptamers for Small Molecules. Front. Chem. 2016, 4, 25. 10.3389/fchem.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


