Abstract
We describe useful vectors to select double-crossover events directly in site-directed marker exchange mutagenesis in gram-negative bacteria. These vectors contain the gusA marker gene, providing colorimetric screens to identify bacteria harboring those sequences. The applicability of these vectors was shown by mapping the 3′ end of the Xanthomonas campestris gum operon, involved in biosynthesis of xanthan.
Gene disruption and marker exchange techniques are well-established tools for genetic engineering of bacteria. Site-directed marker exchange methods include several steps: (i) cloning of a gene region into a suicide vector, (ii) disruption of the cloned gene by insertion of a transposon or an interposon cassette, (iii) mobilization of the hybrid plasmid to the recipient strain, (iv) selection of recombinant strains, and (v) identification of double-recombinant strains. In many cases the last step involves tedious work, since the ratio of double-homologous-crossover to single-crossover events can be very low, depending on the organism used. This difficulty can occasionally be overcome by incorporating a copy of the sacB gene coding for the levansucrase enzyme (16–18), whose expression in a sucrose-containing medium leads to a lethal phenotype in some organisms. Unfortunately, in many gram-negative bacteria the expression of this gene in a sucrose-containing medium is not lethal or leads to the growth of small colonies in prolonged incubations (18). To solve this problem, we developed two plasmid vectors containing a marker gene that enables us to discern between single- and double-crossover events without any experimental step.
The gusA gene, originally identified in Escherichia coli (11), is a widespread marker mainly used to study plant gene expression. It encodes the β-glucuronidase enzyme (GUS). The advantages of GUS over other reporter systems include the robustness of the enzyme and the simplicity of the assays. Compared to the products of other reporter genes, GUS is a very stable gene product. This enzyme accumulates over time, and minute quantities of GUS activity can be accurately measured (12). A key advantage of GUS is the generalized absence of this activity within the taxon Eubacteria (25), with the exception of E. coli and some species of Shigella, Streptococcus, Staphylococcus, Corynebacteria, and Clostridium. The incorporation of the gusA gene into the mobilizable suicide vectors pK18mob and pK19mob facilitates discerning between gene replacement and plasmid integration by simply observing the color of the colonies.
Construction of pKmobGII vectors.
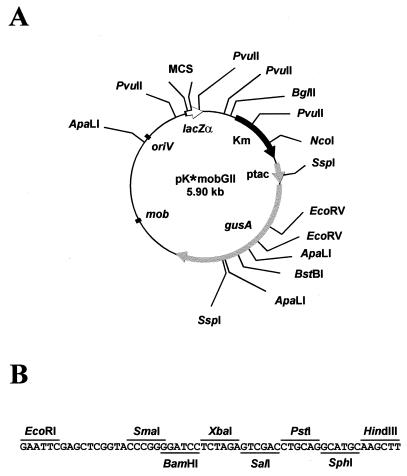
A restriction map of the resulting pKmobGII vectors and the nucleotide sequence of their multiple cloning site (MCS) are shown in Fig. 1. For the construction of the vectors, the gusA gene together with the polyadenylation signal from the nopaline synthase gene (NOS-ter) from the low-copy-number plasmid pBI101 (GenBank accession no. U12639 [12]) were cloned between the HindIII and EcoRI sites of pUC19, giving rise to pDNA4. The tac promoter (GenBank accession no. K01728) was released from plasmid pKK223-3 (5) by digestion with EcoRI and BamHI, and the EcoRI end was filled in. This fragment was cloned into pDNA4, which was previously digested with BamHI and SmaI, originating pTACDNA4. The final plasmids were constructed by cloning the BamHI-SacI fragment, containing the gusA gene from pTACDNA4, into BstBI-digested pK18mob and pK19mob plasmids (GenBank accession no. AF012346 [17]) upon filling in and removing the 3′ and 5′ recessed ends. The additional BamHI site regenerated at the BamHI-BstBI junction was removed by filling in with the Klenow enzyme. These high-copy-number plasmids are 5.9 kb in length and contain the origin of replication of pUC vectors; hence, their host range is restricted to E. coli and a few other species of the Enterobacteriaceae family. The presence of an origin of transfer (mobRK2) enables their transference by conjugation into a wide range of bacteria (17). They also carry the Tn5 kanamycin resistance gene. Selection for this antibiotic resistance does not lead to the formation of satellite colonies and results in a high yield of DNA from standard plasmid preparations (15). The pUC multiple cloning site located within the gene encoding the LacZα peptide enables the application of these vectors as standard cloning vectors.
FIG. 1.
(A) Physical map of pKmobGII vectors. The asterisk symbolizes the plasmid label 18 or 19. (B) Nucleotide sequence of the MCS. Unique restriction sites for cloning purposes are indicated. The enzyme recognition sites and the orientation of the MCS correspond to those in plasmids pK18mob and pK19mob. Km, kanamycin resistance gene (black arrow); mob, origin of transfer; oriV, origin for vegetative replication; lacZα, lac fragment for α-complementation (open arrow); ptac, tac promoter (small gray arrow); gusA, β-glucuronidase gene (large gray arrow).
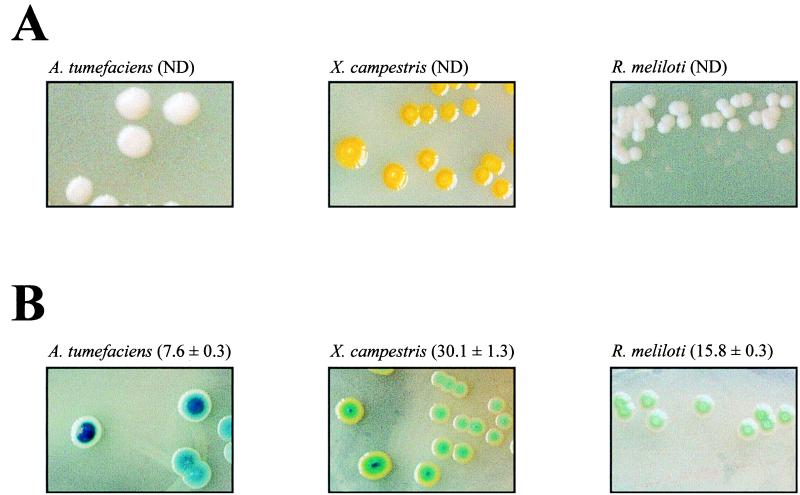
These plasmids were successfully tested in Xanthomonas campestris pv. campestris FC2 (14), Rhizobium meliloti 1021 (1), and Agrobacterium tumefaciens A348 (7). For this purpose, DNA fragments of X. campestris, A. tumefaciens, and R. meliloti were subcloned into pK19mobGII and the resulting hybrid plasmids were transferred to their correspondent wild-type host from the broad-host-range mobilizing strain E. coli S17-1, as described previously (19). Integration of hybrid plasmids into their correspondent host genomes was selected by the vector-encoded antibiotic resistance. Cells carrying pKmobGII sequences integrated into their chromosomes can be easily distinguished from those lacking these sequences in plates containing 50 mg of 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide (X-gluc)/liter or 25 mg of 5-bromo-6-chloro-3-indoxyl-β-d-glucuronide (magenta-gluc)/liter. Phenotypes of X. campestris, R. meliloti, and A. tumefaciens cells carrying or lacking pKmobGII sequences are shown in Fig. 2. GUS activities were assayed by following a protocol originally designed for plants (10). Relative units of GUS for a strain were calculated by using the following formula: (picomoles of methyl-umbelliferone × reaction volume [in milliliters])/(time [in minutes] × OD600 of culture × cell culture [in milliliters]), where OD600 is the optical density at 600 nm.
FIG. 2.
Phenotypes of different strains grown on X-gluc-containing medium. (A) X. campestris FC2, R. meliloti Rm1021, and A. tumefaciens A348 wild-type cells grown for 2 days at 28°C in tryptone-yeast plates (22) containing 50 μg of X-gluc/ml. (B) The same cells harboring pK19mobGII sequences integrated into their genomes were grown as described for panel A in the presence of 50 μg of kanamycin/ml. The GUS activity of each strain is indicated in parentheses. The GUS activities were assayed by employing five independent samples. ND, not detected. The photographs of X. campestris cells were taken after storing the plates for 7 days at 4°C.
Although GUS activity that results from the expression of pKmobGII sequences varies among different hosts (Fig. 2), a correlation between GUS activity and cell color development was not observed. Whereas blue colonies of A. tumefaciens and R. meliloti strains could be observed 2 days after plating, X. campestris cells harboring a copy of pKmobGII needed to be stored at 4°C for a week to become green (Fig. 2). This might be due to differences in the cell wall composition that could make X. campestris cells more impermeable to chromogenic substrates.
The only requisite for the usefulness of these vectors is that the synthesized GUS protein should become accessible to its substrate (X-gluc). There is no need for any specific sugar consumption by the cell for color development. The catalysis of X-gluc might proceed either in the cytoplasm, if the substrate gains access to it, or outside the cell, if the enzyme is released by spontaneous lysis. To test whether the presence of the glucuronide-specific permease gene gusB (11) facilitated color development, this gene was cloned into the broad-host-range vector pRK293 and introduced into a X. campestris mutant with pKmobGII sequences integrated into its chromosome. The presence of the gusB-containing plasmid did not shorten the time in which cells developed a green color.
Mapping of the 3′ end of the gum operon in X. campestris.
The plant-pathogenic bacterium X. campestris produces an acidic exopolysaccharide termed xanthan gum, which is composed of d-glucosyl, d-mannosyl, and d-glucuronyl acid residues in a molar ratio of 2:2:1 and variable proportions of O-acetyl and pyruvyl residues (9, 21). Because of its physical properties, xanthan is widely used as a thickener or viscosifier in both the food and nonfood industries (reviewed in reference 3). The biosynthesis of xanthan mainly consists of a stepwise assembly and decoration of pentasaccharide subunits attached to a polyprenol phosphate carrier, which are subsequently polymerized and exported (8). The enzymes required for these processes are encoded by a 16-kb genome region named xpsI, or gum (14). This region is composed of 12 genes designated gumB to gumM (GenBank accession no. U22511). Transcription of the X. campestris gum gene cluster was proved to be directed by a promoter located upstream of the first gene, gumB (13), but the 3′ end of the operon has not been mapped.
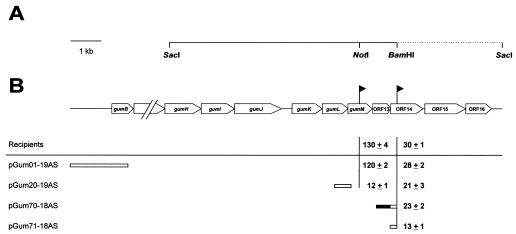
In order to assess whether this operon continues downstream of gumM, we sequenced a 3.6-kb DNA fragment adjacent to the gum region contained in the plasmid pJC372 (J. M. Cleary, Kelco Biopolymers Group of Monsanto Company) (Fig. 3A). This plasmid was obtained from a X. campestris genomic library constructed by cloning total DNA digested with SacI into pJC106, a derivative of the broad-host-range vector RSF1010. DNA sequences were obtained from overlapping nested deletion clones generated by exonuclease III digestion and processed by using an ALFexpress DNA sequencer (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer’s instructions. The nucleotide and amino acid sequences were analyzed by using the MacVector sequence analysis software (Oxford Molecular Ltd., United Kingdom). The amino acid sequences deduced from the nucleotide sequence were compared to the GenBank database with the gapped BLAST algorithm (2). A coding region analysis, using a X. campestris codon usage table, suggested the presence of three open reading frames, which were named, from 5′ to 3′, ORF14, ORF15, and ORF16 (Fig. 3B). While a comparison of the deduced amino acid sequences of ORF14 and ORF16 revealed no homologies in the GenBank database, the deduced sequence of ORF15 was found to be homologous to those of a large family of 3-ketoacyl-acyl carrier proteins involved in the biosynthetic pathway of fatty acids (24).
FIG. 3.
Mapping of the 3′ end of the gum operon. (A) Physical map of the plasmid pJC372. DNA sequences that overlap the gum region (GenBank accession no. U22511) are indicated by a solid line. The dotted line represents the nucleotide sequence of a 3.6-kb DNA fragment determined in this work (GenBank accession no. U70053). The relevant restriction sites are indicated. (B) Genetic map of the cosmid pJC372. The position and orientation of lacZ transcriptional fusions are indicated by solid flags. The relative activities (± standard deviations) of lacZ transcriptional fusions are given in β-Gal units (Miller units) above the line. Below the line, the restriction fragments used for integration mutagenesis are presented. A potential promoter is indicated by a solid box. The relative β-Gal activities (± standard deviations) of the lacZ transcriptional fusions located downstream from the vector integration sites are listed; the activities were calculated from five independent measurements.
To test whether ORF14 is cotranscribed with gumM, we employed a method consisting of the application of lacZ transcriptional fusions combined with plasmid integration (13). A promoterless lacZ-aacC1 interposon (4) was inserted into the unique BamHI site of the SmaI-ClaI fragment (nucleotides 14056 [GenBank accession no. U22511] and 2071 [GenBank accession no. U70053], respectively), which was previously subcloned into pK19mobGII. The hybrid plasmid was transferred from the broad-host-range mobilizing strain E. coli S17-1 (21) to wild-type X. campestris FC2 (14), as described previously (19). Exconjugants were selected in agar medium containing gentamicin, rifampin, and X-gluc. Yellow colonies appeared with a frequency of 10−4 and were sensitive to kanamycin. Correct gene replacement was verified by Southern hybridization. The strain mutated in ORF14 (XcORF14) produced normal amounts of xanthan, as judged by precipitation of the polymer from the broth as previously described (6) (not shown). Also, the biochemical characterization of their lipid-linked intermediates, following established protocols (14), revealed that XcORF14 is capable of both synthesizing and decorating the xanthan lipid-linked repeating unit (not shown). A second mutant in ORF14 was built up by plasmid integration. An internal fragment of ORF14 (nucleotides 763 to 1170; GenBank accession no. U70053) was cloned into the mobilizable suicide vector pK18mob. The resulting plasmid was transferred to X. campestris FC2, and the integration of the hybrid plasmid into the X. campestris genome was selected by the vector-encoded antibiotic resistance and verified by Southern hybridization. The phenotype of this mutant was identical to that of XcORF14, reinforcing the hypothesis that ORF14 might not be related to xanthan biosynthesis. Further studies are required to determine the function of the ORF14 gene product.
To investigate whether ORF14 belongs to the gum operon, subfragments of the gum region were cloned into pK18mob or pK19mob cloning vectors. The gum genes encoded by the cloned fragments were transcribed in the orientation opposite to that of the lacZα promoter located on the vector. The hybrid plasmids were transferred to XcORF14. Integration of these plasmids, by a single-crossover event, caused interruption of the gum region. Transconjugants were verified by Southern hybridization and assayed for their β-galactosidase (β-Gal) activities (Fig. 3B). Integration of pGum01-19AS resulted in strains that displayed β-Gal activities similar to those of the recipient strains without the integrated plasmid (13) (Fig. 3B). Whereas the integration of pGum20-19AS reduced the β-Gal activity mediated by the gumM transcriptional fusion to 10% (13) (Fig. 3B), the β-Gal activity of the XcORF14 strain was not significantly affected by the integration of pGum20-19AS or pGum70-18AS. However, the integration of pGum71-18AS reduced the β-Gal activity of XcORF14 more than 50%. These results suggest that transcription of the gum operon may terminate within a region of 589 bp between the stop codon of gumM and the start codon of ORF14. No clear transcriptional terminators were located in this fragment, although a short open reading frame, not described in the original gum sequence, was found. This open reading frame, whose deduced amino acid sequence displayed no homology to those of proteins of known function, was termed ORF13, and it partially overlaps sequences with weak promoter activity located immediately upstream of ORF14, responsible for the reduced β-Gal activity due to integration of pGum71-18AS. These sequences display no clear homology to the proposed X. campestris consensus promoter sequences (13) or to E. coli ς70 promoters.
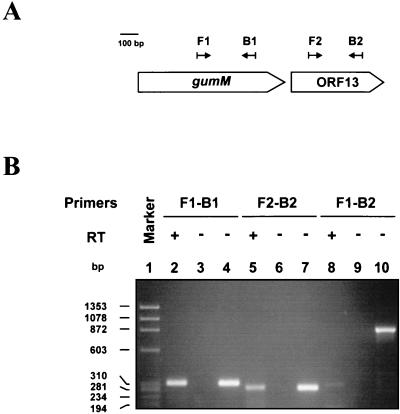
To determine whether ORF13 and gumM belong to the same transcriptional unit, reverse transcriptase (RT) PCR experiments were performed as described previously (23), applying a single amplification round. Whole-cell RNA was isolated from strain FC2 as described previously (13), with a further treatment with RNase-free DNase I (GIBCO BRL, Gaithersburg, Md.). The nucleotide sequences of the primers used were as follows: F1 (5′-CCGACCTGATTCCGTACCTTTG-3′), B1 (5′-GAGAGAAAATCCAGCAAGGCG-3′), F2 (5′-CGCCAGGTGCTGGATGACAG-3′), and B2 (5′-CGAAACCGTGCAGCCCAC-3′). A diagram showing the position and orientation of each primer is presented in Fig. 4A. The right primer of each correspondent pair was employed for reverse transcription. The results are shown in Fig. 4B. While internal sequences of gumM and ORF13 could be RT-PCR amplified (Fig. 4B, lanes 2 and 5, respectively), the expected 893-bp RT-PCR product encompassing both sequences could not be obtained (Fig. 4B, lane 8). These results indicate that ORF13 is transcribed separately from gumM. Therefore, the gum transcript terminates within 418 bp downstream of gumM. These results, in conjunction with previous data (13), show that the gum gene cluster is expressed as a 12-cistron operon, from gumB to gumM.
FIG. 4.
RT-PCR analysis of the 3′ end of the gum operon. (A) Scheme showing the position and orientation of the primers (black arrows) in respect to gumM and ORF13. (B) Ethidium bromide-stained agarose gel (1.8%) of whole-cell RNA-based RT-PCR (lanes 2, 3, 5, 6, 8, and 9) and genome-based PCR (lanes 4, 7, and 10) products amplified with the following primers: F1 and B1 (lanes 2 to 4), F2 and B2 (lanes 5 to 7), and F1 and B2 (lanes 8 to 10). Controls without RT added were included (lanes 3, 6, and 9). DNA size standards (in base pairs) are indicated (lane 1). +, present; −, absent.
Nucleotide sequence accession number.
The GenBank accession number of the DNA fragment determined in this work is U70053.
Acknowledgments
We are very grateful to Jürgen Brosius, Andreas Schäfer, Joseph M. Cleary, and Mario Aguilar for providing pKK223-3, the pKmob vectors, pJC372, and the strain Rm1021, respectively. We thank Pablo Cerdán for assistance in GUS assays and A. J. Parodi for critically reading the manuscript. Photographs were kindly taken by Eduardo J. Rattner.
F.K. acknowledges Ph.D. grants from Universidad de Buenos Aires. Work at the L.I. laboratory was partly supported by grants from Universidad de Buenos Aires and from CONICET. L.I. is a member of Carrera del Investigador (CONICET, Buenos Aires, Argentina).
REFERENCES
- 1.Aguilar O M, Kapp D, Pühler A. Characterization of a Rhizobium meliloti fixation gene (fixF) located near the common nodulation region. J Bacteriol. 1985;164:245–254. doi: 10.1128/jb.164.1.245-254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker A, Katzen F, Pühler A, Ielpi L. Xanthan gum biosynthesis and application: a biochemical-genetic perspective. Appl Microbiol Biotechnol. 1998;50:145–152. doi: 10.1007/s002530051269. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Schmidt M, Jäger W, Pühler A. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene. 1995;162:37–39. doi: 10.1016/0378-1119(95)00313-u. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadmus M C, Rogovin S P, Burton K A, Pittsley J E, Knutson C A, Jeanes A. Colonial variation in Xanthomonas campestris NRRL B-1459 and characterization of the polysaccharide from a variant strain. Can J Microbiol. 1976;22:942–948. doi: 10.1139/m76-136. [DOI] [PubMed] [Google Scholar]
- 7.Castro O A, Zorreguieta A, Ielmini V, Vega G, Ielpi L. Cyclic β-(1,2)-glucan synthesis in Rhizobiaceae: roles of the 319-kilodalton protein intermediate. J Bacteriol. 1996;178:6043–6048. doi: 10.1128/jb.178.20.6043-6048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ielpi L, Couso R O, Dankert M A. Sequential assembly and polymerization of the polyprenol-linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris. J Bacteriol. 1993;175:2490–2500. doi: 10.1128/jb.175.9.2490-2500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson P E, Kenne L, Lindberg B. Structure of extracellular polysaccharide from Xanthomonas campestris. Carbohydr Res. 1975;45:275–282. doi: 10.1016/s0008-6215(00)85885-1. [DOI] [PubMed] [Google Scholar]
- 10.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Reporter. 1987;5:387–405. [Google Scholar]
- 11.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jefferson R A, Kavanagh T A, Bevan M W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzen F, Becker A, Zorreguieta A, Pühler A, Ielpi L. Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J Bacteriol. 1996;178:4313–4318. doi: 10.1128/jb.178.14.4313-4318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katzen F, Ferreiro D U, Oddo C G, Ielmini M V, Becker A, Pühler A, Ielpi L. Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol. 1998;180:1607–1617. doi: 10.1128/jb.180.7.1607-1617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 16.Sarker M R, Cornelis G R. An improved version of suicide vector pKNG101 for gene replacement in gram-negative bacteria. Mol Microbiol. 1997;23:410–411. doi: 10.1046/j.1365-2958.1997.t01-1-00190.x. [DOI] [PubMed] [Google Scholar]
- 17.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 18.Selbitschka W, Niemann S, Pühler A. Construction of gene replacement vectors for Gram− bacteria using a genetically modified sacRB gene as a positive selection marker. Appl Microbiol Biotechnol. 1993;38:615–618. [Google Scholar]
- 19.Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196:413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- 20.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 21.Stankowski J, Mueller B, Zeller S. Location of a second O-acetyl group in xanthan gum by the reductive-cleavage method. Carbohydr Res. 1993;241:321–326. doi: 10.1016/0008-6215(93)80123-v. [DOI] [PubMed] [Google Scholar]
- 22.Steinmann D, Wiggerich H G, Klauke B, Schramm U, Pühler A, Priefer U B. Saturation mutagenesis in Escherichia coli of a cloned Xanthomonas campestris DNA fragment with the lux transposon Tn4431 using the delivery plasmid pDS1, thermosensitive in replication. Appl Microbiol Biotechnol. 1993;40:356–360. doi: 10.1007/BF00170392. [DOI] [PubMed] [Google Scholar]
- 23.Swartley J S, Ahn J H, Liu L J, Kahler C M, Stephens D S. Expression of sialic acid and polysialic acid in serogroup B Neisseria meningitidis: divergent transcription of biosynthesis and transport operons through a common promoter region. J Bacteriol. 1996;178:4052–4059. doi: 10.1128/jb.178.14.4052-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai H, Post-Beittenmiller D, Jaworski J G. Cloning of a cDNA encoding 3-ketoacyl-acyl carrier protein synthase III from Arabidopsis. Plant Physiol. 1994;106:801–802. doi: 10.1104/pp.106.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson K J, Hughes S G, Jefferson R A. The Escherichia coli gus operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In: Gallagher S R, editor. GUS protocols: using the gus gene as a reporter of gene expression. San Diego, Calif: Academic Press; 1992. pp. 7–22. [Google Scholar]