Abstract
A culture-independent phylogenetic survey for an anaerobic trichlorobenzene-transforming microbial community was carried out. Small-subunit rRNA genes were PCR amplified from community DNA by using primers specific for Bacteria or Euryarchaeota and were subsequently cloned. Application of a new hybridization-based screening approach revealed 51 bacterial clone families, one of which was closely related to dechlorinating Dehalobacter species. Several clone sequences clustered to rDNA sequences obtained from a molecular study of an anaerobic aquifer contaminated with hydrocarbons and chlorinated solvents (Dojka et al., Appl. Env. Microbiol. 64:3869–3877, 1998).
Due to their widespread application in industry and agriculture and their chemical stability, chlorobenzenes (CB) are ubiquitous pollutants in soil, sediments, and aquifers (17). Since the toxicity of CBs increases with the number of chlorine substituents (14), microbial dechlorination of CBs is of major interest. In contrast to di- and monochlorobenzenes (DCB and MCB), more highly chlorinated benzenes are more resistant to aerobic dechlorination. However, for these compounds, reductive dechlorination by different anaerobic microbial communities could be demonstrated (1, 8, 15, 16, 19). Bioreactors inoculated with complex dechlorinating anaerobic microbiota seem to be promising technologies for bioremediation of CB-contaminated aquifers. Several studies showed the efficiency of such bioreactors in treating chloroaromatic-contaminated wastewater (6, 26). However, due to the unknown species diversity, microbial activity had to be treated as a “black box” and direct optimization was hampered. To determine the microbial diversity of an anaerobic consortium (16) in a fluidized bed reactor (FBR) used for dechlorination of trichlorobenzene (TCB), we employed comparative sequence analysis of 16S rRNA genes after direct PCR amplification and cloning from community DNA, since culture-based methods have been shown to be strongly biased (2, 7). Here we report the successful application of a new hybridization-based screening approach for bacterial 16S rDNA clone libraries and describe numerous novel 16S rDNA sequences representing yet-uncultured microorganisms of the anaerobic TCB-transforming consortium.
A 5-liter FBR was inoculated with the anaerobic TCB-transforming consortium (16) immobilized on polyurethane foam cubes (1 cm3) and was supplied with a mineral salt medium (21) containing TCB (10 mg/liter), sodium acetate (100 mg/liter), and methanol (100 mg/liter). Transformation of TCB to DCB, MCB, and benzene was analyzed by gas chromatography three times per week. The FBR was operated for several months under stable conditions (pH 7.0; redox potential, −300 mV), transforming about 80 to 90% of supplied TCB. For extraction of community DNA, one foam cube was removed from the bioreactor. Cells were broken by lysozyme treatment (3 mg of lysozyme/ml in 6 ml of 0.15 M NaCl–0.01 M EDTA at 37°C for 3 h) and by the addition of 6 ml of a sodium dodecyl sulfate solution (0.5 M Tris-HCl–0.1 M NaCl–10% SDS [pH 8]) followed by six cycles of freezing on dry ice and thawing at 65°C for 15 min. Remaining intact cells were lysed by incubation at 50°C after 160 μl of a proteinase K solution (20 mg/ml) was added. The lysate was extracted with phenol and chloroform, and nucleic acids were subsequently ethanol precipitated. PCR inhibitors and fragmented DNA were removed from high-molecular-weight DNA by using preparative agarose gel electrophoresis (13) and QiaQuick spin columns (Qiagen, Düsseldorf, Germany). To overcome some of the reported pitfalls in the determination of microbial diversity by PCR-mediated direct analysis of rRNA (for a review, see reference 23), a total of four 16S rDNA clone libraries were generated from the same batch of community DNA, which differed in PCR and cloning conditions. In PCRs A and B, 16S rDNA was amplified with Bacteria-specific primers TPU1 (AGAGTTTGATCMTGGCTCAG; Escherichia coli positions 8 to 27) and RTU8 (AAGGAGGTGATCCANCCRCA; E. coli positions 1522 to 1541) by using the following cycling conditions (parameters for reaction B are given in brackets): 98°C for 3 [2] min, 93°C for 3 [1] min, addition of AmpliTaq polymerase (Perkin-Elmer, Weiterstadt, Germany), 28 cycles of 93°C for 1 min, 53°C for 1 min, and 72°C for 3 [5] min, and final extension at 72°C for 7 min. PCR products were cloned either by TA cloning (Invitrogen, de Schelp, Netherlands), resulting in clone library A, or by ligation-independent cloning (3), resulting in clone library B. In PCRs C and D, 16S rDNA was amplified with Euryarchaeota-specific reverse primer 1390Ra (CGGTGTGTGCAAGGAGC; E. coli positions 1385 to 1401 [18]) and Euryarchaeota-specific forward primer 10-30Fa (TCCGGTTGATCCTGCC; E. coli positions 8 to 23 [18]) (reaction C) or 357Fa (ACGGGGCGCAGCAGGC; E. coli positions 344 to 359 [18]) (reaction D) using a tricine buffer with 5% acetamide (4) and the following cycling conditions (parameters for reaction D are given in brackets): 94°C for 6 min, addition of AmpliTaq polymerase, 30 [27] cycles of 92°C for 1.5 min, 50°C for 1.5 min, and 72°C for 2 [4] min, and final extension at 72°C for 7 min. PCR products of both reactions were cloned by TA cloning, resulting in clone libraries C and D. In all PCRs, the amount of template DNA for maximum synthesis of amplification products was empirically determined. DNA sequencing of 16S rDNA clone inserts was performed by using the Thermo Sequenase Fluorescent Labelled Primer Cycle Sequencing kit (Amersham, Braunschweig, Germany) and an automated LI-COR 4000 or 4000L DNA sequencer (MWG-Biotech, Ebersberg, Germany).
Euryarchaeotal rDNA clone libraries C and D were dominated by clone sequences nearly identical to the 16S rDNA of Methanosaeta concilii (97.7 to 99.7%). Hybridization of 153 clones of library C (SJC clones) and of 56 clones of library D (SJD clones) with the novel digoxigenin (DIG)-labeled oligonucleotide probe MCONC (GAGTACGCTGGCAACTGT; E. coli positions 1120 to 1135) specific for M. concilii and closely related clone sequences revealed 136 (88.8%) and 32 (57.1%) positive clones, respectively. Due to the higher diversity of bacterial clone libraries A and B, the design of clone-specific oligonucleotides seemed not to be appropriate to enable a rapid comparison of both libraries. Consequently, a new screening approach to avoid sequencing of identical or closely related clone sequences was developed; it was based on hybridization with DIG-labeled 16S rDNA fragments (a detailed protocol is available on request). Briefly, DIG-labeled rDNA fragments of single clones were generated by nested PCR with primers TPU1 and RTU2 (TGCCTCCCGTAGGAGTYTGG; E. coli positions 334 to 353) from full-length 16S rDNA clone inserts, which had been previously PCR amplified from plasmid DNA with insert flanking primers M13(−40)F (GTTTTCCCAGTCACGAC) and M13(−24)R (AACAGCTATGACCATG). Resulting DIG-labeled rDNA fragments (about 350 bp in length) were used for filter hybridization of purified plasmid DNA by using a hybridization buffer containing 50% deionized formamide and the DIG Luminescent Detection kit (Boehringer, Mannheim, Germany). A total of 145 full-length rDNA inserts of clone library A (SJA clones) and of 163 rDNA inserts of clone library B (SJB clones) was hybridized with 29 different DIG-labeled 16S rDNA fragments of SJA clones. All SJA clones not identified by hybridization were sequenced. This combined approach allowed the detection of 52 distinct clone families in clone library A (Fig. 1). Hybridization results revealed that four clone families showed significantly different distributions in both clone libraries and seven unique SJA clones could not be detected in library B. However, clone families SJA-36, SJA-102, and SJA-186 represented more than one-fourth of all SJA and SJB clones (28.8% of SJA clones, 26.4% of SJB clones), and 16 clone families of library A were present in both libraries with similar low frequencies (between 0.6 and 4.9%), corresponding to one to eight positive clones. We therefore concluded that the different cloning approaches (TA versus ligation-independent cloning) and slight variations of PCR conditions had no major impact on the composition of clone libraries A and B and that consequently SJA clone families roughly represented bacterial diversity of the TCB-transforming consortium. By using both fractional treeing based on overlapping datasets of about 400 nucleotides and the CHECK_CHIMERA tool (12), one full-length SJA sequence (SJA-7) was found to be chimeric. For phylogenetic analysis, the remaining 51 representative full-length SJA sequences were initially compared to the available databases by using the BlastN tool of the HUSAR software package (DKFZ, Heidelberg, Germany) to determine their approximate phylogenetic affiliations. SJA sequences and phylogenetically related sequences were incorporated into a dendrogram (tree_1400_feb97) of the ARB database (22) by using the ARB parsimony insertion tool. Prior to phylogenetic analyses, highly variable regions or those of uncertain alignment were excluded by omitting, among others, the first 104 positions (E. coli numbering) by using a consensus mask generated by the ARB program (22). The phylogenetic tree shown in Fig. 2 was constructed from 1,228 positions by applying the Jukes and Cantor correction (11) and the neighbor-joining method (20) with 1,000 bootstrap resamplings.
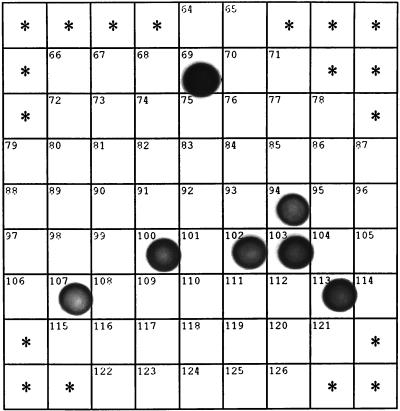
FIG. 1.
Dot blot hybridization with the DIG-labeled rDNA fragment of clone SJA-103 of purified plasmid DNA from SJA clones. Inserts of clones SJA-69, SJA-100, and SJA-102 were sequenced and showed similarity values between 96.5 and 98.1%, indicating that all SJA-103-positive clones belonged to the same clone family.
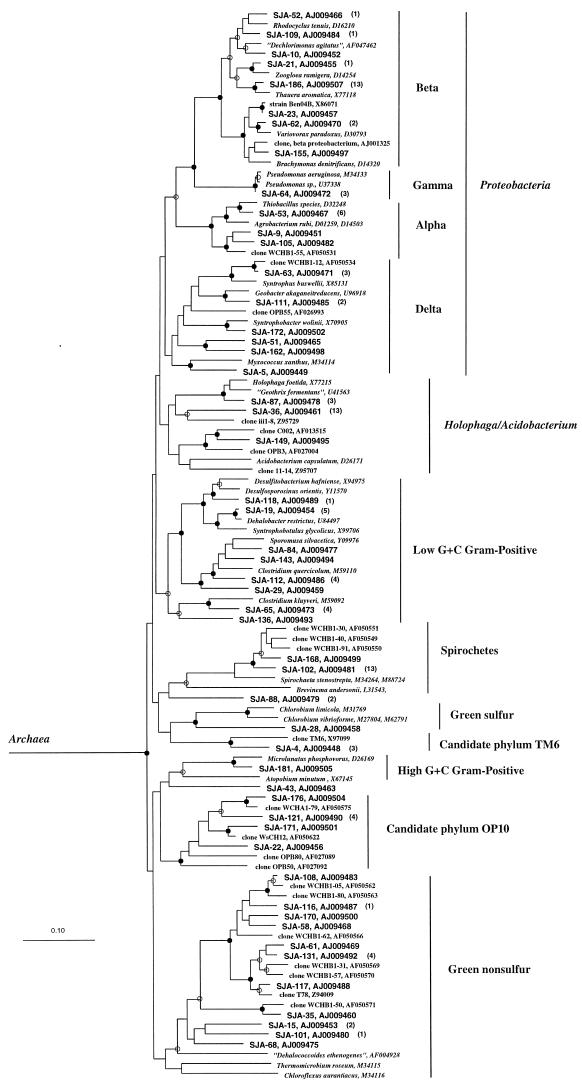
FIG. 2.
Phylogenetic tree showing the relationships of SJA sequences and bacterial 16S rDNA sequences. M. concilii (X16932) and Thermoplasma acidophilum (M20822) were used as an outgroup. The number of additional clone sequences belonging to the individual SJA clone families is given in brackets. Environmental rDNA clone sequences obtained from a contaminated aquifer in an independent study (5) are indicated by the prefixes WCHB1, WCHA1, or WsCH. Branching points supported by bootstrap values >74% are indicated by filled circles. Open circles indicate branching points, which are supported by bootstrap values between 50 to 74%. Branching points without circles were not resolved (bootstrap values, <50%). The scale bar represents 10% sequence divergence.
In this study, a new screening approach for bacterial 16S rDNA clone libraries derived from the TCB-transforming consortium was developed. Hybridization with the DIG-labeled 16S rDNA fragments turned out to be a powerful method for the detection of identical or closely related clone sequences (clone families) within bacterial rDNA clone libraries. Compared to other screening procedures, this novel approach had the following advantages. (i) In contrast to the application of group-specific oligonucleotide probes (13) or restriction fragment length polymorphism analysis of 16S rDNA inserts (24), amplified inserts detected only identical or closely related clones. (ii) Amplicons were specific for the respective inserts without the need of prior sequencing of targeted and nontargeted rDNA inserts, in contrast to the design of insert-specific oligonucleotide probes. (iii) Hybridization and washing conditions were almost the same for all 29 16S rDNA fragments, thus avoiding the laborious optimization effort required for oligonucleotide probes.
Comparative sequence analysis revealed that none of the SJA sequences was identical to 16S rRNA sequences of cultured microorganisms. However, several SJA sequences were closely related to cultivated representatives of well-known bacterial genera (homologies ranging from 96.7 to 99.7%) (Fig. 2). Of these, rDNA sequences of clone family SJA-19 showed similarities between 98.8 and 99.4% to the 16S rDNA of Dehalobacter restrictus. This indicates that these sequences may represent yet-uncultivated members of the genus Dehalobacter. Both known members of this genus are strictly anaerobic bacteria that grow by reductive dechlorination of chlorinated ethene (9, 25). This suggests a similar respiratory process for the yet-uncultivated bacteria described here. Since rDNA clone libraries are not suited to show the quantitative distribution of microorganisms in an ecosystem, direct methods, such as whole-cell hybridization with specific 16S rRNA-targeted oligonucleotides or quantitative hybridization of extracted rRNA, will help to elucidate the functional role of the SJA-19 organisms by correlating cell numbers with the TCB-dechlorination activity in the bioreactor.
In a recent study, Dojka et al. (5) investigated the microbial diversity of an anaerobic aquifer contaminated with hydrocarbons and chlorinated solvents. Interestingly, as in our study, an archaeal rDNA clone library obtained from community DNA was dominated by clone sequences nearly identical to M. concilii (81% of all clones). Furthermore, several SJA sequences affiliated with the green nonsulfur phylum, the Proteobacteria, the candidate phylum OP10 (10), and the spirochetes were closely related to bacterial rDNA sequences retrieved from the contaminated aquifer (Fig. 2). This correspondence might reflect an important role of these yet-uncultivated microorganisms in the anaerobic transformation or degradation of chlorinated organic compounds in a polluted environment, possibly making these bacteria useful indicator organisms. The rDNA sequences obtained in this study represent a solid framework for quantitative, 16S rRNA-based investigations of our TCB-transforming community, which will be necessary to verify this hypothesis.
Nucleotide sequence accession numbers.
Sixty full-length SJA sequences and eight SJC and SJD sequences were deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AJ009448 to AJ009515.
Acknowledgments
We thank Andreas Zapf (TU-Berlin, Germany) for analysis of chlorobenzenes and Wolfgang Ludwig and Oliver Strunk (TU-München, Germany) for providing the ARB package and for many helpful instructions. F.v.W. is especially grateful to Erko Stackebrandt (DSMZ, Braunschweig, Germany) for his extended help and to Fred Rainey (Lousiana State University) for providing primer sequences.
This work was supported by grants of the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 193, “Biological Treatment of Industrial Wastewaters,” given to W.H. and to U.B.G., and by a grant of the Studienstiftung des Deutschen Volkes given to F.v.W.
REFERENCES
- 1.Adrian L, Manz W, Szewzyk U, Görisch H. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl Environ Microbiol. 1998;64:496–503. doi: 10.1128/aem.64.2.496-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslanidis C, de Jong P J, Schmitz G. Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl. 1994;4:172–177. doi: 10.1101/gr.4.3.172. [DOI] [PubMed] [Google Scholar]
- 4.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flora J R V, Suidan M T, Wuellner A M, Boyer T K. Anaerobic treatment of a simulated high-strength industrial wastewater containing chlorophenols. Water Environ Res. 1994;66:21–31. [Google Scholar]
- 7.Göbel U B. Phylogenetic amplification for the detection of uncultured bacteria and the analysis of complex microbiota. J Microbiol Methods. 1995;23:117–128. [Google Scholar]
- 8.Holliger C, Schraa G, Stams A J M, Zehnder A J B. Enrichment and properties of an anaerobic mixed culture reductively dechlorinating 1,2,3-trichlorobenzene to 1,3-dichlorobenzene. Appl Environ Microbiol. 1992;58:1636–1644. doi: 10.1128/aem.58.5.1636-1644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder A J B. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- 10.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 12.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malle K-G. Die Bedeutung der 129 Stoffe der EG-Liste für den Gewässerschutz. Zeitung Wasser-Abwasser-Forsch. 1984;17:75–81. [Google Scholar]
- 15.Middeldorp P J M, De Wolf J, Zehnder A J B, Schraa G. Enrichment and properties of a 1,2,4-trichlorobenzene-dechlorinating methanogenic microbial consortium. Appl Environ Microbiol. 1997;63:1225–1229. doi: 10.1128/aem.63.4.1225-1229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak J, Kirsch N H, Hegemann W, Stan H-J. Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture enriched from Saale river sediment. Appl Microbiol Biotechnol. 1996;45:700–709. [Google Scholar]
- 17.Oliver B G, Nicole K D. Chlorobenzenes in sediments, water, and selected fish from Lakes Superior, Huron, Erie, and Ontario. Environ Sci Technol. 1982;16:532–536. doi: 10.1021/es00114a014. [DOI] [PubMed] [Google Scholar]
- 18.Rainey, F. A. Personal communication.
- 19.Ramanand K, Balba M T, Duffy J. Reductive dehalogenation of chlorinated benzenes and toluenes under methanogenic conditions. Appl Environ Microbiol. 1993;59:3266–3272. doi: 10.1128/aem.59.10.3266-3272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Shelton D R, Tiedje J M. General method for determining anaerobic biodegradation potential. Appl Environ Microbiol. 1984;48:850–857. doi: 10.1128/aem.47.4.850-857.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strunk O, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 23.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 24.Weidner S, Arnold W, Pühler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wild A, Hermann R, Leisinger T. Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichlorethene. Biodegradation. 1996;7:507–511. doi: 10.1007/BF00115297. [DOI] [PubMed] [Google Scholar]
- 26.Woods S L, Ferguson J F, Benjamin M M. Characterization of chlorophenol and chloromethoxybenzene biodegradation during anaerobic treatment. Environ Sci Technol. 1989;23:62–68. [Google Scholar]