Abstract
Chemotherapeutic agents, such as 5-fluorouracil (5-FU) and oxaliplatin (Oxi), can not only kill the cancer cell but also influence the proliferation of gut microbiota; however, the interaction between these drugs and gut microbiota remains poorly understood. In this study, we developed a powerful framework for taxonomy composition and genomic variation analysis to investigate the mutagenesis effect and proliferation influence of chemotherapeutic agents, such as 5-FU and Oxi, on gut microbiota and the interaction between these drugs and gut microbiota during chemotherapy. Using the gut microbiome data, we detected 1.45 million variations among the chemotherapy groups and found the drugs significantly affected mutation signatures of gut microbiota. Oxi notably increased transversion rate, whereas 5-FU reduced the rate. Traits related to cell division and nutrient mobilization showed evidence of strong selection pressure from chemotherapeutic agents. In addition, drug-associated bacteriome shift patterns and functional alterations were found: the metabolism changes in the 5-FU group implied that gut microbiota could provide additional nicotinamide adenine dinucleotide (NAD+) to inhibit cancer cell autophagy; in the Oxi group, the ribosome and lysine biosynthesis genes were obviously enriched. Our study provides a blueprint for characterizing the role of microbes and drug–microbe interaction in the gut microbiota response to chemotherapy.
Keywords: chemotherapeutic agent, gut microbiota, microbes, gene mutation, genomic variation analysis
Introduction
Chemotherapeutics have long been used to treat a variety of human tumors (Kemp and Kwon, 2021). As one of the commonly used chemotherapy protocols, the FOLFOX regimen contains different chemotherapeutic agents [e.g., 5-fluorouracil (5-FU) and oxaliplatin (Oxi)] with a broad range of cytotoxicity (de Gramont et al., 2000). These drugs can effectively inhibit DNA replication of cancer cells via different mechanisms of action. For example, 5-FU produces FdUMP, which directly inhibits thymidylate synthase and thus causes thymine-less cell death (Pinedo and Peters, 1988). In addition, the products of 5-FU may be converted into FUTP or FdUTP, leading to RNA or DNA damage, respectively. An understanding of the interaction of chemotherapeutics and gut microbial populations will probably trigger applications toward improving the effectiveness of chemotherapy (Yuan et al., 2018).
Chemotherapeutic agents may affect gut microbial communities by disrupting the homeostatic balance among resident microorganisms; meanwhile, they can accelerate the microbe evolution at a molecular level (Montassier et al., 2015; Baym et al., 2016; Hong et al., 2019). Many recent studies have explored the interaction between microbiota and anticancer drugs along with interventions aimed at shaping microbiota to optimize drug efficacy and reduce side effects (Cheng et al., 2020). However, previous studies mainly focused on the taxonomic characterization and functional compositions of different cohorts at the genus or species level (Lloyd-Price et al., 2017). Variations in taxonomic abundance as well as functions encoded by these gut microbiota have been described in the cohorts of inflammatory bowel disease (IBD) (Halfvarson et al., 2017), type 2 diabetes (T2D) (Qin et al., 2012), hypertension (Li et al., 2017), and liver cirrhosis (Qin et al., 2014). However, genomic variations within species, which lead to their phenotypic diversity and adaptations to chemotherapeutic agents, have been studied in only a few taxa (Morowitz et al., 2011). For example, in the common gut commensal bacteria Escherichia coli (Bagel et al., 1999), only few point mutations can confer clinically relevant antibiotic resistance, and the natural variation in a single gene can lead to pathogenic adaptation.
Given the importance of the gut microbiota in human health and a growing number of studies reporting associations between gut microbiota and diseases (Hauser et al., 2015), a better understanding of the interaction between gut microbiota and chemotherapeutic drugs during chemotherapy helps to improve the efficacy of the anticancer chemotherapeutic agents. We recruited 37 cancer patients and collected prechemotherapy and postchemotherapy stool samples to study the taxonomic composition, metabolic capacity, and molecular evolution during the chemotherapy. In order to avoid the bias caused by distinct diet, regional, and genetic differences, we collected healthy samples from well-known studies about Han Chinese with the standard clinical assessments and same sequence platform to build the basic of microbiome structure and mutational pattern. We analyzed our cohort and stool samples from T2D (Qin et al., 2012), liver cirrhosis (Qin et al., 2014), and hypertension cohorts (Li et al., 2017) with the same bioinformatics pipeline and parameters. Our goal was to build a power framework for metagenomics analysis to gather basic knowledge on the genomic variation landscape and taxonomic abundance shift in gut microbiota metagenomes during chemotherapy.
Materials and Methods
Cohort Recruitment and Sample Collection
Participants were recruited in the Oncology Department of Beijing Hospital, Beijing, China, in 2018. This study was approved by the Beijing Hospital Ethics Committee. Written informed consent was obtained from all of the participants. The exclusion criteria included the following: with a history of IBDs; having been exposed to probiotics, prebiotics, or broad-spectrum antibiotics within 30 days; or having received nasal-tube feeding or parenteral nutrition in the month prior to initiation of the study. Detailed information about all patients is listed in Supplementary Table 1. We collected two fecal samples from each participant. A fecal sample was collected on hospital inpatient admission (day 0), prior to administration of chemotherapy, and 30 days later immediately prior to chemotherapy (day 30), respectively. For each fecal sample, 1 g of stool was collected into a sterile tube and then subsequently stored at –80°C for molecular analysis. Total genomic DNA was extracted from 200 to 500 mg of fecal sample using a QIAamp DNA Stool Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. The concentration of the extracted DNA was measured by a NanoDrop2000 (Thermo Fisher Scientific, United States), and then the DNA was stored at –80°C for the next studies.
16S Ribosomal RNA Sequencing and Bioinformatics Analysis
To develop the 16S rRNA amplicon libraries, the V3–V4 region of 16S rRNA gene was polymerase chain reaction–amplified with the primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′), modified by adding barcodes for multiplexing (Caporaso et al., 2012). Pooled amplicons were paired-end sequenced [PE 2 × 250 base pairs (bp)] by using an Illumina HiSeq 2500 platform according to the manufacturer’s protocol. Paired-end reads were merged and quality filtered using FLASH and QIIME (Bokulich et al., 2013). Chimeras were detected and removed against the Gold reference database using the UCHIME algorithm (Edgar, 2013). Sequences with ≥ 97% similarity were assigned to the same OTUs. For each representative sequence, the GreenGenes Database was used based on the RDP classifier algorithm to annotate. α Diversity analysis including Shannon and Chao1 was calculated with QIIME. Phylogenetic β diversity distances, including unweighted and weighted UniFrac distances, were measured using QIIME. Principal component analysis was performed to visualize by ggplot2 package in R software (version 3.5.0). Further, diversity analyses, such as Adonis and Anosim, were performed by running a workflow on QIIME.
Shotgun Metagenomics Sequencing Library Construction
For shotgun metagenomics sequencing, adaptive focused acoustics (Covaris) was used to shear a standard volume of 50 μL of the extracted DNA. DNA libraries were prepared using Illumina TruSeq Sample Preparation Kit (Illumina, United States) according to the manufacturer’s protocol. The DNA libraries were quantified using an Agilent Bioanalyzer 2100 (Agilent Technologies, United States). Paired-end sequencing (PE 2 × 150 bp) was performed on successful DNA libraries using an Illumina HiSeq X-Ten instrument at the Annoroad Genome Biotech in Beijing. Finally, we obtained 4.27–8.9 GB of raw data for each sample (average, 5.98 GB; Supplementary Table 2).
Metagenomics Bioinformatics Analysis
Concatenated and human DNA sequences were removed using KneadData with default parameter.1 The remaining high-quality reads were assembled with IDBA-UD (version 2.04, parameters: -pre_correction -min_contig 200) (Peng et al., 2012). We mapped the clean reads against scaffolds using Bowtie2 (version 2.3.4.2) (Langmead and Salzberg, 2012). Genes (minimum length of 100 nucleotides) were predicted on contigs longer than 500 bp and annotated using Prokka (Parameters: -metagenome) (Seemann, 2014). The gene abundance was determined by using a method similar to RPKM (reads per kilo bases per million reads) used for quantifying gene expression from RNA sequencing data. In brief, high-quality reads were counted with HTSeq-count (Anders et al., 2015). For each gene, Gi, the number of reads that aligned to it divided by the length of the gene was calculated as NGi, and the relative abundance, RNGi, of each gene in each sample (n genes) was computed using the following formula:
Species-level quantitative taxonomic profiling was performed using MetaPhlAn2 (version 9760413b180f) on the KneadData-filtered reads (Truong et al., 2015). Community composition was calculated with MetaPhlan2 using the default settings. Taxonomic profiles including bacteria, archaea, microbial eukaryotes, and viruses were inferred by MetaPhlAn2 using the 1M unique clade-specific marker genes identified from 17,000 reference genomes (13,500 bacterial and archaeal, 3,500 viral, and 110 eukaryotic).
Functional Annotation
Two different methods were used for functional alteration and pathway composition analysis. Pathway-level composition was calculated with HUMANn2 using the UniRef90 database with default settings, which was followed by further statistical analysis and visualization in STAMP (Parks et al., 2014). In order to obtain the detailed information of each metabolic pathway, all genes in our genomes were aligned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using DIAMOND (version 0.7.9.58, parameter: -k 50 -sensitive -e 0.00001) and KOBAS (Xie et al., 2011). Each protein was assigned to the KEGG Orthology (KO) families by the highest-scoring annotated hits containing at least one HSP scoring over 60 bits. The abundance of KO/module was calculated using the methods as mentioned previously. For KO enrichment analysis, the significant enriched KEGG pathway was defined as the adjusted p-value (< 0.05) of the hypergeometric test as the previous study (Jiang et al., 2016).
Generation of a Reference Genome Set
We followed the previous method to build the reference genomes to study the genomic variation of gut microbiota (Schloissnig et al., 2013). Approximately 3,934 prokaryotic genomes were downloaded from NCBI2 on August 1, 2018. A set of 40 universal single copy marker genes was identified in these genomes using HMM profiles made for each marker gene from the corresponding orthologous group from the eggnog database (Ciccarelli et al., 2006; Sorek et al., 2007). For each marker gene, pairwise DNA sequence identities among all genomes were calculated using BLASTn (version 2.2.25, parameter: e < 10–5 -F F) (Altschul et al., 1990). For each genome pair, the median identity of all marker genes was used as a proxy for average nucleotide identity (ANI) between the two genomes. Using an operational 95% ANI recommended for identifying species, we generated 929 clusters of genomes. In order to select a reference genome from each cluster, high-quality reads from a subset of shotgun metagenomes (bgi-BGI-06A, bgi-DLF001, bgi-DOF002 from T2B cohort; nHF611710, nHF411719 from HTN; HD-2, HD-31 from liver; 1-1, 4-1, 5-1 from this study) were mapped to the 3934 genomes using Bowtie2 with the options “–very-fast.” Then, the genome with the highest read coverage was selected, resulting in a set of 958 reference genomes, each likely representing a unique species (Supplementary Table 7).
Mapping to Reference Genomes and Variant Calling
Illumina reads from 718 fecal samples, including the T2D, liver, and hypertension cohorts were quality controlled using KneadData. KneadData-filtered reads were mapped to reference genomes using Bowite2 (version 2.3.4.2) with default parameter. The number of reads mapping to reference genomes was counted and normalized by the genome size in order to obtain quantitative relative abundances of each genome in every sample.
To improve the accuracy of genomic variation, we modified the protocol of the previous study (Schloissnig et al., 2013). The SAMtools/BCFtools suite was used for calling high-quality InDel supported by more than five reads and at least one read on each strand (Li et al., 2009). We then performed local realignment of the aligned reads with those high-quality InDels using the Genome Analysis Toolkit (GATK) (McKenna et al., 2010) to minimize the mismatch bases. GATK’s HaplotypeCaller was used to call variants with default parameter. BreakDancer was used to detect the structural variation with at least three reads supporting the event (Fan et al., 2014).
Positive Selection Analysis
Coding DNA sequences (CDSs) were annotated based on their protein family membership using KOBAS (Xie et al., 2011). With MUSCLE (Edgar, 2004), a multiple sequence alignment (MSA) of the protein sequences was created using each KO. Based on the MSA and the CDS nucleotide sequences, a codon-based alignment was constructed for each KO module with PAL2NAL using default parameters (Suyama et al., 2006). We then used FastTree, a relaxed neighbor joining algorithm, to reconstruct a phylogenetic tree for each protein family from the obtained MSA (Price et al., 2009). We excluded low-confidence positions in the alignment with a large number of gaps with Gblocks (Castresana, 2000). Dn/Ds was calculated with PAML (Yang, 2007). A one-sided Fisher test was performed to identify protein families with a significant enrichment of Dn vs. Ds changes in comparison to the entire sample. The false discovery rate (FDR) was controlled using the Benjamini and Hochberg procedure and α set to 5%.
In vitro Experiment to Validate the Mutational Pattern Shift
A filter-sterilized stock solution of 100 mM 5-FU, Oxi, and 5-FU + Oxi (1:1) (Sigma–Aldrich) was prepared in dimethyl sulfoxide and further diluted to 50, 75, and 100 mM. Stock solutions were further diluted (1:1,000) in culture medium for the experiments. Four mono-isolated gut microorganisms were obtained from the stools of recruited patients. Luria broth (LB) agar plates used for passaging were prepared by adding stock 5-FU, Oxi, and 5-FU + Oxi. Subsequently, cultures were transferred into fresh plates and allowed to grow 48 h for each batch. After 20 batches, single colony on each plate was then scraped off and transferred to Eppendorf tubes with liquid LB medium and cultured overnight at 37°C. Total DNA was then extracted using the QIAamp UCP Pathogen Mini Kit (Qiagen). The concentration of the extracted DNA was measured by a NanoDrop2000 (Thermo Fisher Scientific, United States). Illumina DNA sequence libraries were prepared as mentioned previously. JSpeciesWS was used to assign the taxonomy of each isolated strain (Richter et al., 2016). Reads were then mapped to the closest reference genomes with BWA, and variant calling was performed as mentioned previously.
Statistical Analysis
The Wilcoxon signed-ranks test was performed to evaluate differences in richness and diversity for KEGG and microbial taxa at various taxonomic ranks. Mutation rate comparisons were conducted to determine statistical significance between two groups for incidences where differences were declared significant at p < 0.05 with the Student t-test.
Data Availability
All sequences generated in this study have been available in the NCBI sequence read archive under the accession number PRJNA551354.
Results
Overall Composition of Gut Microbiota During Chemotherapy
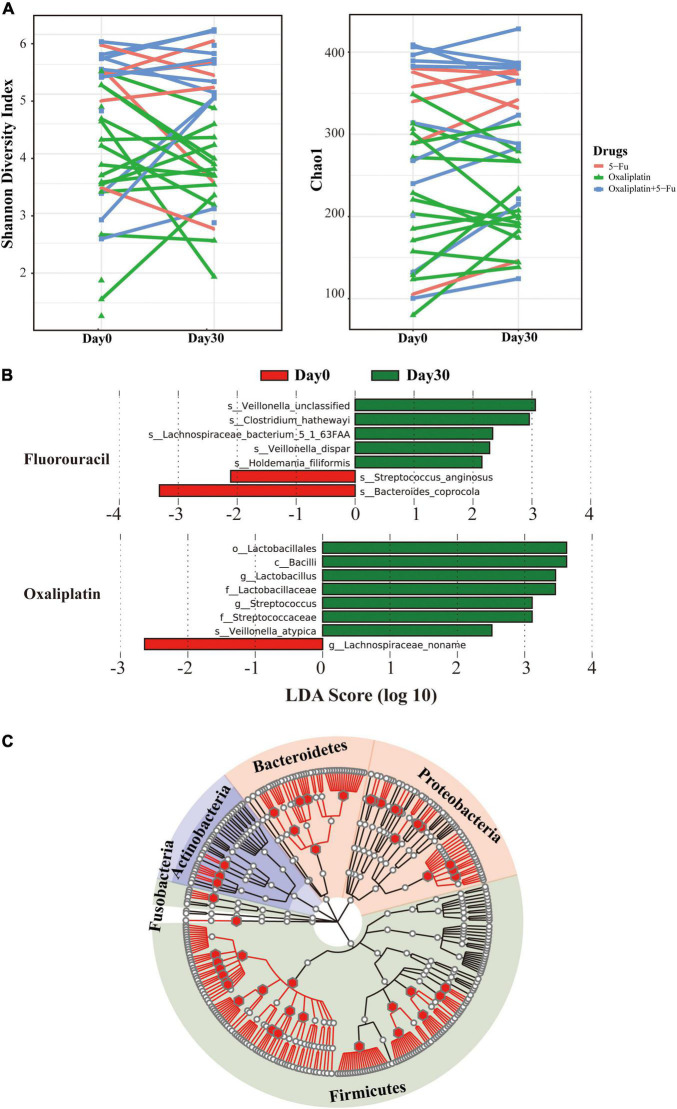
To characterize the diversity and composition of gut microbiome during the chemotherapy of the patients, we characterized the gut microbial composition of 68 fecal samples from 37 patients (six samples were excluded because of DNA extraction failure) using 16S rRNA gene sequencing. Detailed information about stool samples and patients is listed in Supplementary Tables 1, 2. This cohort consisted of 7 patients using 5-FU, 13 patients using 5-FU + Oxi, and 17 patients using Oxi. All the participants were from a cohort study in Beijing Hospital. Using the samples collected at the start of chemotherapy (day 0) and 30 days later (day 30), we traced the dynamic changes of gut microbial species by ecological α and β diversity measures. The chemotherapeutic drugs caused modest changes in the gut microbiome. Shannon diversity and Chao1 index were calculated to estimate the within-sample (α) diversity (Figure 1A), and there was no significant alteration pattern. Weighted UniFrac principal coordinate analysis was used to compare community phylogenetic composition among samples, and the results revealed a separation between days 0 and 30 along axis 1 (explaining 41.5% of the variation in the data, Supplementary Figure 1). β Diversity represented how much the community changed in comparison to the baseline (day 0) and was calculated with weighted UniFrac distance between days 0 and 30 (Supplementary Table 3). ANOSIM with permutations confirmed significant separation of samples (p = 0.02, r = 0.24). No significant differences in microbial communities were observed when the Oxi and 5-FU + Oxi groups were analyzed separately. Although the adjusted p (adjusted p = 0.06) was not significant, it indicated that there was a difference pattern in the microbial composition between days 0 and 30 with 5-FU. Next, we used linear discriminant analysis (LDA) effect size (LEfSe) to analyze the microbial communities of different chemotherapeutic drugs. For all day 30 samples, we identified gut microbiome signatures with a higher abundance of bacilli (specifically Streptococcaceae and Lactobacillaceae, Figure 1B). For 5-FU, seven species were discovered as biomarkers for separating gut microbiota between days 0 and 30 (Supplementary Figure 2). Five of these species were higher, and two were lower on day 0 than on day 30. For example, the abundances of Bacteroides coprocola and Streptococcus anginosus were lower on day 30 (p = 0.03, two-sided Wilcoxon signed-ranks test). Contrary to 5-FU, in the Oxi group, eight of these species were higher, and one was lower on day 0 than on day 30. No significant changes were observed in the 5-FU + Oxi group. To gain further insights into the interaction between chemotherapeutic drugs and gut microbiota, we used the same gut microbiota DNA preparations for independent Illumina shotgun sequencing. The yielded sequences belonged to 10 phyla and 381 species, including several DNA viruses (Supplementary Table 4 and Supplementary Figure 3). Firmicutes was the most abundant phylum, accounting for 40.9% ± 22.3% of the total reads, followed by Bacteroidetes (36.3% ± 23.7%) and Proteobacteria (15.7% ± 23.4%), similar to previous studies (Qin et al., 2014). Two commensal species including Streptococcus salivarius and Ligilactobacillus salivarius were significantly enriched after the chemotherapy (Supplementary Figure 4). Correlation tests between the abundance estimates for bacterial taxa at genus level were performed by using the two methods (Pearson correlation coefficient: 0.84; p < 2.2 × 10–16; Figure 1C and Supplementary Figure 5), highlighting a highly positive correlation between 16S rRNA and shotgun metagenomics sequencing.
FIGURE 1.
Gut microbial diversity before and after the chemotherapy. (A) Shannon diversity (left) and Chao (right) based on 16S rRNA sequencing. Different colors stand for drugs: red for 5-fluorouracil, green for oxaliplatin, and blue for 5-fluorouracil and oxaliplatin. The y-axis stands for the collection time. (B) Cladogram derived from LEfSe analysis of metagenomic sequences based on the shotgun sequencing comparing day 0 with day 30. Green shaded areas indicate microbe orders that more consistently describe the fecal microbiome from day 30; red shaded areas indicate microbe orders that more consistently describe from day 0. The prefixes “c,” “o,” “f,” “g,” and “s” represent the annotated level of class, order, family, genus, and species. (C) Comparison of 16S rRNA and metagenome abundances. The tree represents all taxonomically classified species from the shotgun metagenome survey as well as 16S rRNA sequence. The branches of the tree do not reflect evolutionary distances. The position of the dots in the tree corresponds to the taxonomic placement of the representative sequences in the NCBI taxonomy. Empty dots represent the phylotypes found in the shotgun metagenome classification; red dots were identified from both methods.
Functional Alterations in Gut Microbiota During the Chemotherapy
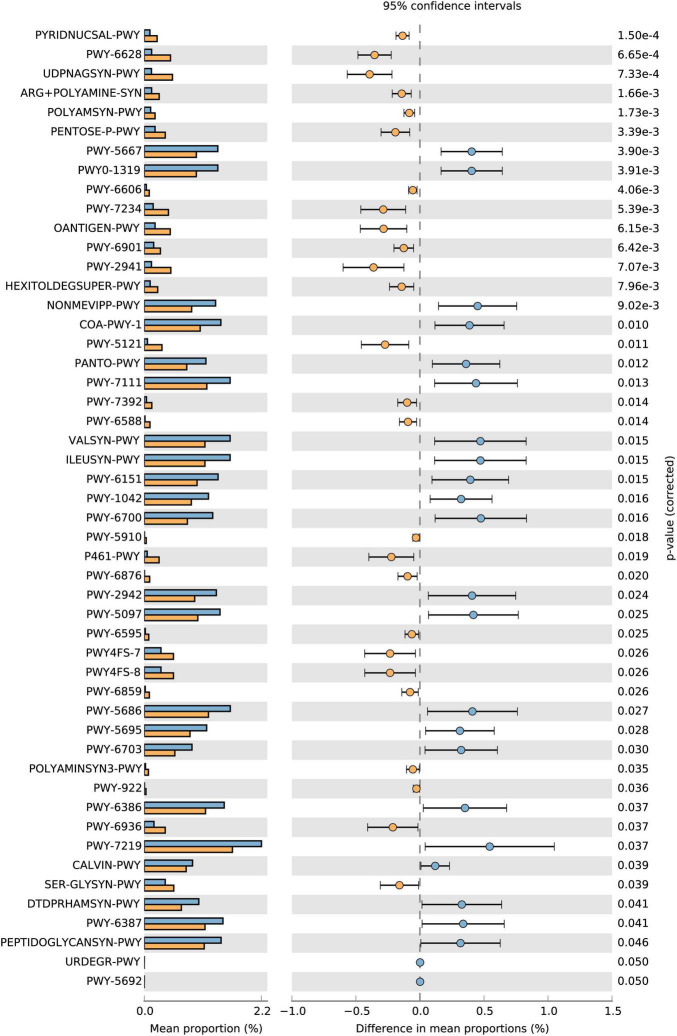
To determine the functional alterations with the chemotherapeutic agents, we used the HUMAnN2/MinPath (Franzosa et al., 2018) and KEGG database (Kanehisa et al., 2017) to evaluate gut microbial functions across groups in our study cohort. According to the HUMAnN2/STAMP analysis of the metabolic function pathways (Parks et al., 2014), most differences occurred in carbohydrate metabolism and energy-related pathways in the 5-FU group (Figure 2). The microbiome from day 30 included more genes involved in nicotinamide adenine dinucleotide (NAD) salvage pathway (PYRIDNUCSAL-PWY) than that from day 0 in the 5-FU group (adjusted p = 1.4 × 10–4). A previous study suggested an active role for the NAD salvage pathway in modulating cancer cell viability via the replenishing of the NAD reservoir (Sharif et al., 2016), which is a novel strategy to protect the cell against DNA damage. Moreover, amino acid biosynthesis, such as L-phenylalanine and arginine, UDP-N-acetyl-D-glucosamine biosynthesis pathway were more active in the samples in the 5-FU group. In the other two groups, few pathways were found to be significantly different before and after chemotherapy (Supplementary Figure 6).
FIGURE 2.
Extended error bar plots showing the abundance of pathways differing significantly between days 0 and 30 in the 5-FU group. Extended error bar plots showing the abundance of pathways differing significantly between days 0 and 30 in the 5-FU group. Corrected p-values are shown on the right. Adjusted p < 0.05 was considered significant. The yellow color stands for day 30 and blue for day 0.
In order to deeply explore the functional role of the gut microbiota during the chemotherapy, all the genes were aligned to the KEGG database, and the abundance of KO groups and pathways for each sample were estimated using a method similar to RPKM (Hu et al., 2013). Approximately 277 KOs were significantly enriched among the three groups (p < 0.05, two-sided Wilcoxon rank sum test; Supplementary Table 5). Overrepresentation of three KEGG pathways involved in metabolism was observed in the 5-FU group with hypergeometric distribution, which is involved in the categories “galactose metabolism” and “phosphotransferase system (PTS)” (Supplementary Table 6). Enriched genes encoding PTS, which reflected increased representation of multiple carbohydrate transporters, were an important mechanism to confer resistance to selection pressure, such as antibiotic, as reported in the previous studies (Stogios et al., 2016). In the Oxi group, two KEGG pathways “ribosome” and “lysine biosynthesis” were significantly enriched. Although Oxi, a platinum-based drug, is generally considered to be DNA-damaging agents, recent studies have unexpectedly shown that Oxi did not alter DNA integrity but instead inhibited rDNA transcription, leading to p53 induction, most likely through the “impaired ribosome biogenesis checkpoint” (Pelletier et al., 2018).
Genomic Variation in Intestinal Microorganisms
To enable comparative analyses in multiple metagenomes and to identify the mutation signatures in the different groups, we modified the previous method (Schloissnig et al., 2013) to identify single-nucleotide polymorphisms (SNPs), short insertions/deletions (InDels; range, 1–50 bp), and structural variations (>50 bp) in each sample. We used 3,934 prokaryotic genomes to generate a set of reference genomes (Supplementary Table 7) for the analysis of genomic variation in gut microbial species in 786 samples. Next, local realignment around high-quality InDels, which was a frequently used strategy in genomic variation analysis, was performed to correct mapping errors to improve the accuracy of variant calling (DePristo et al., 2011). We only considered variants with allele frequency larger than 1% and supported by more than five reads. To validate our SNP calling procedure, we used the approaches to calculate error rates in 40 essential single-copy marker genes, following the method mentioned previously (Schloissnig et al., 2013). False-positive rates were estimated at 3.6% (Supplementary Table 8).
We identified 1.45 million SNPs from 343 genomes (at least three SNPs for one genome), of which 1.38 million SNPs (95%) in 47 genomes (0.88% of the total 156 Mb) across 68 stool samples from 37 subjects (Supplementary Figure 7). We also identified 26,662 InDels and 56,820 structural variants. Subsequent analyses were restricted to SNPs because of their orders-of-magnitude higher count over other variation types. Examination of the SNP distributions of the protein coding genes of the selected 47 gut bacteria revealed that 4,189 genes with at least one non-synonymous mutation in the 47 microbe species had valid coverage (≥ 10 × depth) with sufficient prevalence (Supplementary Table 9). Among them, we identified 22 genes (0.5%) with significantly differentiated SNP densities between days 0 and 30 samples (Wilcoxon signed-ranks test, p < 0.05). Most of these genes (17/22) were found in Bacteroides thetaiotaomicron VPI-5482, the second most common human commensal bacteria, whose reference genome contains 4,779 protein-coding genes (Xu et al., 2003; Supplementary Table 10).
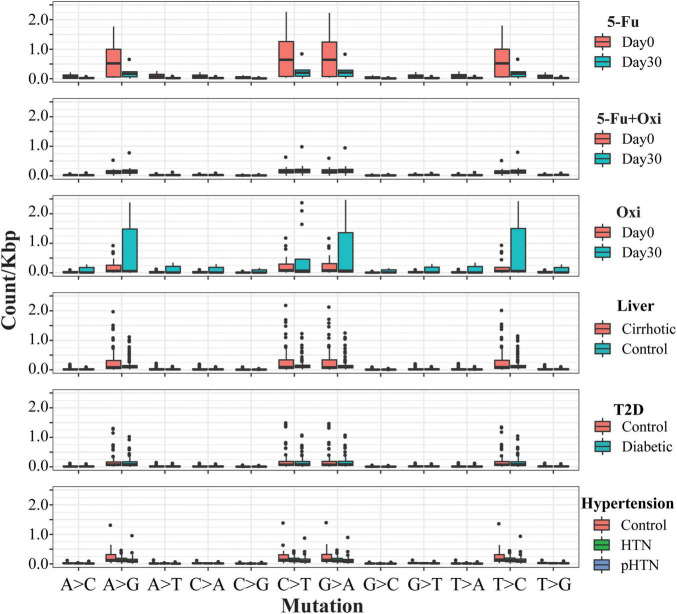
Then, we checked the substitution pattern shift between days 0 and 30. A large portion of base substitutions was attributable to C > T substitution, which ranged from 29.5 to 43.0% on day 0 and from 35.0 to 42.3% on day 30. Comparisons of the mutation rates in the other Chinese cohorts, including liver cirrhosis (Qin et al., 2014), hypertension (Li et al., 2017), and T2D (Qin et al., 2012; Figure 3 and Supplementary Table 11), showed a significantly reduced transversion (Student’s t-test, adjusted p = 0.0003) and transition rate (Student’s t-test, adjusted p = 0.01) in the 5-FU group (Supplementary Table 12). By contrast, we found an increased ratio of transversion (Student t-test, adjusted p = 5.4 × 10–4) and transition (Student’s t-test, adjusted p = 8.2 × 10–7) in the group of Oxi. The similar mutational pattern was also observed in the studies about the effect of antibiotic on the genome (Long et al., 2016). Thus, we conclude that genome-wide substitutions rate of gut microbiota were influenced by chemotherapeutic drugs. To directly test whether selection might have biased the mutation rate of different regions in the genomes, we examined the synonymous and non-synonymous mutation from the coding region from all cohorts. Non-synonymous/synonymous mutation ratio from day 30 was not significantly different from day 0 (Student’s t-test, p > 0.05 in all comparison), indicating that the vast majority of acquired amino acid–altering mutations was not selectively promoted by chemotherapeutic agents but simply accumulated in a neutral fashion.
FIGURE 3.
Mutational signatures found in all fecal samples. Each cohort is displayed according to the 12 substitutions. The different groups are displayed in different colors as the legends show. The mutation types are on the horizontal axes, whereas vertical axes depict the number of mutations detected in the kilobase attributed to a specific mutation type.
In order to validate the shift of mutational pattern, we explored how stool-isolated microorganism mutation rates change in vitro when treated with 5-FU and Oxi. We exposed four gut species (two E. coli strains, Citrobacter sp. MGH104 and Enterococcus faecium) from four different patients, all of which were with significantly low growth rate limited by the chemotherapeutic agents, in different concentrations of 75 μM 5-FU and Oxi (Supplementary Figure 8). Most notably, the transition/transversion ratio of mutations covaried with agents concentrations (Supplementary Figure 9).
Dn/Ds Across Gut Species and Individuals
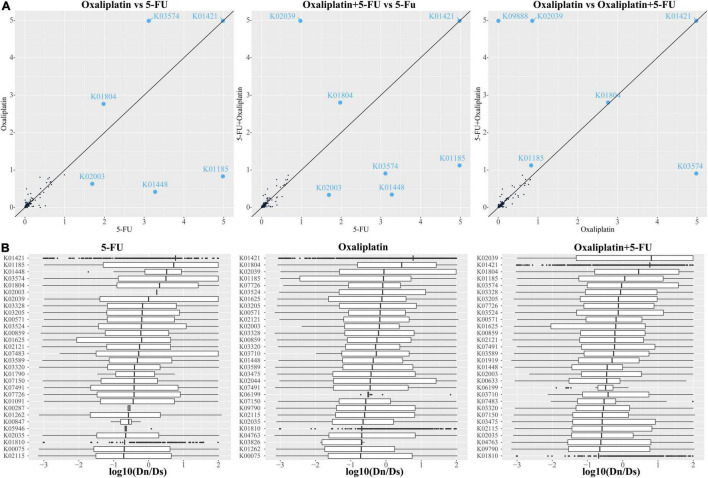
To gain further insights on the molecular mechanisms driving the functional diversification of the gut microbiota, the gene families identified in the assembled metagenome were annotated based on the KEGG, and we calculated, for each KO, the ratio between the number of non-synonymous (Dn) and synonymous (Ds) changes, a proxy for evolutionary pressure (Supplementary Figure 10). Our analyses showed that the average Dn/Ds was 0.063, and the median was 0.016 from 939 KO (Supplementary Table 13 and Supplementary Figure 10). Approximately 17.9% (17.6% in the 5-FU group, 17.7% in the 5-FU + Oxi group, and 18.3% in the Oxi group) of the gene families had significantly higher Dn values and lower Ds values than the mean value calculated over all annotated sequences (one-sided Fisher test, FDR < 0.05), suggesting that they might be under positive selection (Supplementary Table 14 and Supplementary Figure 11). A closer investigation of these gene families revealed that positive selection signatures markedly characterized diverse proteins involved in the cell division, as well as proteins essential for amino acid biosynthesis (Figure 4 and Supplementary Figure 12). According to the abundance analysis of KO items between days 0 and 30, we found that genes were under significant positive selection pressure, such as secY, which is a key in the protein secretion system and important for critical cell functions, such as pathogens and virulence. In addition, as in the previous study (Bulgarelli et al., 2015), we also found the coding sequence of phage infection protein (yhgE) and the genes related to transport system were under positive selection. Thus, the genes under positive selection might have a key role in the interaction between bacteria and chemotherapeutic drugs and provide the bacteria additional survival advantage during the chemotherapy.
FIGURE 4.
KEGG modules under positive selection in different chemotherapeutic drugs. (A) The correlation of average Dn/Ds ratio of all KO modules between the drugs. KO modules with average Dn/Ds ratio larger than 1 were marked by blue points. Maximum ratio was limited at 5. (B) Boxplot of top 20 ranked KO modules under positive selection with significantly increased Dn/Ds statistic, with sorted by their median log transformed Dn/Ds in descending order. The row stand for different KO modules.
Discussion
The human gut microbiota is highly complex and exists in a dynamic balance between symbiosis and pathogenesis, which can influence almost any aspect of host physiology (Sommer and Backhed, 2013). Growing evidences suggest that the gut microbiota not only plays a key role in carcinogenesis but also influences the efficacy and toxicity of anticancer therapy (Panebianco et al., 2018; Pothuraju et al., 2021). The microbiota modulates the host response to chemotherapy via numerous mechanisms, such as alteration of community structure and immune microenvironment. Furthermore, exploitation of the microbiota offers opportunities for the personalization of chemotherapeutic regimens and the development of novel therapies.
Chemotherapeutic agents, Oxi and 5-FU, exert their cytotoxic effect mostly through DNA damage. When DNA damage is caused by chemotherapeutic drugs, the microbiota composition also changes, which can further affect drug efficacy and overall health. It is very important to systematically explore the interaction between chemotherapeutic drugs and gut microbiota not only from microbiome composition (Yu et al., 2017; Hakim et al., 2018) but also genomic variation (Schloissnig et al., 2013; Zeevi et al., 2019). In this study, we first used two different methods (16S rRNA and shotgun metagenomics sequencing) to investigate the disruption of the intestinal microbiome in terms of taxonomic composition. Correlation tests between the abundance estimates for bacterial taxa showed that the two methods highlighted a highly positive correlation. According to the 16S rRNA sequence, we found that different chemotherapeutic agents had a distinct effect on the gut microbiota. 5-FU–associated bacteriome shifts included depletion of common health-associated commensals from the genera Streptococcus and Bacteroides and enrichment of Gram-negative bacteria such as Clostridium hathewayi and Lachnospiraceae bacterium. A previous study had shown that 5-FU could improve T cell–dependent antitumor immunity (Vincent et al., 2010), and Clostridia strains play a key role in enhancing regulatory T cell abundance and inducing important anti-inflammatory molecules, such as interleukin 10 (Atarashi et al., 2013). In contrast, Oxi-associated bacteriome showed different shift patterns including depletion of commensals from the genus Lachnospiraceae noname and enrichment of genus Lactobacillus and Streptococcus.
Subsequently, we used shotgun metagenomics with a quantitative metagenomic species approach to identify which species significantly changed during the chemotherapy with different agents. Two commensal species including S. salivarius and L. salivarius were significantly enriched after the chemotherapy. According to the previous studies, these two species appeared to be highly resistant to 5-FU (Vanlancker et al., 2016), which might be the main driver for a community dysbiosis. We hypothesized that if 5-FU was responsible for the dysbiosis changes seen during chemotherapy, then depleted taxa should be susceptible to the drug whereas the enriched species would be resistant to 5-FU. The resultant damage to the intestinal barrier increases the risk of colitis, bacterial translocation, and infection. Probiotic supplementation may have extra beneficial effects to correct dysbiosis of gut microbiota after the chemotherapy.
Concomitant with the alteration of gut microbial composition, we also observed a dysbiosis in bacterial gene functions, and different chemotherapeutic agents showed distinct influence patterns. Microbial metabolism may cause side effects severe enough to necessitate cessation of chemotherapy. The metagenome of 5-FU patients was enriched in genes associated with NAD salvage pathway. As NAD+ plays central roles in a variety of biological processes ranging from cellular metabolism and energy production in both human and microbiology, it was reasonable to hypothesize that gut microbiota could provide additional NAD+ to inhibit cancer cell autophagy and enhance survival of cancer cells (Sharif et al., 2016). By contrast, the metagenome of gut microbiota in Oxi patients was enriched in the genes associated with ribosome. Oxi, unlike cisplatin and carboplatin, kills cancer cells not only through the DNA-damage response, but also by inducing ribosome biogenesis stress (Bruno et al., 2017). In order to adapt the selection pressure from Oxi, only the species with a high copy number of the genes related to ribosome can survive during the chemotherapy similar to cancer cells, which may be why the genes involved with ribosomes were significantly enriched.
Next, in order to study the effects of DNA damage caused by chemotherapeutic agents, we collected the metagenomics sequencing data of 768 stool samples from 755 Chinese human individuals with the standard clinical assessments and reanalyzed the shift of mutational pattern with unified bioinformatics pipeline. Consistent with our expectation, we found obvious shift in the mutation signatures. A significantly reduced transversion rate (G:C- > T:A) was observed in the 5-FU group. In contrast, we found an increased ratio of the transversion in the Oxi group. In addition, the genome-wide substitution rates of gut microbiota were influenced by chemotherapeutic drugs without bias among different genomic regions. To further confirm that the shifts of mutation signatures were due to strong and diverse environmental selection, we analyzed the patterns of correlation between gene function and the Dn/Ds ratio across all the KO modules. The stable Dn/Ds ratios of most KEGG modules (80.1%) between days 0 and 30 suggest that the core microbiome was evolutionarily conserved. However, some modules, such as the genes involved in cell division, protein export, and phage-related, showed a high-level selection pressure. A previous study has shown that phage-related genes usually have fast mutation rate and are under positive selection (Petersen et al., 2007). However, the other genes, such as zapA and mutT, which are housekeeping genes involved in cell amplification and previously are thought to be with slow rate of amino acid evolution, have remarkably high Dn/Ds ratio and are under positive selection pressure. On the other hand, in order to be adaptive to the pressure of chemotherapeutic agents, bacteria take different strategies to obtain the survival advantage during the therapy, for example, by secreting an amount of protein to modify the host microenvironment with Sec system, which was a potential chemotherapeutic target to many human pathogens.
Conclusion
We have described the disordered profiles of gut microbiota in cancer patients treated with different chemotherapeutic agents, explored the mutational pattern of different drugs, and provided a new clue for the interaction between drugs and microbe. Our findings suggest that a distinct intestinal microbiome pattern of gut dysbiosis during the chemotherapy is dominated by bacilli and a lack of other bacteria, and the potential restorative influence of probiotic supplements should be investigated in the chemotherapeutic research. Positive selection on the protein export system provides an additional survival advantage for gut microbiota and could be a potential therapy target for gut dysbiosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Beijing Hospital Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
FX and SH conceived the study and designed the experiments. FS and LW performed the experiments. HL, GS, and HX made clinical samples validation. LZ, FS, HL, and GS collected clinical samples and analyzed the clinical data. FS, LZ, and SH analyzed the high throughput sequencing data. LW, FS, and FX wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Key Research and Development Program of China (Grant Nos. 2018YFC2000505 and 2020YFC2009003), the CAMS Innovation Fund for Medical Sciences (Grant No. 2021-I2M-1-050), the National Natural Science Foundation of China (Grant No. 81902618), the Beijing Hospital Nova Project (Grant No. BJ-2020-083), and the Beijing Hospital Project (Grant No. BJ-20 19-153).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.841458/full#supplementary-material
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., et al. (2013). “Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota.”. Nature 500 232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Bagel S., Hullen V., Wiedemann B., Heisig P. (1999). Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43 868–875. 10.1128/AAC.43.4.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M., Lieberman T. D., Kelsic E. D., Chait R., Gross R., Yelin I., et al. (2016). Spatiotemporal microbial evolution on antibiotic landscapes. Science 353 1147–1151. 10.1126/science.aag0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10 57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno P. M., Liu Y., Park G. Y., Murai J., Koch C. E., Eisen T. J., et al. (2017). A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 23 461–471. 10.1038/nm.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Garrido-Oter R., Munch P. C., Weiman A., Droge J., Pan Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microb. 17 392–403. 10.1016/j.chom.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Cheng W. Y., Wu C. Y., Yu J. (2020). The role of gut microbiota in cancer treatment: friend or foe? Gut 69 1867–1876. 10.1136/gutjnl-2020-321153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F. D., Doerks T., von Mering C., Creevey C. J., Snel B., Bork P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311 1283–1287. 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- de Gramont A., Figer A., Seymour M., Homerin M., Hmissi A., Cassidy J., et al. (2000). “Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer.”. J. Clin. Oncol. 18 2938–2947. [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., et al. (2011). “A framework for variation discovery and genotyping using next-generation DNA sequencing data.”. Nat. Genet. 43 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Fan X., Abbott T. E., Larson D., Chen K. (2014). “BreakDancer: Identification of Genomic Structural Variation from Paired-End Read Mapping.”. Curr. Protoc. Bioinform. 45 15.16.11–11. 10.1002/0471250953.bi1506s45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E. A., McIver L. J., Rahnavard G., Thompson L. R., Schirmer M., Weingart G., et al. (2018). Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15 962–968. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim H., Dallas R., Wolf J., Tang L., Schultz-Cherry S., Darling V., et al. (2018). “Gut Microbiome Composition Predicts Infection Risk During Chemotherapy in Children With Acute Lymphoblastic Leukemia.”. Clin. Infect. Dis. 67 541–548. 10.1093/cid/ciy153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfvarson J., Brislawn C. J., Lamendella R., Vazquez-Baeza Y., Walters W. A., Bramer L. M., et al. (2017). “Dynamics of the human gut microbiome in inflammatory bowel disease.”. Nat. Microbiol. 2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser P. J., VanGordon S. B., Seavey J., Sofinowski T. M., Ramadan M., Abdullah S., et al. (2015). Abnormalities in Expression of Structural, Barrier and Differentiation Related Proteins, and Chondroitin Sulfate in Feline and Human Interstitial Cystitis. J. Urol. 194 571–577. 10.1016/j.juro.2015.01.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B. Y., Sobue T., Choquette L., Dupuy A. K., Thompson A., Burleson J. A., et al. (2019). Chemotherapy-induced oral mucositis is associated with detrimental bacterial dysbiosis. Microbiome 7:66. 10.1186/s40168-019-0679-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., et al. (2013). “Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota.”. Nat. Commun. 4:2151. 10.1038/ncomms3151 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Xiong X., Danska J., Parkinson J. (2016). Metatranscriptomic analysis of diverse microbial communities reveals core metabolic pathways and microbiome-specific functionality. Microbiome 4:2. 10.1186/s40168-015-0146-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2017). “KEGG: new perspectives on genomes, pathways, diseases and drugs.”. Nucleic Acids Res. 45 D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp J. A., Kwon Y. J. (2021). Cancer nanotechnology: current status and perspectives. Nano. Converg. 8:34. 10.1186/s40580-021-00282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao F., Wang Y., Chen J., Tao J., Tian G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension.”. Microbiome 5:14. 10.1186/s40168-016-0222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A. B., et al. (2017). “Strains, functions and dynamics in the expanded Human Microbiome Project.”. Nature 550 61–66. 10.1038/nature23889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Miller S. F., Strauss C., Zhao C., Cheng L., Ye Z., et al. (2016). “Antibiotic treatment enhances the genome-wide mutation rate of target cells.”. Proc. Natl. Acad. Sci. U S A 113 E2498–E2505. 10.1073/pnas.1601208113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genom. Res. 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montassier E., Gastinne T., Vangay P., Al-Ghalith G. A., Bruley S., des Varannes S., et al. (2015). “Chemotherapy-driven dysbiosis in the intestinal microbiome.”. Aliment. Pharmacol. Ther. 42 515–528. 10.1111/apt.13302 [DOI] [PubMed] [Google Scholar]
- Morowitz M. J., Denef V. J., Costello E. K., Thomas B. C., Poroyko V., Relman D. A., et al. (2011). Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl. Acad. Sci. U S A 108 1128–1133. 10.1073/pnas.1010992108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panebianco C., Andriulli A., Pazienza V. (2018). Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 6:92. 10.1186/s40168-018-0483-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Thomas G., Volarevic S. (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer 18 51–63. 10.1038/nrc.2017.104 [DOI] [PubMed] [Google Scholar]
- Peng Y., Leung H. C., Yiu S. M., Chin F. Y. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28 1420–1428. 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- Petersen L., Bollback J. P., Dimmic M., Hubisz M., Nielsen R. (2007). Genes under positive selection in Escherichia coli. Genom. Res. 17 1336–1343. 10.1101/gr.6254707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo H. M., Peters G. F. (1988). Fluorouracil: biochemistry and pharmacology. J. Clin. Oncol. 6 1653–1664. 10.1200/JCO.1988.6.10.1653 [DOI] [PubMed] [Google Scholar]
- Pothuraju R., Chaudhary S., Rachagani S., Kaur S., Roy H. K., Bouvet M. (2021). Mucins, gut microbiota, and postbiotics role in colorectal cancer. Gut Microb. 13:1974795. 10.1080/19490976.2021.1974795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). “A metagenome-wide association study of gut microbiota in type 2 diabetes.”. Nature 490 55–60. [DOI] [PubMed] [Google Scholar]
- Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513 59–64. [DOI] [PubMed] [Google Scholar]
- Richter M., Rossello-Mora R., Oliver Glockner F., Peplies J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A., et al. (2013). Genomic variation landscape of the human gut microbiome. Nature 493 45–50. 10.1038/nature11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sharif T., Ahn D. G., Liu R. Z., Pringle E., Martell E., Dai C., et al. (2016). The NAD(+) salvage pathway modulates cancer cell viability via p73. Cell Death Differ. 23 669–680. 10.1038/cdd.2015.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Backhed F. (2013). The gut microbiota - masters of host development and physiology. Nat. Rev. Microbiol. 11 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Sorek R., Zhu Y., Creevey C. J., Francino M. P., Bork P., Rubin E. M. (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318 1449–1452. 10.1126/science.1147112 [DOI] [PubMed] [Google Scholar]
- Stogios P. J., Cox G., Spanogiannopoulos P., Pillon M. C., Waglechner N., Skarina T., et al. (2016). Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nat. Commun. 7:11343. 10.1038/ncomms11343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). “PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments.”. Nucleic Acids Res. 34 W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D. T., Franzosa E. A., Tickle T. L., Scholz M., Weingart G., Pasolli E., et al. (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12 902–903. 10.1038/nmeth.3589 [DOI] [PubMed] [Google Scholar]
- Vanlancker E., Vanhoecke B., Smet R., Props R., Van de Wiele T. (2016). 5-Fluorouracil sensitivity varies among oral micro-organisms. J. Med. Microbiol. 65 775–783. 10.1099/jmm.0.000292 [DOI] [PubMed] [Google Scholar]
- Vincent J., Mignot G., Chalmin F., Ladoire S., Bruchard M., Chevriaux A., et al. (2010). 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 70 3052–3061. 10.1158/0008-5472.CAN-09-3690 [DOI] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). “KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases.”. Nucleic Acids Res. 39 W316–W322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299 2074–2076. 10.1126/science.1080029 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., et al. (2017). Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 170(3), 54e516. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zhang S., Li H., Yang F., Mushtaq N., Ullah S., et al. (2018). The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. 108 184–193. 10.1016/j.biopha.2018.08.165 [DOI] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Godneva A., Bar N., Kurilshikov A., Lotan-Pompan M., et al. (2019). Structural variation in the gut microbiome associates with host health.”. Nature 568 43–48. 10.1038/s41586-019-1065-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated in this study have been available in the NCBI sequence read archive under the accession number PRJNA551354.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.