Abstract
Background
Caudatin is extracted from radix cynanchi bungei and has an inhibitory effect on cancer progression. The study aims to reveal the impacts of hsa_circ_0060927 on Caudatin‐mediated colorectal cancer (CRC) development and the underneath mechanism.
Methods
The expression levels of hsa_circ_0060927, microRNA‐421 (miR‐421) and miR‐195‐5p were detected by quantitative real‐time reverse transcription‐polymerase chain reaction. The protein expression was analyzed by Western blot or immunohistochemistry assay. Cell viability and proliferation were analyzed by 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide or 5‐Ethynyl‐29‐deoxyuridine assay. Cell apoptosis was quantified by flow cytometry analysis. Cell migration and invasion were investigated by transwell assay. The putative associations among hsa_circ_0060927, miR‐421 and miR‐195‐5p were predicted by the starbase online database, and identified by dual‐luciferase reporter, RNA pull‐down and RNA immunoprecipitation (RIP) assays. The impacts of Caudatin treatment on tumor growth in vivo were revealed by a xenograft tumor model assay.
Results
Hsa_circ_0060927 expression was significantly upregulated, whereas miR‐421 and miR‐195‐5p were downregulated in CRC tissues and cells compared with control groups. Hsa_circ_0060927 expression was closely associated with lymph node metastasis and tumor‐node‐metastasis stage. Caudatin treatment significantly decreased hsa_circ_0060927 expression but increased miR‐421 and miR‐195‐5p expression. Caudatin exposure suppressed CRC cell proliferation, migration and invasion, and induced cell apoptosis; however, hsa_circ_0060927 overexpression hindered these impacts. Additionally, hsa_circ_0060927 was associated with miR‐421/miR‐195‐5p. Depletion of miR‐421 or miR‐195‐5p attenuated the influences of hsa_circ_0060927 silencing on CRC development. Furthermore, Caudatin treatment repressed tumor growth in vivo.
Conclusion
Caudatin inhibited CRC cell malignancy through the hsa_circ_0060927/miR‐421/miR‐195‐5p pathway, which provided a potential therapeutic agent for CRC.
Keywords: Caudatin, colorectal cancer, hsa_circ_0060927, miR‐195‐5p, miR‐421
Caudatin treatment increased miR‐421/miR‐195‐5p expression by inhibiting hsa_circ_0060927, which induced CRC cell proliferation, migration and invasion but suppressed apoptosis, further hindering CRC cell malignancy.

1. INTRODUCTION
Colorectal cancer (CRC) is an aggressive carcinoma that occurs in the colon or rectum and is the common cause of cancer‐related death. 1 Despite much progress in CRC pathogenesis, the survival rates of CRC cases are still low. 2 , 3 At present, more and more people are diagnosed with CRC, which poses a heavy financial burden to the sufferers, particularly to those from poor households. Caudatin is extracted from cynanchum auriculatum and plays a repressive role in cancer progression. 4 , 5 Thus, the study on the effects and mechanism of Caudatin in CRC progression is worthy and important to develop therapeutic strategies for CRC.
Circular RNA (circRNA) is an endogenous transcript with a closed‐loop structure. 6 , 7 , 8 CircRNA commonly acts as a competing endogenous RNA to sponge microRNA (miRNA). 9 , 10 Considerable evidence has demonstrated that circRNA is a vital regulator in the development of CRC. 11 , 12 For example, hsa_circ_103948 inhibited CRC cell autophagy. 13 Hsa_circ_002144 contributed to CRC cell growth and metastasis by sponging miR‐615‐5p. 14 , 15 Hsa_circ_0060927 is a novel circRNA that is significantly upregulated in CRC tissues and cells and has the potential as a diagnostic marker for CRC. 16 However, whether Caudatin‐mediated CRC development involves hsa_circ_0060927 remains unclear.
MiRNA is a noncoding RNA with approximately 20 nucleotides in size and can interact with the 3′‐untranslated region of the target gene to regulate its expression. 17 Multiple researchers demonstrated that miRNAs functioned as oncogenes or tumor repressors in cancer process. 18 , 19 , 20 Given the miRNA sponge effect of circRNA, 21 hsa_circ_0060927‐associated miRNAs were predicted by the starbase online database (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA). As a result, miR‐421 and miR‐195‐5p contained the binding sites of circ_0060927. MiR‐421 repressed cell proliferative capacity and metastasis in CRC progression, 22 and miR‐195‐5p hindered CRC progression. 23 It was unknown whether Caudatin regulated CRC progression through the hsa_circ_0060927/miR‐421/miR‐195‐5p axis.
Herein, we analyzed the role of Caudatin in CRC cell malignancy and the underlying mechanism. We found that hsa_circ_0060927 participated in Caudatin‐reduced CRC cell malignancy by binding to miR‐421 or miR‐195‐5p. Our findings might provide novel evidence for Caudatin as an anti‐CRC compound.
2. MATERIALS AND METHODS
2.1. Tissue acquisition and storage
Colorectal cancer tissues and paracancerous non‐cancerous colorectal tissues (NC) (N = 68, respectively) were collected from CRC patients who underwent a surgical operation in Nanjing Hospital of Chinese Medicine Affiliated to Nanjing University of Chinese Medicine. The normal tissues were obtained 5 cm from the tumor margin. These tissues were stored at −80°C in a freezer. CRC patients signed the written informed consents before surgery. The Ethics Committee of Nanjing Hospital of Chinese Medicine Affiliated to Nanjing University of Chinese Medicine approved the study (approval number 2020NJK R‐013‐03).
2.2. Cell purchase and treatment
BNCC provided human CRC cell lines (HCT116 and SW480) and human colorectal mucous membrane cell line (FHC). Cells were cultivated in Roswell Park Memorial Institute‐1640 (RPMI‐1640; Procell) supplemented with 10% fetal bovine serum (FBS; Procell) and antibiotics (100 μg/ml penicillin, 100 μg/ml streptomycin) (Procell) at 37°C in an incubator with 5% CO2.
Caudatin (Sigma) (0, 25, 50 and 100 μM) was used to stimulate HCT116 and SW480 cells for 12, 24 and 36 h to determine its effect on CRC progression.
2.3. Cell transfection
The overexpression plasmid of hsa_circ_0060927 (hsa_circ_0060927) and its control (vector) were provided by Geneseed Co., Ltd. The small interfering RNA against hsa_circ_0060927 (si‐circ_0060927) and its control (si‐NC) were synthesized by GENEWIZ Co., Ltd. MiR‐421 mimic (miR‐421), miR‐421 inhibitor (anti‐miR‐421), miR‐195‐5p mimic (miR‐195‐5p), miR‐195‐5p inhibitor (anti‐miR‐195‐5p) and controls (miR‐NC and anti‐NC) were synthesized by Ribobio Co., Ltd. Lipofectamine 2000 (Thermo Fisher) was employed for cell transfection as instructed. 24 The sequences are si‐circ_0060927 5′‐ATACGCCTCAGGGAAGGGGAA‐3′, miR‐421 5′‐AUCAACAGACAUUAAUUGGGCGC‐3′, anti‐miR‐421 5′‐GCGCCCAAUUAAUGUCUGUUGAU‐3′, miR‐195‐5p 5′‐UAGCAGCACAGAAAUAUUGGC‐3′, anti‐miR‐195‐5p 5′‐GCCAAUAUUUCUGUGCUGCUA‐3′, si‐NC 5′‐GCTAGACCCGTAACAGTAT‐3′, miR‐NC 5′‐UUUGUACUACACAAAAGUACUG‐3′ and anti‐NC 5′‐CAGUACUUUUGUGUAGUACAAA‐3′.
2.4. 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide assay
HCT116 and SW480 cells were diluted in RPMI‐1640 medium containing 10% FBS (Procell) and added into 96‐well plates (5 × 103 per well). The cells were treated with Caudatin, plasmids or oligonucleotides according to the defined purposes when cell confluence reached about 60%. Cells were then cultured for 12, 24 and 36 h, respectively. After cells were incubated with 10 μl 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide (MTT) solution (5 mg/ml; Beyotime), 100 μl dimethyl sulfoxide (Sigma) was placed into the culture well to dissolve formazan. Cell viability was determined by assessing the output of optical density (OD) of samples at wavelength 490 nm with a Varioskan LUX multimode microplate reader (Thermo Fisher).
2.5. 5‐Ethynyl‐29‐deoxyuridine assay
Cell proliferation was detected using an 5‐Ethynyl‐29‐deoxyuridine (EDU) staining kit (Ribobio) following the guidebook. 50 μM EDU that was diluted in RPMI‐1640 (Procell) was pre‐incubated with 96‐well plates for 2 h. HCT116 and SW480 cells were grown in 6‐well plates and treated with test compounds. Forty‐eight hours later, the cells were digested using trypsin (Thermo Fisher), and passaged in 96‐well plates (4 × 103 per well). Then, the cells were fixed using 50 μl paraformaldehyde (Beyotime) for 30 min, and incubated with 50 μl glycine (2 mg/ml) for 5 min, followed by destaining using 0.5% TritonX‐100 for 10 min. After being stained with Apollo® solution and 2‐(4‐Amidinophenyl)‐6‐indolecarbamidine dihydrochloride (DAPI), the cells were observed with an IX71fluorescence microscope (Olympus).
2.6. Flow cytometry analysis
The apoptotic rate of CRC cells was analyzed according to the guidebook of Annexin V‐fluorescein isothiocyanate (Annexin V‐FITC) apoptosis detection kit (Solarbio). 1 × 105 cells for each experiment were seeded in 12‐well plates and treated with 50 μM Caudatin for 24 h or plasmids for 48 h. The cells were collected and washed using a cold phosphate buffer solution (PBS; Solarbio). Then, the cells were suspended in binding buffer (Solarbio), and incubated with 5 μl Annexin V‐FITC (Solarbio) for 10 min and 5 μl propidium iodide (PI; Solarbio) for 5 min in the dark. Samples were assessed with an Attune NxT flow cytometer (Thermo Fisher).
2.7. Western blot analysis
For cell assay, 5 × 104 cells for each experiment were seeded in 24‐well plates and treated with 50 μM Caudatin for 24 h or plasmids for 48 h. Tissues and cells were lysed using RIPA buffer (Beyotime). The lysates were mixed with loading buffer (Thermo Fisher) and boiled in boiling water for 10 min. The protein samples were loaded onto 12% bis‐tris‐acrylamide gels (Thermo Fisher), electrotransferred onto polyvinylidene fluoride membranes (Sigma), and immersed in fat‐free milk (Solarbio) that was diluted with Tris‐buffered saline Tween (TBST; Sigma). Then, the polyvinylidene fluoride membranes were incubated with anti‐proliferating cell nuclear antigen (anti‐PCNA) (AF0239; 1:1500; Affinity), anti‐cleaved‐caspase 3 (anti‐Cleaved‐caspase 3) (AF7022; 1:1500; Affinity), anti‐E‐cadherin (AF0131; 1:2500; Affinity), anti‐N‐cadherin (SAB5700641; 1:1000; Sigma) and anti‐glyceraldehyde 3‐phosphate dehydrogenase (anti‐GAPDH) (AF7021; 1:5000; Affinity), respectively. After that, a horseradish peroxidase‐labeled secondary antibody (A9169; 1:80,000; Sigma) was utilized to incubate the membranes. Protein bands were visualized using RapidStep ECL Reagent (Sigma). GAPDH was employed as an internal reference.
2.8. Transwell assay
The migration and invasion of HCT116 and SW480 cells were analyzed by transwell chambers without or with Matrigel (Corning), respectively. 5 × 104 cells diluted in FBS‐free RPMI‐1640 (Procell) were added into the upper chambers, and treated with 50 μM Caudatin or plasmids. RPMI‐1640 containing 12% FBS (Procell) was placed into the lower chambers. Twenty‐four hours later, the medium was removed, and 4% paraformaldehyde (Beyotime) and 0.1% crystal violet (Beyotime) were used to incubate the cells. Results were assessed by calculating the number of cells on the top surface of the lower chambers under a CKX53 inverted microscope (Olympus) with a 100(×) magnification.
2.9. RNA extraction and quantitative real‐time polymerase chain reaction
TransZol (TransGen) was used to lyse tissues and cells. RNA was extracted from the lysates with an RNAsimple kit (Tiangen). cDNA was synthesized according to the instruction of FastKing RT Kit (Tiangen) and MicroRNA RT Kit (Thermo Fisher). Then, SuperReal PreMix Color Kit (Tiangen) was utilized to detect the amount of circRNA/miRNA/mRNA as per the guidebook. The quantitative real‐time polymerase chain reaction (qRT‐PCR) was cycled at 95°C for 15 min, 40 cycles at 95°C for 10 s, and 60°C for 20 s. Obtained data were assessed using the 2−∆∆Ct method. U6 and GAPDH were used as internal controls. The primer sequences are displayed in Table S2.
2.10. RNase R treatment assay
This assay was performed to confirm the stability of hsa_circ_0060927. Two μg of RNA from HCT116 and SW480 cells were incubated with 6 U RNase R (Geneseed) at 37°C for 20 min. After being purified using Qiagen Cleaning Kit, RNA samples were analyzed by qRT‐PCR with CYP24A1 as a control gene reference.
2.11. Cytoplasmic and nuclear RNA analysis
The cytoplasmic and nuclear RNA of hsa_circ_0060927 were analyzed by qRT‐PCR with PARIS™ Kit (Thermo Fisher) as instructed. HCT116 and SW480 cells were collected and then washed using PBS (Thermo Fisher). Then, the cells were suspended in cold cell fractionation buffer (Thermo Fisher) for 8 min. Cell samples were centrifuged at 400 rpm for 3 min. After that, cytoplasmic fraction and nuclear pellet were divided, and lysed using lysis buffer (TransGen). The amount of hsa_circ_0060927 was determined by qRT‐PCR. U6 and GAPDH were employed as control gene references.
2.12. Target prediction and dual‐luciferase reporter assay
The binding relationship between hsa_circ_0060927 and miR‐421 or miR‐195‐5p was predicted by the starbase online database (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA). The wild‐type (WT) plasmid of hsa_circ_0060927 (circ_0060927‐WT) was built by inserting the sequence of hsa_circ_0060927 containing the binding sites of miR‐421 or miR‐195‐5p into the pmirGLO (Promega). The mutant sequence of hsa_circ_0060927 was synthesized by GENEWIZ Co., Ltd., and introduced into the pmirGLO vector to generate the mutant reporter plasmid of hsa_circ_0060927 (circ_0060927‐WT). The constructed plasmids were transfected into HCT116 and SW480 cells with Lipofectamine 2000 (Thermo Fisher) according to the published methods. 25 Luciferase activities were detected by Dual‐Lucy Assay Kit (Solarbio) with Renilla luciferase activity as a control.
2.13. RNA pull‐down assay
Biotinylated hsa_circ_0060927 (Bio‐circ_0060927; Geneseed) and control (Bio‐NC; Geneseed) were transfected into HCT116 and SW480 cells for 48 h. The cells were lysed using lysis buffer (Sigma) for 10 min. Then, the lysates were incubated with Streptavidin‐coupled Dynabeads (Invitrogen) at room temperature for 1 h. RNA was isolated, and the expression of miR‐421 and miR‐195‐5p was detected by qRT‐PCR.
2.14. RNA immunoprecipitation assay
RIP assay was conducted with a Magna RNA immunoprecipitation (RIP) kit (Millipore). HCT116 and SW480 cells were lysed using RIP buffer (Sigma) containing RNase inhibitor (Sigma). Then, the lysates were incubated with magnetic beads coated with anti‐immunoglobulin G (anti‐IgG; ab109489; Abcam) or anti‐argonaute2 (anti‐ago2; ab186733; Abcam). At 24 h after co‐incubation, the beads were washed, and proteinase K (Millipore) was added to digest proteins. The expression of hsa_circ_0060927, miR‐421 and miR‐195‐5p in complexes was determined by qRT‐PCR.
2.15. Xenograft tumor model assay
This assay was carried out under the agreement of the Animal Care and Use Committee of Nanjing Hospital of Chinese Medicine Affiliated to Nanjing University of Chinese Medicine. Male BALB/c nude mice (5‐week old) were purchased from Vital River Laboratories. All mice lived in a pathogen‐free environment were randomly divided into 2 groups (con group and Caudatin group, N = 6 per group). 5 × 106 SW480 cells were diluted in 200 μl PBS (Thermo Fisher) and then inoculated into the flank of every mouse. On the 7th day of inoculation, the mice were treated with 50 mg/kg caudatin every other day for 21 days, administered by caudal vein injection, and tumor volume was measured every 3 days based on the formula that volume (mm3) = width2 × length × 0.5. Twenty‐eight days later, mice were killed by cervical dislocation after being anesthetized with xylazine (10 mg/kg). Tumors were excised for tumor weight and gene expression analysis.
2.16. Immunohistochemistry assay
Immunohistochemistry (IHC) assay was designed to analyze the expression of nuclear proliferation marker (Ki‐67), Cleaved‐caspase 3, E‐cadherin and N‐cadherin in the transplanted tumors from xenograft tumor model assay as instructed. 26 The tumors were cut into the 4‐µm‐thick sections, and embedded into paraffin, followed by the heating at 60°C. Twenty minutes later, the sections were deparaffinized and hydrated with xylene (Millipore) as well as ethanol (Millipore). After that, the sections were dipped in the sodium citrate (Millipore) and heated to conduct the antigen retrieval. After being immersed in Hydrogen Peroxide (Millipore) for 10 min, the sections were incubated with the primary antibodies against Ki‐67 (AF0198; 1:100; Affinity), Cleaved‐caspase 3 (AF7022; 1:100; Affinity), E‐cadherin (AF0131; 1:100; Affinity) and N‐cadherin (AF4039; 1:100; Affinity) as well as secondary antibodies (S0001; 1:200; Affinity), respectively. Hematoxylin (Millipore) was incubated with the tissues, and protein expression was analyzed under a CX31‐LV320 microscope (Olympus).
2.17. Statistical analysis
SPSS 21.0 software (IBM) was employed to analyze the data from three independent duplicate tests. Data were presented as means ± standard deviations (SD). Spearman's rank correlation coefficient test was utilized to compare the significance in Spearman correlation analysis. Significant differences were compared with two‐tailed Student's t‐tests or Wilcoxon rank‐sum test between the two groups, or were analyzed with one‐way analysis of variance between the multiple groups. p value <0.05 was considered statistically significant.
3. RESULTS
3.1. Caudatin treatment inhibited cell proliferation, migration and invasion whereas induced cell apoptosis in HCT116 and SW480 cells
Results showed that Caudatin exposure repressed the viability of HCT116 and SW480 cells in a dose‐dependent manner (p value <0.05; Figure 1A,B). HCT116 and SW480 cells were treated with 50 μM Caudatin for 24 h in subsequent Caudatin‐related studies as the 50% cell viability under the condition. Then, the results showed that Caudatin treatment repressed cell proliferation but induced cell apoptosis in HCT116 and SW480 cells (p value <0.0001; Figure 1C,D). In support, Caudatin treatment decreased the expression of proliferation‐related PCNA and increased the expression of apoptosis‐related Cleaved‐caspase 3 (p value <0.0001; Figure 1E). Additionally, the migration and invasion of HCT116 and SW480 cells were suppressed under Caudatin exposure (p value <0.001; Figure 2A,B). As presented in Figure 2C, Caudatin treatment significantly downregulated N‐cadherin protein level but upregulated E‐cadherin expression (p value <0.0001; Figure 2C). These results demonstrated that Caudatin suppressed CRC cell malignancy.
FIGURE 1.

Caudatin exposure suppressed cell proliferation while induced cell apoptosis in CRC cells. (A and B) The effects of Caudatin (0, 25, 50 and 100 μM) treatment on the viability of HCT116 and SW480 cells were determined by MTT assay. (C) The impact of 50 μM Caudatin on HCT116 and SW480 cell proliferation was determined by EDU assay. (D) Flow cytometry analysis was performed to analyze the influence of 50 μM Caudatin on the apoptosis of HCT116 and SW480 cells. (E) Western blot analysis was employed to detect the effects of 50 μM Caudatin treatment on the protein levels of PCNA and Cleaved‐caspase 3 in HCT116 and SW480 cells. Con: 0 μM. *p < 0.05, **p < 0.01 and ***p < 0.001. CRC, Colorectal cancer; EDU, 5‐Ethynyl‐29‐deoxyuridine; MTT, 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide; PCNA, proliferating cell nuclear antigen
FIGURE 2.

Caudatin treatment hindered the metastasis of HCT116 and SW480 cells. (A and B) The impacts of 50 μM Caudatin on the migration and invasion of HCT116 and SW480 cells were disclosed by transwell assay. (C) The influences of 50 μM Caudatin on the protein levels of E‐cadherin and N‐cadherin were revealed by Western blot in HCT116 and SW480 cells. **p < 0.01 and ***p < 0.001
3.2. Caudatin exposure significantly downregulated hsa_circ_0060927 expression
This study continued to analyze the effect of Caudatin treatment on hsa_circ_0060927 expression. As shown in Figure 3A,B, hsa_circ_0060927 expression was upregulated in CRC tissues and CRC cells (HCT116 and SW480) compared with paracancerous non‐cancerous colorectal tissues and FHC cells (p value <0.0001). Also, we found that hsa_circ_0060927 expression was closely associated with lymph node metastasis and tumor‐node‐metastasis (TNM) stage (Table S1). Hsa_circ_0060927 expression had no significant difference after treatment of RNase R, although linear CYP24A1 expression was reduced (p value <0.0001; Figure 3C,D), which suggested that hsa_circ_0060927 was a circular RNA. We confirmed the head‐to‐tail splicing of hsa_circ_0060927 via Sanger sequencing (Figure S1). Subsequently, Caudatin treatment downregulated hsa_circ_0060927 expression in HCT116 and SW480 cells (p value <0.0001; Figure 3E,F).
FIGURE 3.

Caudatin treatment decreased hsa_circ_0060927 expression. (A and B) Hsa_circ_0060927 expression was detected by qRT‐PCR in CRC tissues (n = 68), paracancerous non‐cancerous colorectal tissues (n = 68) and FHC, HCT116 and SW480 cells. (C and D) The stability of hsa_circ_0060927 was determined by RNase R treatment assay. (E and F) The effect of 50 μM Caudatin treatment on hsa_circ_0060927 expression was determined by qRT‐PCR in HCT116 and SW480 cells. ***p < 0.001. CRC, Colorectal cancer
3.3. Hsa_circ_0060927 overexpression attenuated the effects of Caudatin treatment on CRC cell malignancy
Whether Caudatin inhibited CRC cell processes via regulating hsa_circ_0060927 was further investigated. The efficiency of hsa_circ_0060927 overexpression was detected, and the results were presented in Figure 4A (p value <0.0001). Subsequently, Caudatin treatment inhibited cell viability and proliferation, but the effects were relieved after hsa_circ_0060927 overexpression (p value <0.0001; Figure 4B,C). Caudatin exposure induced the apoptosis of HCT116 and SW480 cells, which was remitted by the enforced expression of hsa_circ_0060927 (p value <0.0001; Figure 4D). A similar pattern of data among different groups was also found from the Transwell assay (p value <0.001; Figure 4E–G). Caudatin‐induced inhibition of PCNA and N‐cadherin production and promotion of Cleaved‐caspase 3 and E‐cadherin were counteracted after hsa_circ_0060927 overexpression (p value <0.0001; Figure 4H). These findings demonstrated that Caudatin repressed CRC cell malignancy via regulating hsa_circ_0060927.
FIGURE 4.
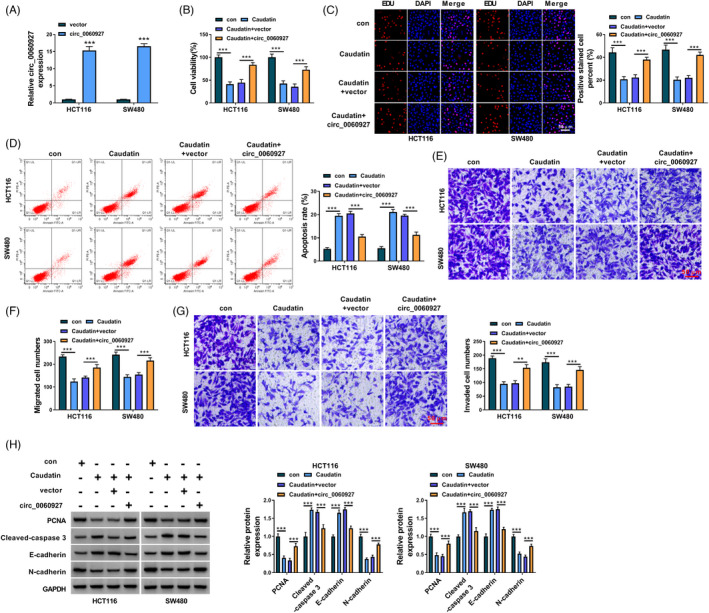
Caudatin inhibited CRC progression by modulating hsa_circ_0060927 expression. (A) The overexpression efficiency of hsa_circ_0060927 was detected by qRT‐PCR in HCT116 and SW480 cells. (B and C) MTT and EDU assays were performed to investigate the impacts of Caudatin treatment and hsa_circ_0060927 overexpression on the viability and proliferation of HCT116 and SW480 cells. (D) Flow cytometry analysis was employed to reveal the impacts of Caudatin treatment and hsa_circ_0060927 overexpression on the apoptosis of HCT116 and SW480 cells. (E–G) The influences of Caudatin exposure and ectopic hsa_circ_0060927 expression on HCT116 and SW480 cell migration and invasion were revealed by transwell assay. (H) Western blot was carried out to determine the impacts of Caudatin treatment and hsa_circ_0060927 overexpression on the protein levels of PCNA, Cleaved‐caspase 3, E‐cadherin and N‐cadherin in HCT116 and SW480 cells. **p < 0.01 and ***p < 0.001. CRC, Colorectal cancer; EDU, 5‐Ethynyl‐29‐deoxyuridine; MTT, 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide; PCNA, proliferating cell nuclear antigen
3.4. Hsa_circ_0060927 acted as a sponge of miR‐421 and miR‐195‐5p in CRC cells
The mechanism of hsa_circ_0060927 mediating CRC cell malignancy under Caudatin treatment was further analyzed. Results first showed that hsa_circ_0060927 expression was higher in the cytoplasm than in the cell nucleus (Figure 5A), suggesting that hsa_circ_0060927 mainly participated in post‐transcriptional regulation. The qRT‐PCR data showed that the small interfering RNA against hsa_circ_0060927 was effective in reducing hsa_circ_0060927 expression (p value <0.0001; Figure 5B). Then, we predicted potential target miRNAs of hsa_circ_0060927 by starbase and circBANK online databases. As presented in Figure 5C, we found that there were 10 miRNAs containing the putative binding sites of hsa_circ_0060927. The data from Figure 5D showed that only miR‐421 and miR‐195‐5p were upregulated after hsa_circ_0060927 silencing (p value <0.0001), which suggested that miR‐421 and miR‐195‐5p might be associated with hsa_circ_0060927. Subsequently, the putative binding sites between hsa_circ_0060927 and the 2 miRNAs were predicted and shown in Figure 5E. The high overexpression efficiency of miR‐421 and miR‐195‐5p mimic was displayed in Figure 5F (p value <0.0001). Dual‐luciferase reporter assay showed that miR‐421 or miR‐195‐5p overexpression significantly inhibited the luciferase activity of wild‐type reporter plasmid of hsa_circ_0060927 (p value <0.0001) but not that of mutant hsa_circ_0060927 (Figure 5G). A comparative analysis of RNA pull‐down assay showed that miR‐421 and miR‐195‐5p were significantly enriched by the biotinylated hsa_circ_0060927 (p value <0.0001; Figure 5H). Meanwhile, the RIP assay showed the ago2 antibody could significantly enrich miR‐421, miR‐195‐5p and hsa_circ_0060927 as compared with the anti‐IgG group (p value <0.0001; Figure 5I). As shown in Figure 5J, miR‐421 and miR‐195‐5p were downregulated in CRC tissues as compared with paracancerous non‐cancerous colorectal tissues (p value <0.0001). Furthermore, miR‐421 and miR‐195‐5p expression were negatively related to hsa_circ_0060927 expression in CRC tissues (p value <0.01; Figure 5K). The above data demonstrated that hsa_circ_0060927 directly interacted with miR‐421 and miR‐195‐5p.
FIGURE 5.

Hsa_circ_0060927 was associated with miR‐421 and miR‐195‐5p in CRC cells. (A) Cytoplasmic and nuclear RNA analysis was performed to detect the expression levels of GAPDH, U6 and hsa_circ_0060927. (B) The efficiency of si‐circ_0060927 in reducing has_circ_0060927 expression was determined by qRT‐PCR in HCT116 and SW480 cells. (C) Starbase and circBANK online databases were employed to predict the possible target miRNAs of hsa_circ_0060927. (D) The effects of hsa_circ_0060927 silencing on the expression levels of the 10 miRNAs possessing the binding sites of hsa_circ_0060927 were analyzed by qRT‐PCR. (E) Starbase online database was performed to predict the putative binding sites of hsa_circ_0060927 for miR‐421 and miR‐195‐5p. (F) The efficiency of miR‐421 and miR‐195‐5p overexpression was determined by qRT‐PCR in HCT116 and SW480 cells. (G) Dual‐luciferase reporter assay was employed to detect luciferase activity in HCT116 and SW480 cells. (H and I) RNA pull‐down and RIP assays were conducted to analyze the direct binding relationships between hsa_circ_0060927 and miRNAs (miR‐421 and miR‐195‐5p). (J) The expression levels of miR‐421 and miR‐195‐5p were detected by qRT‐PCR in 68 pairs of CRC tissues and paracancerous non‐cancerous colorectal tissues. (K) The linear relationship between hsa_circ_0060927 and miR‐421 or miR‐195‐5p expression was revealed by Spearman correlation analysis. ***p < 0.001. CRC, Colorectal cancer
3.5. Hsa_circ_0060927 silencing repressed CRC cell malignancy by sponging miR‐421 or miR‐195‐5p
Given the binding relationship between hsa_circ_0060927 and miR‐421/miR‐195‐5p, whether hsa_circ_0060927 regulated CRC cell processes by sponging miR‐421/miR‐195‐5p was revealed in this part. The results first showed the high knockdown efficiency of both miR‐421 and miR‐195‐5p (Figure 6A,B). Subsequently, hsa_circ_0060927 silencing repressed the viability and proliferation of HCT116 and SW480 cells (p value <0.0001), whereas miR‐421 or miR‐195‐5p inhibitor attenuated these effects (Figure 6C,D). Hsa_circ_0060927 knockdown also induced cell apoptosis (p value <0.0001), but depletion of miR‐421 or miR‐195‐5p hindered the influence (Figure 6E). The migration and invasion of HCT116 and SW480 cells were suppressed after hsa_circ_0060927 silencing (p value <0.0001); however, knockdown of miR‐421 or miR‐195‐5p relieved these impacts (Figure 6F–H). Additionally, hsa_circ_0060927 depletion downregulated the protein levels of PCNA and N‐cadherin but upregulated Cleaved‐caspase 3 and E‐cadherin (p value <0.0001); however, these impacts were attenuated by decreasing miR‐421 or miR‐195‐5p (Figure 6I). These data manifested that hsa_circ_0060927 regulated CRC cell processes by binding to miR‐421/miR‐195‐5p.
FIGURE 6.

Hsa_circ_0060927 silencing inhibited CRC progression by sponging miR‐421 or miR‐195‐5p. (A and B) The efficiency of miR‐421 and miR‐195‐5p knockdown was detected by qRT‐PCR in HCT116 and SW480 cells. (C and D) The effects of hsa_circ_0060927 silencing and miR‐421/miR‐195‐5p inhibitor on HCT116 and SW480 cell viability and proliferation were determined by MTT and EDU assays. (E) The impacts of hsa_circ_0060927 silencing and miR‐421/miR‐195‐5p inhibitor on the apoptosis of HCT116 and SW480 cells were explained by flow cytometry analysis. (F–H) Transwell assay was performed to analyze the influences of si‐circ_0060927 and anti‐miR‐421 or anti‐miR‐195‐5p on the migration and invasion of HCT116 and SW480 cells. (I) Western blot was conducted to investigate the effects of circ_0060927 silencing and miR‐421 inhibitor or miR‐195‐5p inhibitor on the protein levels of PCNA, Cleaved‐caspase 3, E‐cadherin and N‐cadherin in HCT116 and SW480 cells. ***p < 0.001. EDU, 5‐Ethynyl‐29‐deoxyuridine; MTT, 3‐(4,5)‐dimethylthiahiazo (‐z‐y1)‐3,5‐di‐phenytetrazoliumromide; PCNA, proliferating cell nuclear antigen
3.6. Caudatin increased the expression levels of miR‐421 and miR‐195‐5p by regulating hsa_circ_0060927
Based on the above result, the study continued to investigate whether Caudatin modulated miR‐421 and miR‐195‐5p expression through hsa_circ_0060927. Results first showed that miR‐421 and miR‐195‐5p expression were significantly downregulated in HCT116 and SW480 cells compared with FHC cells (p value <0.0001; Figure 7A,B). Subsequently, Caudatin upregulated the expression levels of miR‐421 and miR‐195‐5p in HCT116 and SW480 cells (p value <0.0001; Figure 7C,D). Furthermore, hsa_circ_0060927 overexpression attenuated the promoting effects of Caudatin exposure on the expression of miR‐421 and miR‐195‐5p (p value <0.0001; Figure 7E,F), suggesting that Caudatin upregulated miR‐421 and miR‐195‐5p by inhibiting hsa_circ_0060927 expression.
FIGURE 7.

Caudatin regulated miR‐421 and miR‐195‐5p expression by repressing hsa_circ_0060927. (A and B) MiR‐421 and miR‐195‐5p expression were detected by qRT‐PCR in FHC, HCT116 and SW480 cells. (C and D) The impacts of Caudatin treatment on the expression levels of miR‐421 and miR‐195‐5p were determined by qRT‐PCR in HCT116 and SW480 cells. (E and F) The impacts of Caudatin treatment and hsa_circ_0060927 overexpression on the expression levels of miR‐421 and miR‐195‐5p were revealed by qRT‐PCR in HCT116 and SW480 cells. ***p < 0.001. CRC, Colorectal cancer
3.7. Caudatin repressed tumor formation in vivo
The impacts of Caudatin treatment on the tumorigenesis of CRC cells were further analyzed using a xenograft tumor model assay. Results showed that Caudatin exposure reduced the volume and weight of the primary tumors from SW480 cells (p value <0.001; Figure 8A,B). We then analyzed the protein expression of proliferation‐related proteins (Ki‐67 and PCNA), apoptosis‐linked protein (Cleaved‐caspase 3) and metastasis‐related proteins (E‐cadherin and N‐cadherin) using IHC assay and Western blot analysis. As shown in Figure 8C, the cells expressing Ki‐67 or N‐cadherin were fewer, but the cells expressing Cleaved‐caspase 3 or E‐cadherin were more in the Caudatin group than in the control group. Then, we found that Caudatin exposure decreased hsa_circ_0060927 expression but increased miR‐421 and miR‐195‐5p expression (p value <0.0001; Figure 8D), suggesting that Caudatin regulated the expression of hsa_circ_0060927, miR‐421 and miR‐195‐5p in the transplanted tumors. Western blot analysis also confirmed that Caudatin treatment significantly downregulated the protein expression of PCNA and N‐cadherin whereas upregulated Cleaved‐caspase 3 and E‐cadherin expression in tumor xenografts (p value <0.0001; Figure 8E). Collectively, these findings suggested that Caudatin inhibited CRC cell tumorigenesis by downregulating hsa_circ_0060927 expression and upregulating miR‐421 and miR‐195‐5p expression in vivo.
FIGURE 8.

Caudatin treatment suppressed tumor growth in vivo. (A–B) The impacts of Caudatin treatment on the volume and weight of tumors were revealed in vivo. (C) The protein expression of Ki‐67 and Cleaved‐caspase 3 was analyzed by IHC assay. (D) qRT‐PCR was employed to investigate the effects of Caudatin treatment on the expression levels of hsa_circ_0060927, miR‐421 and miR‐195‐5p in the forming tumors from SW480 cells. (E) Western blot analysis was conducted to determine the influences of Caudatin on the protein levels of Ki‐67, PCNA, Cleaved‐caspase 3, E‐cadherin and N‐cadherin in the forming tumors from SW480 cells. **p < 0.01 and ***p < 0.001. PCNA, proliferating cell nuclear antigen
4. DISCUSSION
The present work was organized to analyze the function of Caudatin on CRC progression and the underlying mechanism. Herein, we reported that Caudatin repressed CRC cell processes by regulating circRNA (hsa_circ_0060927) for the first time. Our results showed that Caudatin treatment repressed CRC cell proliferation, migration and invasion but induced cell apoptosis through the hsa_circ_0060927/miR‐421/miR‐195‐5p pathway. The results from the xenograft tumor model assay showed that Caudatin injection restrained the volume and weight of the primary tumors from CRC cells by regulating hsa_circ_0060927, miR‐421, and miR‐195‐5p. Our findings demonstrated that Caudatin acted as a suppressor in CRC malignant progression.
As reported, Caudatin inhibits the development of breast cancer, 27 gastric cancer 28 and hepatoma. 29 However, the effects and mechanism of Caudatin in CRC progression were unknown. Our data showed that Caudatin repressed CRC cell malignancy. In particular, Caudatin treatment significantly decreased hsa_circ_0060927 expression in vitro and in vivo. Hsa_circ_0060927 was closely associated with lymph node metastasis and TNM stage of CRC patients. Cytoplasmic and nuclear RNA analysis showed that hsa_circ_0060927 was mainly located in the cytoplasm, suggesting that hsa_circ_0060927 principally functioned at the post‐transcriptional level. Additionally, increasing hsa_circ_0060927 expression attenuated the anti‐cancer effects of Caudatin on CRC. We also observed that hsa_circ_0060927 was augmented in CRC tissues and HCT116 and SW480 cells, which was consistent with the published data. 16 These results explained that hsa_circ_0060927 contributed to CRC cell malignancy and that Caudatin repressed CRC development by regulating hsa_circ_0060927.
It has been well documented that circRNAs commonly modulate cancer evolution by sponging miRNAs. 30 , 31 Thus, the miRNAs that bound to hsa_circ_0060927 were screened by the starbase online database. Through analysis of qRT‐PCR, miR‐421 and miR‐195‐5p were employed as follow‐up subjects. We found that hsa_circ_0060927 expression was negatively related to miR‐421/miR‐195‐5p. Additionally, miR‐421 and miR‐195‐5p expression were downregulated in CRC tissues and cells. MiR‐421/miR‐195‐5p inhibitor hindered the impacts of hsa_circ_0060927 silencing on CRC cell processes, implicating that miR‐421 and miR‐195‐5p served as repressors in CRC progression. Xue et al. 22 have explained that miR‐421 restrained CRC cell proliferation, migration and invasion, and our findings were consistent with these results. The studies focusing on miR‐195‐5p have revealed that miR‐195‐5p can repress cell proliferation, migration and invasion but facilitate cell apoptosis in CRC in vitro. 23 , 32 Our data were identical to their results. Thus, the above evidence suggested that hsa_circ_0060927 regulated CRC progression by sponging miR‐421/miR‐195‐5p. Furthermore, hsa_circ_0060927 overexpression impaired the promoting effects of Caudatin on the expression levels of miR‐421 and miR‐195‐5p, which demonstrated that Caudatin inhibited miR‐421/miR‐195‐5p expression by regulating hsa_circ_0060927.
The disadvantage of this research is lacking data about the influences of hsa_circ_0060927 on Caudatin‐mediated CRC growth in vivo, which will be explored in the subsequent study.
Summarily, Caudatin exposure inhibited CRC cell proliferation and metastasis whereas facilitated apoptosis. In terms of mechanism, Caudatin upregulated miR‐421/miR‐195‐5p expression via regulating hsa_circ_0060927, which acted as a promoter in CRC progression (Figure 9). These data provide a theoretical basis for the study of CRC therapy with Caudatin.
FIGURE 9.
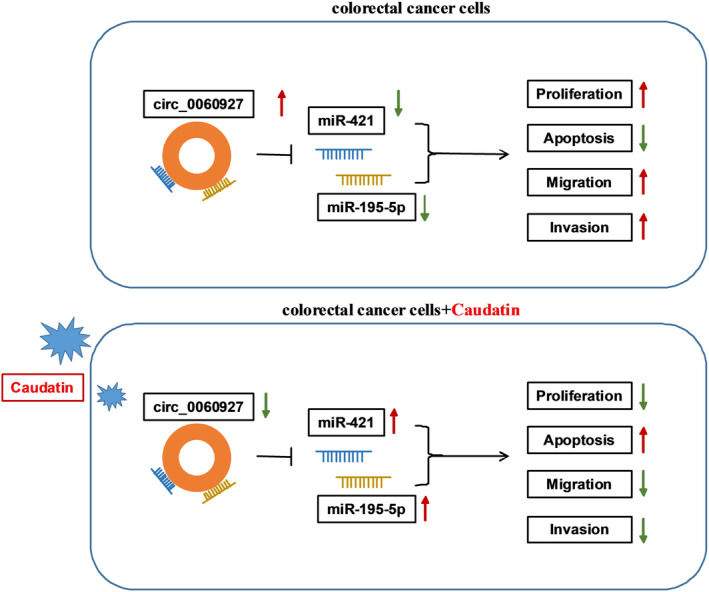
The schematic illustration showed the mechanisms underlying Caudatin regulating CRC malignant progression. CRC, Colorectal cancer
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Figure S1
Table S1
Table S2
Chen J, Xu L, Fang M, Xue Y, Cheng Y, Tang X. Hsa_circ_0060927 participates in the regulation of Caudatin on colorectal cancer malignant progression by sponging miR‐421/miR‐195‐5p. J Clin Lab Anal. 2022;36:e24393. doi: 10.1002/jcla.24393
Funding information
None
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Global Burden of Disease Cancer C , Fitzmaurice C, Allen C, et al. Global, Regional, and National Cancer Incidence, mortality, years of life lost, years lived with disability, and disability‐adjusted life‐years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3(4):524‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peng HX, Yang L, He BS, et al. Combination of preoperative NLR, PLR and CEA could increase the diagnostic efficacy for I‐III stage CRC. J Clin Lab Anal. 2017;31(5):e22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. [DOI] [PubMed] [Google Scholar]
- 4. Zhen X, Choi HS, Kim JH, et al. Caudatin isolated from Cynanchum auriculatum inhibits breast cancer stem cell formation via a GR/YAP signaling. Biomolecules. 2020;10(6):925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song J, Ding W, Liu B, et al. Anticancer effect of caudatin in diethylnitrosamine‐induced hepatocarcinogenesis in rats. Mol Med Rep. 2020;22(2):697‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z, Ruan Y, Zhang H, et al. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588‐5600. [DOI] [PubMed] [Google Scholar]
- 9. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐691. [DOI] [PubMed] [Google Scholar]
- 10. Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co‐expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7(30):47186‐47200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li A, Wang WC, McAlister V, Zhou Q, Zheng X. Circular RNA in colorectal cancer. J Cell Mol Med. 2021;25(8):3667‐3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vakhshiteh F, Hassani S, Momenifar N, Pakdaman F. Exosomal circRNAs: new players in colorectal cancer. Cancer Cell Int. 2021;21(1):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang N, Zhang X, Xu W, Zhang X, Mu Z. CircRNA_103948 inhibits autophagy in colorectal cancer in a ceRNA manner. Ann N Y Acad Sci. 2021;1503(1):88‐101. [DOI] [PubMed] [Google Scholar]
- 14. Wu M, Kong C, Cai M, et al. Hsa_circRNA_002144 promotes growth and metastasis of colorectal cancer through regulating miR‐615‐5p/LARP1/mTOR pathway. Carcinogenesis. 2021;42(4):601‐610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen T, Cheng X, Liu X, et al. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR‐183. Artif Cells Nanomed Biotechnol. 2019;47(1):4038‐4045. [DOI] [PubMed] [Google Scholar]
- 16. Sadeghi H, Heiat M. A novel circular RNA hsa_circ_0060927 may serve as a potential diagnostic biomarker for human colorectal cancer. Mol Biol Rep. 2020;47(9):6649‐6655. [DOI] [PubMed] [Google Scholar]
- 17. Rupaimoole R, Calin GA, Lopez‐Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Z, Sha HH, Li HJ. Functions and mechanisms of miR‐186 in human cancer. Biomed Pharmacother. 2019;119:109428. [DOI] [PubMed] [Google Scholar]
- 20. Li T, Pan H, Li R. The dual regulatory role of miR‐204 in cancer. Tumour Biol. 2016;37(9):11667‐11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. OncoTargets Ther. 2020;13:2067‐2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xue L, Yang D. MiR‐421 inhibited proliferation and metastasis of colorectal cancer by targeting MTA1. J BUON. 2018;23(6):1633‐1639. [PubMed] [Google Scholar]
- 23. Cheng H, Malhotra A. Evaluation of potential of long noncoding RNA NEAT1 in colorectal cancer. J Environ Pathol Toxicol Oncol. 2020;39(2):101‐111. [DOI] [PubMed] [Google Scholar]
- 24. Pena‐Philippides JC, Gardiner AS, Caballero‐Garrido E, et al. Inhibition of MicroRNA‐155 supports endothelial tight junction integrity following oxygen‐glucose deprivation. J Am Heart Assoc. 2018;7(13):e009244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu P, Jia S, Wang K, et al. MiR‐140 inhibits classical swine fever virus replication by targeting Rab25 in swine umbilical vein endothelial cells. Virulence. 2020;11(1):260‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang D‐W, Huang M, Lin X‐S, Huang Q. CD155 expression and its correlation with clinicopathologic characteristics, angiogenesis, and prognosis in human cholangiocarcinoma. OncoTargets Ther. 2017;10:3817‐3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fei HR, Yuan C, Wang GL, et al. Caudatin potentiates the anti‐tumor effects of TRAIL against human breast cancer by upregulating DR5. Phytomedicine. 2019;62:152950. [DOI] [PubMed] [Google Scholar]
- 28. Li X, Zhang X, Liu X, et al. Caudatin induces cell apoptosis in gastric cancer cells through modulation of Wnt/β‐catenin signaling. Oncol Rep. 2013;30(2):677‐684. [DOI] [PubMed] [Google Scholar]
- 29. Luo Y, Sun Z, Li Y, et al. Caudatin inhibits human hepatoma cell growth and metastasis through modulation of the Wnt/β‐catenin pathway. Oncol Rep. 2013;30(6):2923‐2928. [DOI] [PubMed] [Google Scholar]
- 30. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. [DOI] [PubMed] [Google Scholar]
- 31. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bai J, Xu J, Zhao J, Zhang R. lncRNA SNHG1 cooperated with miR‐497/miR‐195‐5p to modify epithelial‐mesenchymal transition underlying colorectal cancer exacerbation. J Cell Physiol. 2020;235(2):1453‐1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


