Abstract
The abortive infection system AbiG is encoded by the lactococcal plasmid pCI750. The abiG locus (consisting of two genes, abiGi and abiGii) was examined by Northern blot analysis, revealing two transcripts of approximately 2.8 and 1.5 kb which were homologous to the two gene-specific probes. A transcriptional start site was mapped upstream of abiGi, and it appeared that the two genes were cotranscribed, resulting in the 2.8-kb transcript. The smaller transcript may be the result of independent transcription of abiGii within abiGi or of the presence of a weak terminator within abiGii. The locus was shown to be constitutively expressed. Evidence is presented for the possible existence of a second Abi mechanism on pCI750. Examination of phage sk1 RNA synthesis demonstrated that both the subcloned AbiG and, to a greater extent, pCI750 inhibited this process. pCI750 also severely inhibited synthesis of both early and late phage c2 transcripts, while the presence of the subclone resulted in a reduction in late transcript synthesis only.
The ongoing demand of the dairy industry for lactococcal starter cultures with high levels of phage resistance has stimulated the search for natural resistance systems which are abundant in these hosts (for a recent review, see reference 9). This has resulted in the identification of a myriad of abortive infection (Abi) systems, 13 of which have been sequenced to date. It has become apparent that the genetic flexibility of phages has allowed them to evolve in response to the selective pressure of resistance mechanisms (6, 13, 19, 21), and it has been proposed that the best defensive approach is that of rotating a number of different mechanisms within an isogenic strain (12). To optimize this strategy, it is useful to combine systems targeting different stages of the phage life cycle. Therefore, there is a need not only to accumulate a large number of resistance systems but also to characterize them so that they may be used optimally. In addition, it is of interest to understand how these systems are controlled. The aim of this study was to examine the expression and regulation of the two genes of the lactococcal AbiG system (23), abiGi and abiGii, and to determine the effect of this defense mechanism on intracellular phage development.
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Lactococcus lactis was grown at 30°C in M17 medium (28) supplemented with 0.5% glucose (GM17). Chloramphenicol was added at 5 μg/ml where necessary. Phages were propagated by the method of Terzaghi and Sandine (28).
TABLE 1.
Bacterial strains, plasmids, and bacteriophages
| Strain, plasmid, or phage | Relevant characteristics | Source or reference |
|---|---|---|
| L. lactis subsp. cremoris | ||
| LM0230 | Plasmid-free derivative of C2 | 14 |
| AB002 | MG1363(pCI750) | 22 |
| LOC758 | LM0230(pCI758) | 23 |
| LOC735 | LM0230(pCI735) | 23 |
| Plasmids | ||
| pCI750 | 65-kb plasmid from L. lactis subsp. cremoris UC653 | 3 |
| pCI784 | 8.3-kb SnaBI-SalI pCI724 fragment cloned into pCI372 | 23 |
| pCI758 | BbsI-SalI deletion derivative of pCI784 | 23 |
| pCI735 | 3.5-kb BspHI fragment cloned into pAM401 | 23 |
| Phages | ||
| sk1 | Small isometric-headed, phage species 936, lytic for strains LM0230 and MG1363 and sensitive to AbiG-containing cells (i.e., no plaques formed) | 26 |
| c2 | Prolate-headed, phage species c2, lytic for strains LM0230 and MG1363 and sensitive to AbiG-containing cells (i.e., reduced plaque diam) | UCCa phage collection |
UCC, University College Cork.
Northern blot analysis of abiG.
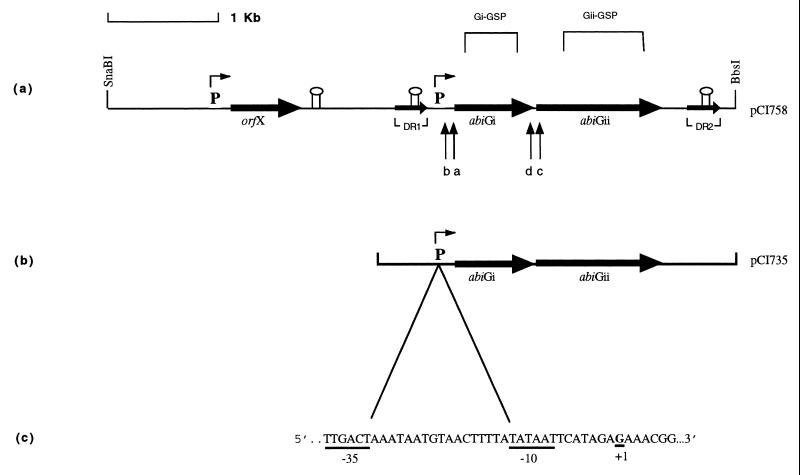
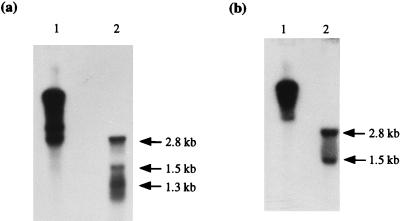
The organization of the abiG locus is diagrammed in Fig. 1. To determine if the two abiG genes were coordinately or discoordinately transcribed, RNA isolated (2) from Lactococcus lactis subsp. cremoris LOC758 (containing an abiG subclone, pCI758) was probed with PCR products Gi-GSP (generated by using primers GiL [5′ TATATTCCTATGACCTG 3′] and GiR [5′ AATTCAAAAGACCATTC 3′]) and Gii-GSP (generated by using primers GiiL [5′ TTGTATTAGATGAAACC 3′] and GiiR [5′ ACTCTATCTTGTAAATCC 3′]), specific for abiGi and abiGii, respectively (Fig. 1a) (Taq polymerase was supplied by Promega, Madison, Wis.). In order to eliminate any difficulties in comparing the estimated transcript sizes obtained, duplicate RNA samples were electrophoresed on the same gel and then transferred separately by capillary blotting to nylon membranes. These were probed with the two different gene-specific probes. Analysis with Gi-GSP identified three transcripts of approximately 2.8, 1.5, and 1.3 kb based on the Promega RNA markers (Fig. 2a). The 1.3-kb transcript was within a smear, and in subsequent analysis of RNA isolated from LOC735 (harboring the smallest functional subclone, pCI735 [Fig. 1b]), only a region of smearing with no distinctive band was observed (see Fig. 3a). The Gii-GSP probe hybridized to the same two transcripts identified by using Gi-GSP, i.e., the 2.8- and 1.5-kb bands (Fig. 2b). It therefore appears that the two genes are cotranscribed, resulting in the 2.8-kb transcript (abiGi is 750 bp and is separated by 2 bp from the 1,194-bp abiGii). The smaller transcript may be the result of independent transcription of abiGii from within abiGi or of the presence of a weak terminator structure within abiGii.
FIG. 1.
(a) Molecular organization of the abiG locus on a 5.8-kb segment of pCI758 (adapted from reference 23). The two open reading frames of abiG and orfX (of unknown function) are indicated by horizontal arrows.  indicates a 309-bp direct repeat with 5 mismatches.
indicates a 309-bp direct repeat with 5 mismatches.  and
and  represent putative promoter sequences and terminator structures, respectively. Gi-GSP and Gii-GSP are PCR-generated probes specific for abiGi and abiGii, respectively. Lowercase letters a through d represent primers. (b) A 3.5-kb insert of pCI735. (c) The mapped promoter and transcriptional start site (+1) upstream of abiGi.
represent putative promoter sequences and terminator structures, respectively. Gi-GSP and Gii-GSP are PCR-generated probes specific for abiGi and abiGii, respectively. Lowercase letters a through d represent primers. (b) A 3.5-kb insert of pCI735. (c) The mapped promoter and transcriptional start site (+1) upstream of abiGi.
FIG. 2.
Northern analysis of abiGi and abiGii. RNA isolated from L. lactis subsp. cremoris LOC758 (lanes 2) or pCI784 DNA used as a positive control (lanes 1) was hybridized with 32P-labelled Gi-GSP (a) or 32P-labelled Gii-GSP (b). Arrows indicate the sizes of the transcripts, estimated by using RNA size standards.
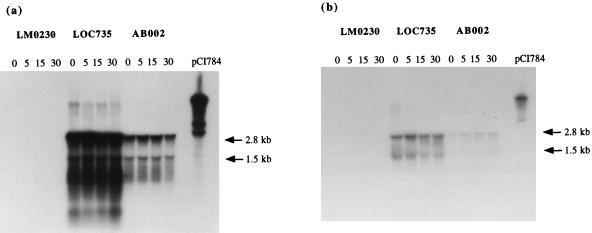
FIG. 3.
Hybridization of total RNA isolated from phage sk1-infected L. lactis subsp. cremoris LM0230, LOC735, or AB002 at 0, 5, 15, and 30 min postinfection with 32P-labelled Gi-GSP (a) or 32P-labelled Gii-GSP (b). The last lanes contain pCI784 DNA used as a positive control.
Mapping of the 5′ ends of the abiG transcripts.
As sequence analysis had identified a putative promoter upstream of abiGi (23) (Fig. 1a), this region was first examined in order to determine the 5′ end of the abiG mRNA by using the 5′/3′ RACE (rapid amplification of cDNA ends) kit (Boehringer Corporation Ltd., East Sussex, United Kingdom). Primer a (5′ ATATCGTGGTAAGAAGGC 3′) (Fig. 1a) was used as outlined by the manufacturers to make cDNA from LOC758 total RNA, while the nested primer b (5′ TTAGCACTAAGCTCCGAA 3′) and primers from the kit were used to generate a PCR fragment. A PCR product of approximately 100 bp was obtained. Sequence analysis of this product demonstrated its homology to the region upstream of abiGi and identified a G as the first base of the original cDNA (Fig. 1c). Due to the close proximity (i.e., separation by 7 bp) of this base to the −10 box of the previously specified putative promoter, it was concluded that this G was the transcriptional start site. This site and the accompanying promoter were typical of those found in lactococci by Van de Guchte et al. (29), who most frequently observed an A residue at the start point, with G residues being the next most commonly encountered. The −10 box did not possess the TGy extension (reported to be a feature of strong promoters [20]) which has been observed in the abiD1 (1) promoter, the putative promoters of abiF (16) and abiI (27), and other lactococcal promoters.
In order to determine if the smaller transcript (1.5 kb) started upstream of abiGii (within abiGi), even though no putative promoter was evident based on sequence analysis (20), attempts were made to map the 5′ end of an abiGii-specific message by using primer c (5′ TGTTGCTTTATCATTAG 3′) and the nested primer d (5′ TCTTCCTTTATTTCATCG 3′) (Fig. 1a). While PCR products of approximately 310 and 400 bp were obtained, sequence analysis demonstrated no homology with the abiG locus, suggesting that they were nonspecific products. The distance between primer c and the transcriptional start site upstream of abiGi is 954 bp; assuming that this is also the transcriptional start site for abiGii, giving rise to the 2.8-kb transcript, it is likely that the cDNA reaction of the 5′ RACE protocol did not successfully extend the 954 bp.
Previously, examination of the DNA sequence revealed a putative terminator downstream of abiGii (23). The distance between this and the mapped transcriptional start site is 2.539 kb, which is in reasonable agreement with the estimated size of the larger transcript (2.8 kb). Given that the smaller transcript was estimated at 1.5 kb and is recognized by both the Gi-GSP and Gii-GSP probes, it seems most likely that it begins at the same site as the larger transcript and results from the presence of a weak terminator within abiGii (although no obvious candidate was detected in the sequence).
Transcriptional regulation of the abiG genes.
To determine whether the abiG genes were constitutively transcribed or induced upon phage infection, we used the AbiG-sensitive phage sk1, which is unable to plaque in the presence of AbiG. Total RNA was isolated from phage sk1-infected cells of LM0230 (plasmid free), LOC735 (containing a 3.5-kb subclone), and AB002 (containing the native plasmid) and was probed with Gi-GSP and Gii-GSP (Fig. 1a). Eighty-milliliter volumes of culture were grown to an optical density at 600 nm of 0.3. A 20-ml sample was removed and harvested by centrifugation in a Beckman JA-14 rotor at 10,000 rpm for 1 min, after which the pellet was resuspended in ice-cold RNA extraction buffer (2) and stored on ice until all the samples were collected. CaCl2 was added to a final concentration of 5 mM, and phage was added at a multiplicity of infection of 5 to 10; samples were harvested as before. Total RNA was extracted as described by Arnau et al. (2); samples were DNase treated and run on a 1.1% formaldehyde gel.
The Gi-GSP probe recognized the previously identified 2.8- and 1.5-kb transcripts in both LOC735 and AB002 (Fig. 3a). Surprisingly, an additional faint band of approximately 7 kb was seen in the subclone containing LOC735, and after extended exposure it was also visible in AB002 (data not shown). This band may represent plasmid DNA contamination, even though DNase treatment was performed, or it may be the result of low-level read-through of the terminator of the abiG locus. The Gi-GSP probe demonstrated that abiGi was not induced by phage infection (Fig. 3a). While the 2.8-kb transcript was very concentrated and thus a slight increase in transcription might be difficult to detect, examination of the 1.5-kb transcript indicated clearly that it was not induced. Similarly, the Gii-GSP probe hybridized to the 2.8- and 1.5-kb transcripts and, as expected, demonstrated that abiGii also was not induced (Fig. 3b).
Two other abi genes have been examined for induction. In the case of abiA, the ability of its promoter to confer chloramphenicol resistance when cloned upstream of a promoterless cat68 gene suggested that it was constitutively expressed (18), while Anba et al. (1) observed no alteration in the level of transcription of abiD1 upon phage infection. Northern analysis of the latter abi gene revealed that transcription was initiated at a strong promoter but stopped at a terminator located 48 bp downstream. This observation, together with expression studies of the AbiD1 protein, strongly suggested that AbiD1 overproduction was toxic to the lactococcal cells. No such stringent control was observed for abiG transcription, although it is interesting that cloning of intact abiGi was lethal in Escherichia coli cells (data not shown).
It was noted that a much higher level of transcription was observed in LOC735- than in AB002-containing cells, by using either Gi-GSP or Gii-GSP. As care was taken to load the same amount of RNA in each well, it is most improbable that such a concentration effect could be attributed to experimental error. Moreover, the samples used in Fig. 3a and b were isolated on separate occasions, further supporting the reproducibility of this observation. A possible explanation is that the copy number of the subclone is higher than that of the original plasmid, and indeed this subclone is based on pAM401, a high-copy-number plasmid (31). It is widely accepted that increasing the copy number of an abi gene results in increased phage resistance (24, 10); however, in this instance the opposite would appear to be the case, given that pCI750 offers more protection to cells against phage infection than pCI735 (23). It therefore seems very plausible that pCI750 encodes a second Abi mechanism.
Influence of AbiG on bacteriophage RNA synthesis.
Previously, we reported that AbiG-containing cells did not inhibit intracellular phage DNA replication (23). In this study we examined the ability of AbiG to interfere with phage RNA synthesis, using pCI750 and the 3.5-kb subclone pCI735. Transcription of phages c2 and sk1 was investigated as restriction data, and temporal transcription maps, describing the order in which parts of the phage genome are expressed, were available for both phages (5, 8). Phage sk1 is unable to plaque in the presence of either the original plasmid or the subclone, while c2 forms plaques of reduced size in the presence of both and has an efficiency of plating (EOP) of 2.5 × 10−3 on AB002 but an EOP of 1.0 in the presence of AbiG subclones (23).
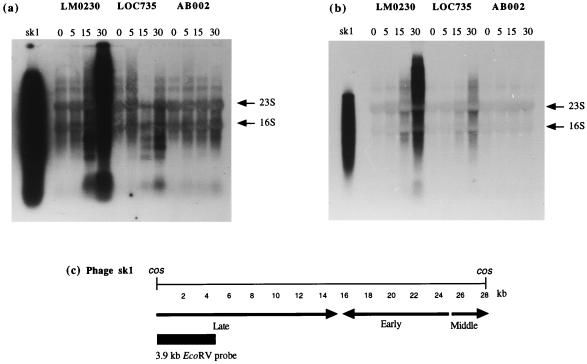
The results obtained following probing of total-RNA extracts from sk1-infected cells (obtained as described in the previous section) with sk1 total DNA are presented in Fig. 4a. The bands visible at 0 and 5 min after phage infection (T0 and T5, respectively) for all three strains appear to represent nonspecific hybridization to two host transcripts in addition to the 23S and 16S rRNA bands. A comparison of the signal from the T15 samples clearly demonstrated that less RNA is being synthesized in the pCI750- and pCI735-containing cells. A comparison of the T30 values suggests that virtually no phage RNA synthesis occurred in the presence of pCI750 and only a very limited amount occurred in the presence of the subcloned abiG. These findings are substantiated by a 3.9-kb EcoRV probe specific for the late-expressed region of phage sk1 (8). This probe revealed considerably reduced synthesis of late-expressed RNA in the presence of pCI735 and an apparent complete inhibition of synthesis in cells containing the original plasmid (Fig. 4b). Whether early and middle sk1 transcripts are also affected is difficult to determine from experiments using the total phage DNA probe (Fig. 4a). The sk1-specific signal (obtained by using either the total-DNA probe or the late-specific probe) was a smear rather than discrete bands. Chandry et al. (8) also observed this smearing and suggested that it may be the result of nonspecific processing of transcripts, transcript degradation, the presence of fragments of increasing size with a common 5′ end, or a combination of these factors.
FIG. 4.
Hybridization of total RNA isolated from phage sk1-infected L. lactis subsp. cremoris LM0230, LOC735, or AB002 at 0, 5, 15, and 30 min postinfection with 32P-labelled sk1 total DNA digested with EcoRV (a) or a 32P-labelled 3.9-kb EcoRV fragment from the late-expressed region (b). The first lanes contain EcoRV-digested phage sk1 DNA used as a positive control. Arrows indicate the 16S and 23S rRNA bands. (c) Temporal transcription map of phage sk1 with early-, middle-, and late-expressed regions indicated; adapted from Chandry et al. (8) with permission from the publisher.
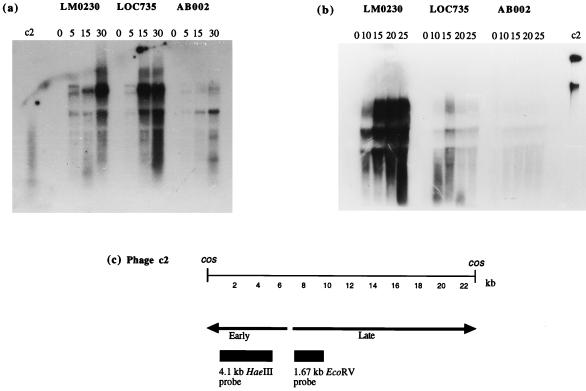
In the case of early phage c2 RNA synthesis, the signal obtained was more specific, with no signal observed prior to phage infection (i.e., T0 [Fig. 5a]). The probe used was a 4.1-kb HaeIII fragment specific for the early-expressed region of c2 (5). A comparison of the amount of RNA synthesized in plasmid-free and pCI735-containing cells suggested that early transcription was normal in the presence of the abiG subclone. Indeed, there appeared to be slightly more RNA present at T15 and T30 in the presence of Abi. This presumably reflects the experimental conditions, although care was taken to harvest RNA from cells at the same optical density and to subsequently load the same total amount of RNA in each well. In contrast to the influence of the subclone, the presence of pCI750 dramatically inhibited phage c2 early transcription. Late c2 transcription was examined by using a 1.67-kb EcoRV fragment as a probe (Fig. 5b). The signal obtained was similar to late sk1 in that no discrete bands were visible (see above). In this instance the abiG subclone resulted in reduced levels of phage transcription, while the presence of pCI750 severely inhibited the transcription process. The latter result was expected, given the previously observed effect of pCI750 on early c2 transcription.
FIG. 5.
(a) Hybridization of total RNA isolated from phage c2-infected L. lactis subsp. cremoris LM0230, LOC735, or AB002 at 0, 5, 15, and 30 min postinfection with a 32P-labelled 4.1-kb HaeIII fragment from the early-expressed region. (b) Hybridization of total RNA isolated from phage c2-infected L. lactis subsp. cremoris LM0230, LOC735, or AB002 at 0, 10, 15, 20, and 25 min postinfection with a 32P-labelled 1.67-kb EcoRV fragment from the late-expressed region. In both panels, lanes labeled c2 contain phage c2 DNA used as a positive control. (c) Temporal transcription map of phage c2 with early- and late-expressed regions indicated; adapted from Beresford et al. (5).
Notably, the level of interference in sk1 RNA synthesis was similar for both pCI750 and pCI735 when total and late-specific transcription was examined. In the case of c2 RNA synthesis, the subclone had no effect on early transcription, while pCI750 did. Thus, it may be concluded that abiG does not interfere with phage c2 early transcription. However, when the effect of pCI750 on late c2 transcription was determined, there was clearly a degree of inhibition greater than that mediated by the subclone. This raises the question whether there is another mechanism(s) on pCI750, which is active against c2, particularly affecting early transcription, while having no significant effect on sk1.
Interestingly, since c2 DNA replication appeared to be unaffected by the presence of pCI750 and yet early c2 transcription was inhibited, it seems possible that early c2 genes are not necessary for c2 DNA replication to occur. This concurs with the findings of Waterfield et al. (30), who had previously found that a noncoding fragment containing the ori was capable of directing DNA replication in Lactococcus but not in E. coli and concluded that host factors alone were sufficient to initiate DNA replication at ori.
In conclusion, AbiG inhibits phage sk1 and late c2 RNA synthesis; however, whether this process of transcription is the primary target of the system or a secondary effect remains to be determined. The AbiB mechanism also interferes with phage RNA but has been shown to result in rapid degradation of sensitive phage transcripts 10 to 15 min after infection (25). Geis et al. (17) reported that the presence of the Abi-encoding lactococcal plasmids pBU1-8 and 1149-3 resulted in complete inhibition of RNA synthesis and proposed that phage transcription was the primary target of these systems. However, it should be noted that these native lactococcal plasmids may encode other resistance mechanisms, which together may have contributed to this observed inhibition. Deficiencies in transcription have been observed in some of the better-characterized Abi systems of gram-negative bacteria, such as the λ-encoded Rex system (15), the ColIb-encoded Abi (11), and the F-encoded Pif (4); however, all these systems also severely inhibited DNA replication, a feature not observed for AbiG. Deficiencies in φ29 transcription and translation in the course of its aborted infection of Vibrio cholerae biotype el tor cells were shown to be due to destabilization of phage DNA concatamer intermediates (7). AbiG most closely resembles the el tor-encoded resistance in terms of its effect on phage DNA replication and RNA transcription.
Acknowledgments
This work was supported by European Community BRIDGE and BIOTECHNOLOGY grants to G. F. Fitzgerald (contracts BIO-CT91-0263 and BIO4-CT96-0402, respectively).
REFERENCES
- 1.Anba J, Bidnenko E, Hillier A, Erlich D, Chopin M-C. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J Bacteriol. 1995;177:3818–3823. doi: 10.1128/jb.177.13.3818-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnau J, Sorensen K I, Appel K F, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. doi: 10.1099/13500872-142-7-1685. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner A, Murphy M, Daly C, Fitzgerald G F. Conjugative co-transfer of lactose and bacteriophage resistance plasmids from Streptococcus cremoris UC653. FEMS Microbiol Lett. 1986;35:233–237. [Google Scholar]
- 4.Beck P J, Molineux I J. Defective transcription of the right end of bacteriophage T7 DNA during an abortive infection of F plasmid-containing Escherichia coli. J Bacteriol. 1991;173:947–954. doi: 10.1128/jb.173.3.947-954.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beresford T P J, Ward L J H, Jarvis A W. Temporally regulated transcriptional expression of the genomes of lactococcal bacteriophages c2 and sk1. Appl Environ Microbiol. 1993;59:3708–3712. doi: 10.1128/aem.59.11.3708-3712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidnenko, E., J. Anba, D. Ehrlich, and M.-C. Chopin. 1996. A phage-encoded nuclease essential for phage development is depleted in AbiD1 cells. FEMS Microbiol. Rev. (Abstract F22.)
- 7.Biswas S K, Chowdhury R, Das J. A 14-kilodalton inner membrane protein of Vibrio cholerae biotype el tor confers resistance to group IV choleraphage infection to classical vibrios. J Bacteriol. 1992;174:6221–6229. doi: 10.1128/jb.174.19.6221-6229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandry P S, Davidson B E, Hillier A J. Temporal transcription map of the Lactococcus lactis bacteriophage sk1. Microbiology. 1994;140:2251–2261. doi: 10.1099/13500872-140-9-2251. [DOI] [PubMed] [Google Scholar]
- 9.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteriophage resistance. Antonie Leeuwenhoek. 1996;70:99–110. doi: 10.1007/BF00395928. [DOI] [PubMed] [Google Scholar]
- 10.Dinsmore P K, Klaenhammer T R. Phenotypic consequences of altering the copy number of abiA, a gene responsible for aborting bacteriophage infections in Lactococcus lactis. Appl Environ Microbiol. 1994;60:1129–1136. doi: 10.1128/aem.60.4.1129-1136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duckworth D H, Glenn J, McCorquodale D J. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol Rev. 1981;45:52–71. doi: 10.1128/mr.45.1.52-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durmaz E, Klaenhammer T R. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defenses in a single Lactococcus lactis strain. Appl Environ Microbiol. 1995;61:1266–1273. doi: 10.1128/aem.61.4.1266-1273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durmaz, E., and T. R. Klaenhammer. 1996. Characterization and sequencing of recombinant lactococcal phage φ31.3. FEMS Microbiol. Rev. (Abstract F27.)
- 14.Efstathiou J D, McKay L L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977;130:257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garen A. Physiological effects of rII mutations in bacteriophage T4. Virology. 1961;14:151–163. doi: 10.1016/0042-6822(61)90190-8. [DOI] [PubMed] [Google Scholar]
- 16.Garvey P, Fitzgerald G F, Hill C. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4160–4166. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geis A, Janzen T, Teuber M, Wirsching F. Mechanism of plasmid-mediated bacteriophage resistance in lactococci. FEMS Microbiol Lett. 1992;94:7–14. [Google Scholar]
- 18.Hill C, Miller L A, Klaenhammer T R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990;56:2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill C, Miller L A, Klaenhammer T R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991;173:4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Malloch R A, Fujita N, Simillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an extended minus 10 promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 21.Moineau S, Pandian S, Klaenhammer T R. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol. 1994;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy M C. Ph.D. thesis. Cork, Ireland: National University of Ireland; 1988. [Google Scholar]
- 23.O’Connor L, Coffey A, Daly C, Fitzgerald G F. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol. 1996;62:3075–3082. doi: 10.1128/aem.62.9.3075-3082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Sullivan D J, Hill C, Klaenhammer T R. Effects of increasing the copy number of bacteriophage origins of replication, in trans, on incoming phage proliferation. Appl Environ Microbiol. 1993;59:2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parreira R, Ehrlich S D, Chopin M-C. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol Microbiol. 1996;19:221–230. doi: 10.1046/j.1365-2958.1996.371896.x. [DOI] [PubMed] [Google Scholar]
- 26.Pillidge C J, Jarvis A W. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. N Z J Dairy Sci Technol. 1988;23:411–416. [Google Scholar]
- 27.Su P, Harvey M, Im H J, Dunn N W. Isolation, cloning and characterisation of the abiI gene from Lactococcus lactis subsp. lactis M138 encoding abortive phage infection. J Biotechnol. 1997;54:95–104. doi: 10.1016/s0168-1656(97)01692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophage. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van de Guchte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Rev. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 30.Waterfield N R, Lubbers M W, Polzin K M, Le Page R W F, Jarvis A W. An origin of DNA replication from Lactococcus lactis bacteriophage c2. Appl Environ Microbiol. 1996;62:1452–1453. doi: 10.1128/aem.62.4.1452-1453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]