Abstract
Spirochetes are a large group of prokaryotes that originated from Gram‐negative bacteria and are capable of causing a variety of human and animal infections. However, the pathogenesis of spirochetes remains unclear, as different types of spirochetes play pathogenic roles through different pathogenic substances and mechanisms. To survive and spread in the host, spirochetes have evolved complicated strategies to evade host immune responses. In this review, we aimed to provide a comprehensive overview of immune evasion strategies in spirochetes infection. These strategies can be explained from the following points: (i) Antigenic variation: random, unidirectional, and segmental conversion of the gene to evade immune surveillance; (ii) Overcoming the attack of the complement system: recruitment of host complement regulators, cleavage of complement components and inhibition of complement activation to evade immune defenses; (iii) Interfering with immune cells to regulating the immune system; (iv) Persistent infection: invading and colonizing the host cell to escape immune damage.
Keywords: immune evasion, immune response, persistent infection, spirochetes
In this review, we aimed to provide a comprehensive overview of immune evasion strategies in spirochetes infection. These strategies can be explained from the following points: (i) Antigenic variation: random, unidirectional, and segmental conversion of the gene to evade immune surveillance; (ii) Overcoming the attack of the complement system: recruitment of host complement regulators, cleavage of complement components and inhibition of complement activation to evade immune defenses; (iii) Interfering with immune cells to regulating the immune system; (iv) Persistent infection: invading and colonizing the host cell to escape immune damage.
1. INTRODUCTION
Spirochetes are prokaryotic microorganisms that are slender, flexible, spiral‐shaped, and highly motile. They are widely distributed in nature and are commonly found in water, soil, and other putrefactive organic matter, as well as in the human mouth and animal body. Spirochetes are mainly divided into five genera of Spirochaetaceae based on differences in size, regularity, space structure, physiological characteristics, and host cells. There are namely Cristispira, 1 Spirochaeta, 2 Treponema, 3 Borrelia, 4 and Leptospira. 5 Only Treponema, Borrelia, and Leptospira are known to cause diseases in humans (Table 1). Epidemiological studies have shown that the incidences of syphilis, 6 , 7 Lyme disease, 8 , 9 and leptospirosis 10 , 11 have increased rapidly worldwide and posed major threats to public health.
TABLE 1.
The taxonomy and pathogenicity of spirochetes
| Spirochaetaceae | Morphology | Length (μm) | Type species | Diseases | Pathogens | Medium |
|---|---|---|---|---|---|---|
| Cristispira | Ridged spiral | 30–180 | Cristispira pectinis | No | ‐‐‐‐‐ | Mud, sewage |
| Spirochaeta | Curved spiral | 5–250 | Spirochaeta plicatilis | No | ‐‐‐‐‐ | Mollusk shell |
| Treponema | Tight spiral | 1–20 | Treponema pallidum | Syphilis | Treponema pallidum | Genital |
| Periodontal disease | Treponema denticola | Oral cavity | ||||
| Yaws | Treponema pertenue | Mucous membrane | ||||
| Pinta | Treponema carateum | Mucous membrane | ||||
| Borrelia | Sparsely wavy | 10–35 | Borrelia anserina | Relapsing fever | Borrelia recurrentis | Ixodes |
| Lyme disease | Borrelia burgdorferi | Body louse | ||||
| Leptospira | Hooked spiral | 6–12 | Leptospira interrogans | Leptospirosis | Leptospira interrogans | Rodents, mammals |
Like with other pathogenic bacteria, a series of immune reactions occur when pathogenic spirochetes enter the body. The immune responses can be divided into innate and acquired immune responses. When pathogen‐associated molecular patterns (PAMPs) interact with pattern‐recognition receptors (PRRs), these PRRs include Toll‐like receptors (TLRs), nucleotide‐binding oligomerization domain (Nod), RIG‐like receptors (RLRs), C‐type lectin receptors (CLRs), and so on. 12 , 13 The innate immune system is activated, with macrophages, dendritic cells (DCs), and neutrophils helping to identify the pathogen and subsequently activating the complement system to eliminate nonspecific pathogens. In addition, specific pathogens are then killed by the acquired immune system. Although there is an obvious immune response after spirochete infection, the pathogenic bacteria cannot be completely cleared, which is why it leads to persistent infection in vivo. In this review, we will focus on the immune evasion mechanism of spirochetes in terms of antigen variation, complement inhibition, and immune interference, and summarize how pathogenic spirochetes escape from the strictly controlled immune system and colonize the host to affect humans and animals alike. A deeper understanding of the immune escape mechanisms of spirochetes will help us to better understand why it is so challenging to eradicate spirochete infections and ultimately provide new insights for developing effective vaccines against them in the future.
2. ANTIGENIC VARIATION
It is generally accepted that antigenic variation is a common pathogenic mechanism adopted by bacterial, protozoan, and fungal pathogens to cope with host identification and defense. 14 It is commonly found in an evolutionary variety of obligate parasites, such as Neisseria gonorrhoeae (gonorrhea), Giardia lamblia (giardiasis or beaver fever), Treponema pallidum (syphilis), Mycoplasma pulmonis (mycoplasma pneumonia), Borrelia recurrentis (relapsing fever), or Borrelia burgdorferi (Lyme disease). In this part, we will consider B. burgdorferi as an example to illustrate the mechanism of antigen variation.
Lyme disease is a multisystem infectious disease caused by B. bugdorferi and is the most common vector‐borne disease affecting humans in North America and temperate Eurasia. In the United States, Lyme disease is transmitted by hard‐bodied ticks of the genus Ixodes. Infected ticks will spread B. burgdorferi to humans during feeding, which can lead to a local infection (erythema migrans) at the site of the tick bite. 15 The clinical manifestation is a chronic infection characterized by neuritis, arthritis, and myocarditis. B. burgdorferi has a highly evolved variable major protein (VMP)‐like sequence (vls) antigenic variation system, which possesses the ability to establish persistent infection by continuous variation of vlsE, a surface‐bound lipoprotein. 16 The vls locus of B31 is located on the 28‐kb linear plasmid (lp28‐1). The vls locus comprises an expression site encoding the 35 kDa lipoprotein vlsE and a consecutive array of 15 silent vls cassettes (474–594 bp in length). The direction of these silent cassettes is opposite to the vlsE locus (Figure 1A). 17 The vlsE locus and the silent vls cassettes are separated by a short intergenic region, and this intergenic space includes a 51‐bp inverted repeat (IR) sequence. 18 Furthermore, a part of the promoter required for vlsE expression is located within this inverted repeat. The expression region of vlsE is composed of a central variable cassette, and the constant region (CR) is located on both sides of the central variable cassette. There are 17‐bp direct repeats (DR) at the junction of the variable and CRs and both ends of most silent cassettes (Figure 1B). 19 The silent cassettes show high homology with the central variable cassette region of vlsE (90–96.1% nucleotide sequence identity). There are six variable regions and six invariant regions located in the central variable cassette region of vlsE, and most of the antigenic variation sequence differences are found in these six variable regions (Figure 1C).
FIGURE 1.
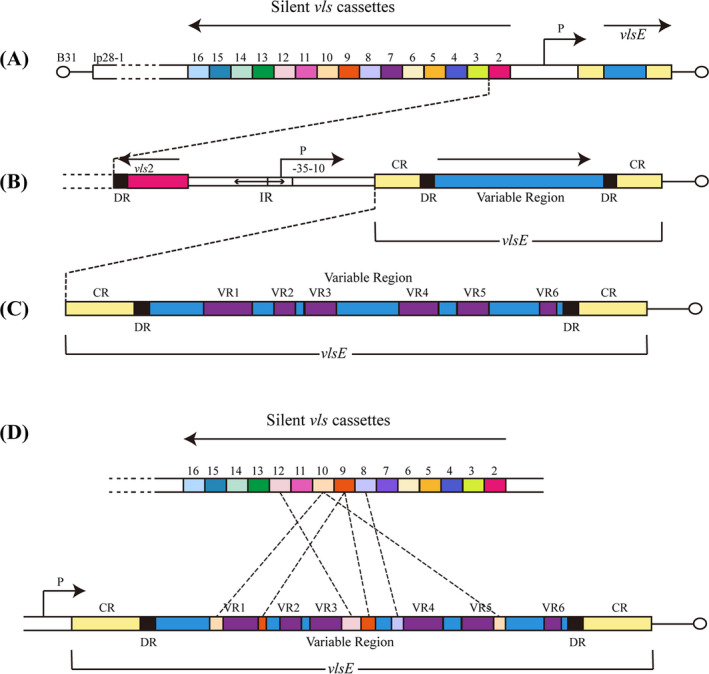
Schematic diagram of the vls system structure and recombination pattern. (A) In the B. burgdorferi of B31‐type strain, the vls locus is located on lp28‐1 and the vlsE gene is located near the telomere of the hairpin, 15 silent cassettes are located near and upstream of vlsE, but in the opposite direction (arrow). (B) The vlsE region is described in more detail. It is composed of the variable region (VR) and the constant region (CR), the variable region is flanked by 17‐bp direct repeats (DRs). To the left of vlsE is a part of the promoter (P), with the −10 and −35 inverted repeat sequences (about 100‐bp in length). (C) The central variable cassette region of vlsE is comprised of six variable regions (VR1–VR6) and six invariant regions (IRs). (D) In infected mammalian hosts, a random, unidirectional, and segmental combination of the vlsE protein‐coding site and the silent cassettes leads to differences in the expression of vlsE variants. Modified from Ref 16, 19, 30
Antigenic variation is a random combination of the vlsE protein‐coding site and the silent cassettes that lead to a difference in the expression of vlsE variants. In the vls system, B. burgdorferi can evade the host's acquired immunity via random, unidirectional, and segmental gene conversion. 16 , 20 Previous studies in infected mice 21 or rabbits 22 showed that while vlsE sequence variation occurs within 4 days and continues throughout infection, they could not be detected in vitro or the tick vector. 23 This suggests that vlsE variants only exist in mammals. Although vlsE through segmental gene conversion provides a large number of diversity variable sequences during antigenic variation to evade immune surveillance, more about its variation mechanism is still unclear. The proposed model for vlsE antigenic conversion indicated 24 that the vlsE central cassette regions are replaced by fragments of varied length and location in the silent cassettes. Bankhead 19 performed an in‐depth analysis of the vlsE sequence changes and found that the vls antigenic variation system promotes varying length (short or long) recombination events in each cassette region. With the development of gene‐conversion events, other template‐independent changes also resulted in the amplification of vlsE variant sequences. The result of these cumulative changes is the generation of a new vlsE sequence with a mosaic structure (Figure 1D). Thus, these mutated antigens prevent recognition by the host immune system. Moreover, it has been well documented that the lack of vlsE or vls genes residing on lp28‐1 will cause the spirochete to lose its persistent infection. Other lp28‐1 non‐vls genes are not involved in the virulence, persistence, and recombination of vlsE. In other words, the vls locus and vlsE protein are key factors for immune escape and persistence of pathogens. 25 , 26 , 27 Similarly, T. pallidum has a corresponding protein gene (TprK) that causes persistent infection through antigenic variation, 28 but the mechanism is not yet known.
In recent years, some technical progress has been made in the study of the vls locus. These include the development of a new‐generation sequencing method 29 for the analysis of vlsE recombination switches and the mini‐vls system 30 for the genetic manipulation of the vls locus. Through this information, we have deepened our understanding of the antigenic variation mechanism of spirochetes. Although vlsE is flexible and variable in evolution, the locus has strictly conservative structural characteristics. These structures are indispensable in the process of antigenic variation. Therefore, whether we can inhibit the persistent infection by destroying the structure of the vls locus or using some genetic tools to achieve the effect of disease treatment remains to be further discussed. Understanding the definite mechanism behind the antigenic variation of spirochetes may lay the foundation for intervention measures to inhibit infection. For now, however, we still have a long way to go to overcome the obstacles of gene manipulation.
3. OVERCOMING THE ATTACK OF THE COMPLEMENT SYSTEM
The complement system is comprised of more than 50 glycoproteins, including plasma complement components, soluble proteins, membrane‐bound proteins, and complement receptors. In innate immunity, the complement system forms an important line of defense against microbial invasion. Actually, the complement system can be activated through three relatively independent and interrelated pathways to exert various biological effects such as regulating phagocytosis, cracking cells, mediating inflammation, regulating immune and clearing immune complexes. These three pathways are the classical pathway (CP), alternative pathway (AP), and lectin pathway (LP). All three pathways lead to the production of C3 invertase and C5 invertase that cleave C3 and C5, respectively, and pass through a common terminal pathway to form a membrane attack complex (MAC) (Figure 2). After activation of complement molecules, the complement components and surface substance of the pathogens form a complex, which ultimately promotes bacterial dissolution. 31 In order to overcome the attack of the complement system, spirochetes take some evasion strategies that include acquiring host complement regulators, cracking complement components by binding of surface protein and Plg, and inhibiting complement activation by the interaction between the surface proteins of spirochetes and complement substances are introduced.
FIGURE 2.
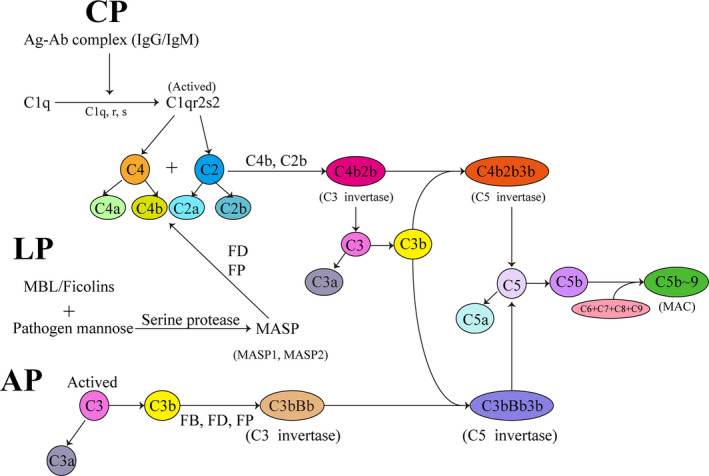
Schematic diagram of three activation pathways of the complement. There are three pathways to activate the complement system: classical pathway (CP), lectin pathway (LP), and alternative pathway (AP). All three pathways lead to the production of C3 invertase and C5 invertase, which cleave C3 and C5 respectively, and the C5 is then cleaved to form C5a and C5b, in that C5b combines with C6 and C7 to form a C5bC6C7 complex which is inserted into the cell membrane. The complex binds to C8 and joins with C9 to form a membrane attack complex (MAC). Finally, the cell starts to lyse. MBL, mannose‐binding lectin; MASP, MBL‐associated serine protease; FB, Factor B; FD, Factor D; FP, Factor D; MAC, membrane attack complex (C5b‐9)
3.1. Recruitment of host complement regulators
The activation of the complement system requires complement factors to perform normal physiological functions. If complement regulation is out of control, a large number of the complement factors will be consumed, which will lead to a decrease in the body's resistance to infection and also cause severe inflammation or damage to its tissues and cells. Therefore, the role of complement regulators is particularly important. Common complement regulators are soluble regulators and membrane‐binding proteins. The former mainly includes C1 inhibitor (C1‐INH), C4b‐binding protein (C4BP), Factor I (FI), Factor H (FH), and factor H‐like‐1 (FHL‐1). The latter mainly includes complement receptor 1 (CR1), membrane cofactor protein (MCP), and decay‐accelerating factor (DAF). In addition, the regulators of the terminal activation sequence comprise vitronectin (Vn), clusterin (Cn), and some of the factor H‐related (FHR) proteins. 32 Serum‐resistant strains of Borrelia, 33 , 34 Leptospira, 35 , 36 and Treponema 37 , 38 recruit the Factor H family, FHL‐1, FHR, and C4BP to inhibit the CP and AP. Different types of spirochetes have different surface‐binding proteins, which accelerate the inactivation of C3b and C4B by binding to these complement regulators.
Several Borrelia proteins such as CspA (CRASP‐1) 39 , 40 and CspZ (CRASP‐2) 41 , 42 can recruit FH/FHL‐1 to accelerate the inactivation of C3b. In addition, CspA binds the complement components C7 and C9 of the terminal pathway and blocks the assembly and membrane insertion of the terminal complement complex (TCC), thereby inhibiting the terminal complement pathway. 43 ErpP (CRASP‐3), ErpC (CRASP‐4), and ErpA (CRASP‐5) have been discovered which combined with FHR‐1, FHR‐2, and partly with FHR‐5 to inhibit complement activation on the cell surface. 44 Moreover, P43 (an outer membrane protein) can bind to C4bp and activate cofactor activity of factor I‐mediated cleavage of C4b. 34 Among the Leptospira proteins, such as L. interrogans endostatin‐like outer membrane proteins A and B (LenA/LenB), 45 Leptospira immunoglobulin‐like proteins A and B (LigA and LigB), 35 Lsa33 (LIC11834), and Lsa25 (LIC12253) 46 can also bind to FH, FHL‐1, FHR‐1, and C4BP at different sites in vitro and regulate the complement activation pathway. Furthermore, Leptospira complement regulator‐acquiring protein A (LcpA) is capable of binding vitronectin via regulating the terminal pathway to reduce MAC deposition. 47 Different from Borrelia and Leptospira, Factor H binding protein B (FhbB) of Treponema can reduce complement factor activity by interacting with FH. 48 Taken together, all of the above examples illustrate the mechanism of immune evasion mediated by spirochetes through the recruitment of complement regulators.
3.2. Cleavage of complement components
When pathogenic microorganisms invade the body, the inactivated complement components in human serum will be activated in the complement system by antigen–antibody complexes. The activated complement components possess enzymatic activity, which can mediate the immune response and inflammatory response, lyse C3b to combine with bacteria to promote phagocytosis of phagocytes, or form membrane attack complex (MAC) to lyse target cells and cause cell death. Spirochete surface proteins, as a kind of important factor to resist innate defense, can cleave the complement factors by binding to plasmin (Pla).
Based on published literature, we know that several plasminogen/plasmin (Plg/Pla) receptors of Borrelia can degrade the complement components C3, C3b, C4b, and C5 via binding to surface proteins such as the Erp family, 49 BBA70 protein, 50 CspA, 39 and CspZ 51 proteins. Furthermore, the HcpA protein, as a surface protein of B. recurrentis can also reduce the deposition of C3b via Pla binding. 52 Similar to Borrelia, the surface Plg/Pla of Leptospira results in downregulation of C3b, C4, and C5 by capturing LigA and LigB, 53 Lsa23 (LIC11360), 54 Lsa30, 55 and elongation factor thermo unstable (EF‐Tu), 56 among other proteins. However, thus far, it is still a challenge to determine the biological correlation between the surface protein of spirochetes and Plg/Pla‐binding protein.
3.3. Inhibition of complement activation
The complement system can be activated by three pathways. Each of the complement components from these three pathways is critical, and the absence or downregulation of even one component will inhibit complement activation. Expect the above two points about the recruitment of complement regulators and the cleavage of complement components, the direct effect of spirochete surface protein on complement factors can also inhibit activation of the complement system.
There is a new mechanism for downregulating the CP of complement. The surface protein BBK32 expressed by Borrelia can block C1r to inhibit the C1 complex composed of C1q, C1r, and C1s. BBK32 acts directly on C1r and inhibits the autocatalysis of C1r in the C1 complex, which fails activation of the CP. However, the LP and AP are not affected, because they lack the targeting factor for BBK32. 57 Therefore, BBK32 can be used as an effective inhibitor of the CP by interfering with C1r. At the same time, BGA66, BGA71, and CspA inhibit complement activation by interfering with the terminal pathway. They interact directly with C7, C8, and C9, and may affect MAC assembly by preventing c5b‐8 complexes from inserting into the target cell membrane correctly and also inhibit the ability of C9 to aggregate with subsequent factors. 43 , 58 Besides, early work by Caine et al. 59 established that the outer surface protein C (OspC) from B. burgdorferi has complement resistance, which can inhibit CP and LP by competing with complement protein C2 for C4B binding and survive in the bloodstream.
Recently, two new hypothesized proteins of Leptospira encoded by the LIC12587 60 and LIC13259 61 genes have been reported. Recombinant protein LIC12587 interacts with C7, C8, and C9 components of the complement system in a dose‐dependent manner, reducing the sterilization effect of the complement. Binding to C9 may result in inhibition of C9 polymerization and interfere with the formation of MAC. Similarly, recombinant protein LIC13259 can recruit and interact with vitronectin, C7, C8, and C9 from normal human serum. Cavenague et al. showed that the binding of rLIC13259 with C8 and vitronectin was inhibited gradually with the increase of heparin concentration, indicating that the interaction with vitronectin occurred through the heparin domain. Most interestingly, the interaction between rLIC13259 and C9 can prevent C9 polymerization from inhibiting MAC formation. To summarize, by degrading or downregulating the complement components, the pathogenic spirochetes can limit activation of the complement system, and finally achieve the goal of immune escape.
4. INTERFERENCE WITH IMMUNE REGULATION
The immune response is that immune cells recognize, activate, proliferate, and produce immune substances under the stimulation of antigens, so as to mediate specific immune effects and finally eliminate invading pathogens. The body can maintain a relatively stable state through appropriate immune regulation. As is well known, immune responses can be divided into innate immunity and adaptive immunity. First, how does the immune system detect the existence of pathogens? There are multiple receptors on the surface of mammalian cells, which can recognize specific molecular characteristics of pathogens. These characteristics are called pathogen‐associated molecular patterns (PAMPs), 13 including bacterial lipopolysaccharides (LPS), lipoproteins, peptidoglycan, and flagellar proteins. The host receptors are called pattern recognition receptors (PRRs) that interact with PAMPs to initiate a series of intracellular signal cascades that trigger an innate immune response and subsequent adaptive immune responses. This process initiates the production of a series of cytokines, including polymorphonuclear neutrophils, monocytes, macrophages, dendritic cells, and natural Killer T Cells, which lead to inflammation in the body. Toll‐like receptors (TLRs) are the main recognition receptors of spirochetes. TLRs can recognize spirochetal membrane components, which play a major role in the inflammatory induction of spirochete infections. Interestingly, spirochetes (Treponema, Borrelia) as gram‐negative bacteria, despite the lack of surface LPS, can still be attacked by TLRs recognition and the immune system. 62 Next, we will focus on the interference of innate immune cells and their surface lipoproteins on immune regulation.
4.1. Impairment of bactericidal function of polymorphonuclear neutrophils (PMNs)
Human PMNs are one of the fastest and earliest immune cells to react during acute infection and possess a variety of microbicide mechanisms. After TLRs recognize and activate neutrophils, hydrolases and strong oxidizing bactericidal substances, such as H2O2, myeloperoxidase, and antimicrobial peptides will be released, which will eventually lead to the elimination of bacteria through the phagocytosis, hydrolysis, and oxidative burst of neutrophils. Furthermore, a study 63 has proposed a novel pathogen‐killing mechanism of neutrophils. When bacterial or fungal species are activated, the nuclear DNA of neutrophils will be released outside of the cell. These DNA structures are called neutrophils extracellular traps (NETs) that can kill pathogens without relying on phagocyte uptake and degranulation.
These mechanisms may be important for early infection and transmission of spirochetes. Surprisingly, Leptospira is hardly phagocytized in neutrophils and can be killed only in the presence of specific antibodies. Vieira et al. proved that Leptospiral outer membrane protein LipL21 can be used as a myeloperoxidase inhibitor to inhibit the oxidation and chlorination activity of myeloperoxidase without interfering with neutrophil degranulation, which is conducive to the survival of Leptospira in the host. 64 Likewise, in the B. burgdorferi lipoprotein BBA57 experiment, infected mice with wild‐type or BBA57 mutant were genetically analyzed that BBA57 mutant mice reduced the expression levels of various pro‐inflammatory factors such as IL‐1β, IL‐6, TNF, or the chemokines Ccl3. On the other hand, the transcription level mediated by PRRS and downstream signaling molecules is also downregulated including Tlr2, Myd88, and Nlrp3. The results suggest that BBA57 can inhibit the activation of innate immune cells. 65 In addition, spirochetes seem to release significant nucleases to degrade the nuclear DNA of neutrophils and prevent them from being trapped and killed by NETs, which helps to spread in the host. 66 However, these specific mechanisms remain to be further studied.
4.2. Anti‐phagocytosis and pro‐apoptosis effect on monocytes and macrophages (M/M)
Other than neutrophils, M/M also provides innate immune protection to the host, especially in the early stages of infection. Spirochete lipoproteins bind to CD14 on the M/M membrane and activate the nuclear transcription factor‐κB (NF‐κB) pathway. 67 , 68 Spirochetes stimulate M/M to produce cytokines such as IL‐1, IL‐10, IL‐6, and TNF‐α, which is one of the main immune responses under TLRs mediation. 69 , 70 , 71 Thus, we speculate that spirochetes may evade the immune response by resisting phagocytosis or promoting apoptosis of immune cells.
In recent years, the mechanism by which leptospirosis induces apoptosis of macrophages has generated much discussion. Two likely mechanisms are discussed below to explain the apoptotic process. Hu et al. 72 demonstrated that Leptospira induced apoptosis through mitochondrial damage in macrophages. The release of apoptosis‐inducing factor (AIF) and endonuclease G (EndoG) from the mitochondria and subsequent nuclear translocation can lead to nuclear DNA breakage and apoptosis. During infection, caspase‐8 and Bid protein were activated, and highly reactive oxygen species (ROS) led to Akt (or protein kinase B, PKB) dephosphorylation. More specifically, Bid‐mediated mitochondrial release of AIF and/or EndoG and nuclear translocation are both major mechanisms of Leptospira‐induced apoptosis in macrophages, and this process is regulated by both caspase‐8 and ROS‐Akt signal pathways. Another new study reported that LPS on the surface of Leptospira promotes the expression of Fas and FasL in macrophages and cell membrane migration. The newly discovered recombinant protein L. interrogans LB047 gene is the only protein captured by mouse and human Fas proteins, which is significantly upregulated in macrophage infection. With the participation of transcription factors c‐Jun and ATF2, c‐Jun N‐terminal kinase (JNK) and p38 mitogen‐activated protein kinase (MAPK) signal pathways are activated by TLR2, and finally, macrophage apoptosis is induced by Fas/FasL‐caspase‐8/‐3 pathway. 73 On the other hand, by recognizing CD14 and/or TLR2 on the cell surface, the outer membrane protein Tp92 of T. pallidum induces apoptosis of THP‐1 cells via the pro‐caspase‐1 pathway. At the same time, the protein induces apoptosis of THP‐1 cells by the protein kinase 1/caspase‐8/caspase‐3 pathway under the receptor interaction. 74
Results from these related studies have helped us to define the anti‐phagocytosis and pro‐apoptotic mechanism of spirochetes and provide a new perspective.
4.3. Inhibition of dendritic cells (DCs) and natural killer T (NKT) cells
Dendritic cells and NKT cells are also essential cells in the innate immune system and play an irreplaceable role in eliminating bacterial infection. DCs are the most powerful antigen‐presenting cells, which can induce naive T cells and act as the sentinel of immune responses. NKT cells can nonspecifically modulate Th1/Th2. They provide a bridge between innate and adaptive immunity.
Inhibiting the function of these cells may be one of the mechanisms of spirochete immune escape. TLR‐mediated pro‐inflammatory effects can produce a large number of cytokines, 75 the most special of which is IL‐10. Because it is different from other cytokines such as IL‐1 and IL‐12, IL‐10 76 is thought to have the ability to downregulate the inflammatory response via the TLR pathway. It can help to fight against spirochetal cell wall infection and any possible chronic effects such as arthritis. 77 In a mouse experiment, Zhang et al. 78 found that when Leptospira and TLR2 agonist Pam3csk4 were injected into hamsters, the IL‐10 produced in the tissues of mice injected with Leptospira and Pam3csk4 was increased as compared with the group injected with Leptospira alone. Similarly, the IL‐10 level of TLR2‐deficient mice was lower than that of wild‐type mice. This suggests the use of TLR2 agonists to induce IL‐10 production, IL‐10 can downregulate the inflammatory system, thereby weakening the inflammatory response of the body. LipL32, as a major outer membrane protein of pathogenic Leptospira, is a TLR2 agonist and can induce a strong antibody response. 79 Like Leptospira, B. burgdorferi can also induce IL‐10 to inhibit the production of inflammatory mediators by M/M and/or DCs in mice, and reduce the inflammatory response. 76 , 80
Regarding NKT cells, an important example is that spirochetes can directly interfere with NKT cells, which respond to CD1d glycolipids on the surface of spirochetes such as B. burgdorferi. 81 Although the exact mechanism of interference is still unclear, further studies are needed to understand the possible interactions between spirochetes and NKT cells.
5. PERSISTENT INFECTION
The fibrinolytic (or Plg/Pla) system is an enzyme cascade consisting of many proteases and inhibitors that are involved in the production and regulation of Pla. Plg is transformed into Pla by tissue‐type Plg activator (tPA) or urokinase‐type Plg activator (uPA) in the fibrinolytic system. Pla, a broad‐spectrum serine protease is the core component of the fibrinolysis system, and its main function is the degradation of fibrinolytic proteins. Several studies have proved that spirochetes such as Borrelia, Leptospira, and T. denticola can bind Plg on their outer surface. Plg appears to combine with protein receptors through its kringle domains. As a Plg/Pla binding site, the lysine residue of spirochete receptors can induce the expression and/or release of Plg activators (tPA or uPA), thereby favoring conversion of surface‐bound Plg into Pla. 82 These spirochetes with Pla activity will cause proteolysis of fibronectin and laminin, which are important parts of ECM and basal membranes. The result is to promote bacterial invasion of cells and transmission. In addition to binding Plg, spirochetes can also stimulate human monocytes to secrete Pla activators that will be helpful to form the Pla on the bacterial surface. 83 On the other hand, pathogens can also indirectly promote the fibrinolytic system by stimulating endothelial cells to secrete matrix metalloproteinases (MMPs). 84
There are different types of outer membrane proteins in spirochetes that can combine with Plg to produce Pla. For instance, Borrelia species have the outer surface proteins A 85 and C 86 (OspA and OspC). These two proteins attach to the intestinal tract of ticks and infect mammalian hosts to colonize and survive. Likewise, in relapsing fever disease, CbiA showed the ability to promote complement binding and inhibit the activation of regulators through interaction with Pla, which would prevent the complement system from attacking pathogens. 87 Some Leptospira Plg receptors such as the major outer membrane protein LenA, 88 LipL46, 89 OmpL1, 90 and OmpA (Lsa66), 91 or T. denticola chymotrypsin‐like protease were also discovered that invades host cells by degrading the ECM and basement membrane components. 92
From the above conclusion, we found that interaction between spirochetes and the host fibrinolytic system provides the bacterial membrane‐related proteins with hydrolytic activity. This property contributes to the degradation of ECM or basal membrane components and epithelial or endothelial tissue penetration, thereby promoting the bacterial invasion of the host, immune escape, and transmission. Recently, a new study reported that Leptospira has two newly developed recombinant proteins, the gene loci of which are LIC11711 60 and LIC13259. 61 Both these recombinant proteins are capable of acquiring Plg from normal human serum and translating them into enzyme‐active Pla under the effect of the Plg activator. The discovery of these new Plg‐binding proteins will play an important role in the invasion and colonization of hosts. The introduction of more new recombinant proteins may deepen our understanding of the fibrinolytic system and the immunopathogenesis of spirochetes.
6. CONCLUSION
Spirochetes are prokaryotic microorganisms between bacteria and protozoa. It not only has the similar structure and biological characteristics of bacteria, including cell wall, binary fission, amorphous nucleus, and sensitivity to antibiotics (penicillin), but also are soft as protozoa. It can move flexibly by bending and contracting the elastic filaments between the cell wall and the cell membrane, but it differs from other pathogens such as fungi, viruses, and parasites in that it does not have a complete cell structure, nor is it a strictly intra‐host parasitic organism. However, these pathogens all have a similar set of self‐protection and independent immune evasion mechanisms that interact with the immune capacity of the host organism to form a certain balance, which is the result of their long‐term co‐evolution. Under the surveillance of the powerful and hostile immune system, spirochetes have developed many strategies to resist the detrimental effects of immune factors. In this work, we summarized several escape mechanisms of spirochetes, including antigenic variation, complement inhibition, and immune interference to subvert the immune response. All the above‐mentioned measures increase the possibility of spirochete survival in the host and lead to a persistent chronic infection. As we know, the vls locus has strictly conservative structural characteristics. With the successful construction of a mini‐vls system, we speculate whether we can block the recombination switch or destroy the structure of the vls site or use other genetic tools to inhibit persistent infection, to achieve the effect of successful disease treatment that remains to be further investigated. There are still several challenges concerning gene manipulation as a potential therapy. Moreover, we found that many escape mechanisms of spirochetes are closely associated with the spirilla surface proteins. In recent years, scientists have also developed some candidate vaccines for spirochetes using surface proteins. Our future studies are aimed at identifying more immune‐related proteins to contribute to the development of vaccines against spirochetes.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
Jielite Huang drafted the manuscript. Jinlin Chen and Yafeng Xie modified the manuscript. Zhuoran Liu conceived the idea.
Huang J, Chen J, Xie Y, Liu Z. Subversion of the immune response of human pathogenic spirochetes. J Clin Lab Anal. 2022;36:e24414. doi: 10.1002/jcla.24414
Funding information
This work was supported by the National Natural Science Foundation of China (82002182 and 81702046), the Natural Science Foundation of Hunan Province (2021JJ40479 and 2019JJ50535), the Hunan Province Clinical Medical Technology Innovation Guidance Project (S2018SFYLJS0025), the Hunan Science and Technology Innovation Project (2018SK51504), Postgraduate Research and Innovation Project of Hunan Province (CX20190770), General Funding of Hunan Provincial Health Commission (202201063177 and 202211003920), Key Funding of Hunan Provincial Education Department (21A0282), and Hengyang Science and Technology Bureau Guiding Plan Project (hyzdxjh202103)
Contributor Information
Yafeng Xie, Email: xieyafeng1989@163.com.
Zhuoran Liu, Email: LZR9656@163.com.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Paster BJ, Breznak JA. Cristispira. Bergey's manual of systematics of archaea and bacteria. 2015. doi: 10.1002/9781118960608.gbm01247 [DOI]
- 2. Leschine S, Paster BJ. Spirochaeta. Bergey's manual of systematics of archaea and bacteria. 2015. doi: 10.1002/9781118960608.gbm01248 [DOI]
- 3. Norris SJ, Paster BJ, Smibert RM. Treponema. Bergey's manual of systematics of archaea and bacteria. 2015. doi: 10.1002/9781118960608.gbm01249 [DOI]
- 4. Barbour AG, Schwan TG. Borrelia. Bergey's manual of systematics of archaea and bacteria. 2018. doi: 10.1002/9781118960608.gbm01246.pub2. [DOI]
- 5. Picardeau M.. Leptospira. Bergey's manual of systematics of archaea and bacteria. 2017. doi: 10.1002/9781118960608.gbm01244.pub2 [DOI]
- 6. Peeling RW, Mabey D, Kamb ML, et al. Syphilis. Nat Rev Dis Primers. 2017;3:17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghanem KG, Ram S, Rice PA. The modern epidemic of syphilis. N Engl J Med. 2020;382(9):845‐854. [DOI] [PubMed] [Google Scholar]
- 8. Mead PS. Epidemiology of Lyme disease. Infect Dis Clin North Am. 2015;29(2):187‐210. [DOI] [PubMed] [Google Scholar]
- 9. Bacon RM, Kugeler KJ, Mead PS. Surveillance for Lyme disease–United States, 1992–2006. MMWR Surveill Summ. 2008;57(10):1‐9. [PubMed] [Google Scholar]
- 10. Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12(4):351‐357. [DOI] [PubMed] [Google Scholar]
- 11. Vijayachari P, Sugunan AP, Shriram AN. Leptospirosis: an emerging global public health problem. J Biosci. 2008;33(4):557‐569. [DOI] [PubMed] [Google Scholar]
- 12. Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science (New York, NY). 2010;327(5963):291‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16‐34. [DOI] [PubMed] [Google Scholar]
- 14. Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7(7):493‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stanek G, Wormser GP, Gray J, et al. Lyme borreliosis. Lancet (London, England). 2012;379(9814):461‐473. [DOI] [PubMed] [Google Scholar]
- 16. Norris SJ. The vls antigenic variation systems of Lyme disease Borrelia: eluding host immunity through both random, segmental gene conversion and framework heterogeneity. Microbiol Spectr. 2014;6(2). doi: 10.1128/microbiolspec.mdna3-0038-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang JR, Hardham JM, Barbour AG, et al. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP‐like sequence cassettes. Cell. 1997;89(2):275‐285. [DOI] [PubMed] [Google Scholar]
- 18. Coutte L, Botkin DJ, Gao L, et al. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5(2):e1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bankhead T. Role of the VlsE lipoprotein in immune avoidance by the Lyme disease spirochete Borrelia burgdorferi . For Immunopathol Dis Therap. 2016;7(3‐4):191‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette‐specific, segmental gene conversion. Infect Immun. 1998;66(8):3698‐3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang JR, Norris SJ. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi . Infect Immun. 1998;66(8):3689‐3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Embers ME, Liang FT, Howell JK, et al. Antigenicity and recombination of VlsE, the antigenic variation protein of Borrelia burgdorferi, in rabbits, a host putatively resistant to long‐term infection with this spirochete. FEMS Immunol Med Microbiol. 2007;50(3):421‐429. [DOI] [PubMed] [Google Scholar]
- 23. Indest KJ, Howell JK, Jacobs MB, et al. Analysis of Borrelia burgdorferi vlsE gene expression and recombination in the tick vector. Infect Immun. 2001;69(11):7083‐7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palmer GH, Bankhead T, Lukehart SA. ‘Nothing is permanent but change’‐ antigenic variation in persistent bacterial pathogens. Cell Microbiol. 2009;11(12):1697‐1705. doi: 10.1111/j.1462-5822.2009.01366.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bankhead T, Chaconas G. The role of VlsE antigenic variation in the Lyme disease spirochete: persistence through a mechanism that differs from other pathogens. Mol Microbiol. 2007;65(6):1547‐1558. [DOI] [PubMed] [Google Scholar]
- 26. Magunda PR, Bankhead T. Investigating the potential role of non‐vls genes on linear plasmid 28–1 in virulence and persistence by Borrelia burgdorferi . BMC Microbiol. 2016;16(1):180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rogovskyy AS, Bankhead T. Variable VlsE is critical for host reinfection by the Lyme disease spirochete. PLoS One. 2013;8(4):e61226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giacani L, Molini BJ, Kim EY, et al. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J Immunol. 2010;184(7):3822‐3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verhey TB, Castellanos M, Chaconas G. Analysis of recombinational switching at the antigenic variation locus of the Lyme spirochete using a novel PacBio sequencing pipeline. Mol Microbiol. 2018;107(1):104‐115. [DOI] [PubMed] [Google Scholar]
- 30. Castellanos M, Verhey TB, Chaconas G. A Borrelia burgdorferi mini‐vls system that undergoes antigenic switching in mice: investigation of the role of plasmid topology and the long inverted repeat. Mol Microbiol. 2018;109(5):710‐721. [DOI] [PubMed] [Google Scholar]
- 31. Barbosa AS, Isaac L. Complement immune evasion by spirochetes. Curr Top Microbiol Immunol. 2018;415:215‐238. [DOI] [PubMed] [Google Scholar]
- 32. Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9(10):729‐740. [DOI] [PubMed] [Google Scholar]
- 33. Kraiczy P, Skerka C, Kirschfink M, et al. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL‐1/reconectin and Factor H. Eur J Immunol. 2001;31(6):1674‐1684. [DOI] [PubMed] [Google Scholar]
- 34. Pietikäinen J, Meri T, Blom AM, et al. Binding of the complement inhibitor C4b‐binding protein to Lyme disease Borreliae. Mol Immunol. 2010;47(6):1299‐1305. [DOI] [PubMed] [Google Scholar]
- 35. Castiblanco‐Valencia MM, Fraga TR, Silva LB, et al. Leptospiral immunoglobulin‐like proteins interact with human complement regulators factor H, FHL‐1, FHR‐1, and C4BP. J Infect Dis. 2012;205(6):995‐1004. [DOI] [PubMed] [Google Scholar]
- 36. Meri T, Murgia R, Stefanel P, et al. Regulation of complement activation at the C3‐level by serum resistant leptospires. Microb Pathog. 2005;39(4):139‐147. [DOI] [PubMed] [Google Scholar]
- 37. McDowell JV, Frederick J, Miller DP, et al. Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola . Mol Oral Microbiol. 2011;26(2):140‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDowell JV, Lankford J, Stamm L, et al. Demonstration of factor H‐like protein 1 binding to Treponema denticola, a pathogen associated with periodontal disease in humans. Infect Immun. 2005;73(11):7126‐7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hammerschmidt C, Koenigs A, Siegel C, et al. Versatile roles of CspA orthologs in complement inactivation of serum‐resistant Lyme disease spirochetes. Infect Immun. 2014;82(1):380‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kraiczy P, Hellwage J, Skerka C, et al. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP‐1, a novel linear plasmid‐encoded surface protein that interacts with human factor H and FHL‐1 and is unrelated to Erp proteins. J Biol Chem. 2004;279(4):2421‐2429. [DOI] [PubMed] [Google Scholar]
- 41. Marcinkiewicz AL, Dupuis AP 2nd, Zamba‐Campero M, et al. Blood treatment of Lyme borreliae demonstrates the mechanism of CspZ‐mediated complement evasion to promote systemic infection in vertebrate hosts. Cell Microbiol. 2019;21(2):e12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartmann K, Corvey C, Skerka C, et al. Functional characterization of BbCRASP‐2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL‐1. Mol Microbiol. 2006;61(5):1220‐1236. [DOI] [PubMed] [Google Scholar]
- 43. Hallström T, Siegel C, Mörgelin M, et al. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. MBio. 2013;4(4):e00481‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haupt K, Kraiczy P, Wallich R, et al. Binding of human factor H‐related protein 1 to serum‐resistant Borrelia burgdorferi is mediated by borrelial complement regulator‐acquiring surface proteins. J Infect Dis. 2007;196(1):124‐133. [DOI] [PubMed] [Google Scholar]
- 45. Stevenson B, Choy HA, Pinne M, et al. Leptospira interrogans endostatin‐like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One. 2007;2(11):e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Domingos RF, Vieira ML, Romero EC, et al. Features of two proteins of Leptospira interrogans with potential role in host‐pathogen interactions. BMC Microbiol. 2012;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. da Silva LB, Miragaia Ldos S, Breda LC, et al. Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA. Infect Immun. 2015;83(3):888‐897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tegels BK, Oliver LD Jr, Miller DP, et al. Plasminogen binding and degradation by Treponema denticola: identification of the plasminogen binding interface on the FhbB protein. Mol Oral Microbiol. 2018;33(3):249‐256. [DOI] [PubMed] [Google Scholar]
- 49. Brissette CA, Haupt K, Barthel D, et al. Borrelia burgdorferi infection‐associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect Immun. 2009;77(1):300‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koenigs A, Hammerschmidt C, Jutras BL, et al. BBA70 of Borrelia burgdorferi is a novel plasminogen‐binding protein. J Biol Chem. 2013;288(35):25229‐25243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siegel C, Schreiber J, Haupt K, et al. Deciphering the ligand‐binding sites in the Borrelia burgdorferi complement regulator‐acquiring surface protein 2 required for interactions with the human immune regulators factor H and factor H‐like protein 1. J Biol Chem. 2008;283(50):34855‐34863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grosskinsky S, Schott M, Brenner C, et al. Borrelia recurrentis employs a novel multifunctional surface protein with anti‐complement, anti‐opsonic and invasive potential to escape innate immunity. PLoS One. 2009;4(3):e4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Castiblanco‐Valencia MM, Fraga TR, Pagotto AH, et al. Plasmin cleaves fibrinogen and the human complement proteins C3b and C5 in the presence of Leptospira interrogans proteins: a new role of LigA and LigB in invasion and complement immune evasion. Immunobiology. 2016;221(5):679‐689. [DOI] [PubMed] [Google Scholar]
- 54. Siqueira GH, Atzingen MV, de Souza GO, Vasconcellos SA, Nascimento ALTO. Leptospira interrogans Lsa23 protein recruits plasminogen, factor H and C4BP from normal human serum and mediates C3b and C4b degradation. Microbiology. 2016;162(2):295‐308. [DOI] [PubMed] [Google Scholar]
- 55. Souza NM, Vieira ML, Alves IJ, et al. Lsa30, a novel adhesin of Leptospira interrogans binds human plasminogen and the complement regulator C4bp. Microb Pathog. 2012;53(3‐4):125‐134. [DOI] [PubMed] [Google Scholar]
- 56. Wolff DG, Castiblanco‐Valencia MM, Abe CM, et al. Interaction of Leptospira elongation factor Tu with plasminogen and complement factor H: a metabolic leptospiral protein with moonlighting activities. PLoS One. 2013;8(11):e81818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garcia BL, Zhi H, Wager B, et al. Borrelia burgdorferi BBK32 inhibits the classical pathway by blocking activation of the C1 complement complex. PLoS Pathog. 2016;12(1):e1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hammerschmidt C, Klevenhaus Y, Koenigs A, et al. BGA66 and BGA71 facilitate complement resistance of Borrelia bavariensis by inhibiting assembly of the membrane attack complex. Mol Microbiol. 2016;99(2):407‐424. [DOI] [PubMed] [Google Scholar]
- 59. Caine JA, Lin YP, Kessler JR, et al. Borrelia burgdorferi outer surface protein C (OspC) binds complement component C4b and confers bloodstream survival. Cell Microbiol. 2017;19(12). doi: 10.1111/cmi.12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kochi LT, Fernandes LGV, Souza GO, et al. The interaction of two novel putative proteins of Leptospira interrogans with E‐cadherin, plasminogen and complement components with potential role in bacterial infection. Virulence. 2019;10(1):734‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cavenague MF, Teixeira AF, Filho AS, et al. Characterization of a novel protein of Leptospira interrogans exhibiting plasminogen, vitronectin and complement binding properties. Int J Med Microbiol. 2019;309(2):116‐129. [DOI] [PubMed] [Google Scholar]
- 62. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783‐801. [DOI] [PubMed] [Google Scholar]
- 63. Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science (New York, NY). 2004;303(5663):1532‐1535. [DOI] [PubMed] [Google Scholar]
- 64. Vieira ML, Teixeira AF, Pidde G, et al. Leptospira interrogans outer membrane protein LipL21 is a potent inhibitor of neutrophil myeloperoxidase. Virulence. 2018;9(1):414‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bernard Q, Smith AA, Yang X, et al. Plasticity in early immune evasion strategies of a bacterial pathogen. Proc Natl Acad Sci USA. 2018;115(16):E3788‐E3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scharrig E, Carestia A, Ferrer MF, et al. Neutrophil extracellular traps are involved in the innate immune response to infection with Leptospira. PLoS Negl Trop Dis. 2015;9(7):e0003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sellati TJ, Bouis DA, Kitchens RL, et al. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytic cells via a CD14‐dependent pathway distinct from that used by lipopolysaccharide. J Immunol. 1998;160(11):5455‐5464. [PubMed] [Google Scholar]
- 68. Wooten RM, Morrison TB, Weis JH, et al. The role of CD14 in signaling mediated by outer membrane lipoproteins of Borrelia burgdorferi. J Immunol. 1998;160(11):5485‐5492. [PubMed] [Google Scholar]
- 69. Hirschfeld M, Kirschning CJ, Schwandner R, et al. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll‐like receptor 2. J Immunol. 1999;163(5):2382‐2386. [PubMed] [Google Scholar]
- 70. Guerau‐de‐Arellano M, Huber BT. Chemokines and Toll‐like receptors in Lyme disease pathogenesis. Trends Mol Med. 2005;11(3):114‐120. [DOI] [PubMed] [Google Scholar]
- 71. Radolf JD, Arndt LL, Akins DR, et al. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytes/macrophages. J Immunol. 1995;154(6):2866‐2877. [PubMed] [Google Scholar]
- 72. Hu WL, Dong HY, Li Y, et al. Bid‐induced release of AIF/EndoG from mitochondria causes apoptosis of macrophages during infection with Leptospira interrogans . Front Cell Infect Microbiol. 2017;7:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Du P, Li SJ, Ojcius DM, et al. A novel Fas‐binding outer membrane protein and lipopolysaccharide of Leptospira interrogans induce macrophage apoptosis through the Fas/FasL‐caspase‐8/‐3 pathway. Emerg Microbes Infect. 2018;7(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74. Luo X, Zhang X, Gan L, et al. The outer membrane protein Tp92 of Treponema pallidum induces human mononuclear cell death and IL‐8 secretion. J Cell Mol Med. 2018;22(12):6039‐6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ma Y, Seiler KP, Tai KF, et al. Outer surface lipoproteins of Borrelia burgdorferi stimulate nitric oxide production by the cytokine‐inducible pathway. Infect Immun. 1994;62(9):3663‐3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gautam A, Dixit S, Philipp MT, et al. Interleukin‐10 alters effector functions of multiple genes induced by Borrelia burgdorferi in macrophages to regulate Lyme disease inflammation. Infect Immun. 2011;79(12):4876‐4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lochhead RB, Zachary JF, Dalla Rosa L, et al. Antagonistic interplay between MicroRNA‐155 and IL‐10 during Lyme carditis and arthritis. PLoS One. 2015;10(8):e0135142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang W, Zhang N, Xie X, et al. Toll‐like receptor 2 agonist Pam3CSK4 alleviates the pathology of leptospirosis in hamster. Infect Immun. 2016;84(12):3350‐3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hsu SH, Lo YY, Tung JY, et al. Leptospiral outer membrane lipoprotein LipL32 binding on toll‐like receptor 2 of renal cells as determined with an atomic force microscope. Biochemistry. 2010;49(26):5408‐5417. [DOI] [PubMed] [Google Scholar]
- 80. Chung Y, Zhang N, Wooten RM. Borrelia burgdorferi elicited‐IL‐10 suppresses the production of inflammatory mediators, phagocytosis, and expression of co‐stimulatory receptors by murine macrophages and/or dendritic cells. PLoS One. 2013;8(12):e84980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang J, Li Y, Kinjo Y, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107(4):1535‐1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vieira ML, Nascimento AL. Interaction of spirochetes with the host fibrinolytic system and potential roles in pathogenesis. Crit Rev Microbiol. 2016;42(4):573‐587. [DOI] [PubMed] [Google Scholar]
- 83. Fuchs H, Simon MM, Wallich R, et al. Borrelia burgdorferi induces secretion of pro‐urokinase‐type plasminogen activator by human monocytes. Infect Immun. 1996;64(10):4307‐4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vieira ML, Alvarez‐Flores MP, Kirchgatter K, et al. Interaction of Leptospira interrogans with human proteolytic systems enhances dissemination through endothelial cells and protease levels. Infect Immun. 2013;81(5):1764‐1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fuchs H, Wallich R, Simon MM, et al. The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc Natl Acad Sci USA. 1994;91(26):12594‐12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Önder Ö, Humphrey PT, McOmber B, et al. OspC is potent plasminogen receptor on surface of Borrelia burgdorferi. J Biol Chem. 2012;287(20):16860‐16868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nguyen NTT, Röttgerding F, Devraj G, et al. The complement binding and inhibitory protein CbiA of Borrelia miyamotoi degrades extracellular matrix components by interacting with plasmin(ogen). Front Cell Infect Microbiol. 2018;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Verma A, Brissette CA, Bowman AA, et al. Leptospiral endostatin‐like protein A is a bacterial cell surface receptor for human plasminogen. Infect Immun. 2010;78(5):2053‐2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Santos JV, Pereira PRM, Fernandes LGV, et al. Binding of human plasminogen by the lipoprotein LipL46 of Leptospira interrogans. Mol Cell Probes. 2018;37:12‐21. [DOI] [PubMed] [Google Scholar]
- 90. Fernandes LG, Vieira ML, Kirchgatter K, et al. OmpL1 is an extracellular matrix‐ and plasminogen‐interacting protein of Leptospira spp. Infect Immun. 2012;80(10):3679‐3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oliveira R, de Morais ZM, Gonçales AP, Romero EC, Vasconcellos SA, Nascimento ALTO. Characterization of novel OmpA‐like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS One. 2011;6(7):e21962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Grenier D, Uitto VJ, McBride BC. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect Immun. 1990;58(2):347‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


