Abstract
Background
Circular RNAs (circRNAs) serve as critical regulators in the chemoresistance of human cancers, including non‐small cell lung cancer (NSCLC). We aimed to explore the role of hsa_circ_0011298 (circ_0011298) and its mechanism in Taxol resistance of NSCLC.
Methods
Circ_0011298, microRNA‐486‐3p (miR‐486‐3p), and CRABP2 mRNA expression were determined using qRT‐PCR. EdU and MTT assays were used to detect cell proliferation. Cell cycle distribution and cell apoptosis were detected by flow cytometry. Cell migratory and invasive abilities were detected using transwell assay. Cellular glycolysis was determined by specific kits. Protein levels were examined by western blot. Dual‐luciferase reporter and RIP assays were performed to confirm the relationship between miR‐486‐3p and circ_0011298 or CRABP2. Xenograft mice model was established to confirm the function of circ_0011298 in vivo.
Results
Circ_0011298 was overexpressed in Taxol‐resistant NSCLC cells and tissues. Circ_0011298 knockdown enhanced Taxol sensitivity by decreasing cell proliferation, migration, invasion, and glycolysis and inducing apoptosis and cell cycle arrest in Taxol‐resistant NSCLC cells. Circ_0011298 was a sponge of miR‐486‐3p, and the impact of circ_0011298 silencing on Taxol resistance was rescued by miR‐486‐3p inhibition. Moreover, miR‐486‐3p directly targeted CRABP2, and miR‐486‐3p inhibited Taxol resistance by targeting CRABP2. Furthermore, circ_0011298 regulated CRABP2 expression through targeting miR‐486‐3p. Importantly, circ_0011298 interference elevated Taxol sensitivity of NSCLC in vivo.
Conclusion
Circ_0011298 elevated Taxol resistance of NSCLC by sponging miR‐486‐3p and upregulating CRABP2, providing a possible circRNA‐targeted therapy for NSCLC.
Keywords: circ_0011298, CRABP2, miR‐486‐3p, non‐small cell lung cancer, Taxol resistance
Circ_0011298 and CRABP2 were upregulated while miR‐486‐3p was downregulated in taxol resistance of non‐small cell lung cancer. Circ_0011298 promotes the cell viability, DNA synthesis, migration, invasion, and glycolysis, while reduces cell cycle arrest and apoptosis in taxol‐resistant non‐small cell lung cancer cells by sponging miR‐486‐3p and upregulating CRABP2, thereby accelerating taxol resistance.
1. INTRODUCTION
Lung cancer is a common and aggressive cancer, and non‐small cell lung cancer (NSCLC) is the major type of lung cancer. 1 , 2 Although tremendous efforts have been made in NSCLC treatment, the survival rate for patients remains poor. 3 Taxol (also known as paclitaxel) is identified as an effective first‐line chemotherapy drug for many cancers, including NSCLC. 4 Nevertheless, many NSCLC patients have developed Taxol resistance or even multiple‐drug resistance during chemotherapy, leading to treatment failure. 5 Hence, it is essential to clarify the mechanism of Taxol resistance in NSCLC and identify a new therapeutic method that can sensitize Taxol‐resistant NSCLC cells to Taxol.
Circular RNAs (circRNAs) are featured by covalent closed loops with neither 5′‐end cap nor 3′‐end poly (A) tail 6 . Recently, many circRNAs are involved in the regulation of pathological processes and multiple cellular activities. 7 , 8 Growing evidence has revealed that some circRNAs have essential roles in NSCLC by acting as tumor suppressors or promoters. 9 , 10 Moreover, circRNAs play key roles in Taxol resistance of many cancers. For example, circCELSR1 promoted tumor growth and Taxol resistance in breast cancer. 11 Circ‐PVT1 knockdown inhibited paclitaxel resistance in gastric cancer cells via increasing cell apoptosis and suppressing proliferation and invasion. 12 A previous study has shown that hsa_circ_0011298 (circ_0011298; chr1:32050751–32051086) is derived from tubulointerstitial nephritis antigen like 1 (TINAGL1) gene and overexpressed in Taxol‐resistant NSCLC cells, 13 suggesting that circ_0011298 might be associated with the resistance of NSCLC to Taxol. Nevertheless, the precise function of circ_0011298 and its regulatory mechanism in Taxol resistance of NSCLC have not been reported.
CircRNAs can serve as microRNA (miRNA) sponges or competitive endogenous RNAs (ceRNAs) by competitively binding to miRNA response elements, thereby regulating target gene expression. 14 , 15 Dysregulation of miRNAs is closely related to cancer development and chemoresistance. 16 , 17 MiR‐486‐3p serves as a tumor‐suppressive miRNA in lung cancer. 18 Nevertheless, the function of miR‐486‐3p and the connection between miR‐486‐3p and circ_0011298 in Taxol resistance of NSCLC are still unclear. It is widely acknowledged that miRNAs can regulate various physiological and biological processes via binding with target mRNAs, resulting in the expression suppression and function restriction of mRNAs. 19 CRABP2 plays a promotional role in lung cancer progression. 20 However, whether CRABP2 can be directly targeted by miR‐486‐3p and its function in Taxol resistance of NSCLC have not been studied.
In this study, we detected circ_0011298 expression in Taxol‐resistant NSCLC cells and tissues and explored its role in Taxol resistance of NSCLC. Additionally, the relationships among circ_0011298, miR‐486‐3p, and CRABP2 were explored in Taxol resistance of NSCLC. The aim of our work was to provide promising strategies for the Taxol‐resistant NSCLC patients.
2. MATERIALS AND METHODS
2.1. Specimen collection
NSCLC tissues (n = 65) and corresponding normal tissues (n = 65) were acquired from NSCLC patients at Dongguan Hospital of Guangzhou University of Chinese Medicine. Based on the response of patients with NSCLC to Taxol, these tissues were divided into two groups: Taxol‐sensitive (n = 35) and Taxol‐resistant groups (n = 30), and were promptly frozen in liquid nitrogen. Informed consent was acquired from all enrolled SCLC patients. This research was permitted by the Research Ethics Committee of Dongguan Hospital of Guangzhou University of Chinese Medicine.
2.2. Cell culture and transfection
NSCLC cells (A549 and H1299) were commercially provided by COBIOER. All cells were grown in RPMI‐1640 medium (Invitrogen) containing 10% FBS (Invitrogen) and maintained in standard conditions (37, 5% CO2). The corresponding Taxol‐resistant NSCLC cells (A549/Taxol and H1299/Taxol) were generated via exposing the parental cells (A549 and H1299) to increasing doses of Taxol (Sigma‐Aldrich). Then, Taxol (50 nM) was added into the culture medium to maintain a resistant phenotype.
Short‐hairpin RNA targeting circ_0011298 (si‐circ_0011298#1 and si‐circ_0011298#2) and shRNA control (sh‐NC), mimics or inhibitor of miR‐486‐3p (miR‐486‐3p and anti‐miR‐486‐3p) and matched control (miR‐NC or anti‐NC), CRABP2‐overexpressing vector (CRABP2), and corresponding control (vector) were commercially provided by GenePharma. Cells were introduced with the plasmid or/and oligonucleotide by Lipofectamine 3000 reagent (Invitrogen).
2.3. Quantitative real‐time reverse transcription–polymerase chain reaction (qRT‐PCR)
After extraction of RNA with Trizol reagent (Invitrogen), the synthesis of first‐strand cDNA was conducted using M‐MLV Reverse Transcriptase kit (Promega) or miScript II RT kit (Invitrogen). Next, qRT‐PCR reaction was manipulated with SYBR Green PCR Master Mix kit (Invitrogen). Primer sequences: circ_0011298 (F, 5′‐CACTGCCCCAACAGCTATGT‐3′; R, 5′‐GAAGGGGTAGCAGTGGTCAG‐3′); TINAGL1 (F, 5′‐CCTCTTGACCAAGGCAACTGTG‐3′; R, 5′‐GGTGTCACAAGACAGCAGGTTC‐3′); miR‐486‐3p (F, 5′‐GTATGACGGGGCAGCTCAGTA‐3′; R, 5′‐CAGTGCGTGTCGTGGAGT‐3′); miR‐616 (F, 5′‐GCCGAGAGTCATTGGAGGGT −3′; R, 5′‐CAGTGCGTGTCGTGGAGT‐3’); CRABP2 (F, 5′‐TTGAGGAGCAGACTGTGGATGG‐3′; R, 5′‐GTTCTCTGGTCCACGAGGTCTT‐3′); GAPDH (F, 5′‐GTCTCCTCTGACTTCAACAGCG‐3′; R, 5′‐ACCACCCTGTTGCTGTAGCCAA‐3′); and U6 (F, 5′‐CTCGCTTCGGCAGCACATATACT‐3′; R, 5′‐ACGCTTCACGAATTTGCGTGTC‐3′). The levels of genes were evaluated with 2−ΔΔCt method and normalized to U6 (for miRNA) or GAPDH (for circRNA/mRNA).
2.4. 3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyl‐tetrazolium bromide (MTT) assay
MTT assay was applied to analyze cell viability and IC50 value of Taxol. Cell suspension (100 uL) was inoculated into 96‐well plates. After treatment with Taxol or transfection with oligonucleotide or/and vector, MTT (5 mg/ml; 10 µl; Solarbio) reagent was placed into per well and further incubated for 3–4 h. DMSO (200 µl; Sigma‐Aldrich) was added into per well after removing the medium. The absorbance was detected with a microplate reader at 490 nm. IC50 value of Taxol was calculated using the relative survival curve.
2.5. CircRNA identification
Circ_0011298 and TINAGL1 were amplified with oligo (dT)18 or random primers. At last, qRT‐PCR was conducted to confirm whether circ_0011298 contained poly (A) tail.
Total RNA was treated with or without RNase R (Seebio) for 0.5 h at 37℃. Next, circ_0011298 and TINAGL1 mRNA levels were assessed by qRT‐PCR.
For Actinomycin D assay, Actinomycin D (Sigma‐Aldrich) was added to cells for blocking transcription. Thereafter, qRT‐PCR was used to detect circ_0011298 and TINAGL1 mRNA expression.
2.6. Subcellular fraction assay
Cytoplasm and nucleus fractions were isolated with PARIS™ kit (Invitrogen). Circ_0011298, U6 (as a nucleus control) and GAPDH (as a cytoplasm control) levels were assessed by qRT‐qPCR.
2.7. 5‐ethynyl‐20‐deoxyuridine (EdU) assay
KFluor488 EdU kit (KeyGene, Nanjing, China) was used to test DNA synthesis and cell proliferation. In short, A549/Taxol and H1299/Taxol cells were seeded into a 12‐well plate. After transfection, cells were then treated with EdU solution (50 μM) for 2 h and fixed with 4% paraformaldehyde for 20 min. Next, 0.5% TritonX‐100 was sued to permeabilize cells, and then cells were stained with Click‐It reaction in a dark place for 0.5 h. Afterwards, cell nucleus was stained with DAPI. Finally, cells were observed using a fluorescence microscope (Leica).
2.8. Flow cytometry analysis
For cell cycle assay, A549/Taxol and H1299/Taxol cells were collected after transection and fixed in ethanol (70%; Beyotime) for 12 h at −20℃. After fixation, the cells were harvested via centrifugation, followed by addition of propidium iodide (PI; KeyGene) and RNase A (Beyotime). FACScan® flow cytometry was employed to detect cell cycle distribution.
For detection of cell apoptosis, the transfected cells were harvested. After suspending in binding buffer (0.5 ml), cells were labeled with Annexin V‐FITC and PI (Beyotime), followed by measurement of apoptotic cells with FACScan® flow cytometry.
2.9. Transwell assay
The transwell chambers were employed to evaluate cell invasive and migratory capacities. The only difference was that the top chambers were coated with Matrigel for detecting cell invasion. In brief, serum‐free medium (200 µl) containing transfected cells (A549/Taxol and H1299/Taxol) was seeded into the upper chamber. In the bottom compartment, complete medium (600 µl) was used as a chemoattractant. Twenty‐four hours later, the invaded/migrated cells were fixed in methanol, dyed using crystal violet, and examined by microscopy (Leica).
2.10. Glycolysis detection
Apo SENSOR ADP/ATP Ratio Assay kit (BioVision), Glucose Assay kit (BioVision), and Lactate Assay kit (BioVision) were used to detect ATP/ADP ratio, glucose uptake, and lactate production, respectively, following the manufacturer's protocol.
2.11. Western blot assay
Total protein was extracted by RIPA lysis buffer (Keygen). After detection of protein concentration, extracted protein was separated with SDS‐PAGE (Beyotime). After transferring onto PVDF membranes (Millipore), the membranes were blocked in non‐fat dried milk (Yili). After that, the membranes were probed with unique primary antibodies: KI67 (ab181255; 1:5000; Abcam), Bax (ab32503; 1:3000; Abcam), multidrug resistance protein (MRP1; ab76464; 1:1000; Abcam), CRABP2 (ab181255; 1:3000; Abcam), and β‐actin (ab8226; 1:1000; Abcam), and then secondary antibody was used for combination with primary antibodies. At last, the combined signals were detected by ECL reagent (Abcam).
2.12. Dual‐luciferase reporter assay
The relationship between miR‐486‐3p and circ_0011298 or CRABP2 was predicted by Circular RNA Interactome or TargetScan. The fragments of circ_0011298 or CRABP2 containing miR‐486‐3p‐binding sequence or not were inserted into pmirGLO vector (GenePharma) to form wild‐type reporter vector (circ_0011298 WT and CRABP2 3′UTR WT) and mutated reporter vector (circ_0011298 MUT, CRABP2 3′UTR MUT). Next, cells were introduced with reporter vector and miR‐NC/miR‐486‐3p for 48 h. The luciferase activities were detected using Dual‐Luciferase Reporter Assay System (Promega).
2.13. RNA immunoprecipitation (RIP) assay
EZ‐Magna RIP kit (Millipore) was utilized to conduct RIP assay. Briefly, cells were harvested and lysed using RIP lysis buffer, and cell extracts were then treated with magnetic beads previously binding to human antibody against immunoglobulin G (Anti‐IgG; as a control) or Argonaute2 (Anti‐Ago2). Subsequently, the RNA in the immunoprecipitates was extracted for detection of miR‐486‐3p and circ_0011298 enrichment by qRT‐PCR.
2.14. Tumor formation assay in vivo
The following work was approved by the Animal Care and Use Committee of Dongguan Hospital of Guangzhou University of Chinese Medicine. BALB/c nude mice (6‐week‐old, male, n = 12, weighing 20–25 g) were commercially provided by Vital River. Briefly, A549/Taxol cells (1 × 106, circ_0011298 downregulating or control) were inoculated into nude mice. Seven days later, these mice were intraperitoneally administrated with 3 mg/kg of Taxol every 3 days. The tumor volume was measured using a caliper every 7 days. Thirty‐five days later, the mice were killed for subsequent analysis.
2.15. Immunohistochemistry (IHC)
Tumor tissues were fixed in formalin solution (10%) and embedded in paraffin, followed by cutting into 4‐μm‐thick section. Afterwards, these sections were probed with the primary antibody: CRABP2 (ab181255; 1:100; Abcam) or Ki67 (ab15580; 1:200; Abcam). Next, these sections were incubated with secondary antibody (ab171870; 1:2000; Abcam). After staining with diaminoaniline (DAB; Maxim, Fuzhou, China) and counterstaining with hematoxylin (Maxim), the sections were observed using a microscopy (Leica).
2.16. Statistical analysis
All data in our research were expressed as the mean ± standard deviation from at least three independent experiments and analyzed by GraphPad Prism. Student's t‐test or ANOVA was used for calculating significant differences. The correlations among circ_0011298, miR‐486‐3p, and CRABP2 were analyzed using spearman's correlation coefficient analysis. The significance was determined when p< 0.05.
3. RESULTS
3.1. Circ_0011298 was upregulated in Taxol‐resistant NSCLC
Circ_0011298 level was increased in Taxol‐resistant NSCLC cells (A549/Taxol and H1299/Taxol) compared to parental cells (A549 and H1299) (Figure 1A). Next, cells were exposed to different doses of Taxol and then MTT was used to measure IC50. As expected, IC50 value of Taxol in Taxol‐resistant NSCLC cells was higher than that in parental cells (Figure 1B), confirming the production of Taxol resistance. Similarly, circ_0011298 level was demonstrated to be increased in Taxol‐sensitive NSCLC tissues relative to normal tissues, and circ_0011298 level was higher in Taxol‐resistant NSCLC tissues than that in Taxol‐sensitive NSCLC tissues (Figure 1C). Next, we analyzed the characteristics of circ_0011298 in A549/Taxol and H1299/Taxol cells. Circ_0011298 level was decreased when oligo(dT)18 primers were used (Figure 1D,E), indicating the absence of a poly (A) tail for circ_0011298. Circ_0011298 was resistant to RNase R (Figure 1F,G), suggesting the cyclic structure of circ_0003998. The half‐life of TINAGL1 mRNA transcript was about 8 h, while the half‐life of circ_0011298 transcript exceeded 24 h (Figure 1H,I), implying that circ_0011298 transcript was more stable. Subsequently, the localization of circ_0011298 in Taxol‐resistant NSCLC cells was detected. Circ_0011298 was demonstrated to be primarily located in the cytoplasm (Figure 1J).
FIGURE 1.
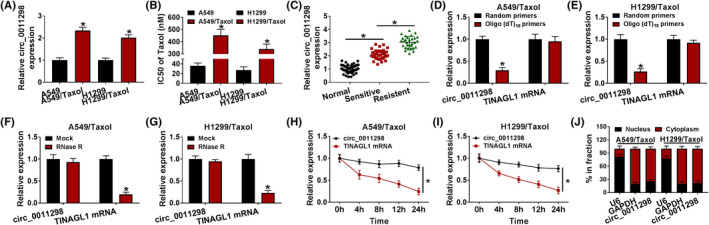
High expression of circ_0011298 was detected in Taxol‐resistant NSCLC cells and tissues. A, Circ_0011298 expression was detected using qRT‐PCR in Taxol‐resistant NSCLC cells (A549/Taxol and H1299/Taxol) and parental cells (A549 and H1299). B, IC50 of Taxol in Taxol‐resistant NSCLC cells was measured by MTT assay. C, Circ_0011298 level in normal, Taxol‐sensitive NSCLC, and Taxol‐resistant NSCLC tissues was determined. D and E, Crc_0011298 and TINAGL1 mRNA levels were detected by qRT‐PCR after reverse transcription with oligo (dT)18 primers and random primers in Taxol‐resistant NSCLC cells. F‐I, After treatment with RNase R or Actinomycin D, qRT‐PCR was used to measure circ_0011298 and linear TINAGL1 levels in Taxol‐resistant NSCLC cells. J, The localization of circ_0011298 was determined in the nuclear and cytosolic fractions. *p < 0.05
3.2. Circ_0011298 knockdown elevated Taxol sensitivity in Taxol‐resistant NSCLC cells
Lose‐of‐function experiments were performed in Taxol‐resistant NSCLC cells to explore the role of circ_0011298. Circ_0011298 level was reduced in cells by transfection with sh‐circ_0011298#1 and sh‐circ_0011298#2, and circ_0011298 expression was lower after transfection with sh‐circ_0011298#1 than si‐circ_0011298#2 group; however, transfection of sh‐circ_0011298#1 or sh‐circ_0011298#2 could not affect the mRNA expression of TINAGL1 (Figure 2A,B). Therefore, we choose sh‐circ_0011298#1 for further research. MTT and EdU analyses demonstrated that circ_0011298 silencing decreased cell viability and DNA synthesis in Taxol‐resistant NSCLC cells (Figure 2C‐E), revealing that circ_0011298 interference suppressed cell proliferative ability. Circ_0011298 downregulation increased G0/G1 phase and reduced S phase (Figure 2F,G). In addition, circ_0011298 knockdown caused significant apoptosis in Taxol‐resistant NSCLC cells (Figure 2H). A549/Taxol and H1299/Taxol cell migration and invasion were repressed by downregulation of circ_0011298 (Figure 2I,J). Moreover, IC50 value of Taxol was decreased in A549/Taxol and H1299/Taxol after deficiency of circ_0011298 (Figure 2K). Glycolysis can promote cancer progression. Next, the effect of circ_0011298 on glycolysis was studied. Downregulation of circ_0011298 suppressed glycolysis via reducing glucose uptake, lactate production, and ATP/ADP ratio (Figure 2L‐N). The protein expression of Ki67 (a proliferation marker), Bax (a pro‐apoptotic molecule), and MRP1 (drug resistance‐associated protein) was then measured. As presented in Figure 2O,P, circ_0011298 silencing inhibited Ki67 and MRP1 protein levels and increased Bax protein abundance.
FIGURE 2.

Circ_0011298 knockdown sensitized Taxol‐resistant NSCLC cells to Taxol. A and B, Circ_0011298 and TINAGL1 mRNA levels were detected in Taxol‐resistant NSCLC cells introduced with sh‐NC, sh‐circ_0011298#1, or sh‐circ_0011298#2. C‐P, Taxol‐resistant NSCLC cells were introduced with sh‐NC or sh‐circ_0011298#1. C and D, MTT assay was used to measure cell viability. (E) DNA synthesis was determined by EdU analysis. F‐H, Flow cytometry analysis was performed to analyze cell cycle process and apoptosis. I and J, Transwell assay was performed to detect cell migratory and invasive abilities (×100). K, IC50 of Taxol was assessed using MTT assay. L‐N, Glucose uptake, lactate consumption, and cellular ATP/ADP ratio were assessed using corresponding kits. O and P, Western blot was conducted to analyze Ki67, Bax, and MRP1 protein levels. *p < 0.05
3.3. Circ_0011298 was a sponge of miR‐486‐3p
CircRNA can act as a sponge for miRNA. 21 MiR‐486‐3p and miR‐616 were found to have higher prediction scores by using Circular RNA Interactome scores. Next, miR‐486‐3p and miR‐616 levels were explored in normal tissue samples and resistant/sensitive NSCLC tissues specimens, and we demonstrated that miR‐486‐3p level was reduced in Taxol‐sensitive NSCLC tissue samples in contrast to normal tissue samples and miR‐486‐3p level further decreased in Taxol‐resistant NSCLC tissues; however, miR‐616 expression presented an opposite effect (Figure 3A). Hence, miR‐486‐3p was chosen for further study. The predicted binding sequence between circ_0011298 and miR‐486‐3p was displayed in Figure 3B. MiR‐486‐3p expression was remarkably elevated after introduction with miR‐486‐3p and reduced after introduction with anti‐miR‐486‐3p (Figure 3C), suggesting that transfection efficiency was high. As shown in Figure 3D,E, upregulation of miR‐486‐3p led to a reduction in the luciferase intensity of circ_0011298 WT, but this change was not found in circ_0011298 MUT group. MiR‐486‐3p and circ_0011298 were obviously enriched in Ago2 group with respect to IgG group (Figure 3F,G). Additionally, miR‐486‐3p expression was found to be reduced in Taxol‐resistant NSCLC cells relative to parental cells (Figure 3H). Likewise, miR‐486‐3p expression was declined in Taxol‐sensitive NSCLC tissues, and its expression was lower in Taxol‐resistant NSCLC tissues (Figure 3I). An inverse correlation between circ_0011298 and miR‐486‐3p expression was observed in Taxol‐resistant NSCLC tissues (Figure 3J). Furthermore, we observed that circ_0011298#1 transfection increased miR‐486‐3p level, which was reversed by anti‐miR‐486‐3p co‐transfection (Figure 3K).
FIGURE 3.
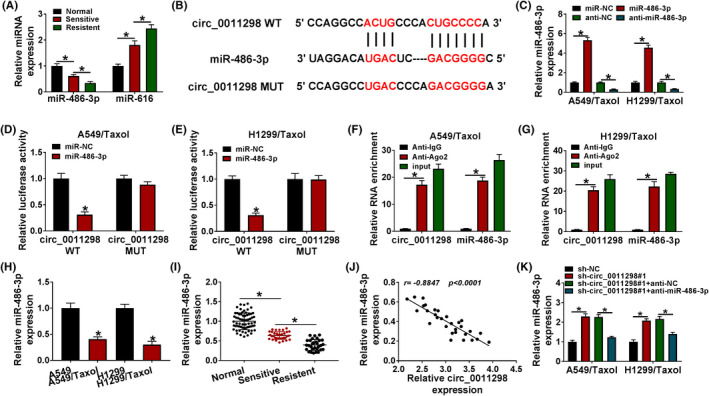
MiR‐486‐3p was sponged by circ_0011298. A, The expression of miR‐486‐3p and miR‐616 (potential target miRNAs of circ_0011298) was determined in normal, Taxol‐sensitive NSCLC, and Taxol‐resistant NSCLC tissues. B, The putative binding sites of miR‐486‐3p and circ_0011298 were exhibited. C, MiR‐486‐3p expression was measured in Taxol‐resistant NSCLC cells introduced with miR‐NC, miR‐486‐3p, anti‐NC, or anti‐miR‐486‐3p. D and E, Taxol‐resistant NSCLC cells were co‐introduced with circ_0011298 WT/MUT and miR‐NC/miR‐486‐3p, and the luciferase activity was detected. F and G, Circ_0011298 and miR‐486‐3p enrichment were determined by RIP assay in Taxol‐resistant NSCLC cells. H, Detection of miR‐486‐3p level in Taxol‐resistant NSCLC cells and parental cells. I, MiR‐486‐3p level was tested in normal, Taxol‐sensitive NSCLC, and Taxol‐resistant NSCLC tissues. J, The correlation between circ_0011298 and miR‐486‐3p in Taxol‐resistant NSCLC tissues was analyzed. K, MiR‐486‐3p level was measured in Taxol‐resistant NSCLC cells introduced with sh‐NC, sh‐circ_0011298#1, sh‐circ_0011298#1+anti‐NC, or sh‐circ_0011298#1+anti‐miR‐486‐3p. *p < 0.05
3.4. Circ_0011298 silencing repressed Taxol resistance in Taxol‐resistant NSCLC cells via upregulating miR‐486‐3p
To explore the association between circ_0011298 and miR‐486‐3p in regulating Taxol resistance, rescue assays were performed. MTT and EdU assays indicated that miR‐486‐3p suppression abrogated the repressive effect of circ_0011298 interference on proliferation (Figure 4A‐C). Moreover, circ_0011298 downregulation‐caused cell cycle arrest and apoptosis were relieved via anti‐miR‐486‐3p reintroduction (Figure 4D‐F). Also, the inhibiting effects of circ_0011298 downregulating on migration and invasion were counteracted by decreasing miR‐486‐3p (Figure 4G,H). Furthermore, the influence of circ_0011298 interference on decrease in IC50 value of Taxol, glucose uptake, lactate production, and ATP/ADP ratio was neutralized via inhibiting miR‐486‐3p (Figure 4I‐L). MiR‐486‐3p reduction reversed the effects of circ_0011298 deficiency on decreasing Ki67 and MRP1 protein expression and increasing Bax protein expression (Figure 4M,N).
FIGURE 4.
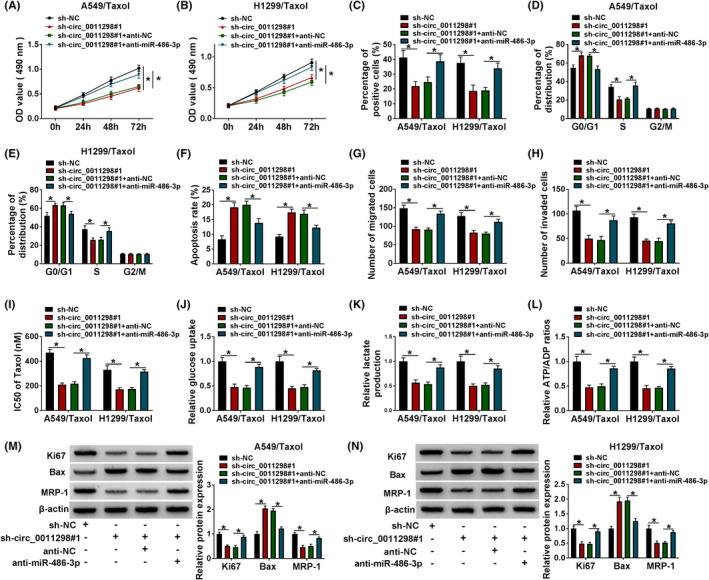
Circ_0011298 knockdown enhanced Taxol sensitivity in Taxol‐resistant NSCLC cells by targeting miR‐486‐3p. A‐N, A549/Taxol and H1299/Taxol cells were transfected with sh‐NC, sh‐circ_0011298#1, sh‐circ_0011298#1+anti‐NC, or sh‐circ_0011298#1+anti‐miR‐486‐3p. A‐C, Cell proliferation was analyzed. D‐F, Cell cycle distribution and cell apoptosis were detected. (G and H) Cell migratory and invasion capacities were evaluated. I, IC50 value of Taxol was examined. (J‐L) Glucose uptake, lactate consumption, and cellular ATP/ADP ratio were determined. M and N, Ki67, Bax, and MRP1 protein levels were analyzed. *p < 0.05
3.5. CRABP2 was targeted by miR‐486‐3p
By searching for TargetScan, the downstream target for miR‐486‐3p was explored. As presented in Figure 5A, CRABP2 possessed the possible binding sequence for miR‐486‐3p. MR‐486‐3p overexpression showed a dramatic suppression in the luciferase activity of CRABP2 3’UTR WT but not CRABP2 3’UTR MUT (Figure 5B,C). GEPIA showed that CRABP2 expression was upregulated in both LUAD and LUSC tissues relative to normal tissues, and patients in high CRABP2 expression group showed poor survival (Figure 5D,E). The levels of CRABP2 mRNA and protein were increased in Taxol‐sensitive NSCLC tissues; moreover, its expression was higher in Taxol‐resistant NSCLC tissues (Figure 5F,I). In addition, CRABP2 mRNA expression was inversely associated with miR‐486‐3p expression and positively correlated with circ_0011298 level in Taxol‐resistant tissues (Figure 5G,H). Moreover, an enhancement in CRABP2 protein level was detected in Taxol‐resistant NSCLC cells relative to parental cells (Figure 5J). Upregulation of miR‐486‐3p could reduce CRABP2 protein expression in Taxol‐resistant NSCLC cells (Figure 5K). Downregulation of circ_0011298 reduced the protein expression of CRABP2, which was neutralized by decreasing miR‐486‐3p expression (Figure 5L,M), suggesting that circ_0011298 sponged miR‐486‐3p to positively modulate CRABP2 expression.
FIGURE 5.
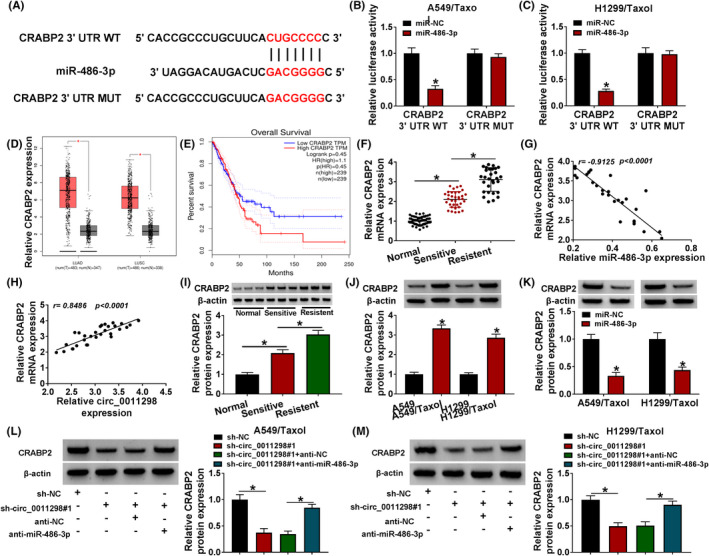
MiR‐486‐3p directly interacted with CRABP2. A, The predicted binding sites between CRABP2 and miR‐486‐3p were presented. B and C, The relationship between CRABP2 and miR‐486‐3p was verified by dual‐luciferase reporter assay. D and E, GEPIA database showed CRABP2 expression in tumor and normal tissue samples, and the survival rate for patients with high and low CRABP2 expression. F, CRABP2 mRNA expression was determined in normal, Taxol‐sensitive NSCLC, and Taxol‐resistant NSCLC tissues. G and H, The correlation between CRABP2 and miR‐486‐3p or circ_0011298 in Taxol‐resistant NSCLC tissues was analyzed. I and J, CRABP2 protein expression was examined in normal tissues, Taxol‐sensitive NSCLC tissues, Taxol‐resistant NSCLC tissues, Taxol‐resistant NSCLC cells, and parental cells. K, The protein expression of CRABP2 was examined in Taxol‐resistant NSCLC cells after transfection with miR‐NC/miR‐486‐3p. L and M, CRABP2 protein expression was measured in Taxol‐resistant NSCLC cells after introduction with sh‐NC, sh‐circ_0011298#1, sh‐circ_0011298#1+anti‐NC, or sh‐circ_0011298#1+anti‐miR‐486‐3p. *p < 0.05
3.6. MiR‐486‐3p targeted CRABP2 to increase Taxol sensitivity in Taxol‐resistant NSCLC cells
As illustrated in Figure 6A, CRABP2 protein expression was increased by transfection of CRABP2 in Taxol‐resistant NSCLC cells, indicating a high transfection efficiency. To elucidate whether miR‐486‐3p exerted its roles via targeting CRABP2 in Taxol‐resistant NSCLC cells, rescue experiments were performed. Enhancement of miR‐486‐3p reduced cell proliferative ability in Taxol‐resistant NSCLC cells, which was abated by elevating CRABP2 (Figure 6B‐D). Enforced expression of miR‐486‐3p arrested cells in G0/G1 phase and increased cell apoptosis as well as inhibited migratory and invasive abilities, which could be reversed by overexpression of CRABP2 (Figure 6E‐I). MiR‐486‐3p overexpression reduced the IC50 value of Taxol and glycolysis in Taxol‐resistant NSCLC cells, but these changes were counteracted by CRABP2 elevation (Figure 6J‐M). Meanwhile, restoration of miR‐486‐3p led to reduction in the protein levels of Ki67 and MRP1 and promotion in Bax protein expression, which was counteracted by upregulating CRABP2 (Figure 6N).
FIGURE 6.
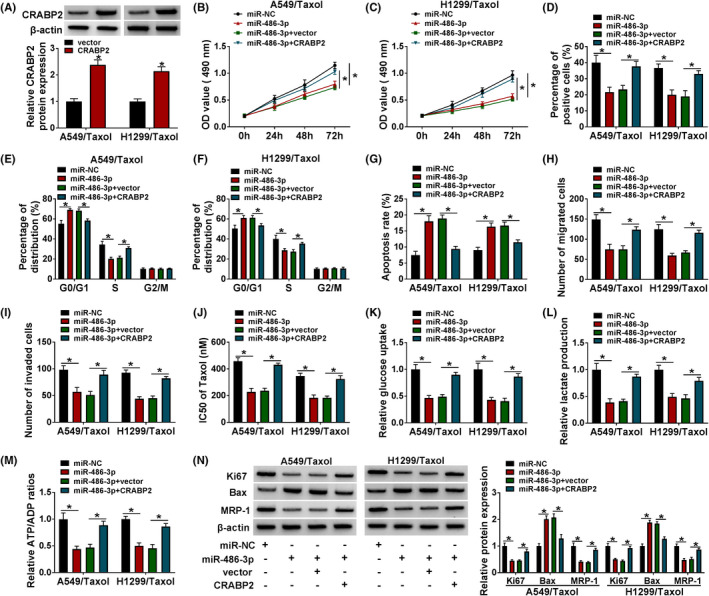
Upregulation of miR‐486‐3p inhibited Taxol resistance by downregulating CRABP2 in Taxol‐resistant NSCLC cells. A, The protein expression of CRABP2 was examined in Taxol‐resistant NSCLC cells introduced with vector or CRABP2. (B‐N) Taxol‐resistant NSCLC cells were introduced with miR‐NC, miR‐486‐3p, miR‐486‐3p+vector, or miR‐486‐3p+CRABP2. B‐D, Cell proliferation was evaluated. E‐G, Cell cycle distribution and cell apoptosis were detected. H and I, Cell migration and invasion were measured. J, The IC50 of Taxol was calculated by MTT assay. K‐M, Glucose uptake, lactate consumption, and ATP/ADP ratio were examined. N, Ki67, Bax, and MRP1 protein levels were determined. *p < 0.05
3.7. Circ_0011298 downregulation enhanced Taxol sensitivity of NSCLC in vivo
The volume of tumor in Taxol+sh‐circ_0011298#1 group grew slower than that in Taxol+sh‐NC group (Figure 7A). Figure 7B showed the representative photographs of the tumors. In addition, Taxol treatment and circ_0011298 knockdown reduced tumor volume compared with Taxol treatment alone (Figure 7C). Moreover, circ_0011298 expression and CRABP2 mRNA and protein expression were reduced and miR‐486‐3p expression was enhanced in Taxol+sh‐circ_0011298#1 group relative to Taxol+sh‐NC group (Figure 7D,E). IHC analysis showed that Taxol treatment and circ_0011298 downregulation decreased CRABP2 and Ki67 expression compared to Taxol+sh‐NC group (Figure 7F). Collectively, circ_0011298 deficiency repressed Taxol resistance of NSCLC in vivo.
FIGURE 7.
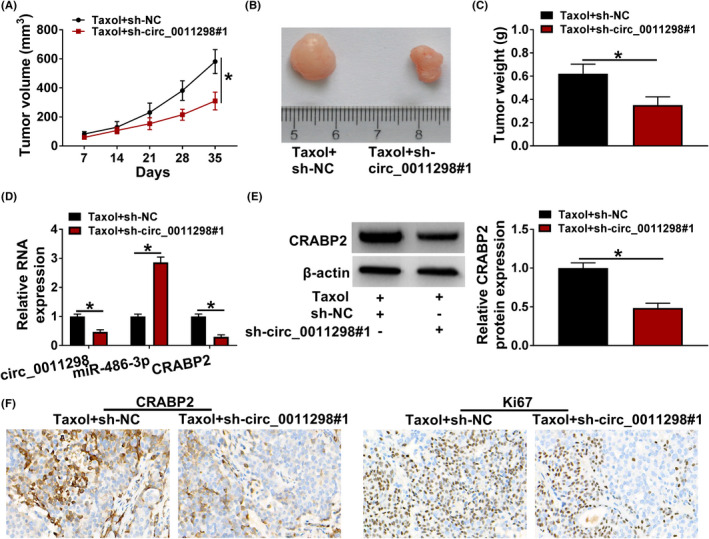
Circ_0011298 interference increased Taxol sensitivity of NSCLC in vivo. A, Tumor volume was recorded every 7 days. B, Images of the tumors were displayed. C, After 35 days inoculation, tumor weight was examined. D, Circ_0011298, miR‐486‐3p, and CRABP2 mRNA levels were determined. E, CRABP2 protein abundance was detected. F, IHC analysis was used to examine CRABP2 and Ki67 expression. *p < 0.05
4. DISCUSSION
Although Taxol‐based chemotherapy is particularly effective in most cancer cases, the acquisition of Taxol resistance is a major obstacle for cancer treatment. 22 , 23 In this research, we confirmed that circ_0011298 interference reduced the resistance of Taxol‐resistant NSCLC cells through regulating miR‐486‐3p/CRABP2 axis.
Many circRNAs have been gradually identified in various diseases, including cancers. 24 Several circRNAs are dysregulated and involved in regulation of Taxol resistance in NSCLC. For example, circ_ZFR augmented the Taxol resistance and progression of NSCLC by modulating KPNA4/miR‐195‐5p axis. 25 Circ_0011292 was upregulated in NSCLC, and accelerated tumorigenesis and Taxol resistance in NSCLC by regulating TRIM65 and sponging miR‐379‐5p. 26 In addition, hsa_circ_0002874 silencing could reverse Taxol resistance in NSCLC. 27 In this research, the influence of circ_0011298 on Taxol resistance was explored in Taxol‐resistant NSCLC cells. Herein, circ_0011298 was overexpressed in Taxol‐resistant NSCLC cells and tissues, which was in line with former research. 13 Functionally, circ_0011298 knockdown sensitized Taxol‐resistant NSCLC cells to Taxol through inhibiting cell growth, cell cycle progression, invasion, migration, and glycolysis and inducing apoptosis. Importantly, circ_0011298 downregulation could block tumor growth to improve Taxol sensitivity in NSCLC in vivo. Our data revealed that circ_0011298 increased the resistance of NSCLC to Taxol.
Many studies have suggested that circRNAs located in the cytoplasm can exert their functions by interacting with miRNAs to release mRNAs that are targeted by miRNAs. 28 Here, circ_0011298 was demonstrated to be primarily located in the cytoplasm. Hence, circ_0011298 might promote Taxol resistance of NSCLC through directly binding to miRNAs. In our study, circ_0011298 was first verified to interact with miR‐486‐3p. MiR‐486‐3p is able to function as a suppressor in some cancers and is related to drug resistance. For instance, Ji et al. disclosed that miR‐486‐3p expression was reduced in sorafenib‐resistant hepatocellular carcinoma cells and miR‐486‐3p‐targeted FGFR4 could overcome sorafenib resistance. 29 Wu et al. declared that miR‐486‐3p could improve temozolomide sensitivity through targeting MGMT in glioblastoma. 30 Moreover, Pan et al. claimed that miR‐486‐3p showed a low abundance in lung cancer, and upregulation of miR‐486‐3p suppressed lung cancer cell viability, invasion, and migration. 18 We wondered whether miR‐486‐3p could regulate Taxol sensitivity of NSCLC. Low abundance of miR‐486‐3p in Taxol‐resistant NSCLC cell lines and tissue samples was observed. MiR‐486‐3p reduction could revere the suppressing influence of circ_0011298 downregulation on Taxol resistance, indicating that circ_0011298 regulated Taxol sensitivity through targeting miR‐486‐3p.
Next, we further explored the possible targets for miR‐486‐3p. Our data revealed that CRABP2 was directly targeted by miR‐486‐3p, and circ_0011298 sponged miR‐486‐3p to upregulate CRABP2 expression. Many researches have suggested that CRABP2 is involved in regulation of the drug sensitivity in several cancers. 31 , 32 , 33 In addition, Favorskaya et al. reported that CRABP2 mRNA level was elevated in NSCLC samples. 34 Moreover, Kim et al. proved that plasma CRABP2 level was increased in NSCLC patients. 35 More importantly, Wu et al. suggested that CRABP2 silencing limited the metastasis and growth of lung cancer cells. 20 In our research, CRABP2 was overexpressed in Taxol‐resistant NSCLC tissue samples and cell lines. CRABP2 overexpression neutralized miR‐486‐3p‐mediated improvement of Taxol sensitivity in Taxol‐resistant NSCLC cells, implying that miR‐486‐3p exerted its function in Taxol resistance of NSCLC cells by targeting CRABP2.
In conclusion, circ_0011298 was overexpressed in Taxol‐resistant NSCLC, and circ_0011298 enhanced Taxol resistance by sponging miR‐486‐3p and upregulating CRABP2. Our research may provide a new molecular mechanism for Taxol resistance and a possible circRNA‐targeted therapy for NSCLC patients with Taxol resistance.
5. COMPLIANCE WITH ETHICAL STANDARDS
This research was permitted by the Research Ethics Committee of Dongguan Hospital of Guangzhou University of Chinese Medicine.
The following work was approved by the Animal Care and Use Committee of Dongguan Hospital of Guangzhou University of Chinese Medicine.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Wu Y, Xie J, Wang H, Hou S, Feng J. Circular RNA hsa_circ_0011298 enhances Taxol resistance of non‐small cell lung cancer by regulating miR‐486‐3p/CRABP2 axis. J Clin Lab Anal. 2022;36:e24408. doi: 10.1002/jcla.24408
Funding information
None
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10‐29. [DOI] [PubMed] [Google Scholar]
- 3. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non‐small‐cell lung cancer. Lancet. 2016;388(10048):1012‐1024. [DOI] [PubMed] [Google Scholar]
- 4. Zhang D, Qiu L, Jin X, Guo Z, Guo C. Nuclear factor‐kappaB inhibition by parthenolide potentiates the efficacy of Taxol in non‐small cell lung cancer in vitro and in vivo. Mol Cancer Res. 2009;7(7):1139‐1149. [DOI] [PubMed] [Google Scholar]
- 5. Pu J, Shen J, Zhong Z, Yanling M, Gao J. KANK1 regulates paclitaxel resistance in lung adenocarcinoma A549 cells. Artif Cells Nanomed Biotechnol. 2020;48(1):639‐647. [DOI] [PubMed] [Google Scholar]
- 6. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu P, Wang L, Xie X, et al. Hsa_circ_0001869 promotes NSCLC progression via sponging miR‐638 and enhancing FOSL2 expression. Aging (Albany NY). 2020;12(23):23836‐23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen T, Yang Z, Liu C, et al. Circ_0078767 suppresses non‐small‐cell lung cancer by protecting RASSF1A expression via sponging miR‐330‐3p. Cell Prolif. 2019;52(2):e12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang W, Gong P, Yang Y, et al. Circ‐ABCB10 contributes to paclitaxel resistance in breast cancer through Let‐7a‐5p/DUSP7 axis. Cancer Manag Res. 2020;12:2327‐2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu YY, Zhang LY, Du WZ. Circular RNA circ‐PVT1 contributes to paclitaxel resistance of gastric cancer cells through the regulation of ZEB1 expression by sponging miR‐124‐3p. Biosci Rep. 2019;39(12):BSR20193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu N, Chen S, Liu Y, et al. Profiles and bioinformatics analysis of differentially expressed circrnas in taxol‐resistant non‐small cell lung cancer cells. Cell Physiol Biochem. 2018;48(5):2046‐2060. [DOI] [PubMed] [Google Scholar]
- 14. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67‐79. [DOI] [PubMed] [Google Scholar]
- 16. Liu T, Guo J, Zhang X. MiR‐202‐5p/PTEN mediates doxorubicin‐resistance of breast cancer cells via PI3K/Akt signaling pathway. Cancer Biol Ther. 2019;20(7):989‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Lu M, Jin J, et al. miR‐449a suppresses tamoxifen resistance in human breast cancer cells by targeting ADAM22. Cell Physiol Biochem. 2018;50(1):136‐149. [DOI] [PubMed] [Google Scholar]
- 18. Pan J, Huang G, Yin Z, et al. Circular RNA FLNA acts as a sponge of miR‐486‐3p in promoting lung cancer progression via regulating XRCC1 and CYP1A1. Cancer Gene Ther. 2022;29(1):101‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3’ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu JI, Lin YP, Tseng CW, Chen HJ, Wang LH. Crabp2 Promotes metastasis of lung cancer cells via HuR and Integrin β1/FAK/ERK signaling. Sci Rep. 2019;9(1):845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. [DOI] [PubMed] [Google Scholar]
- 22. Stordal B, Pavlakis N, Davey R. A systematic review of platinum and taxane resistance from bench to clinic: an inverse relationship. Cancer Treat Rev. 2007;33(8):688‐703. [DOI] [PubMed] [Google Scholar]
- 23. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575(7782):299‐309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134‐142. [DOI] [PubMed] [Google Scholar]
- 25. Li J, Fan R, Xiao H. Circ_ZFR contributes to the paclitaxel resistance and progression of non‐small cell lung cancer by upregulating KPNA4 through sponging miR‐195‐5p. Cancer Cell Int. 2021;21(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo C, Wang H, Jiang H, Qiao L, Wang X. Circ_0011292 enhances paclitaxel resistance in non‐small cell lung cancer by regulating miR‐379‐5p/TRIM65 Axis. Cancer Biother Radiopharm. 2020;37(2):84–95. [DOI] [PubMed] [Google Scholar]
- 27. Xu J, Ni L, Zhao F, et al. Overexpression of hsa_circ_0002874 promotes resistance of non‐small cell lung cancer to paclitaxel by modulating miR‐1273f/MDM2/p53 pathway. Aging (Albany NY). 2021;13(4):5986‐6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. [DOI] [PubMed] [Google Scholar]
- 29. Ji L, Lin Z, Wan Z, et al. miR‐486‐3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020;11(4):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H, Li X, Zhang T, et al. Overexpression miR‐486‐3p promoted by Allicin enhances temozolomide sensitivity in glioblastoma via targeting MGMT. Neuromolecular Med. 2020;22(3):359‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X, Li H, Wu ML, et al. Resveratrol reverses retinoic acid resistance of anaplastic thyroid cancer cells via demethylating CRABP2 gene. Front Endocrinol (Lausanne). 2019;10:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Enikeev AD, Komelkov AV, Axelrod ME, et al. CRABP1 and CRABP2 protein levels correlate with each other but do not correlate with sensitivity of breast cancer cells to retinoic acid. Biochemistry (Mosc). 2021;86(2):217‐229. [DOI] [PubMed] [Google Scholar]
- 33. Gupta S, Pramanik D, Mukherjee R, et al. Molecular determinants of retinoic acid sensitivity in pancreatic cancer. Clin Cancer Res. 2012;18(1):280‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Favorskaya I, Kainov Y, Chemeris G, et al. Expression and clinical significance of CRABP1 and CRABP2 in non‐small cell lung cancer. Tumour Biol. 2014;35(10):10295‐10300. [DOI] [PubMed] [Google Scholar]
- 35. Kim DJ, Kim WJ, Lim M, et al. Plasma CRABP2 as a novel biomarker in patients with non‐small cell lung cancer. J Korean Med Sci. 2018;33(26):e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


