Abstract
Environmentally dominant members of the genus Beggiatoa and Thioploca spp. are united by unique morphological and physiological adaptations (S. C. McHatton, J. P. Barry, H. W. Jannasch, and D. C. Nelson, Appl. Environ. Microbiol. 62:954–958, 1996). These adaptations include the presence of very wide filaments (width, 12 to 160 μm), the presence of a central vacuole comprising roughly 80% of the cellular biovolume, and the capacity to internally concentrate nitrate at levels ranging from 150 to 500 mM. Until recently, the genera Beggiatoa and Thioploca were recognized and differentiated on the basis of morphology alone; they were distinguished by the fact that numerous Thioploca filaments are contained within a common polysaccharide sheath, while Beggiatoa filaments occur singly. Vacuolate Beggiatoa or Thioploca spp. can dominate a variety of marine sediments, seeps, and vents, and it has been proposed (H. Fossing, V. A. Gallardo, B. B. Jorgensen, M. Huttel, L. P. Nielsen, H. Schulz, D. E. Canfield, S. Forster, R. N. Glud, J. K. Gundersen, J. Kuver, N. B. Ramsing, A. Teske, B. Thamdrup, and O. Ulloa, Nature [London] 374:713–715, 1995) that members of the genus Thioploca are responsible for a significant portion of total marine denitrification. In order to investigate the phylogeny of an environmentally dominant Beggiatoa sp., we analyzed complete 16S rRNA gene sequence data obtained from a natural population found in Monterey Canyon cold seeps. Restriction fragment length polymorphism analysis of a clone library revealed a dominant clone, which gave rise to a putative Monterey Beggiatoa 16S rRNA sequence. Fluorescent in situ hybridization with a sequence-specific probe confirmed that this sequence originated from wide Beggiatoa filaments (width, 65 to 85 μm). A phylogenetic tree based on evolutionary distances indicated that the Monterey Beggiatoa sp. falls in the gamma subdivision of the class Proteobacteria and is most closely related to the genus Thioploca. This vacuolate Beggiatoa—Thioploca cluster and a more distantly related freshwater Beggiatoa species cluster form a distinct phylogenetic group.
Among the numerous conspicuous sulfur-oxidizing bacteria, the genera Beggiatoa and Thioploca have similar morphological and physiological characteristics, including disk-shaped or cylindrical cells arranged in long filaments, gliding motility, intracellular globules of elemental sulfur, and the occurrence of both freshwater and marine representatives that have a wide range of cell diameters (widths). Despite long-standing interest in both of these genera, there are pure cultures of only the narrowest Beggiatoa strains (width, less than 5 μm), while no Thioploca species has been cultured. Although some ultrastructural differences between certain Thioploca and Beggiatoa strains have been reported at the electron microscopy level (21, 22) and although some differentiation may be possible based on filament widths (Table 1), a single Thioploca filament cannot be reliably differentiated from a Beggiatoa filament by light microscopy. In addition, all wide (cell diameter, 12 to 160 μm) uncultured marine representatives of both genera examined to date have a massive central vacuole and accumulate nitrate, presumably in the vacuole and presumably for use as an electron acceptor that allows anaerobic sulfide oxidation (5, 24). Morphologically, Beggiatoa spp. are distinguished from Thioploca spp. only by the fact that in members of the genus Thioploca up to 100 separate filaments are contained within a single common polysaccharide sheath to form a bundle (22). Within each genus, filament width, which seems to divide natural populations into largely nonoverlapping groups, is the basis of species differentiation (37).
TABLE 1.
Summary of properties of vacuolate Beggiatoa and Thioploca spp.
| Organism(s) | Filament width (μm) | Common sheath | Vacuole vol. (% of biovolume) | Evidence for vacuolea | Length of 16S ribosomal DNA sequence (bp) | References |
|---|---|---|---|---|---|---|
| Beggiatoa sp. (Monterey Canyon) | 65–85 | No | 80 | P/V | 1,493 | 24; this study |
| Beggiatoa spp. (Guaymas Basin vents) | 17–35 | No | 68–85 | P/V, EM | None | 29, 31 |
| 32–50 | No | 83 | P/V, EM | None | ||
| 88–140 | No | 92 | P/V, EM | None | ||
| Thioploca ingrica | 2–4.5 | Yes | 39–42b | EM | 1,491 | 18, 19, 38 |
| Thioploca chileae | 12–20 | Yes | 89b | EM | 550 | 20, 21, 38 |
| Thioploca araucae | 30–43 | Yes | >80 | EM | 620 | 5, 20, 21, 38 |
Beggiatoa and Thioploca filaments have been observed to form dense mats on sediments in estuarine, shelf, seep, and deep-sea hydrothermal vent environments (7, 10, 12). The biomass densities of vacuolate forms of members of these genera can be especially impressive, up to 1 kg (wet weight)/m2 of sediment surface (5, 25). Although narrow nonvacuolate Beggiatoa spp. proliferate in a narrow zone whose vertical dimension is less than 1 mm, where both oxygen and H2S occur (13, 30), the densities of the wider, vacuolate forms of both genera are high over a greater vertical distance (e.g., 10 cm), even in the absence of oxygen. Presumably, these organisms employ internal nitrate as an electron acceptor, which allows anaerobic oxidation of sulfide 10 to 15 cm below the sediment surface.
In a recent study Teske et al. reported that there is a relatively close phylogenetic relationship between Thioploca spp. and Beggiatoa spp. based on 16S rRNA gene sequence data (38), but that study included only freshwater, nonvacuolate, heterotrophic representatives of the genus Beggiatoa. In the current study we focused on a very wide (width, 65 to 85 μm), vacuolate, uncultured Beggiatoa sp. from Monterey Canyon, California. The filaments of this organism are ideal for study because they occur at extraordinary biomass densities and can be harvested with minimal contamination from other prokaryotes (24, 25). The Monterey Canyon Beggiatoa sp. is also among the best-characterized representatives having the vacuolate phenotype. Enzymatic studies have shown that it is a chemoautotrophic sulfide oxidizer with the ability to reduce its internal store of nitrate to ammonia while the nitrate serves as a presumptive electron acceptor (24, 25). The results reported here are the first phylogenetic results obtained for a vacuolate marine Beggiatoa sp. from any environment and provide data for an important comparison with previously published partial sequences (38) attributed to marine Thioploca spp. and finer resolution of the phylogeny of vacuolate, nitrate-accumulating, chemoautotrophic, marine, sulfide-oxidizing filaments. Confirmation by fluorescent in situ hybridization (FISH) that our sequence derives from the vacuolate Beggiatoa sp. sequence strongly supports the tight clustering of the genera Beggiatoa and Thioploca.
MATERIALS AND METHODS
Beggiatoa sampling.
Native filaments of a wide uncultured Beggiatoa sp. were collected at a depth of 900 m in Clam Field Seep (1) in Monterey Canyon in April 1997. Plexiglas cores were used for sediment sampling from this sulfide-rich cold seep; the cores were collected by the remotely operated vehicle Ventana. Samples, including at least 10 cm of overlying water, were transported on ice to Davis, Calif., where they were stored at 4°C for 24 h, a period which allowed filaments to glide through the disturbed surface layer and extend into the overlying water. Filaments were gently removed with a Pasteur pipette and transferred to 1.5-ml Eppendorff tubes. As determined by examination with a microscope, the material collected generally consisted of more than 99% (by biovolume) Beggiatoa filaments having widths ranging from 65 to 85 μm. The filaments were pelleted by centrifugation at 5,000 × g for 10 s and kept at −20°C until they were used. Chromosomal DNA was extracted as described by Wilson (44).
Thiomicrospira strains.
Thiomicrospira sp. strain L-12 was obtained from Holger Jannasch, Woods Hole Oceanographic Institution (32). Thiomicrospira sp. strain XCL-2 was cultured from the Galapagos Rift vents in 1988 (28); the DNA base composition, growth rate at 33°C, and morphology of this strain indicate that it is a Thiomicrospira crunogena strain (11). The culture conditions used for both strains were the conditions described previously.
Clone library construction.
Small-subunit 16S rRNA genes were amplified from the potentially mixed DNA by PCR by using Taq DNA polymerase and standard methods (34). The two universal eubacterial 16S ribosomal DNA primers used were based on primers described by Weisburg et al. (43), primers 8fpl (5′-AGAGTTTGATCCTGGCTCAG-3′, corresponding to Escherichia coli positions 8 to 27) and 1492rpl (5′-GGTTACCTTGTTACGACTT-3′, corresponding to positions 1510 to 1492), and contained added polylinkers. The reaction mixtures were overlaid with mineral oil and were incubated in a Perkin-Elmer model 480 DNA thermal cycler. Three control reaction mixtures (one lacking template DNA, one lacking forward primer, and one lacking reverse primer) were prepared. The amplification conditions were as follows: denaturation at 94°C for 5 min, annealing at 45°C for 1 min, and extension at 70°C for 4 min for 30 cycles. Following the final cycle, the reaction mixture was incubated at 72°C for 10 min. The amplified products were inserted into the TA vector and transformed into INVαF′ cells (Invitrogen Corp.). Positive transformants (white colony morphotype) were streaked for isolation and were screened by using a miniprep kit (Qiagen, Chatsworth, Calif.). A total of 111 clones containing the full-length 16S rRNA gene inserts from the Beggiatoa-enriched DNA were obtained from three independent PCR and subsequent multiple cloning reactions.
RFLP analysis and sequencing.
All of the clones were characterized by an EcoRI restriction fragment length polymorphism (RFLP) analysis in which standard methods were used (34). Restriction fragments were resolved by gel electrophoresis (1% agarose in 1× Tris-acetate-EDTA buffer) and stained with ethidium bromide (0.5 μg ml−1). Three representatives of the dominant restriction pattern, which was produced by 76% of the clones, were partially sequenced (approximately 250 bases), which confirmed the sequence identity. A single representative of the dominant operational taxonomic unit (OTU) was then selected for complete bidirectional sequencing by Sanger’s dideoxynucleotide chain termination method (34), in which a Sequenase, version 2.0, kit (U.S. Biochemicals Corp., Cleveland, Ohio) was used. The following sequencing primers were used: forward primer −40 and reverse primer −21 (U. S. Biochemicals Corp.); universal reverse primers 519r, 907r, and 1392r (16); and custom forward primers MBF1 (positions 346 to 363; GGGAGGCAGCAGTAGGGA), MBF2 (positions 666 to 683; GGGAAGCGGAATTCTTAG), and MBF3 (positions 1174 to 1191; GGAGGAAGGTGGGGATGA). The manually obtained sequence data were also confirmed by an automated sequencing analysis in which we used ABI PRISM dye terminator cycle sequencing with dRhodomine terminator chemistry. Reactions were performed by using an ABI PRISM DNA sequencer (model 377) and a 5% Long Ranger gel. Sequence data were edited and analyzed by using ABI PRISM sequencing 2.1.1 software.
Probe design and labeling.
An 18-mer Monterey Beggiatoa-specific probe, MBSP1RC (Table 2), was targeted to variable region 29 (6, 9, 39) of the dominant 16S rRNA clone sequence (OTU 3) for use in FISH. The probe was obtained from OPERON Technologies with the 5′ amino modifier, 6-(4-monomethoxytritylamino)hexyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphor-amide. The 5′ amino groups of this probe and other probes were labeled with the fluorophore BODIPY-TMR (excitation at 542 nm, emission at 574 nm) by using the instructions provided by the supplier (Molecular Probes Inc.). Unlabeled probe was removed by using a spin column purification kit. Aliquots (50 μl; 25 ng/μl) of labeled probe were distributed into nuclease-free tubes, dried in a SpeedVac apparatus, and stored at −20°C in the dark. The probe’s optimal hybridization parameters were calculated as described by Stahl and Amann (36).
TABLE 2.
Fluorescent rRNA-specific oligonucleotide probes used in this study
| Probe | Target positions | Sequence | Specificity |
|---|---|---|---|
| 16S rRNA probes | |||
| MBSP1RC | 850–833 | 5′-AGGATCAATCTCCCCCAA-3′ | Wide Beggiatoa sp. |
| MBSP1Ca | 833–850 | 5′AACCCCCTCTAACTAGGA-3′ | None |
| Eub-338b | 338–355 | 5′-GCTGCCTCCCGTAGGAGT-3′ | Eubacteria |
| ALF1bc | 19–35 | 5′-CGTTCG(C/T)TCTGAGCCAG-3′ | Alpha subdivision |
| Thioploca-829d | 829–849 | 5′-GGATTAATTTCCCCCAACATC-3′ | Thioploca spp. |
| 23S rRNA probec | |||
| BET42a | 1027–1043 | 5′-GCCTTCCCACTTCGTTT-3′ | Beta subdivision |
| GAM42a | 1027–1043 | 5′-GCCTTCCCACATCGTTT-3′ | Gamma subdivision |
FISH.
Filament samples from Monterey Canyon were fixed, prehybridized, and hybridized with fluorescent probes by previously described methods (38), with the following modifications. Filaments fixed in paraformaldehyde (1.4% in filtered [pore size, 0.2 μm] natural seawater) were consecutively dehydrated in 50, 80, and 100% ethanol. After air drying, the slides were prehybridized with 40 μl of prehybridization buffer at 37°C for 2 h. Then the prehybridization buffer was replaced with 40 μl of hybridization buffer and an appropriate oligonucleotide probe at a final concentration of 5 to 10 ng μl−1.
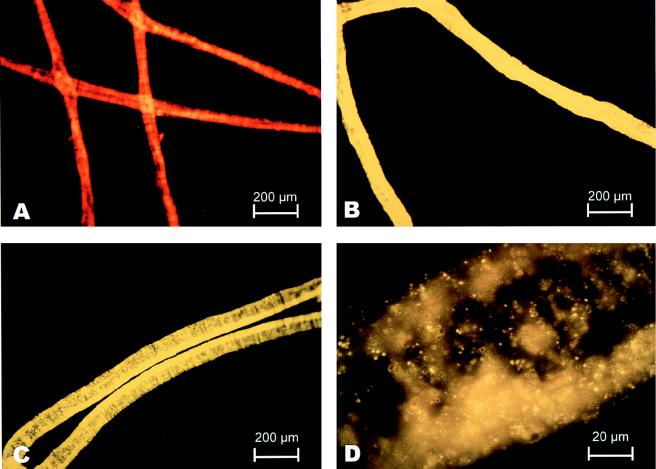
Hybridization was carried out at 37°C (10, 15, or 20% [vol/vol] formamide) or at 37 to 43°C (20% [vol/vol] formamide) for 16 h to determine the conditions for specific binding of probes. The slides were incubated with wash buffer at 37°C for 20 min to remove unbound fluorescent probe, rinsed with water, air dried, and mounted in 100% glycerol. Fluorescence was detected with a Zeiss Axioskop microscope equipped with an epifluorescence filter set (Narrow X HQ 545/565/610; Chroma Technology, Brattleboro, Vt.). Micrographs were taken with a Zeiss model MC 100 camera and Kodak Ektachrome 1600 film. A composite (see Fig. 5) was edited by using Photoshop, version 4.01, and was printed by using a Fujix Pictrography 3000 printer.
FIG. 5.
Photomicrographs of fluorescent FISH results. (C and D) Hybridization (37°C, 20% formamide) of the MBSP1RC probe labeled with the fluorophore BODIPY-TMR in a single wide (width, approximately 75 μm) Beggiatoa filament. The intensity of the hybridization signal was comparable to the intensity of the signal obtained when the universal eubacterial probe (Eub-338) was used as a positive control (B). Nonspecific probe did not hybridize with the Beggiatoa rRNA (A). The orange-red coloration (A) (compared with the bright yellow of the probe [B through D]) appeared to be autoflurorescence and was also detected in unstained filaments.
Phylogenetic analysis.
All of the sequences used for comparison were retrieved from the Ribosomal Database Project (17). Sequences were manually aligned and edited by using the SeqLab program included in the Wisconsin package, version 9.1 (7a). Evolutionary trees were constructed by distance, maximum-parsimony, and maximum-likelihood methods by using programs contained in the phylogeny inference package (PHYLIP, version 3.5c) (4). Two data sets that included only regions in which the alignment was unambiguous were used for phylogenetic analysis. The large set (small mask) consisted of 1,203 aligned positions for the Monterey Beggiatoa sp. and previously published sequence data. The small set (large mask) consisted of only 534 positions that were required to accommodate the partial sequences available for Thioploca araucae and Thioploca chileae. For each alignment, 100 bootstrapped replicate resampling data sets were generated by using the SEQBOOT program with random sequence addition and global rearrangement. We estimated evolutionary distances with the program DNADIST by using the option for Kimura’s two-parameter model for nucleotide change and a transition/transversion ratio of 2.0 (15). We also tested the Jukes-Cantor model (14) for nucleotide substitution. The resulting evolutionary distance matrices were used to reconstruct phylogenetic trees by the neighbor-joining method (33) by using NEIGHBOR. Parsimony and maximum-likelihood trees were reconstructed with the programs DNAPARS and DNAML, respectively. We edited the phylogenetic trees with the program TREECON for Windows 95, version 1.3b (40).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 65- to 85-μm-wide Monterey Beggiatoa sp. has been deposited in the GenBank database under accession no. AF064543. The nucleotide sequences of Thiomicrospira sp. strain L-12 and Thiomicrospira sp. strain XCL-2 have been deposited under accession no. AF064544 and AF064545, respectively.
RESULTS
Collection of environmental sample.
Sediment cores obtained from a sulfide-rich cold seep (Clam Field Seep) off the California coast in Monterey Canyon (depth, 900 m) typically had surface mats consisting of macroscopically visible Beggiatoa sp. filaments that projected 1 to 2 cm above the sediment surface. Microscopic examination of harvested filaments showed that the wide Beggiatoa sp. dominated; all of the unicellular contaminants comprised less than 1% of the biovolume (24). Although the diameters of most of the filaments harvested ranged from 65 to 85 μm, a few of the Beggiatoa sp. filaments were narrower, with diameters ranging from 20 to 30 μm. All of the filaments had sulfur inclusions and moved by gliding.
Extraction of chromosomal DNA.
The initial conventional extraction of bacterial DNA (34) from the wide filaments resulted in a poor yield or degraded DNA. We presumed that a large amount of extracellular polysaccharide in the samples affected the efficiency of cell breakage and separation of DNA from the exopolymers. In order to solve this problem, a modified method in which cetyltrimethylammonium bromide was used (44) allowed removal of cell wall debris and denatured protein and polysaccharide, while the intact nucleic acid remained in solution, as demonstrated by agarose gel electrophoresis (data not shown).
PCR amplification and construction of 16S rRNA gene clone library.
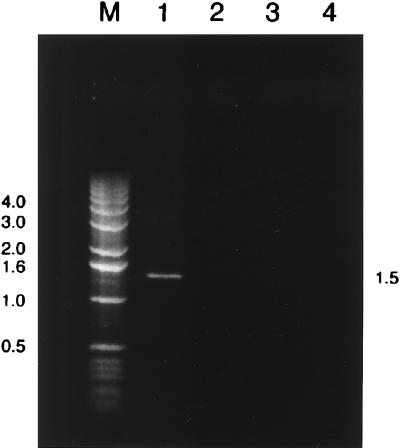
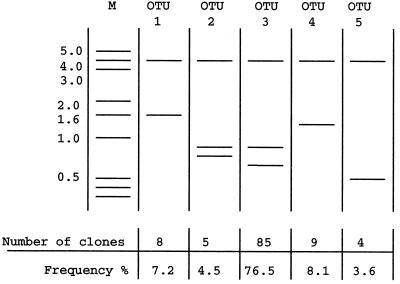
PCR amplification performed with universal eubacterial 16S rRNA gene primers and mixed template DNA from the Beggiatoa-dominated population was successful and yielded a single 1.5-kb DNA fragment (Fig. 1), suggesting that the PCR was specific for the target region. Cloning of the PCR products yielded 111 positive clones having the complete 1.5-kb insert. After positive clones were screened by performing an RFLP analysis, five different OTUs were defined. The dominant restriction pattern (OTU 3) was produced by 76% of the clones screened (Fig. 2).
FIG. 1.
PCR amplification products based on chromosomal DNA extracted from Beggiatoa sp. filaments from Monterey Canyon, California, as revealed by ethidium bromide staining and agarose gel electrophoresis. Each PCR mixture contained a 3-μl sample. Lane M, 1.0-kb DNA ladder; lane 1, PCR amplification product obtained with universal eubacterial 16S rRNA gene primers; lanes 2 through 4, three separate control reactions (lane 2, no DNA template; lane 3, no forward primer; lane 4, no reverse primer). DNA sizes (in kilobase pairs) are indicated on the left and right.
FIG. 2.
Diagram of RFLP patterns (after EcoRI digestion) of cloned 16S rRNA genes, resolved by agarose gel electrophoresis. All five different restriction patterns obtained, defined as OTUs, are shown along with their proportional representation in the 111 clones screened. Molecular weight standards (lane M) were included for comparison. Fragment sizes (in kilobase pairs) are indicated on the left.
Sequencing.
The first 250 bases (5′ to 3′) of three representatives that were picked randomly from OTU 3 were sequenced with primer −40F. All three representatives had identical sequences in this region that included two regions known to be highly variable within the gamma subdivision of the Proteobacteria (7a). The complete 16S rRNA gene sequence of the Monterey Beggiatoa sp. was confirmed by manual and automated sequencing. There were no unresolved mismatches. The 16S rRNA gene sequence of the dominant clone was not a product of chimeric artifacts, as determined by the CHECK_CHIMERA program. To eliminate misincorporation PCR errors as a major source of variations, we used secondary-structure models (9, 45) to examine the nature and position of the sequence variation. In this analysis we assumed that base substitutions caused by DNA polymerase errors should be randomly distributed throughout the sequences. In fact, the secondary-structure analysis confirmed that all substitutions compared to the E. coli sequence were restricted to highly variable regions of the 16S rRNA sequence, were largely compensated for by corresponding substitutions in the complementary stem region, and did not disturb the highly conserved secondary structure.
Database search and alignment of 16S rRNA gene sequence.
Searches of databases were performed with the BLAST program (Wisconsin package, version 9.1 [7a]) in order to identify partial and complete sequences similar to the putative 16S rRNA gene sequence of the Monterey Beggiatoa sp. Beggiatoa sp. strain B1401-13 had the highest score, and the excellent matches included matches with sequences from Thioploca spp., Thiobacillus spp., and a number of free-living and endosymbiotic sulfur-oxidizing bacteria belonging to the gamma subdivision of the Proteobacteria.
Phylogenetic analysis.
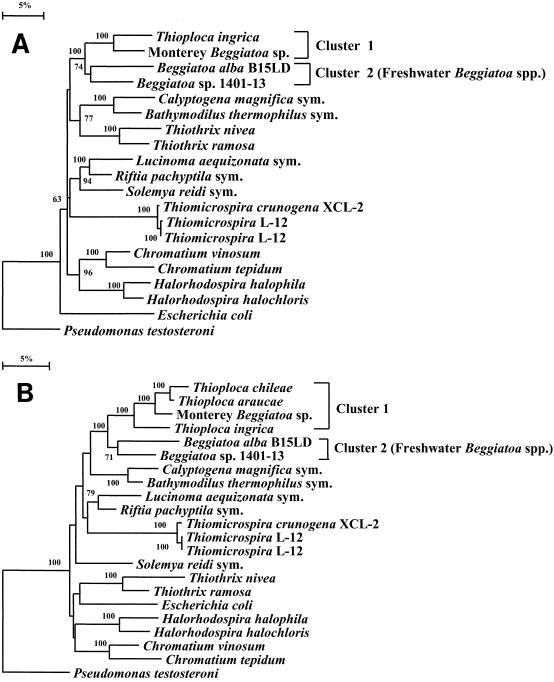
Tree construction analyses performed with both distance and parsimony methods and bootstrapping unambiguously placed the Monterey Beggiatoa sequence in the gamma subdivision of the Proteobacteria. The phylogenetic trees that were inferred from the distance matrix data by neighbor-joining tree reconstruction methods are shown in Fig. 3. Although the number of nucleotide positions analyzed was only 534 when partial sequences of T. araucae and T. chileae were included in the alignment (Fig. 3B), this small number of positions did not result in instability of the overall tree topology or significant changes in the bootstrap values compared to the values obtained for the small mask (1,203 positions) (Fig. 3A).
FIG. 3.
Phylogenetic trees showing the positions of the Monterey Beggiatoa sp. and other representatives of the gamma subgroup of the Proteobacteria, as inferred by the neighbor-joining method. Distances were corrected with Kimura’s two-parameter model. The sequence of Pseudomonas testosteroni (a member of the β subdivision of the Proteobacteria) was used to root the tree. Halorhodospira halophila and Halorhodospira halochloris (9a) sequence data were accessed as data for the corresponding Ectothiorhodospira species. The phylogenetic analyses were performed with programs contained in the PHYLIP package, version 3.5c. There are two main Beggiatoa-Thioploca clusters. Cluster 1 contains the Monterey Beggiatoa sp. sequence and all previously published Thioploca spp. sequences; cluster 2 contains all sequences belonging to freshwater Beggiatoa spp. (A) Small mask tree inferred from 1,203 nucleotide positions. Partial T. araucae and T. chileae sequences were not included. (B) Tree inferred with the full mask by using only 534 positions, which allowed inclusion of partial sequences of T. araucae and T. chileae. All of the sequences used except the new sequences were retrieved from the Ribosomal Database Project (17). Scale bar = 5 substitutions/100 nucleotide positions. Thiomicrospira crunogena XCL-2 and Thiomicrospira sp. strain L-12 were sequenced in this study; the sequence of Thiomicrospira sp. strain L-12 was also determined previously (31). sym, symbiont.
The partial-sequence tree (Fig. 3B) and an evolutionary distance matrix (Table 3) showed that the vacuolate marine thioplocas (T. araucae and T. chileae) are the closest relatives of the Monterey Beggiatoa sp. (Fig. 3B) and that T. araucae is equally close to the Monterey Beggiatoa sp. and T. chileae in terms of evolutionary distance. Narrow, nonvacuolate, freshwater, filamentous bacteria (i.e., Beggiatoa spp. strains B15LD and B1401-13) and Thioploca ingrica are more distantly related to this cluster (Fig. 3). Even with the large mask, the bootstrap values gave complete (100%) support for (i) the three-species cluster that contains all known vacuolate, marine, filamentous sulfur bacteria (i.e., Beggiatoa and Thioploca spp.), (ii) the finding that the freshwater organism T. ingrica is the closest relative of this three-species cluster, and (iii) the monophyletic nature of the Beggiatoa-Thioploca lineage within the gamma subdivision.
TABLE 3.
Evolutionary distances between 16S rRNA sequences of Beggiatoa and Thioploca spp.
| Organism | Estimated no. of substitutions per 100 bases
|
||||||
|---|---|---|---|---|---|---|---|
| Monterey Beggiatoa sp. | Thioploca araucae | Thioploca chileae | Thioploca ingrica | Beggiatoa sp. strain B15LD | Beggiatoa sp. strain B1401-13 | Escherichia coli | |
| Monterey Beggiatoa sp. | 0.00 | 2.60 | 3.74 | 8.73 | 14.89 | 12.48 | 20.67 |
| Thioploca araucae | 0.00 | 2.41 | 9.14 | 14.91 | 12.26 | 20.91 | |
| Thioploca chileae | 0.00 | 10.19 | 15.56 | 12.05 | 20.67 | ||
| Thioploca ingrica | 0.00 | 17.45 | 13.18 | 24.22 | |||
| Beggiatoa sp. strain B15LD | 0.00 | 12.70 | 22.62 | ||||
| Beggiatoa sp. strain B1401-13 | 0.00 | 22.13 | |||||
| Escherichia coli | 0.00 | ||||||
Values were corrected by the Jukes-Cantor model for nucleotide substitution (14).
FISH.
We used FISH to confirm that the sequence which we retrieved is the sequence of the wide vacuolate Beggiatoa sp. from Monterey Canyon. The highlighted target region in the multiple alignment in Fig. 4 shows that the sequence of the 18-mer probe (MBSP1RC), which was designed to be specific for the Monterey Beggiatoa sp., differed from the sequences of the two marine Thioploca spp. by two or three nucleotides. Compared with the sequence of the narrow freshwater organism T. ingrica, there were eight mismatches. The sequence of MBSP1RC was checked, and negative results were obtained with the CHECK_PROBE program of the Ribosomal Database Project (updated 15 June 1997), which allowed two mismatches.
FIG. 4.
16S rRNA target region for the Monterey Beggiatoa sp.-specific probe (MBSP1RC; length 18 nucleotides) aligned with sequences from selected endosymbiotic and free-living sulfur-oxidizing bacteria. The specific target sequence of the Monterey Beggiatoa sp. (in boldface type) differs from the aligned sequences of all of the other sulfur-oxidizing bacteria by at least two nucleotides. The target of the Thioploca-829 probe is also shown in boldface type for T. chileae. The sequences correspond to variable region helix 29 of the E. coli 16S rRNA secondary structure model predicted by Van de Peer et al. (39). All predict a 12-base stem starting at position 829 and ending at position 857 with a five- to seven-base loop beginning at the aligned gap.
MBSP1RC bound specifically to wide Beggiatoa filaments under fairly stringent hybridization conditions (Fig. 5C). The probe did not hybridize to the occasional narrower Beggiatoa filaments (width, 20 to 30 μm) observed in some preparations. The intensity and yellow color of the hybridization signal were comparable to the intensity and color of the signal obtained when universal eubacterial probe was used with the same samples (Fig. 5B). The most intense fluorescence was observed in the presence of 20% formamide with both MBSP1RC and Eub-338. The filaments in these preparations also hybridized with the GAM42a probe, giving a signal that was stronger than the signal observed with either BET42a or ALF1b (data not shown). As a negative control, nonsense probe MBSP1C did not hybridize with the Beggiatoa rRNA (Fig. 5A), and filaments appeared exactly as if no probe had been added. When a mismatched probe or no probe was added, Beggiatoa filaments appeared orange-red due to autofluorescence conferred apparently by abundant cytochromes (data not shown). The Thioploca sp.-specific probe (Thioploca-829) did not hybridize to Monterey Beggiatoa sp. filaments at temperatures above 37°C (Table 4), but MBSP1RC hybridized with the target filaments at temperatures up to 42 or 43°C (Table 4). Since Monterey Beggiatoa cells are very large by bacterial standards, hybridization experiments revealed some internal details of filaments (Fig. 5D); the central vacuole lacking ribosomes appeared as a clear area when the microscope was focused in mid-filament.
TABLE 4.
FISH of preserved Monterey Beggiatoa filaments with various probes at different temperatures
| Probea | Hybridization atb:
|
||||
|---|---|---|---|---|---|
| 37°C | 39°C | 41°C | 42°C | 43°C | |
| MBSP1RC | ++++c | +++ | +++ | ++ | + |
| Thioploca-829 | ++ | − | − | − | − |
| Eub-338 | ++++ | +++ | NDd | +++ | ND |
| MBSP1C | − | − | − | − | − |
Probes MBSP1RC and Thioploca-829 are putatively specific for the Monterey Beggiatoa sp. and Thioploca spp., respectively (Fig. 4).
All hybridizations were performed in the presence of 20% formamide, and the final concentration of the probe in the hybridization buffer was 5 ng/μl.
The hybridization signal intensity ranged from very strong (++++) to no additional signal (−) compared with the background autofluorescence.
ND, not determined.
DISCUSSION
Phenotypic and phylogenetic comparisons of the genera Beggiatoa and Thioploca.
Until recently, massive natural occurrence of filaments of Beggiatoa or Thioploca spp. have been identified based solely on their characteristic morphologies by using (i) the presence (Thioploca spp.) or absence (Beggiatoa spp.) of a single sheath around multiple filaments and (ii) filament widths as the major criteria (Table 1). No strain of a wide marine Beggiatoa or Thioploca sp. has been obtained in pure culture. The physiological properties of these genera can, therefore, be determined only from observations of natural populations. Such studies have revealed several metabolic similarities. These similarities include chemoautotrophic carbon metabolism (20, 24, 29, 31), sulfide oxidation (20, 25), and concentration of nitrate in the vacuolate cells at levels several-thousand-fold above ambient nitrate levels (5, 24). Teske et al. (38) described the phylogenetic position of vacuolate, unusually wide Thioploca filaments and demonstrated that Beggiatoa spp. were their closest relatives. The study of Teske et al. included only two phenotypically similar, nonvacuolate, narrow (width, 2.8 to 3.0 μm), freshwater Beggiatoa isolates that appear to be obligate chemoheterotrophs (26). No marine strains were analyzed.
In the current study we examined the sequence of a marine or vacuolate Beggiatoa sp. for the first time. Phylogenetic trees (Fig. 3) showed that all of the Beggiatoa spp. examined so far fall into a coherent evolutionary cluster (bootstrap value, 100%) that includes as its only other members species of the genus Thioploca. One of the two clusters identified contains only the narrow, nonvacuolate, freshwater, chemoheterotrophic Beggiatoa strains. Somewhat surprisingly, the second cluster contains a narrow freshwater Thioploca strain in addition to all of the vacuolate, marine Beggiatoa and Thioploca strains whose sequences have been determined. Based on the monophyletic association of the wide marine vacuolate bacteria regardless of the presence of a sheath, an association between the freshwater organism T. ingrica and narrow freshwater Beggiatoa spp. might have been anticipated. The search for a common feature to unify the vacuolate Beggiatoa-Thioploca cluster placed emphasis on the presence of vacuoles in all of the members of the cluster (Table 1) and suggested that there should be a search for nitrate accumulation in T. ingrica.
The finer details of the evolutionary relationships between Thioploca spp. and the wide vacuolate Beggiatoa spp. may still be forthcoming because several additional representatives of the latter group have been identified, including strains whose widths overlap the widths of T. ingrica and T. chileae strains (Table 1). Of special interest is understanding whether these strains form two separate well-defined genera or are actually members of more closely related species of a single genus with variable phenotypic responses (e.g., perhaps they form sheaths only in particular environments). The observed transitions from unsheathed forms (Beggiatoa spp.) to sheathed forms (Thioploca spp.) in the Peruvian upwelling (35) and Monterey Canyon seeps (2) support this suggestion. On the other hand, sheathed forms have never been observed among the wide Beggiatoa spp. of the Guaymas Basin vents or Gulf of Mexico seeps (28). In any case, the taxonomy of the Beggiatoa and Thioploca spp. certainly requires revision. The cluster containing the Monterey Beggiatoa sp., T. araucae, and T. chileae (Table 3) is, on the basis of the criteria of Devereux et al. (3), narrow enough to warrant a single genus. In contrast, the distances between any two Beggiatoa spp. or between T. ingrica and the two other Thioploca spp. (Table 3) are greater than the acceptable range of distances for a coherent genus (3).
FISH, gene copy, and ribosome density.
The low number of OTUs observed (see above) suggests that the sample extracted was dominated by DNA from a narrow range of microorganisms. Based on the in situ hybridization results (Fig. 5), the vacuolate genus Beggiatoa is the dominant OTU and has the corresponding 16S rRNA sequence. At first glance, this might have been expected because microscopic examination revealed that the mat material collected was a virtual monoculture of wide Beggiatoa filaments; the volume of all of the other bacterial biomass was equal to less than 1% of the total Beggiatoa biovolume (24). However, because of the huge size of individual Beggiatoa cells (roughly 75 by 20 μm), much higher filament purity or a much high copy number of the Beggiatoa genome seems to be required to account for our findings. For example, if we assumed that unicellular contaminants (1 by 2 μm; same genome copy number and rRNA operon copy number as the Monterey Beggiatoa sp.) were present at a volume that was equivalent to 0.1% of the Beggiatoa biovolume, the contaminants would be expected (assuming no PCR bias) to contribute six times as many rRNA gene copies as the wide vacuolate Beggiatoa sp.
Due to low signal intensity attributed to low ribosome density, FISH signals of individual Thioploca sp. filaments were often difficult to detect (38). Only amplification of a signal emanating from overlying filaments within a bundle made detection straightforward. In contrast, individual Beggiatoa filaments could be readily detected. The presumptive higher density of the Monterey Beggiatoa sp. ribosomes may reflect optimal growth conditions compared to the Chilean sediments, where growth may be restricted for certain periods of time due to the absence of both nitrate and oxygen.
Specificity and fidelity of probe and sequence.
The 16S rRNA sequence reported here appears to reflect the entire population of the 75-μm-wide, vacuolate Beggiatoa sp. from Monterey Canyon. Three independent PCR performed with the mixed DNA showed that the OTU corresponding to this sequence was always dominant, and the partial 16S rRNA sequences of three random representatives of this OTU were identical. A specific probe was designed for a variable region of the 16S rRNA sequence assigned to the Monterey Beggiatoa sp., and in situ hybridization experiments revealed the specificity of this probe for the target species (the probe hybridized only with the Beggiatoa filaments that were 65 to 85 μm wide and did not hybridize with narrow Beggiatoa filaments or unicellular prokaryotes observed in the samples). The MBSP1RC probe hybridized with the Monterey Beggiatoa sp. under conditions stringent enough to eliminate hybridization of the Thioploca-specific probe (Table 4). Suggesting broader specificity, our probe also hybridized with a 70-μm-wide Beggiatoa sp. collected from sulfide-rich seeps (depth, 600 m) in the Gulf of Mexico.
When we sought evidence of a chimera, we observed no abnormalities in the secondary structure when the sequence assigned to the Monterey Beggiatoa sp. was examined for base complementarity within the helical regions of rRNA (8). In addition, a separate phylogenetic analysis of short sequence domains with the CHECK_CHIMERA program of the Ribosomal Database Project (17) gave negative results. We noted, however, that a chimeric sequence resulting from a fusion between two closely related species might go undetected (27). It has been shown (42) that a higher frequency of chimera formation is expected when very complex DNA is used for PCR. Because our environmental sample was dominated by a single 16S rRNA OTU, a low frequency of chimera formation was expected. We believe that the sequence which we obtained is unique and can be assigned to the wide vacuolate Beggiatoa sp. that dominated the Monterey Canyon sample.
Compared to the available partial sequences of closely related Thioploca spp., the complete 16S rRNA sequence of the wide vacuolate Monterey Canyon Beggiatoa sp. retrieved and examined in this study provides a more complete database for comparison with future sequences derived from natural populations of filamentous sulfur bacteria. In addition, the in situ hybridization studies with fluorescent probes which we performed can be extended to establish differences between single Beggiatoa and Thioploca filaments within mixed natural populations, perhaps revealing correlations between subtle sequence differences and morphological differences (e.g., differences in filament width, the presence or absence of a sheath, or niche differences).
ACKNOWLEDGMENTS
This research was supported by grant IBN-9513962 from the National Science Foundation, by grant UAF96-0059 from the NOAA West Coast National Undersea Research Center, and by a grant from the Monterey Bay Aquarium Research Institute.
We are grateful to Patrick Whaling, the crew of the Pt. Lobos, and the pilots of the remotely operated vehicle Ventana for their assistance and perservance in sample collection.
REFERENCES
- 1.Barry J P, Greene H G, Orange D L, Baxter C H, Robinson B H, Kochevar R E, Nybakken J W, Reed D L, McHugh C M. Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep Sea Res Part A Oceanogr Res Pap. 1996;43:1739–1762. [Google Scholar]
- 2.Buck, K. Personal communication.
- 3.Devereux R, He S H, Doyle C L, Orkland S, Stahl D A, LeGall J, Whitman W B. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol. 1990;172:3609–3619. doi: 10.1128/jb.172.7.3609-3619.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 5.Fossing H, Gallardo V A, Jorgensen B B, Huttel M, Nielson L P, Schulz H, Canfield D E, Forster S, Glud R N, Gundersen J K, Kuver J, Ramsing N B, Teske A, Thamdrup B, Ulloa O. Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature (London) 1995;374:713–716. [Google Scholar]
- 6.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 7.Gallardo V A. Large benthic microbial communities in sulphide biota under Peru-Chile subsurface countercurrent. Nature (London) 1977;268:331–332. [Google Scholar]
- 7a.Genetics Computer Group. Wisconsin package, version 9.1. Madison, Wis: Genetics Computer Group; 1997. [Google Scholar]
- 8.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea. Nature (London) 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 9.Gutell R R, Larsen N, Woese C R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Imhoff J F, Suling J. The phylogenetic relationship among Ectothiorhodospiraceae—a reevaluation of their taxonomy on the basis of 16S rDNA analyses. Arch Microbiol. 1996;165:106–113. doi: 10.1007/s002030050304. [DOI] [PubMed] [Google Scholar]
- 10.Jannasch H W, Nelson D C, Wirsen C O. Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature (London) 1989;342:834–836. [Google Scholar]
- 11.Jannasch H W, Wirsen C O, Nelson D C, Robertson L A. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- 12.Jorgensen B B. Distribution of colorless sulfur bacteria (Beggiatoa spp.) in a coastal marine sediment. Mar Biol. 1977;41:19–28. [Google Scholar]
- 13.Jorgensen B B, Revsbech N P. Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp., in O2 and H2S microgradients. Appl Environ Microbiol. 1983;45:1261–1270. doi: 10.1128/aem.45.4.1261-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 15.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Sequencing techniques in bacterial systematics. London, United Kingdom: John Wiley & Sons Ltd.; 1991. pp. 115–174. [Google Scholar]
- 17.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier S. Description of Thioploca ingrica sp. nov., nom. rev. Int J Syst Bacteriol. 1984;34:344–345. [Google Scholar]
- 19.Maier S, Gallardo V A. Thioploca araucae sp. nov. and Thioploca chileae sp. nov. Int J Syst Bacteriol. 1984;34:414–418. [Google Scholar]
- 20.Maier S, Gallardo V A. Nutritional characteristics of two marine thioplocas determined by autoradiography. Arch Microbiol. 1984;139:218–220. [Google Scholar]
- 21.Maier S, Murray R G E. The fine structure of Thioploca ingrica and a comparison with Beggiatoa. Can J Microbiol. 1965;11:645–663. doi: 10.1139/m65-087. [DOI] [PubMed] [Google Scholar]
- 22.Maier S, Volker H, Beese M, Gallardo V A. The fine structure of Thioploca araucae and Thioploca chileae. Can J Microbiol. 1990;36:438–448. [Google Scholar]
- 23.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 24.McHatton S C, Barry J P, Jannasch H W, Nelson D C. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl Environ Microbiol. 1996;62:954–958. doi: 10.1128/aem.62.3.954-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHatton, S. C. Personal communication.
- 26.Mezzino M J, Strohl W R, Larkin J M. Characterization of Beggiatoa alba. Arch Microbiol. 1984;137:139–144. [Google Scholar]
- 27.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, D. C. Unpublished data.
- 29.Nelson D C. The genus Beggiatoa. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3171–3180. [Google Scholar]
- 30.Nelson D C, Jorgensen B B, Revsbech N P. Growth pattern and yield of a chemoautotrophic Beggiatoa sp. in oxygen-sulfide microgradients. Appl Environ Microbiol. 1986;52:225–233. doi: 10.1128/aem.52.2.225-233.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson D C, Wirsen C O, Jannasch H W. Characterization of large autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl Environ Microbiol. 1989;55:2909–2917. doi: 10.1128/aem.55.11.2909-2917.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruby E G, Jannasch H W. Physiological characteristics of Thiomicrospira sp. strain L-12 isolated from deep-sea hydrothermal vents. J Bacteriol. 1982;149:161–165. doi: 10.1128/jb.149.1.161-165.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schulz H N, Jorgensen B B, Fossing H A, Ramsing N B. Community structure of filamentous, sheath-building sulfur bacteria, Thioploca spp., off the coast of Chile. Appl Environ Microbiol. 1996;62:1855–1862. doi: 10.1128/aem.62.6.1855-1862.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl D A, Amann R A. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- 37.Strohl W R. Genus I. Beggiatoa. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 2091–2097. [Google Scholar]
- 38.Teske A, Ramsing N B, Kuver J, Fossing H. Phylogeny of Thioploca and related filamentous sulfide-oxidizing bacteria. Syst Appl Microbiol. 1995;18:517–526. [Google Scholar]
- 39.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 41.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G C-Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 43.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson K. Miniprep of bacterial genomic DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1990. pp. 2.4.1–2.4.2. [Google Scholar]
- 45.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]