Abstract
Objectives: The purpose of this study was to identify the SNP sites and determine the BKV genotype circulating in kidney-transplant Vietnamese recipients based on the VP1 gene region. Methods: 344 samples were collected from post-kidney-transplant recipients at the 103 Vietnam Military Hospital to investigate the number of BKV infections. Positive samples with a sufficient virus concentration were analyzed by nested PCR in the VP1 region, sequencing detected genotyping and single-nucleotide polymorphism. Results: BKV infection was determined in 214 patients (62.2%), of whom 11 (5.1%) were diagnosed with BKV-associated nephropathy. Among the 90 BKV-I strains sequenced, 89 (98.88%) were strains of I/b-1 and 1 (1.12%) was strain I/b-2. The 60 BKV-IV strains had a greater diversity of subgroups, including 40% IV/a-1, 1.66% IV/a-2, 56.68% IV/c-1, and 1.16% IV/c-2. Additionally, of 11 cases diagnosed with BKVN, seven belonged to subgroup I/b-1 (63.6%) and four to subgroup IV/c-1 (36.4%). Moreover, 22 specific SNPs that were genotype I or IV were determined in this Vietnamese population. Specifically, at position 1745, for the Vietnamese BKV-IV strains, the SNP position (A→G) appeared in 57/60 samples (95%). This causes transformation of the amino acid N→S. This SNP site can enable detection of genotype IV in Vietnam. It represents a unique evolution pattern and mutation that has not been found in other international strains. Conclusion: The BKV-I genotype was more common than BKV-IV; however, mutations that occur on the VP1 typing region of BKV-IV strains were more frequent than in BKV-I strains.
Keywords: BK polyomavirus, VP1 region, single nucleotide polymorphism, genotype, amino acid, kidney transplantation, Vietnam
1. Introduction
The human BK polyomavirus (BKV) belongs to the Polyomaviridae family, which is characterized by a double-stranded circular DNA genome that has been isolated from several hosts, including humans, monkeys, rabbits, rodents, and birds [1]. Its genome is approximately 5.3 kb and contains an early region, a late region, and a non-coding control region (NCCR) [2]. BKV is often acquired during early childhood with a seroprevalence of 60–80% in the general population [3]. Following a primary infection, the virus remains latent and mainly asymptomatic within the kidney and urinary tract [4]. It can become active during severe immunosuppressive therapy due to kidney or stem-cell transplantation: reactivation of BKV can then occur, and then replicates massively, resulting in damage to uroepithelial cells [2,5].
In kidney-transplant recipients, the prevalence of BKV reactivation ranges from 10–60%, of which 1–10% develop BKV nephropathy (BKVN), characterized by severe tubulointerstitial nephritis and basement-membrane necrosis [6,7]. Progression of BKVN usually occurs without any clinical signs or symptoms, except for rising serum-creatinine levels. Unless detected promptly, 50% of BKVN cases can cause graft loss within the following months/years [3].
The major capsid component, VP1, can be divided into five outer domains: i.e., BC, DE, EF, GH, and HI, which connect the various β-strands and α-helix of the polypeptide segments [7]. The BC and EF loop regions are most frequently affected by mutations and the BC loop contains a short sequence at positions (1744–1812) used to identify the four main viral genotypes [7,8]. BKV has been classified into genotypes and subgroups based on single-nucleotide polymorphism (SNPs) analysis of the VP1 region and the non-coding control region (NCCR) [9]. Nucleotides of the VP1 coding region show very high conservation (over 95%) in all genotypes of BKV, but the similarity between the amino acid residues from 61E to 83R is only 61–70% [7,10].
Genotype I is the most prevalent and widespread worldwide (about 80% of reported cases), followed by genotype IV (about 15% of reported cases), mainly distributed in Europe and East Asia, while genotypes II and III are rare in all geographic regions (~5%). Genotypes I and IV are further divided into four subgroups of I (I-a, I-b1, I-b2, and I-c) and six of IV (IV-a1, IV-a2, IV-b1, IV-b2, IV-c1, and IV-c2) [11]. Analysis of polymorphism between obtained sequences, especially in the BC loop, is vital in medical research because molecular variation in BKV may entail changes in tropism and influence the clinical manifestations of infection [12]. However, in-depth studies on the molecular genetics of BKV have rarely been reported in Vietnam. Therefore, the purpose of this study was to identify the SNP sites and to determine the BKV genotype circulating in kidney-transplant recipients in Vietnam, based on the VP1 gene region.
2. Materials and Methods
2.1. Sample and DNA Extraction
A total of 344 samples were collected from post-kidney-transplant recipients at the 103 Vietnam Military Hospital. We investigated the number of BKV infections using qPCR. Among the 344 samples, 150 urine or serum samples were selected from patients diagnosed with BKV. Viral DNA was extracted from 200 μL of clinical sample (plasma or urine) using a Geneall DNA viral kit (Geneall, Seoul, Korea), according to the manufacturer’s protocol. The DNA was finally eluted in a final volume of 54 μL of AE buffer and stored at −80 °C until use.
2.2. Quantification of BK Viral Load in Urine and Plasma Samples
We used an in-house quantitative realtime PCR assay developed in our laboratory with a slight modification from a previously published protocol on the Rotor Gene Q5 plex MDx platform (Qiagen, Germany) for quantification of BK virus load in urine and plasma [13]. The BK virus load was expressed in BKV genome copies per milliliter of urine or plasma. The lower limit of BK viral load detection at our center is 250 copies per milliliter.
2.3. Genotyping of BKV
Primers were designed for the VP1 region of BKV, as described previously [14,15]. The external primer pair, BKS + BKAS, and the internal primer pair, BKF + BKR1, amplified a 580-bp and a 327-bp DNA fragment, respectively (Table 1). The PCR reaction was optimized in a total volume of 20 μL using a PCR kit (GoTaq Mastermix, Promega, Madison, WI, USA) containing 5 μL of DNA template, 1 μL primer, 4 μL of molecular-grade water, and 10 μL of 2X Buffer. Cycling conditions were 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 7 min. For the nested PCR, 3 µL of the products from the PCR, using the external primer pair, was added as the DNA template for the second-round of PCR using the internal primer pair. PCR amplifications were performed using the parameters described above. All reactions were implemented on an Eppendorf™ Mastercycler™ pro PCR System. Amplification products were separated by electrophoresis on 1.2% agarose and visualized under UV light after staining with ethidium bromide (10 mg/mL) and sequenced using a 3130xl sequencer.
Table 1.
Primer sequences used in this study.
| Primers | Sequences | Position * | Size | References |
|---|---|---|---|---|
| BKV_S | ATC AAA GAA CTG CTC CTC AAT | 1361–1381 | 580 bp | [15] |
| BKV_AS | GCA CTC CCT GCA TTT CCA AGGG | 1919–1940 | ||
| BKV-F | CAA GTG CCA AAA CTA CTA AT | 1630–1649 | 327 bp | [14] |
| BKV-R1 | TGC ATG AAG GTT AAG CAT GC | 1937–1956 |
Abbreviations: BKV, BK polyomavirus; bp, base pair. * With respect to Genebank accession number V01108 (Dunlop strain).
2.4. Phylogenetic Analysis
All of the obtained sequences were compared with 31 reference sequences retrieved from GenBank using Bioedit 7.0 (http://www.mbio.ncsu.edu/bioedit/bioedit.html, Hall 1999 (accessed on 10 March 2022)) and MEGA 7.0 software (https://www.megasoftware.net/ (accessed on 10 March 2022)) (Table 2). The data were then analyzed by constructing a phylogenetic tree using the neighbor-joining method, and significance level was estimated with 1000 bootstrap replicates.
Table 2.
Reference sequence.
| Access No. | Subgroups | Country |
|---|---|---|
| AB211369 | Ib1 | Japan |
| AB365177 | Ib1 | Japan |
| AB213282 | Ib1 | China |
| JN192433 | Ib1 | Ireland |
| AY628226 | Ib1 | USA |
| AB369236 | Ib2 | USA |
| AB301086 | Ib2 | Japan |
| AB245223 | Ib2 | China |
| AB211370 | Ib2 | Netherland |
| AB369092 | Ia | Japan |
| NC001538 | Ia | USA |
| V01108 | Ia | USA |
| AB263931 | Ic | China |
| AB211381 | Ic | Japan |
| AB211372 | Ic | Japan |
| AB211386 | III | Japan |
| AB263920 | II | English |
| AB269859 | IV a1 | Philippine |
| AB269869 | IV a1 | Vietnam |
| AB268868 | IV a2 | Vietnam |
| AB211389 | IV a2 | Japan |
| AB269867 | IV c1 | Vietnam |
| AB269858 | IV c1 | China |
| AB269863 | IV c1 | China |
| AB211391 | IV b1 | Japan |
| AB217919 | IV b1 | Japan |
| KF468291 | IV c2 | Germany |
| AB269857 | IV c2 | China |
| AB269835 | IV b2 | Japan |
| AB269834 | IV b2 | Japan |
| AB211387 | IV b2 | Japan |
The basis of specific polymorphisms in the portion of the VP1 region, spanning nucleotide (1650–1955) was determined and analyzed in terms of the changes in one or more amino-acid sequences using Bioedit 7 (http://www.mbio.ncsu.edu/bioedit/bioedit.html, Hall 1999 (accessed on 10 March 2022)).
2.5. Renal Allograft Biopsy
Allograft biopsy was performed in patients with acute allograft dysfunction, BK virus nephropathy was characterized by tubular atrophy and fibrosis with a variable inflammatory lymphocytic infiltrate, BK virus nephropathy was confirmed using immunohistochemical nuclear staining with anti-SV40 antibody.
2.6. Statistical Analysis
Differences between categorical variables were analyzed by the χ2 test while independent t-test or Mann-Whitney test were used to compare quantitative variables. All statistical analyses were conducted using the SPSS software version 20.0 (IBM, Armonk, NY, USA). p value less than 0.05 was considered to be statistically significant.
3. Results
3.1. Characteristics of the Kidney-Transplant Recipients
Of the 344 kidney-transplant recipients, BKV was detected in 214 (62.2%), of which 11 (5.1%) developed BKV nephropathy. Among the 214 positive-BKV recipients, 24 (7.0%) only developed BKV viremia, and 50 had BK viruria alone was present (6.98%), whereas 140 (14.5%) had both BKV viremia and BKV viruria. The median values for BKV viremia and viruria were 3.95 ± 1.14 and 6.81 ± 2.82 log copies/mL, respectively (Table 3).
Table 3.
Characteristics of kidney-transplant recipients.
| Variable | Value |
|---|---|
| Age (years) (mean ± SD) | 38.37 ± 10.09 |
| Gender | |
| Male, n, % | 245 (71.2%) |
| Female, n, % | 99 (28.8%) |
| Times of HD (months) | 28.26 ± 41.70 |
| HLA mismatch (mean ± SD) | 3.36 ± 1.18 |
| Serum creatinine at first BKV testing (µmol/L) | 110.40 ± 33.10 |
| eGFR at first BKV testing (mL/min) | 61.28 ± 14.75 |
| BK viremia only, n, % | 24 (7.0%) |
| BK viruria only, n, % | 50 (14.5%) |
| BK viremia and viruria, n, % | 140 (40.7%) |
| BKV, n, % | 214 (62.2%) |
| BK viremia (log copies/mL) | 3.95 ± 1.14 |
| BK viruria (log copies/mL) | 6.81 ± 2.82 |
| CNIs | |
| Tacrolimus, n (%) | 305 (88.66%) |
| Cyclosporine, n (%) | 39 (11.34%) |
Abbreviations: HD, hemodialysis; BKV, BK virus; SD, standard deviation; eGFR, estimated glomerular filtration rate; CNI, calcineurin inhibitor.
We also analyzed for possible associations between the two groups infected with BKV and without BKV. The results showed that there was no significant difference between the two groups with regards to age, gender, HLA mismatch, or types of immunosuppressive drugs. However, the difference between the BKV-negative group and BKV-positive group in serum-creatinine concentration (115.95 ± 36.84 vs. 101.25 ± 23.19, respectively) and estimated glomerular-filtration rate (58.95 ± 14.60 vs. 65.12 ± 14.19, respectively) were determined to be statistically significant (p < 0.001, Table 4).
Table 4.
Comparison between BKV and non-BKV groups.
| Variable | BKV (−) | BKV (+) | p Value |
|---|---|---|---|
| Age (years) (mean ± SD) | 39.01 ± 9.99 | 37.98 ± 10.15 | 0.36 |
| Gender | 0.22 | ||
| Male, n, % | 95 (73.1) | 150 (70.1) | |
| Female, n, % | 35 (26.9) | 64 (29.9) | |
| HLA mismatch (mean ± SD) | 3.33 ± 1.02 | 3.37 ± 1.27 | 0.75 |
| Serum creatinine at first BKV testing (µmol/L) | 101.25 ± 23.19 | 115.95 ± 36.84 | <0.001 |
| eGFR at first BKV testing (mL/min) | 65.12 ± 14.19 | 58.95 ± 14.60 | <0.001 |
| Times after kidney transplantation (months, mean ± SD) | 5.73 ± 5.70 | 10.79 ± 18.40 | 0.003 |
| CNIs | |||
| Tacrolimus, n, % | 118 (90.7) | 187 (87.4) | 0.62 |
| Cyclosporine, n, % | 12 (9.3) | 27 (12.6) |
Abbreviations: SD, standard deviation; eGFR, estimated glomerular filtration rate; CNI, calcineurin inhibitor; BKV, BK virus.
To investigate the possible impacts of the different genotypes, we also compared the differences of patients infected with BKV-I and BKV-IV. However, there were no statistically significant differences in age, sex, HLA mismatch, time after transplantation, serum creatinine, eGFR, BKVN, immunosuppressive therapy, and BKV load between the two groups (Table 5).
Table 5.
Comparison between BKV-I and BKV-IV groups.
| Variable | BKV-I | BKV-IV | p Value |
|---|---|---|---|
| Age (years) (mean ± SD) | 38.44 ± 9.67 | 35.98 ± 10.69 | 0.15 |
| Gender | 0.69 | ||
| Male, n, % | 61 (67.8) | 44 (73.3) | |
| Female, n, % | 29 (32.2) | 16 (26.7) | |
| HLA mismatch (mean ± SD) | 3.40 ± 1.35 | 3.43 ± 1.20 | 0.9 |
| Serum creatinine at first BKV testing (µmol/L) | 113.96 ± 35.40 | 126.42 ± 46.86 | 0.07 |
| eGFR at first BKV testing (mL/min) | 59.07 ± 15.26 | 57.03 ± 16.81 | 0.44 |
| log BK viremia (mean ± SD) | 3.95 ± 1.13 | 4.28 ± 1.19 | 0.11 |
| log BK viruria (mean ± SD) | 7.54 ± 2.65 | 7.19 ± 2.90 | 0.45 |
| BKVN n, % | 7 (7.8%) | 4 (6.7%) | 0.81 |
| CNIs | |||
| Tacrolimus, n, % | 78 (86.7) | 52 (86.7) | 0.59 |
| Cyclosporine, n, % | 12 (13.3) | 8 (13.3) |
Abbreviations: SD, standard deviation; eGFR, estimated glomerular filtration rate; CNI, calcineurin inhibitor; BKV, BK virus.
3.2. Phylogenetic Analysis of BKV Isolates
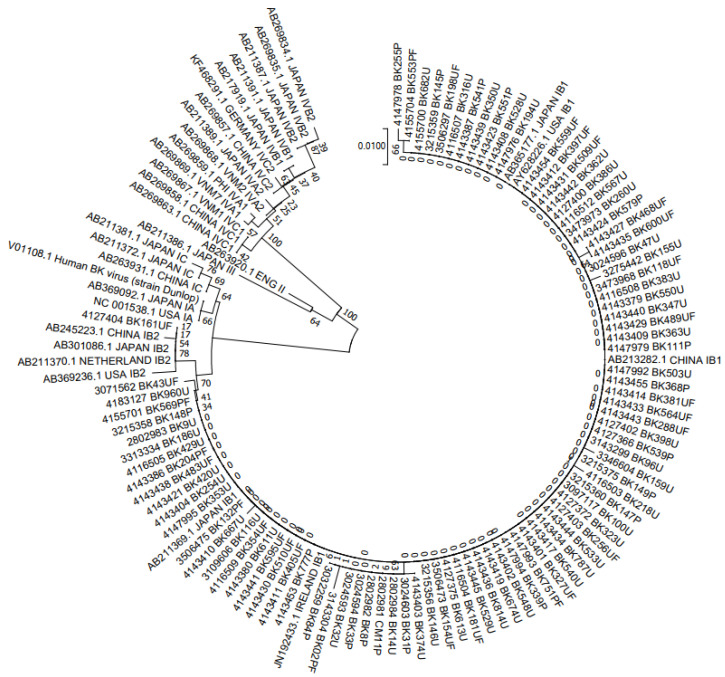
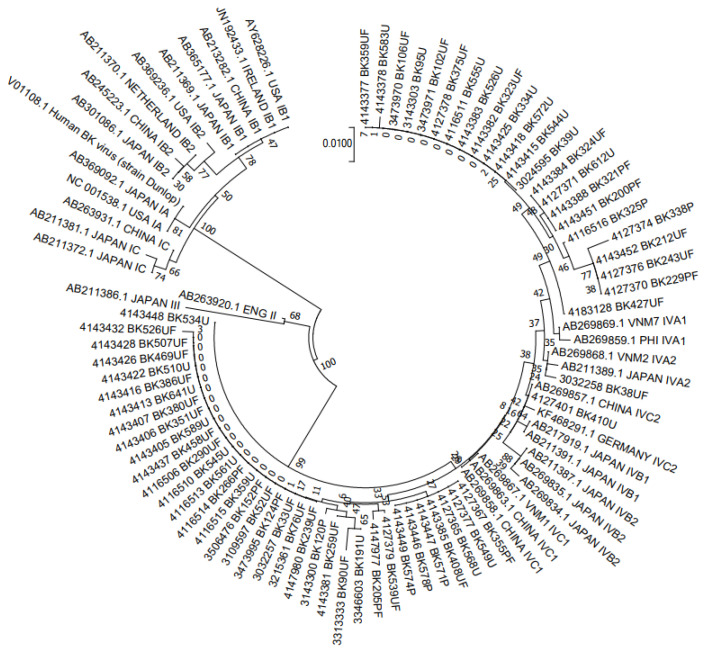
A total of 150 samples were genotyped by constructing a phylogenetic tree from analysis of 305-bp fragments of BKV, aligned with 31 reference sequences retrieved from the GenBank (Figure 1 and Figure 2). The results indicated that 90 (76.6%) belonged to BKV-I and 60 (23.3%) to BKV-IV, with no cases of BKV-II or BKV-III observed. Among the 90 BKV-I strains, 89 (98.9%) strains were of I/b-1 and 1 (1.1%) of I/b-2. BKV-IV had a greater diversity of subgroups, including 40% of IV/a-1, 1.7% of IV/a-2, 56.7% of IV/c-1, and 1.2% of IV/c-2 (Table 6). Of the 11 cases that were diagnosed with BKVN, seven were subgroup I/b-1 (63.6%) and four were subgroup IV/c-1 (36.4%).
Figure 1.
NJ phylogenetic tree clustering of 90 BKV-I polyomavirus sequences to determine subgroups.
Figure 2.
NJ phylogenetic tree clustering 60 BKV-IV polyomavirus sequences to determine subgroups.
Table 6.
Distribution of BKV subgroups.
| BKV-I (n = 90) | BKV-IV (n = 60) | |||||||
|---|---|---|---|---|---|---|---|---|
| I/a | I/b-1 | I/b-2 | I/c | IV/a-1 | IV/a-2 | IV/b-1 | IV/c-1 | IV/c-2 |
| 0 (0%) |
89 (98.9%) |
1 (1.12%) |
0 (0%) |
24 (40%) |
1 (1.7%) |
0 (0%) |
34 (56.7%) |
1 (1.7%) |
The 305-bp typing region sequenced in this study, along with 31 reference sequences, were used to construct a neighbor-joining phylogenetic tree using Kimura’s correction.
3.3. Analysis of SNP
Analysis of 31 reference sequences and 150 sequences that contained the VP1 gene fragment of 305 bp in size (1650–1955) showed 22 different SNP positions between genotypes I and IV (Table 7). Among these, 10 had capsid amino-acid sequence substitution of BKV (61, 62, 66, 69, 71, 74, 75, 77, 82, 117).
Table 7.
SNP difference between BKV-I and BKV-IV.
| SNP Position | Aa Position | BKV-I | BKV-IV | Aa Substitution |
|---|---|---|---|---|
| 1704 | 47 | G | A | |
| 1716 | 51 | C | T | |
| 1722 | 53 | C | T | |
| 1744 | 61 | G | A | E → N/S |
| 1746 | 61 | A | T | |
| 1747 | 62 | A | G | N → D |
| 1760 | 66 | T | A | F → Y |
| 1769 | 69 | A | G/A (3) | K → R |
| 1770 | 69 | G | A | |
| 1775 | 71 | G | C | S → T |
| 1784 | 74 | A | C | N → T |
| 1787 | 75 | A | C | D → A |
| 1792 | 77 | A | G/C | S → D/N/E/Q/H |
| 1793 | 77 | G | A | |
| 1809 | 82 | G/A | C | E → D |
| 1848 | 95 | C | A A |
|
| 1851 | 96 | C | A/G | |
| 1854 | 97 | C | T | |
| 1869 | 102 | C | T | |
| 1890 | 109 | G | A | |
| 1912 | 117 | C | A | Q → K |
| 1938 | 125 | C | T |
Ninety of the BKV-I sequences were further compared with Dunlop strains and international strains of subgroup I in the VP1 gene region (1650–1955) to determine polymorphisms. Sequence analysis showed that 15 SNPs were detected within subgroup I; however, most of these SNP sites appeared singly, except at nucleotide position 1786, as hot spots (Table 8). At this position there was a nucleotide change (G → A) in the amino acid substitution, i.e., Asp → Asn, in a frequency of 9/90 samples (10%).
Table 8.
SNP positions among BKV-I subgroups in the VP1 gene region.
| SNP | Dunlop (I/a) |
AB211369 (I/b-1) |
AB211370 (I/b-2) |
AB211372 (I/c) |
Research Sample | Frequency |
|---|---|---|---|---|---|---|
| 1687 | G | G | C | G | G/C | 89/90, 1/90 |
| 1698 | T | A | A | A | A | 90/90 |
| 1741 | G | G | G | G | G/C | 88/90, 2/90 |
| 1765 | C | G | G | G | C/G | 89/90, 1/90 |
| 1780 | G | G | G | G | G/C | 89/90, 1/90 |
| 1786 | G | G | G | G | A/G | 9/90, 81/90 |
| 1809 | G | A | A | G | A | 90/90 |
| 1848 | C | C | C | C | C/T | 89/90, 1/90 |
| 1863 | T | T | T | T | T/C | 88/90, 2/90 |
| 1908 | T | T | A | T | T/A | 89/90, 1/90 |
| 1919 | A | A | A | A | A/G | 89/90, 1/90 |
| 1923 | T | C | C | T | C | 90/90 |
| 1929 | A | A | A | A | A/G | 89/90, 1/90 |
| 1946 | A | A | A | A | A/C | 89/90, 1/90 |
| 1952–1953 | AT | AT | AT | AT | AT/TG | 89/90, 1/90 |
Sixty BKV-IV sequences were compared with international strains of subgroups IV/a-1 (AB269869), IV/a-2 (AB211389), IV/c-1 (AB269867), and the parent strain (sequence IV, Z19535, Jin L (1993) [3], isolated in the UK) (Table 9). After analysis, there were 15 distinct SNP positions between parent strain IV and isolated strains found in Vietnam, in which many SNPs had changed amino acids. It is notable that most of these SNP sites appeared singly, but all had an altered amino acid sequence (Table 9 and Table 10). Specifically, seven SNP positions that belong to subgroup IV had high frequency, of which 1745 and 1794 had changed amino acid-sequences (95.0% and 61.7%, respectively).
Table 9.
SNP positions among BKV-IV subgroups in the VP1 gene region.
| SNP | IV | IV/a-1 | IV/a-2 | IV/c-1 | IV/c-2 | Research Sample | Frequency |
|---|---|---|---|---|---|---|---|
| 1735 | G | G | G | G | G | G/A | 55/60, 5/60 |
| 1737 | T | T | T | T | T | T/C | 38/60, 23/60 |
| 1741 | G | G | G | G | G | G/C | 59/60, 1/60 |
| 1745 | A | A | A | A | A | A/G | 3/60, 57/60 |
| 1769 | G | G | G | G | G | G/A | 57/60, 3/60 |
| 1780 | G | G | G | G | G | G/T/C | 57/60, 1/60, 2/60 |
| 1781 | A | A | A | A | A | A/C | 59/60, 1/60 |
| 1786 | G | G | G | G | G | G/A | 59/60, 1/60 |
| 1792 | G | G | G | G | G | G/A/C | 51/60, 3/60, 6/60 |
| 1794 | G | G | C | C | C | C/G | 37/60, 23/60 |
| 1851 | A | A | A | A | A | A/G | 38/60, 22/60 |
| 1860 | G | G | A | G | G | A/G | 24/60, 36/60 |
| 1870 | C | C | C | C | C | C/T | 59/60, 1/60 |
| 1905 | G | G | G | A | G | A/G | 35/60, 25/60 |
| 1938 | C | T | T | T | T | T/C | 43/60, 17/60 |
Table 10.
Amino acid substitution between strain IV, Dunlop, and the study strains.
| SNP | AA Position | Dunlop AA | IV AA | Research AA |
|---|---|---|---|---|
| 1735 | 58 | D | D | D(55/60), N(5/60) |
| 1737 | 58 | D | D | |
| 1741 | 60 | D | D | D (59/60), H (1/60) |
| 1745 | 61 | E | N | N (3/60), S (57/60) |
| 1769/1770 | 69 | K | R | R (57/60), K (3/60) |
| 1780/1781 | 73 | E | S | E (57/60), Q (2/60), S (1/60) |
| 1786 | 75 | D | A | A (59/60), T (1/60) |
| 1792 | 77 | S | E | D (33/60), E (18/60), Q (5/60), H (1/60), N (3/60) |
| 1794 | 77 | S | E | |
| 1851 | 96 | L | L | L (60/60) |
| 1860 | 99 | L | L | L (60/60) |
| 1870 | 103 | L | L | L (60/60) |
| 1905 | 114 | V | V | V (60/60) |
| 1938 | 125 | S | S | S (60/60) |
Abbreviations: AA, aminoacid.
4. Discussion
We reported on the prevalence of BKV infection in the blood and urine of 344 kidney-transplant recipients, of which 214 were BKV-positive (62.2%), which was a slightly lower rate than the study of Toan et al. in northern Vietnam (77.4%) [16]. This difference may be due to our larger sample size (344 in our study vs. 82 for Toan et al.’s study). However, the number of cases of BK viruria and BK viremia in kidney-transplant recipients in our study is similar to that reported in other studies [17,18].
Previous studies have reported a number of risk factors for BKV nephropathy, including age, gender, HLA mismatches, deceased-donor transplants, and types of immunosuppressive drugs. Nevertheless, in our study, there was no difference between groups with and without BKV infection regarding any of the risk factors mentioned, except for differences in serum-creatinine and estimated glomerular-filtration rate, which were statistically significant (p < 0.001, Table 4). As BKV infection represents a threat to premature allograft loss and the early monitoring of BKV load using realtime PCR in either plasma or urine can improve the management of patients after kidney transplantation.
The first genotyping scheme for BKV was reported by Jin et al. in 1993 using restriction fragment-length polymorphism (RFLP) based on nucleotide polymorphisms in a very short fragment of the gene of the capsid protein VP1 (nucleotides 1744 to 1812). Sanger sequencing is now a major method of genotyping BKV [19]. In previous studies, the phylogenetic tree was constructed based on a complete genome sequence or any viral genomic region; however, the VP1 gene region contains more polymorphism information that can yield a clearer definition of subgroups [6,20].
BKV can be classified into four genotypes (I → IV) and each genotype is further divided into several subgroups according to geographical distribution [21]. BKV-I was the predominant genotype during the period of study follow at 60% overall (90 of 150 BKV-positive cases), followed by BKV-IV (60 of 150 BKV-positive cases), and no cases of genotypes II or III were found. Intra-genotype diversity was observed in both genotypes; however, BKV-IV revealed more subgroup variety than BKV-I. In genotype I, all sequences belonged to Ib1 except one, which belonged to subgroup Ib2. Subtype Ib-1 has been shown to be highly prevalent in southeast Asia, across Europe, and parts of North Africa, while subtype Ib-2 is common in Europe [16,22].
The only patient in this study with subgroup Ib-2 BKV received a kidney transplant in France. This can be explained because that patient had received a kidney from a donor with BK viruria. Recent studies have concluded that BKV replication in recipients come from donor origin [23,24]. However, the coexisting four subgroups of IV (IV-a1, IV-a2, IV-c1, and IV-c2), and the three subgroups (IV-a1, IV-a2, IV-c1), are the same as reported in previous publications [16,25]. Interestingly, one patient of this study belonged to IV-c2 which was prevalent in Mongolia and Europe [25]. In this situation, the patient or kidney donor could have acquired the BKV infection not from living local community but from geographical regions where BKV-IV/c2 was predominant, which supported the source of BKV after kidney transplantation.
Nukuzuma.S et al. (2006) [26] showed that BKV-I replicates more efficiently than BKV-IV in human renal epithelial cells, while several other reports illustrated the association between genotypes BKV-II, III, IV with a higher risk of BKVN in kidney transplant patients [27,28]. Therefore, the role of particular genotypes or BKV variants in the development of BKVN remains unclear. In this study, we compared two groups of patients, BKV-I and BKV-IV, with some clinical factors. In general, the proportion of patients infected with BKV-I going on to develop BKVN was higher than patients with BKV genotype IV. This can be explained because BKV-I dominates the population of Vietnam as well as by the limited number of BKVN cases in studies. Therefore, a large cohort of BKVN cases should be analyzed to determine the role of BKV genotypes on the pathogenesis and the clinical course of BKVN in patients with a history of kidney transplant.
Of note, suitably selected SNP assays can provide an alternative way for BKV genotyping. A recent study constructed an algorithm in order to determine the 12 BKV subgroups and subtypes based on a region comprising only 100 bp of the VP1 gene (1977 through 2076) with 12 isolated SNPs [29]. Twenty-two specific SNPs for the identification of genotype I or IV in a Vietnamese population were determined (Table 7), of which 12 sites (1760, 1787, 1793, 1848, 1912, 1704, 1760, 1770, 1784,1854, 1809, and 1938) concordant with the results reported by Luo et al. [20]. A subgroup Ib2 sequence has two SNPs (1687 and 1908) showing the remaining known subtype Ib1 strains, but there is no special SNP position to separate the IV subgroups from each other on the VP1 region (1650–1955). Therefore, a study that investigated SNP sites to differentiate subgroups IV should be performed on other genomic regions of BKV, such as LTA or NCCR, because the application of SNP genotyping can remove the dependence on the accuracy of the algorithm when creating a phylogenetic tree.
It is notable that single nucleotide polymorphisms (SNPs) are believed to be vital in the pathogenesis of the virus as well as a mechanism of immune escape. Varella et al. (2017) found differences in viral load between the Ia and Ib1 subgroups, which discriminate in the sequences coding for VP1 [30]. Therefore, changes in the single nucleotide sequence can affect not only its pathogenicity, but also the tropism of the virus or the range of hosts. At position 1786 (corresponding to amino acid 75) of genotype I on some Vietnamese strains, the SNP position (G → A) appeared to change the amino acid Asp → Asn, observed in 9/90 samples (10%). This is a rare SNP site: not within the range of SNPs that Jin et al. described in 1993 to discriminate BKV genotypes [25], but recognized as a rare mutation by Tremolada et al. (2010) and was often present in the urine in patients with BKVN [2]. Most genotype I SNP sites appear singly, whereas, for genotype IV, we observed that 7/15 SNP sites (1737, 1745, 1794, 1851, 1860, 1905, 1938) had high repetition frequencies (28–95%). In particular, position 1745 on Vietnamese strains shows the SNP position (A → G) in 57/60 samples (95%). This causes an amino acid substitution from N → S. To our knowledge, this is a distinct SNP site and can be used for detection of genotype IV in Vietnam, representing a unique evolution pattern and mutation that has not been found in other international strains. However, limitations also exist, because our samples were collected from a particular geographical area. Additional sequences from other areas in Vietnam, including central and south Vietnam, are required to confirm these results. Additionally, this is only a cross-sectional study at a time, therefore the assessment of some related factors has not been really rigorous.
5. Conclusions
The BKV-I genotype was more common than BKV-IV; however, mutations that occur on the VP1 typing region of BKV-IV strains were more frequent than in BKV-I strains. This study has provided more insights into the genotype characteristics and SNP sites of BKV strains among recipients of renal transplant in Vietnam.
Abbreviations
| BKV | BK virus |
| BKVN | BK virus nephropathy |
| eGFR | estimated glomerular-filtration rate |
| HLA | human leucocyte antigen |
| PCR | polymerase chain reaction |
| VP | viral protein |
| SNP | single nucleotide polymorphism |
Author Contributions
T.Q.K., P.B.N., D.T.V., N.V.D., D.M.H., N.T.T.D., N.T.T.H., H.T.V., L.V.T. and B.V.M. recruited the patients and collected the clinical data. T.Q.K. and P.Q.T. designed the study and wrote the manuscript. L.T.B.Q., H.X.S. and T.V.T. performed the virological analyses as well as the statistical analyses. L.R. edited the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The Institutional Review Board of Military Hospital 103, Vietnam Military Medical University in Hanoi (Vietnam) approved this study (protocol code 2885 and date of approval: 7 July 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ambalathingal G.R., Francis R.S., Smyth M.J., Smith C., Khanna R. BK polyomavirus: Clinical aspects, immune regulation, and emerging therapies. Clin. Microbiol. Rev. 2017;30:503–528. doi: 10.1128/CMR.00074-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tremolada S., Delbue S., Castagnoli L., Allegrini S., Miglio U., Boldorini R., Elia F., Gordon J., Ferrante P. Mutations in the external loops of BK virus VP1 and urine viral load in renal transplant recipients. J. Cell. Physiol. 2010;222:195–199. doi: 10.1002/jcp.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta N., Lawrence R.M., Nguyen C., Modica R.F. BK virus in systemic lupus erythematosus (review article) Pediatric Rheumatol. 2015;13:34. doi: 10.1186/s12969-015-0033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavlakis M., Haririan A., Klassen D.K. BK virus infection after non-renal transplantation. Adv. Exp. Med. Biol. 2006;577:185–189. doi: 10.1007/0-387-32957-9_13. [DOI] [PubMed] [Google Scholar]
- 5.Alavi S., Kaji Yazdi M., Parvin M., Zohrehbandian F., Azma R. Haemorrhagic cystitis due to BK virus in a child with ALL on standard chemotherapy without stem cell transplant. Ecancermedicalscience. 2013;7:350. doi: 10.3332/ecancer.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z.Y., Hong W.L., Zhu Z.H., Chen Y.H., Ye W.L., Chu G.Y., Li J.L., Chen B.C., Xia P. Phylogenetic reconstruction and polymorphism analysis of BK virus VP2 gene isolated from renal transplant recipients in China. Exp. Ther. Med. 2015;10:1759–1767. doi: 10.3892/etm.2015.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furmaga J., Kowalczyk M., Zapolski T., Furmaga O., Krakowski L., Rudzki G., Jaroszyński A., Jakubczak A. BK Polyomavirus—Biology, Genomic Variation and Diagnosis. Viruses. 2021;13:1502. doi: 10.3390/v13081502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tremolada S., Delbue S., Larocca S., Carloni C., Elia F., Khalili K., Gordon J., Ferrante P. Polymorphisms of the BK virus subtypes and their influence on viral in vitro growth efficiency. Virus Res. 2010;149:190–196. doi: 10.1016/j.virusres.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma R., Tzetzo S., Patel S., Zachariah M., Sharma S., Melendy T. BK virus in kidney transplant: Current concepts, recent advances, and future directions. Exp. Clin. Transpl. 2016;14:377–384. doi: 10.6002/ect.2016.0030. [DOI] [PubMed] [Google Scholar]
- 10.Krumbholz A., Bininda-Emonds O.R., Wutzler P., Zell R. Evolution of four BK virus subtypes. Infection. Infect. Genet. Evol. 2008;8:632–643. doi: 10.1016/j.meegid.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Wunderink H.F., De Brouwer C.S., Gard L., De Fijter J.W., Kroes A.C.M., Rotmans J.I., Feltkamp M.C.W. Source and relevance of the BK polyomavirus genotype for infection after kidney transplantation. Open Forum Infect. Dis. 2019;6:ofz078. doi: 10.1093/ofid/ofz078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furmaga J., Kowalczyk M., Furmaga O., Rokos C.A., Zapolski T., Krakowski L., Jakubczak A., Rudzki S. Molecular Epidemiology and Variation of the BK Polyomavirus in the Population of Central and Eastern Europe Based on the Example of Poland. Viruses. 2022;14:209. doi: 10.3390/v14020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babel N., Volk H.-D., Reinke P. BK polyomavirus infection and nephropathy: The virus–immune system interplay. Nat. Rev. Nephrol. 2011;7:399–406. doi: 10.1038/nrneph.2011.59. [DOI] [PubMed] [Google Scholar]
- 14.Jin L., Pietropaolo V., Booth J.C., Ward K.H.D., Brown W. Prevalence and distribution of BK virus subtypes in healthy people and immunocompromised patients detected by PCR-restriction enzyme analysis. Clin. Diagn. Virol. 1995;3:285–295. doi: 10.1016/S0928-0197(94)00044-1. [DOI] [PubMed] [Google Scholar]
- 15.Ledesma J., Bouza E., González-Nicolás M.A., García de Viedma D., Rodríguez-Sánchez B., Muñoz P. BK polyomavirus genotyping at inter and intra patient level in Spain. J. Med. Virol. 2013;85:1402–1408. doi: 10.1002/jmv.23612. [DOI] [PubMed] [Google Scholar]
- 16.Toan P.Q., Bao Quyen L.T., Thu Hang D.T., My Anh T.T., Cuong L.M., Lanh N.S., Su H.X. Identification of BK Virus Genotypes in Recipients of Renal Transplant in Vietnam. Transplant. Proc. 2019;51:2683–2688. doi: 10.1016/j.transproceed.2019.03.072. [DOI] [PubMed] [Google Scholar]
- 17.Comerlato J., Souza Campos F., Trindade Oliveira M., Cibulski S.P., Corrêa L., Roos Kulmann M.I., Souza Arantes T., Peretti Hentges L., Rosado Spilki F., Roehe P.M., et al. Molecular detection and characterization of BK and JC polyomaviruses in urine samples of renal transplant patients in Southern Brazil. J. Med. Virol. 2015;87:522–528. doi: 10.1002/jmv.24086. [DOI] [PubMed] [Google Scholar]
- 18.Pape L., Tönshoff B., Hirsch H.H. Perception, diagnosis and management of BK polyomavirus replication and disease in paediatric kidney transplant recipients in Europe. Nephrol. Dial. Transplant. 2016;31:842–847. doi: 10.1093/ndt/gfv392. [DOI] [PubMed] [Google Scholar]
- 19.Jin L., Gibson P.E., Knowles W.A., Clewley J.P. BK virus antigenic variants: Sequence analysis within the capsid VP1 epitope. J. Med. Virol. 1993;39:50–56. doi: 10.1002/jmv.1890390110. [DOI] [PubMed] [Google Scholar]
- 20.Luo C., Bueno M., Kant J., Martinson J., Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J. Virol. 2009;83:2285–2297. doi: 10.1128/JVI.02180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korth J., Anastasiou O.E., Bräsen J.H., Brinkhoff A., Lehmann U., Kribben A., Dittmer U., Verheyen J., Wilde B., Ciesek S., et al. The detection of BKPyV genotypes II and IV after renal transplantation as a simple tool for risk assessment for PyVAN and transplant outcome already at early stages of BKPyV reactivation. J. Clin. Virol. 2019;113:14–19. doi: 10.1016/j.jcv.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Zhong S., Randhawa P.S., Ikegaya H., Chen Q., Zheng H.Y., Suzuki M., Takeuchi T., Shibuya A., Kitamura T., Yogo Y. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J. Gen. Virol. 2009;90:144–152. doi: 10.1099/vir.0.83611-0. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt C., Raggub L., Linnenweber-Held S., Adams O., Schwarz A., Heim A. Donor origin of BKV replication after kidney transplantation. J. Clin. Virol. 2014;59:120–125. doi: 10.1016/j.jcv.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz A., Linnenweber-Held S., Heim A., Framke T., Haller H., Schmitt C. Viral origin, clinical course, and renal outcomes in patients with BK virus infection after living-donor renal transplantation. Transplantation. 2016;100:844–853. doi: 10.1097/TP.0000000000001066. [DOI] [PubMed] [Google Scholar]
- 25.Nishimoto Y., Zheng H.Y., Zhong S., Ikegaya H., Chen Q., Sugimoto C., Kitamura T., Yogo Y. An Asian origin for subtype IV BK virus based on phylogenetic analysis. J. Mol. Evol. 2007;65:103–111. doi: 10.1007/s00239-006-0269-6. [DOI] [PubMed] [Google Scholar]
- 26.Nukuzuma S., Takasaka T., Zheng H.-Y., Zhong S., Chen Q., Kitamura T., Yogo Y. Subtype I BK polyomavirus strains grow more efficiently in human renal epithelial cells than subtype IV strains. J. Gen. Virol. 2006;87:1893–1901. doi: 10.1099/vir.0.81698-0. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda Y., Qazi Y., Iwaki Y. A rapid and efficient method BK polyomavirus genotyping by high resolution melting analysis. J. Med. Virol. 2011;83:2128–2134. doi: 10.1002/jmv.22239. [DOI] [PubMed] [Google Scholar]
- 28.Tremolada S., Akan S., Otte J., Khalili K., Ferrante P., Chaudhury P.R., Gordon J. Rare subtypes of BK virus are viable and frequently detected in renal transplant recipients with BK virus-associated nephropathy. Virology. 2010;404:312–318. doi: 10.1016/j.virol.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Morel V., Martin E., François C., Helle F., Faucher J., Mourez T., Choukroun G., Duverlie G., Castelain S., Brochot E. A simple and reliable strategy for BK virus subtyping and subgrouping. J. Clin. Microbiol. 2017;55:1177–1185. doi: 10.1128/JCM.01180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varella R.B., Zalona A.C.J., Diaz N.C., Zalis M.G., Santoro-Lopes G. BK polyomavirus genotypes Ia and Ib1 exhibit different biological properties in renal transplant recipients. Virus Res. 2018;243:65–68. doi: 10.1016/j.virusres.2017.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request.