Abstract
Background
Lung cancer is one of the most common malignancies globally and a significant component of cancer‐related deaths. The lack of early diagnosis accounts for detecting approximately 75% of cancer patients at an intermediate to an advanced stage, with a low 5‐year survival rate. Therefore, a more comprehensive understanding of the molecular mechanisms of lung cancer development is necessary to find reliable and effective therapeutic and diagnostic biomarkers.
Methods
circ_SAR1A, miR‐21‐5p, and TXNIP in lung cancer tissues, animal xenografts, and cell lines were validated by qRT‐PCR and western blotting analyses. RNase R digestion and nuclear/cytoplasm fractionation experiments were utilized to determine the stability and localization of circ_SAR1A in lung cancer cells. The binding between miR‐21‐5p and circ_SAR1A or TXNIP was confirmed by luciferase reporter, RNA pull‐down, Spearman's correlation, and rescue assays. CCK‐8, colony formation, flow cytometry, Transwell, and western blotting were utilized to illustrate the malignant behavior of lung cancer cells.
Results
circ_SAR1A and TXNIP were down‐regulated while miR‐21‐5p was up‐regulated in lung cancer samples and cells. circ_SAR1A was located predominantly in the cytoplasm; it inhibited lung cancer growth in vitro and in vivo by sponging to miR‐21‐5p. miR‐21‐5p silencing suppressed lung cancer malignancy by targeting TXNIP.
Conclusions
circ_SAR1A is a critical negative regulator of lung carcinogenesis. circ_SAR1A/miR‐21‐5p/TXNIP attenuation inhibited lung cancer progression, presenting an ideal diagnostic and a potential therapeutic target.
Keywords: circ_SAR1A, lung cancer, metastasis, miR‐21‐5p, TXNIP
circ_SAR1A was down‐regulated in lung cancer. circ_SAR1A restrained malignancy properties of lung cancer cells. circ_SAR1A inhibited tumor growth in vivo. circ_SAR1A was a ceRNA for miR‐21‐5p. circ_SAR1A suppressed malignant properties in lung cancer cells by sponging miR‐21‐5p. TXNIP was a direct downstream target of miR‐21‐5p. miR‐21‐5p interference inhibited lung cancer cell growth via targeting TXNIP.
1. INTRODUCTION
Lung cancer is one of the most common malignant tumors globally. 1 According to the World Health Organization (WHO), approximately 9.6 million people died from cancer worldwide in 2018. 2 The cancers with higher mortality rates are lung cancer, colorectal cancer, gastric cancer, hepatocellular carcinoma, and breast cancer. 2 Currently, surgery, radiotherapy, and chemotherapy are the three main lung cancer treatments, which are still relatively conservative. 3 Lung cancer patients cannot be treated accordingly due to the lack of apparent characteristics of the early stage of lung cancer. The primary treatment for patients with advanced lung cancer is radiotherapy combined with chemotherapy. 4 However, radiotherapy and chemoradiotherapy cannot eradicate lung cancer. Studies have confirmed that the 5‐year survival rate of lung cancer patients is only 15%. 5 Hence, it is urgent to identify novel biomarkers for lung cancer diagnosis and treatment.
Circular RNAs (circRNAs) are novel covalently closed continuous‐loop RNAs without 5′‐end caps and 3′‐poly(A) tails. 6 , 7 , 8 circRNAs are not hydrolyzed by nucleic acid exonucleases (RNase R) and are stable in cells, tissues, and body fluids, making circRNAs more stable than linear RNAs. 9 Meanwhile, circRNAs have cell‐specific and tissue‐specific origins. 10 These properties made circRNAs noteworthy molecules in research. Numerous studies have found that circRNAs are involved in the development and progression of various diseases, such as tumors, 11 rheumatic diseases, 12 cardiovascular diseases, 13 neurological diseases, 14 and endocrine diseases. 15 Previous studies have shown that circRNAs are aberrantly expressed in a variety of tumors, such as gastric cancer, 16 hepatocellular carcinoma, 17 and lung cancer. 18 However, the role of circ_SAR1A in lung cancer remains unexplored.
MicroRNAs (miRNAs) are a class of highly conserved endogenous non‐coding single‐stranded RNAs of 18–25 nt in length that is widely found in various organisms. 19 Among a variety of miRNAs, current studies have shown miR‐21‐5p as a crucial oncogenic miRNA (oncomiR) that is highly expressed in various tumors, including gastric cancer, 20 breast cancer, 21 ovarian cancer, 22 cervical cancer, 23 colorectal cancer, 24 hepatocellular carcinoma, 25 glioblastoma, 26 and lung cancer. 27 In addition, tumor growth, apoptosis, metastasis, and other biological phenomenon are regulated by miR‐21‐5p. 28 , 29 However, the regulatory role of circ SAR1A in lung cancer progression remains obscure.
This research aimed to investigate the role of circ_SAR1A and miR‐21‐5p in lung cancer malignant behaviors. Also, the relationship between circ_SAR1A and miR‐21‐5p was investigated.
2. METHODS
2.1. Sample collection
We collected tumor specimens and paired normal paracancerous specimens from 68 lung cancer patients who underwent surgery from December 2017 to November 2020. The samples were obtained from the Nantong First People's Hospital sample bank. Immediately after surgery, tissue specimens were frozen in liquid nitrogen until further use. None of these patients had received radiotherapy or chemotherapy prior to surgery. All samples were pathologically examined, and none were lost during the study period. This study was approved by the Ethics Committee of Nantong First People's Hospital. All patients were aware of the study and provided informed consent.
2.2. Cell culture and transfection
All lung cancer cell lines H1650, H1581, H460, H1299, and A549, and normal human bronchial epithelial cells HBE, were grown in 10% fetal bovine serum (FBS)‐supplemented RPMI1‐1640 complete medium. Cells were typically grown in an incubator at 37°C and 5% CO2. circ‐SAR1A overexpression vector (ov‐SAR1A), negative control (ov‐NC), TXNIP lentiviral expression vector (sh‐TXNIP), negative control sh‐NC, miR‐21‐5p mimics, miR‐21‐5p inhibitor, and negative control (miR‐NC) were purchased from GenePharma. These plasmids were transfected individually or combined into A549 cells for subsequent experiments.
2.3. RNA extraction and quantification
Total RNA was extracted from human tissues and cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The concentration and purity of RNA samples were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher). cDNA was prepared using a commercial reverse transcription kit (Takara). Real‐time quantitative PCR analysis of circRNA, miRNA, and mRNA was performed using an SYBR Green PCR kit (Takara) and the primers listed in Table 1. GAPDH and U6 were used as internal references, and all analyses were performed in triplicates. The relative expression of intracellular circRNA, miRNA, and mRNA was analyzed by the 2−ΔΔCT method.
TABLE 1.
Primer sequences for qRT‐PCR analysis
| Forward | Reverse | |
|---|---|---|
| miR‐21‐5p | 5′‐ CTTACTTCTCTGTGTGATTTCTGTG −3′ | 5′‐ ACAACCTTTCCAAAATCCATGAGGC −3′ |
| U6 | 5′‐TCCGATCGTGAAGCGTTC−3′ | 5′‐GTGCAGGGTCCGAGGT−3′ |
| TXNIP | 5′‐TGTGTGAAGTTACTCGTGTCAAA−3′ | 5′‐GCAGGTACTCCGAAGTCTGT−3′ |
| GAPDH | 5′‐AGAAGGCTGGGGCTCATTTG−3′ | 5′‐AGGGGCCATCCACAGTCTTC−3′ |
| circ_SAR1A | 5′‐ GTTGAACCAGAAAACATTTTC−3 | 5′‐ AGACTTCCTGTTTGGTGATC −3′ |
2.4. Nucleus/cytoplasm fractionation assay
P0028 Extraction Reagent kit (Beyotime) was used to separate the nuclear and cytoplasmic content from tissues or cells. First, P0028 Extraction Reagents A and B were left at room temperature until melted, then homogenized and placed on ice immediately. Cells were digested using trypsin and collected by low‐speed centrifugation until further analysis. 200 μl of Reagent A and 10 μl of Reagent B were added to the cells, followed by vortexing for 5 s, then kept on ice for 1 min. Finally, a Trizol reagent was used to extract total RNA. The mRNA expression was determined by qRT‐PCR; GAPDH or U6 was used as internal inferences.
2.5. RNase R treatment
In the same sample, 2 µg of RNA was taken, of which 1 µg was treated with RNase R, and the other 1 µg was used as a control. The reaction system was prepared and heated at 37°C for 15 min, then 1 ml of Trizol was added to extract the total RNA. The untreated group was used as the control for reverse transcription experiments.
2.6. CCK‐8 analysis
The transfected cells were inoculated in 96‐well plates at a cell density of 1 × 104 per well for cell viability assay. After incubation at 37°C and 5% CO2 for 0, 24, 48, and 72 h, each well has received 10 μl of CCK‐8 reagent (Beyotime). Then, the cells were further incubated at 37°C in the dark for another 4 h. Finally, the optical density was measured at 450 nm using a microplate spectrophotometer (BioTek Instruments).
2.7. Colony formation assay
The colony formation assay was performed by inoculating 1000 cells/well in a 6‐well plate. After 14 days of incubation, the cells were washed three times with PBS, fixed with anhydrous methanol, and stained with 0.1% crystalline violet staining. The cell colonies were subsequently photographed and counted.
2.8. Flow cytometry analysis
According to the manufacturing instructions of AnnexinV‐FITC Apoptosis Kit (Jiamei Biotechnology), the apoptosis rate in A549 cells was detected. Briefly, transfected cells were washed with cold PBS and later suspended in Annexin V binding buffer. Then, the collected cells were placed in a mixture of Annexin V‐FITC and PI away from light exposure at room temperature for 15 min. Finally, the apoptosis rate was measured by flow cytometry.
2.9. Transwell assay
Cell invasion and migration capacity were evaluated using 8 μm pore Transwell chambers (Corning) pre‐coated with or without Matrigel. Briefly, a culture medium (containing 10% fetal bovine serum) was added to the lower chamber as a chemical elicitor. 5 × 104 of transfected cells were incubated in the upper chamber with 200 μl of a serum‐free medium at 37°C and 5% CO2 for 24 h. After incubation, the cells were fixed with 70% ethanol and stained with 0.1% crystal violet. Finally, the invaded or migrated cells were photographed with a Nikon (Nikon Ti‐s) microscope.
2.10. RNA pull‐down analysis
Cells were lysed and transferred to a 2 ml Ep tube. After lysis, cells were centrifuged at 12,000 r/min for 40 min. Streptavidin agarose beads were mixed and rinsed with 1 ml of lysis solution. 10 μl of circ_SAR1A probe was added to the streptavidin agarose beads and incubated for 2 h at 4°C. Lysed cells were mixed with the agarose bead‐probe complex and incubated for 2 h at 4°C. 1 ml of the high salt eluate was added to each sample, followed by 40 µl of the low salt eluate. Samples were mixed gently to obtain the target lysate. The enrichment of miR‐21‐5p was detected using PCR analysis.
2.11. Dual‐luciferase reporter assay
The full‐length sequences of circ_SAR1A and TXNIP 3′‐UTR with or without miR‐21‐5p binding sites were cloned in the psiCHECK2 vector to construct the circ_SAR1A WT, circ_SAR1A MUT, TXNIP WT, and TXNIP MUT vectors (Promega), respectively. Later, miR‐21‐5p mimics or miR‐NC were cotransfected into A549 cells with the luciferase vectors using Lipofectamine 3000 reagent (Invitrogen). After 48 h of transfection, the luciferase activity was assayed with a luciferase reporter kit (Promega) normalized to Renilla luciferase.
2.12. Western blotting assay
Total proteins in tissues and cells were extracted with a protein extraction kit (AmyJet). Then, the proteins were separated on 10% SDS‐PAGE gel and transferred onto the PVDF membrane. The membranes were blocked at room temperature using 5% skimmed milk for 1 h followed by incubation overnight at 4°C with the following primary antibodies: anti‐TXNIP (ab188865; 1:1000; Abcam), anti‐MMP‐2 (ab92536; 1:1000; Abcam), anti‐MMP‐9 (ab76003; 1:1000; Abcam), anti‐N‐cadherin (ab76011; 1:5000; Abcam), anti‐Vimentin (ab92547; 1:1000; Abcam), anti‐E‐cadherin (ab40772; 1:10000; Abcam), and anti‐GAPDH (ab9485; 1:2500; Abcam). Finally, these membranes were incubated with secondary antibodies and visualized using ECL reagents.
2.13. In vivo experiment
Nude mice (4 weeks old, 18~22 g) were divided into two groups (6 mice/group). 200 μl (1 × 108 cells/ml) of control cells (A549 cells stably expressing ov‐NC) or stably expressing ov‐SAR1A A549 cells were injected into the right axilla of each mouse. After successful xenografting, the tumor diameter in all mice was measured and recorded with vernier calipers every day. After 30 days of continuous observation, the nude mice were euthanized, and the subcutaneous tumors were removed, weighed, and photographed. All animal procedures were conducted according to national guidelines and approved by the Institutional Committee of Nantong University for Animal Research.
2.14. Statistical analysis
Data were analyzed and generated by SPSS software (version 23.0) and GraphPad Prism software (version 7.0). p < 0.05 was considered to be statistically significant. All data were expressed as mean ± standard deviation (SD) using the chi‐square test to assess differences among categorical variables. Comparisons between two or more groups were conducted via Student's t test or one‐way ANOVA followed by Tukey's post hoc test. Spearman's correlation analysis was utilized to analyze the correlation between two factors.
3. RESULTS
3.1. circ_SAR1A is down‐regulated in lung cancer
After searching the GEO database (GSM540421‐GSM‐540425), we found that circ_SAR1A was prominently decreased in lung cancer samples resistant to cisplatin compared to non‐resistant lung cancer samples (Figure 1A), suggesting that circ_SAR1A might participate in regulating the progression of lung cancer. After that, we collected paired lung cancer tumor tissues and adjacent non‐tumor counterparts in a cohort of 68 lung cancer patients to detect the ectopic level of circ_SAR1A. Results showed that circ_SAR1A was down‐regulated in tumor tissues compared to adjacent para‐carcinoma tissues (Figure 1B). Meanwhile, compared to human bronchial epithelial cell line HBE, circ_SAR1A was remarkably decreased in lung cancer cell lines H1650, H1581, H460, H1299, and A549 (especially in A549), which was chosen for the following experiments (Figure 1C). After RNase R treatment, we found that circ_SAR1A was more resistant to degradation than linear SAR1A due to its stable structure, suggesting that circ_SAR1A is a circRNA (Figure 1D). Also, the subcellular localization assay further proved that circ_SAR1A was localized in the cytoplasm (Figure 1E). Altogether, circ_SAR1A was down‐regulated in lung cancer and implicated in lung cancer tumorigenesis.
FIGURE 1.
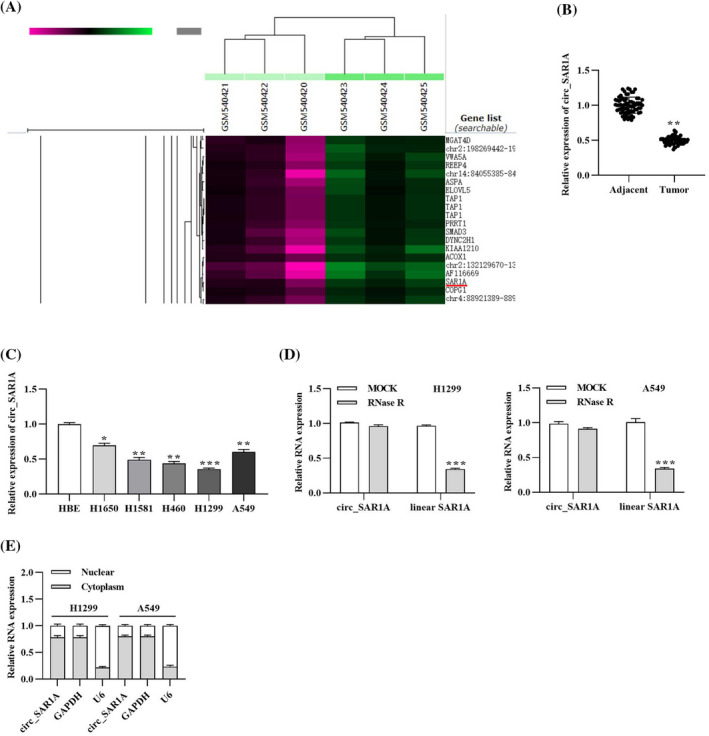
circ_SAR1A is down‐regulated in lung cancer. (A) Expression of circ_SAR1A in lung cancer samples summarized by GEO database. (B) Expression of circ_SAR1A in tumor and non‐tumor adjacent tissues collected from lung cancer patients. **p < 0.01, tumor versus adjacent group. (C) Expression of circ_SAR1A in human bronchial epithelial cells HBE and lung cancer cell lines. *p < 0.05, H1650 versus HBE; **p < 0.01, H1581 versus HBE, H460 versus HBE, A549 versus HBE; ***p < 0.001, H1299 versus HBE. (D) Expression of linear SAR1A and circ_SAR1A after RNase R treatment. ***p < 0.001, RNase R versus MOCK group. (E) Localization of circ_SAR1A in lung cancer cells
3.2. circ_SAR1A inhibited lung cancer malignant properties
To further unravel the molecular mechanisms of circ_SAR1A in lung cancer, we transfected A549 cells with circ_SAR1A overexpression plasmid or its negative control. As depicted in Figure 2A, circ_SAR1A level was significantly enhanced in the ov‐SAR1A group compared to ov‐NC or control group, suggesting successful transfection. Then, a series of cellular experiments were implemented. As demonstrated in Figure 2B‐D, circ_SAR1A overexpression has significantly suppressed A549 cell viability and proliferative capacity. Besides, the flow cytometry analysis in Figure 2E,F suggested that circ_SAR1A could hinder lung cancer cell apoptosis compared to the ov‐NC group.
FIGURE 2.
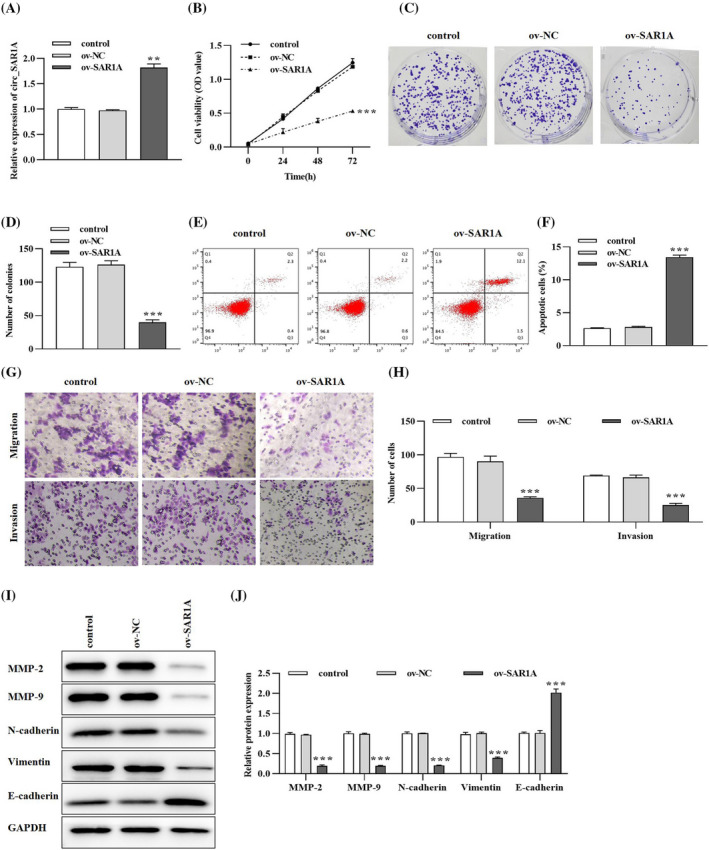
circ_SAR1A inhibits the malignant behavior in lung cancer. (A) Transfection efficacy of circ_SAR1A in lung cancer cells. (B) The effects of circ_SAR1A on lung cancer cell viability were determined by CCK‐8 assay. (C, D) Lung cancer cell proliferation was detected by colony formation assay. (E, F) circ_SAR1A promoted lung cancer cell apoptosis. (G, H) The effects of circ_SAR1A on lung cancer cell migration and invasion were evaluated by Transwell assay. (I, J) Migration‐, invasion‐, and EMT‐related proteins were examined by western blotting analysis. **p < 0.01, ***p < 0.001, ov‐SAR1A versus ov‐NC group
Moreover, the effect of circ_SAR1A on lung cancer cell metastasis was evaluated using Transwell and western blotting analyses. As presented in Figure 2G,H, the migratory and invasiveness of lung cancer cells were notably reduced in the ov‐SARA1 group compared to the ov‐NC or control group. Also, the pro‐metastasis proteins MMP‐2, MMP‐9, N‐cadherin, and Vimentin were decreased while anti‐metastasis protein E‐cadherin was enhanced after overexpressing circ_SAR1A in A549 cells (Figure 2I,J). Collectively, circ_SAR1A might serve as an inhibitory element in lung cancer.
3.3. circ_SAR1A inhibited tumor growth in vivo
The xenograft tumor model was established by injecting A549 cells stably overexpressing circ_SAR1A into nude mice. As shown in Figure 3A, circ_SAR1A level was prominently increased in the ov‐SAR1A group compared to the ov‐NC group. Meanwhile, tumor volume and weight in the ov‐SAR1A group were much lower than the ov‐NC group in a time‐dependent manner (Figure 3B,C). Taken together, the in vivo experiments validated that circ_SAR1A inhibited lung cancer tumor growth.
FIGURE 3.
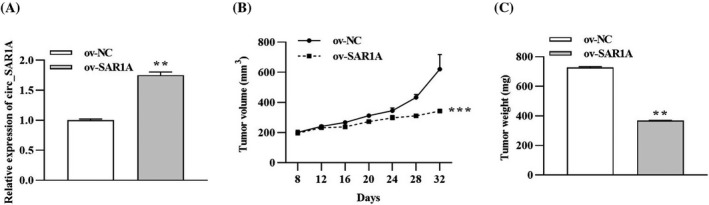
circ_SAR1A suppresses lung cancer tumor growth in vivo. (A) The expression of circ_SAR1A was determined by qRT‐PCR analysis. (B) Tumor volume. (C) Tumor weight. **p < 0.01, ***p < 0.001, ov‐SAR1A versus ov‐NC group
3.4. circ_SAR1A targets miR‐21‐5p
Functional experiments predicted and confirmed the relationship between circ_SAR1A and miR‐21‐5p. First, the Starbase tool predicted the potential binding sites between circ_SAR1A and miR‐21‐5p (https://starbase.sysu.edu.cn/), which are presented in Figure 4A. Then, the dual‐luciferase reporter assay illuminated that miR‐21‐5p mimics decreased the luciferase activity in 3′UTR of circ‐SAR1A WT (Figure 4B) compared to the miR‐NC group. However, there were no apparent differences in the circ‐SAR1A MUT groups. Meanwhile, the RNA pull‐down assay further ascertained that miR‐21‐5p was abundantly enriched in the biotin‐SAR1A group compared to the biotin‐NC group, confirming the binding between circ_SAR1A and miR‐21‐5p (Figure 4C).
FIGURE 4.
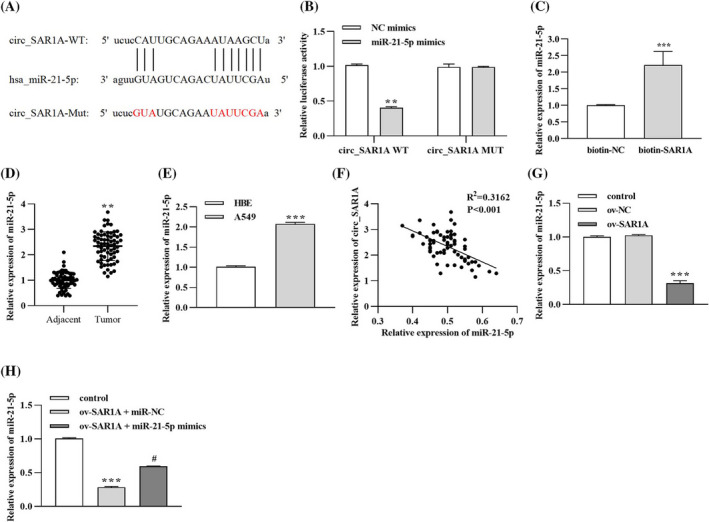
circ_SAR1A sponges miR‐21‐5p in lung cancer. (A) The putative binding sequences between circ_SAR1A and miR‐21‐5p were predicted by Starbase online tool. (B) Dual‐luciferase reporter assay. **p < 0.01, miR‐21‐5p mimics versus NC mimics group. (C) RNA pull‐down assay. ***p < 0.001, biotin‐SAR1A versus biotin‐NC group. (D) Expression of miR‐21‐5p in tumor and non‐tumor para‐carcinoma tissues collected from lung cancer patients. **p < 0.01, tumor versus adjacent group. (E) Expression of miR‐21‐5p in human bronchial epithelial cells HBE and lung cancer cell lines. ***p < 0.001, A549 versus HBE group. (F) Correlation of miR‐21‐5p and circ_SAR1A level in lung cancer patients. (G) Effects of circ_SAR1A overexpressing on miR‐21‐5p expression. **p < 0.01, ov‐SAR1A versus ov‐NC group. (H) Expression of miR‐21‐5p after co‐transfection with circ_SAR1A and miR‐21‐5p in lung cancer cells. ***p < 0.01, ov‐SAR1A + miR‐NC versus control group; # p < 0.05, ov‐SAR1A + miR‐21‐5p mimics versus ov‐SAR1A + miR‐NC group
Moreover, the involvement of miR‐21‐5p in lung cancer onset was studied. As depicted in Figure 4D, miR‐21‐5p was remarkably increased in lung cancer tumor tissues compared to non‐cancerous distal normal tissues. Similarly, miR‐21‐5p was prominently expressed in lung cancer cells line A549 compared to human bronchial epithelial cell line HBE (Figure 4E). Intriguingly, miR‐21‐5p was inversely correlated with circ_SAR1A expression level in a cohort of 68 lung cancer patients (Figure 4F). The transfection experiments suggested that overexpressing circ_SAR1A could decrease the miR‐21‐5p level (Figure 4G), whereas the co‐expression of circ_SAR1A and miR‐21‐5p abated their effects (Figure 4H). Conclusively, the above data implied that circ_SAR1A negatively targets miR‐21‐5p in lung cancer.
3.5. circ_SAR1A suppressed malignant properties in lung cancer cells by sponging miR‐21‐5p
A series of rescue experiments elaborated the regulatory mechanism of the circ_SAR1A/miR‐21‐5p axis in lung cancer. A549 cells were transfected with ov‐NC, ov‐SAR1A, miR‐NC, and miR‐21‐5p mimics. The CCK‐8 and colony formation assays in Figure 5A‐C demonstrated that circ_SAR1A overexpression significantly inhibited lung cancer cell proliferation; however, the inhibitory effect induced by ov‐SAR1A could be partially reversed by co‐transfection with miR‐21‐5p mimics. In addition, flow cytometry analysis depicted that the tumor consolidating effect triggered by ov‐SAR1A in lung cancer cell apoptosis could be partly counteracted by re‐introducing miR‐21‐5p mimics (Figure 5D,E). The migratory and invasive abilities in the ov‐SAR1A group were remarkably hindered compared to the ov‐NC group (Figure 5F,G). Nevertheless, the inhibition of migration and invasion in A549 cells induced by circ_SAR1A overexpression could be reversed by administering miR‐21‐5p mimics. Finally, we identified that pro‐metastasis proteins MMP‐2, MMP‐9, N‐cadherin, and Vimentin levels were prominently declined while anti‐metastasis protein E‐cadherin was promoted in the ov‐SAR1A group; however, the effects of circ_SAR1A on metastasis‐related protein expressions could be reversed by co‐transfection with miR‐21‐5p mimics in part (Figure 5H,I).
FIGURE 5.
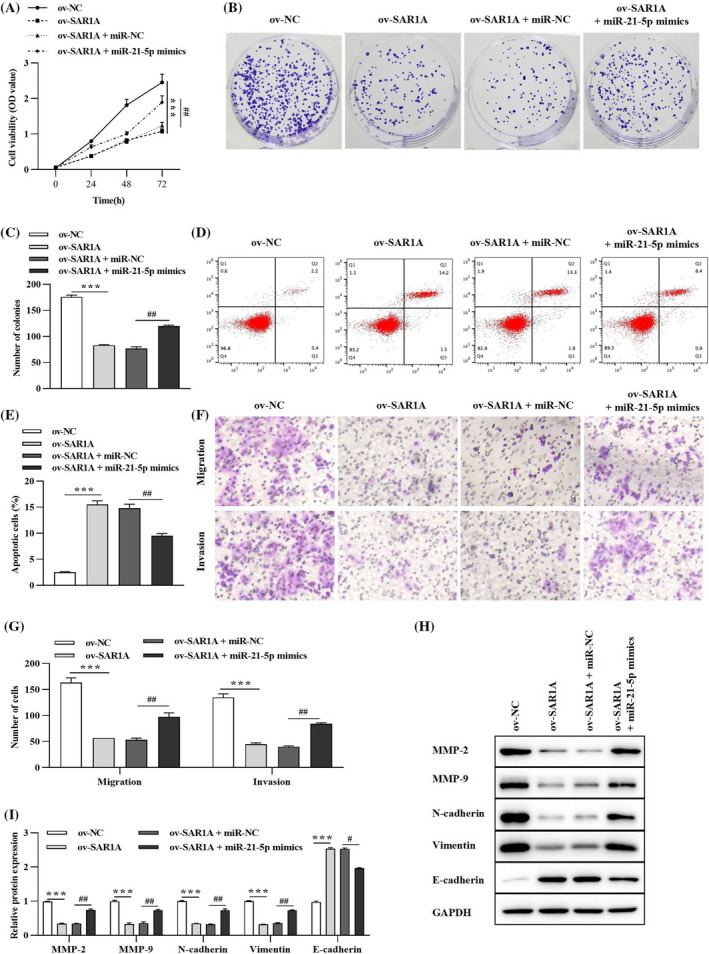
circ_SAR1A functions as a tumor suppressor in lung cancer via sponging miR‐21‐5p. (A) CCK‐8 assay was conducted to measure cell viability. (B, C) Lung cancer cell proliferation was evaluated by colony formation assay. (D, E) Cell apoptosis was verified by flow cytometry analysis. (F, G) Cell migration and invasion were detected by Transwell assay. (H, I) Migration‐, invasion‐, and EMT‐related proteins were examined by western blotting analysis. ***p < 0.001, ov‐SAR1A versus ov‐NC; # p < 0.05, ## p < 0.01, ov‐SAR1A + miR‐21‐5p mimics versus ov‐SAR1A + miR‐NC group
3.6. TXNIP is a direct downstream target of miR‐21‐5p
Starbase software was employed to predict downstream targets of miR‐21‐5p. As shown in Figure 6A, the putative binding sites between miR‐21‐5p and TXNIP 3′UTR are presented. Afterward, dual‐luciferase and RNA pull‐down assays confirmed the target relationship between miR‐21‐5p and TXNIP. From the results in Figure 6B, transfection with miR‐21‐5p mimics significantly decreased luciferase activity in 3′UTR of TXNIP WT. Meanwhile, miR‐21‐5p was abundantly enriched in the biotin‐labeled TXNIP group by contrast with the biotin‐NC group (Figure 6C). Then, the role of TXNIP in lung cancer tumorigenesis was verified. As demonstrated in Figure 6D,E, qRT‐PCR and western blotting assays elaborated that TXNIP was significantly down‐regulated in lung cancer tumor tissues compared to paracancerous non‐tumor tissues.
FIGURE 6.
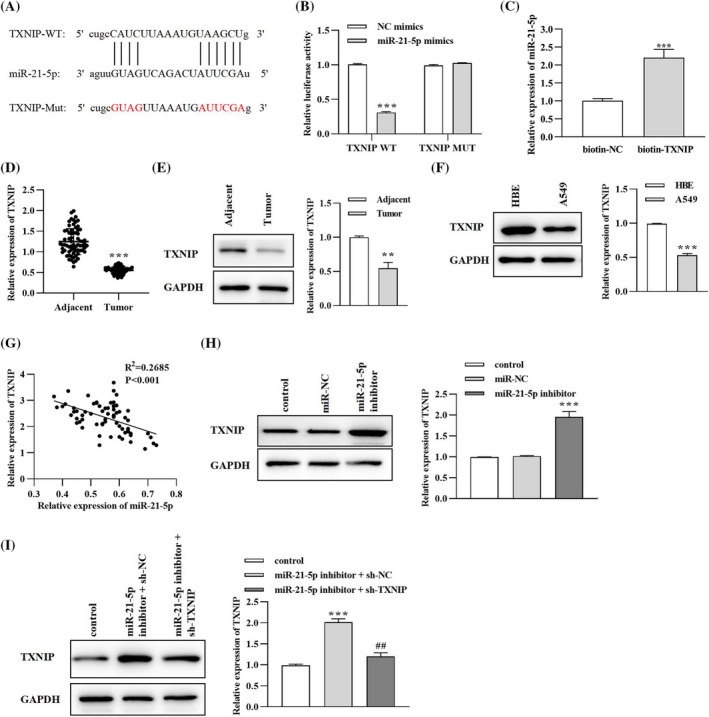
TXNIP is a downstream target of miR‐21‐5p in lung cancer. (A) The putative binding sites between TXNIP and miR‐21‐5p. (B) Dual‐luciferase reporter assay. **p < 0.01, miR‐21‐5p mimics versus NC mimics group. (C) RNA pull‐down assay. ***p < 0.001, biotin‐SAR1A versus biotin‐NC group. (D) Expression of TXNIP in tumor and non‐tumor para‐carcinoma tissues collected from lung cancer patients was evaluated by qRT‐PCR analysis. ***p < 0.001, tumor versus adjacent group. (E) Expression of TXNIP in tumor and non‐tumor para‐carcinoma tissues collected from lung cancer patients was evaluated by western blotting analysis. **p < 0.01, tumor versus adjacent group. (F) Expression of miR‐21‐5p in human bronchial epithelial cells HBE and lung cancer cell lines. ***p < 0.001, A549 versus HBE group. (G) Correlation between miR‐21‐5p and TXNIP level in lung cancer tissues. (H) The effect of miR‐21‐5p level on TXNIP expression was detected by western blotting assay. ***p < 0.001, miR‐21‐5p inhibitor versus miR‐NC group. (I) TXNIP level after co‐transfection with miR‐21‐5p and TXNIP was evaluated by western blotting analysis. ***p < 0.01, miR‐21‐5p inhibitor + sh‐NC versus control group; ## p < 0.05, miR‐21‐5p inhibitor + sh‐TXNIP versus miR‐21‐5p inhibitor + sh‐NC group
Similarly, TXNIP level was declined in A549 lung cancer cells compared to human bronchial epithelial cell line HBE (Figure 6F). The Spearman's correlation analysis in Figure 6G confirmed the inverse correlation between miR‐21‐5p and TXNIP level in lung cancer tissues. After transfection, we found that miR‐21‐5p inhibitor could enhance TXNIP level in lung cancer cells (Figure 6H). However, the enhancement in TXNIP expression induced by miR‐21‐5p inhibitor could be partially counterbalanced with the co‐transfection with lentivirus plasmid sh‐TXNIP (Figure 6I). Generally speaking, TXNIP was found to be a direct target and negatively regulated by miR‐21‐5p in lung cancer.
3.7. Knockdown of miR‐21‐5p inhibited lung cancer cell growth via targeting TXNIP
A549 cells were transfected with miR‐21‐5p inhibitor and sh‐TXNIP to clarify the regulatory role of miR‐21‐5p/TXNIP axis in lung cancer progression. From the results in Figure 7A‐D, miR‐21‐5p inhibitor prominently inhibited lung cancer cell viability, proliferation, migration, and invasion, yet facilitated apoptosis. However, the inhibitory effect of miR‐21‐5p silencing on lung cancer cell malignant behavior could be counterbalanced by silencing TXNIP using sh‐TXNIP (Figure 7A‐D). In addition, western blotting results in Figure 7E illuminated that miR‐21‐5p interference could promote E‐cadherin level but suppresses MMP‐2, MMP‐9, N‐cadherin, and Vimentin expressions; nevertheless, the influence on metastasis‐related proteins aroused by miR‐21‐5p inhibitor could be relieved by co‐transfection with the sh‐TXNIP plasmid (Figure 7E). Ultimately, the above data suggested that miR‐21‐5p inhibition could ameliorate lung cancer malignant properties by targeting TXNIP.
FIGURE 7.
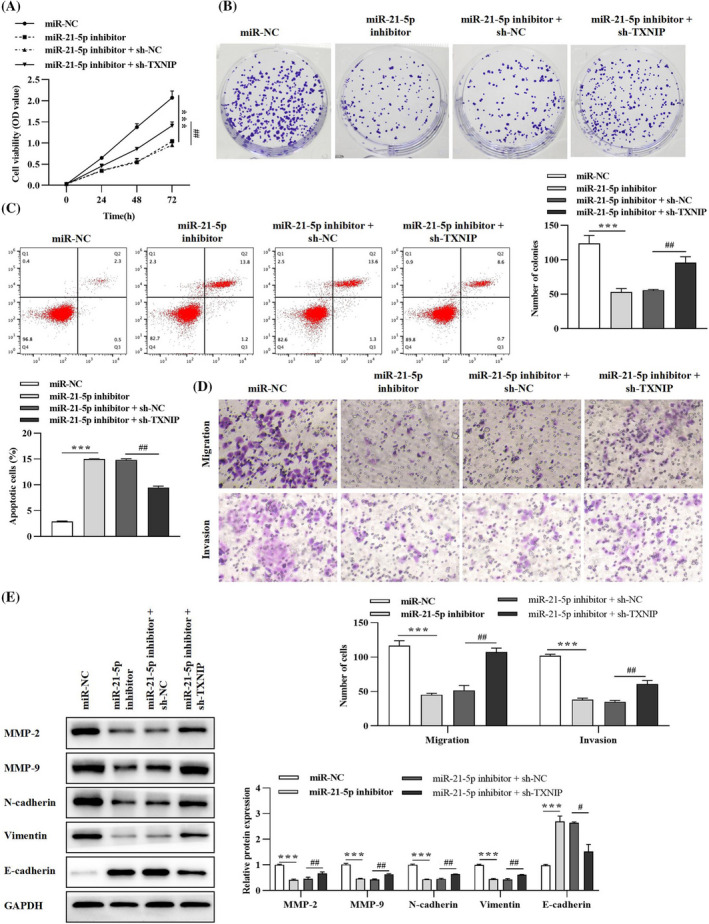
miR‐21‐5p facilitates lung cancer progression via targeting TXNIP. (A) Lung cancer cell viability was examined by CCK‐8 assay. (B) Colony formation was executed to verify cell proliferative ability. (C) Flow cytometry analysis was conducted to evaluate cell apoptosis rate. (D) Transwell assay was performed to ascertain cell migratory and invasive capabilities. (E) Expressions of migration‐, invasion‐, and EMT‐related proteins were detected by western blotting assay. ***p < 0.001, miR‐21‐5p inhibitor versus miR‐NC group; # p < 0.05, ## p < 0.01, miR‐21‐5p inhibitor + sh‐TXNIP versus miR‐21‐5p inhibitor + sh‐NC group
4. DISCUSSION
There is growing evidence that circRNAs have multiple biological functions, including “miRNA sponging,” transcriptional regulators, proteins translators, and templates. 30 , 31 Most molecular mechanisms of circRNAs can be categorized as “miRNA sponges.” Most notably, Hansen et al. first reported the circRNA CDR1as, also known as CIRS‐7, 32 the first circRNA identified as a miRNA sponge. circ_SAR1A was first reported by Salzman et al. 33 in 2013. It was further confirmed by Zhao et al. 34 that circ_SAR1A was highly expressed in renal cell carcinoma (RCC) samples and cells, serving as an oncogene in RCC. In our study, we downloaded information from the GEO database (GSM540421‐GSM‐540425) and the results suggested that circ_SAR1A was declined in samples of lung cancer patients who were resistant to cisplatin. Furthermore, we collected tumor and non‐tumor distal tissues from a cohort of 68 lung cancer patients. The results showed that circ_SAR1A was declined in lung cancer tumor tissues compared to adjacent normal tissues. Further experiments verified that circ_SAR1A was a stable circRNA located in the cytoplasm and is down‐regulated in A549 lung cancer cells, exerting a tumor‐suppressive role in lung cancer. Our research might be the first study that revealed the molecular role of circ_SAR1A in lung cancer.
With the increasing understanding of the biological mechanisms of circRNAs, researchers have found that their functions are primarily through the competitive binding of miRNAs (ceRNAs). For instance, miR‐21‐5p was sponged by circ_PVT1 and participated in laryngeal cancer (LC) development. 35 Han et al. 36 suggested that circ_0027599 could sponge miR‐21‐5p to inhibit gastric cancer progression; Jiang et al. 37 revealed that miR‐21‐5p is up‐regulated in colorectal cancer (CRC) and is sponged by circEPB41L2; Li et al. 38 illuminated that circPUM1 served as a sponge of miR‐21‐5p, promoting papillary thyroid cancer (PTC) tumorigenesis; Ma et al. 39 verified that miR‐21‐5p bound to the 3′UTR of circ_ACAP2 to promote head and neck squamous cell carcinoma (HNSCC) cellular phenotypes. Another research by Yang et al. 40 suggested that miR‐21‐5p is a target of circ‐LARP4, serving as an oncogene in lung cancer. Our study verified that circ_SAR1A sponges and negatively modulates miR‐21‐5p in lung cancer cells. Moreover, miR‐21‐5p is down‐regulated in lung cancer tumor tissues and cell lines. Up‐regulation of circ_SAR1A impeded lung cancer malignant behaviors, and miR‐21‐5p mimics could counteract this inhibitory effect. In conclusion, our study ascertained the carcinogenic role of miR‐21‐5p in lung cancer, which was in concordance with previous studies. 41 , 42 , 43
In a former study, TXNIP was predicted and confirmed to be the downstream target of miR‐21‐5p. 44 Also, TXNIP was revealed to be a tumor suppressor in a wide range of tumors, including cervical cancer, 45 prostate cancer, 46 RCC, 47 and thyroid cancer. 48 In lung cancer, the tumor suppressor role of TXNIP was also scrutinized. For instance, Liang et al. 49 depicted that cirDCUN1D4 could restrain metastasis of lung cancer cells by stabilizing the TXNIP level. Zhang et al. 50 elucidated that TXNIP targets oncogenic miR‐411 in lung cancer carcinogenesis. Consistent with previous studies, our research observed down‐regulation of TXNIP in lung cancer tumor samples and cells. Moreover, TXNIP was a direct target of miR‐21‐5p, which could partially reverse the inhibitory effects of miR‐21‐5p on lung cancer cell growth.
In sum, the expression of circ_SAR1A was found to be significantly decreased in lung cancer tissues and cells. The results of cellular and animal experiments further suggested that circ_SAR1A protects against the development of lung cancer by regulating the miR‐21‐5p/TXNIP axis and may serve as a potential therapeutic target for lung cancer treatment.
CONFLICT OF INTEREST
All authors declare no conflict of interest in this study.
ACKNOWLEDGEMENTS
None.
Zhao Y, Wan Y, He T. Circ_SAR1A regulates the malignant behavior of lung cancer cells via the miR‐21‐5p/TXNIP axis. J Clin Lab Anal. 2022;36:e24366. doi: 10.1002/jcla.24366
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Cheung CHY, Juan HF. Quantitative proteomics in lung cancer. Biomed Sci. 2017;24(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Wakeam E, Acuna SA, Leighl NB, et al. Surgery versus chemotherapy and radiotherapy for early and locally advanced small cell lung cancer: a propensity‐matched analysis of survival. Lung Cancer. 2017;109:78‐88. [DOI] [PubMed] [Google Scholar]
- 4. Fu ZX, Yang X, Wang WQ, et al. Radiotherapy combined with gefitinib for patients with locally advanced non‐small cell lung cancer who are unfit for surgery or concurrent chemoradiotherapy: a phase II clinical trial. Radiat Oncol. 2020;15(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Field EA, Field JK. Lung cancer: a potential role for dentists. Br Dent J. 2020;228(6):413‐414. [DOI] [PubMed] [Google Scholar]
- 6. Huang AQ, Zheng HX, Wu ZY, Chen MS, Huang YL. Circular RNA‐protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503‐3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang HY, Shen YJ, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34(1):e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐691. [DOI] [PubMed] [Google Scholar]
- 11. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang JH, Yan SS, Yang JH, Hu HY, Xu DH, Wang ZY. Non‐coding RNAs in rheumatoid arthritis: from bench to bedside. Front Immunol. 2020;10:3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588‐5600. [DOI] [PubMed] [Google Scholar]
- 14. Li TR, Jia YJ, Wang Q, Shao XQ, Lv RJ. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127(8):726‐734. [DOI] [PubMed] [Google Scholar]
- 15. Chen C, Zhang X, Deng Y, et al. Regulatory roles of circRNAs in adipogenesis and lipid metabolism: emerging insights into lipid‐related diseases. FEBS J. 2021;288(12):3663‐3682. [DOI] [PubMed] [Google Scholar]
- 16. Li R, Jiang JJ, Shi H, Qian H, Zhang X, Xu WR. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661‐1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiu LP, Xu H, Ji MC, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. [DOI] [PubMed] [Google Scholar]
- 18. Wang CD, Tan SY, Li JW, Liu WR, Peng Y, Li WM. CircRNAs in lung cancer ‐ Biogenesis, function and clinical implication. Cancer Lett. 2020;492:106‐115. [DOI] [PubMed] [Google Scholar]
- 19. Zhao YH, Cong L, Lukiw WJ. Plant and animal microRNAs (miRNAs) and their potential for inter‐kingdom communication. Cell Mol Neurobiol. 2018;38(1):133‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P, Guan QY, Zhou DM, Yu Z, Song YB, Qiu WS. miR‐21 inhibitors modulate biological functions of gastric cancer cells via PTEN/PI3K/mTOR pathway. DNA Cell Biol. 2018;37(1):38‐45. [DOI] [PubMed] [Google Scholar]
- 21. Wang H, Tan ZQ, Hu H, et al. microRNA‐21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer. 2019;19(1):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu HY, Zhang YY, Zhu BL, et al. miR‐21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT3. Eur Rev Med Pharmacol Sci. 2019;23(10):4149‐4155. [DOI] [PubMed] [Google Scholar]
- 23. Park S, Eom K, Kim J, et al. MiR‐9, miR‐21, and miR‐155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17(1):658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bautista‐Sánchez D, Arriaga‐Canon C, Pedroza‐Torres A, et al. The promising role of miR‐21 as a cancer biomarker and its importance in RNA‐based therapeutics. Mol Ther Nucleic Acids. 2020;20:409‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao LQ, Yang XW, Chen YB, Zhang DW, Jiang XF, Xue P. Exosomal miR‐21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer. 2019;18(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jesionek‐Kupnicka D, Braun M, Trąbska‐Kluch B, et al. MiR‐21, miR‐34a, miR‐125b, miR‐181d and miR‐648 levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch Med Sci. 2019;15(2):504‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ni KW, Wang DM, Xu HY, et al. miR‐21 promotes non‐small cell lung cancer cells growth by regulating fatty acid metabolism. Cancer Cell Int. 2019;19:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li JC, Huang LX, He ZN, et al. Andrographolide suppresses the growth and metastasis of luminal‐like breast cancer by inhibiting the NF‐κB/miR‐21‐5p/PDCD4 signaling pathway. Front Cell Dev Biol. 2021;9:643525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang R, Xia T. Long non‐coding RNA XIST regulates PDCD4 expression by interacting with miR‐21‐5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51(5):1460‐1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67‐79. [DOI] [PubMed] [Google Scholar]
- 31. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshimoto R, Rahimi J, Hansen TB, Kjems J, Mayeda A. Biosynthesis of circular RNA ciRS‐7/CDR1as is mediated by mammalian‐wide interspersed repeats. iScience. 2020;23(7):101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell‐type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao XL, Zhao ZH, Xu WC, et al. Circ‐SAR1A promotes renal cell carcinoma progression through miR‐382/YBX1 axis. Cancer Manag Res. 2020;12:7353‐7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu F, Lin Y, Ai MM, Tan GJ, Huang JL, Zou ZR. Knockdown of circular RNA hsa_circ_PVT1 inhibited laryngeal cancer progression via preventing wnt4/β‐Catenin signaling pathway activation. Front Cell Dev Biol. 2021;9:658115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han JZ, Yang ZX, Zhao S, Zheng LK, Tian YH, Lv YQ. Circ_0027599 elevates RUNX1 expression via sponging miR‐21‐5p on gastric cancer progression. Eur J Clin Invest. 2021;51(11):e13592. [DOI] [PubMed] [Google Scholar]
- 37. Jiang ZP, Hou ZH, Li L, Liu W, Yu ZM, Chen S. Exosomal circEPB41L2 serves as a sponge for miR‐21‐5p and miR‐942‐5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur J Clin Invest. 2021;51(9):e13581. [DOI] [PubMed] [Google Scholar]
- 38. Li YQ, Qin J, He ZC, Cui G, Zhang K, Wu BQ. Knockdown of circPUM1 impedes cell growth, metastasis and glycolysis of papillary thyroid cancer via enhancing MAPK1 expression by serving as the sponge of miR‐21‐5p. Genes Genomics. 2021;43(2):141‐150. [DOI] [PubMed] [Google Scholar]
- 39. Ma C, Shi TT, Qu ZL, et al. CircRNA_ACAP2 suppresses EMT in head and neck squamous cell carcinoma by targeting the miR‐21‐5p/STAT3 signaling axis. Front Oncol. 2020;10:583682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Z, Xu XX, Song C. Circular RNA la‐related protein 4 inhibits nonsmall cell lung cancer cell proliferation while promotes apoptosis through sponging microRNA‐21‐5p. Cancer Biother Radiopharm. 2020;37(2):111‐118. [DOI] [PubMed] [Google Scholar]
- 41. Zhu YX, Bo H, Chen ZZ, et al. LINC00968 can inhibit the progression of lung adenocarcinoma through the miR‐21‐5p/SMAD7 signal axis. Aging. 2020;12(21):21904‐21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang GA, Zhou YY, Chen WZ, et al. miR‐21‐5p promotes lung adenocarcinoma cell proliferation, migration and invasion via targeting WWC2. Cancer Biomark. 2020;28(4):549‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhong JC, Ren XH, Chen ZH, et al. miR‐21‐5p promotes lung adenocarcinoma progression partially through targeting SET/TAF‐Iα. Life Sci. 2019;231:116539. [DOI] [PubMed] [Google Scholar]
- 44. Zhang HS, Liu MF, Ji XY, Jiang CR, Li ZL, Ouyang B. Gastrodin combined with rhynchophylline inhibits cerebral ischaemia‐induced inflammasome activation via upregulating miR‐21‐5p and miR‐331‐5p. Life Sci. 2019;239:116935. [DOI] [PubMed] [Google Scholar]
- 45. Zhang JH, Tian XB, Yin HF, et al. TXNIP induced by MondoA, rather than ChREBP, suppresses cervical cancer cell proliferation, migration and invasion. J Biochem. 2020;167(4):371‐377. [DOI] [PubMed] [Google Scholar]
- 46. Xie M, Xie RY, Xie S, et al. Thioredoxin interacting protein (TXNIP) acts as a tumor suppressor in human prostate cancer. Cell Biol Int. 2020;44(10):2094‐2106. [DOI] [PubMed] [Google Scholar]
- 47. Chen Q, Liu T, Bao Y, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR‐27a‐3p/TXNIP pathway. Cancer Lett. 2020;469:68‐77. [DOI] [PubMed] [Google Scholar]
- 48. Morrison JA, Pike LA, Sams SB, et al. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer. 2014;13:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang YK, Wang H, Chen B, et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids. 2020;23:355‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang CY, Wang HM, Liu XM, et al. Oncogenic microRNA‐411 promotes lung carcinogenesis by directly targeting suppressor genes SPRY4 and TXNIP. Oncogene. 2019;38(11):1892‐1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


