Abstract
Background
circRNA hsa_circ_0018289‐mediated growth and metastasis of CC cells were investigated, as well as the mechanistic pathway.
Methods
Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) was carried out to examine the expression of hsa_circ_0018289, microRNA (miR)‐1294, and isoprenylcysteine carboxyl methyltransferase (ICMT). CC cell proliferation, migration, and invasion were evaluated by 5‐ethynyl‐2’‐deoxyuridine (EdU) incorporation, colony formation, transwell assays, Western blot analysis of ICMT, and glycolysis‐associated proteins. Dual‐luciferase reporter or RNA pull‐down analysis of the target interaction between miR‐1294 and hsa_hsa_circ_0018289 or ICMT. Xenograft model assay was implemented to assess the role of hsa_circ_0018289 in vivo. Immunofluorescence (IHC) was employed to detect the level of Ki‐67.
Results
Hsa_circ_0018289 was elevated in CC tissues and cells, its deficiency could repress growth, metastasis, and glycolysis of CC cells in vitro, as well as hamper tumor growth in vivo. Hsa_circ_0018289 sponged miR‐1294 while miR‐1294 bound with ICMT, and the inhibition of miR‐1294 or addition of ICMT could partially relieve the effect caused by hsa_circ_0018289 depletion.
Conclusion
Hsa_circ_0018289 contributes to malignant development by regulating the miR‐1294/ICMT axis, affording novel insight into CC therapy.
Keywords: cervical cancer, glycolysis, hsa_circ_0018289, ICMT, miR‐1294
Schematic diagram of circ_0018289 regulating CC cell malignancy behavior.
1. INTRODUCTION
Cervical cancer (CC) ranks as the second most lethal tumor derived from women in the world, only behind breast cancer, seriously threatening female health. 1 According to statistical data, there were exceeding 3.4 × 104 death caused by CC in China only in 2015. 2 Although the application of human papillomavirus (HPV) vaccines has effectively decreased the occurrence of CC, the prognosis of CC patients at advanced stages is still terrible. 3 , 4 Therefore, elucidating the mechanism underlying CC initiation and development is advantageous to develop new strategies of CC therapy.
Circular RNAs (circRNAs) can be divided into non‐coding circRNAs and coding circRNAs. 5 , 6 , 7 They are characterized by a unique closed continuous ring structure and therefore exhibit more stability than their hosting gene under RNase R treatment. 8 A plethora of circRNAs endows potential importance in human malignancies, serving as cancer biomarkers, and acting as cancer promoters or suppressors. 9 Mechanistically, as sponges of microRNAs (miRNAs), circRNAs could participate in cancer progression regulation. 10 Derived from Synaptotagmin 15 (SYT15), circRNA hsa_circ_0018289 was discovered to be upregulated in CC, and it had an oncogenic role in CC development. 11 But the molecular mechanism by which hsa_circ_0018289 functioned in CC is not still entirely elucidated.
MiRNAs are also highly evolutionary conserved non‐coding RNAs, whose length is about 22 nucleotides. 12 A pool of miRNAs was abnormally expressed in both solid and hematopoietic tumors, and they could target certain genes involved in tumorigenesis, thereby affecting tumor development and progression. 13 For instance, miR‐802 was reported to be downregulated in CC, and it could reduce CC cell proliferation via targeting SRSF9. 14 In addition, the gain of miR‐340 targeted EphA3 to repress CC cell malignant behavior. 15 Also, a significant increase in miR‐126 was noticed in CC tissues and cells and exerted a CC‐suppressing role through decreasing ZEB1 expression, suppressing CC cell proliferation, migration, and invasion. 16 Former research reported the downregulation of miR‐1294 in CC and its tumor‐inhibiting function. 17 Whether miR‐1294, as the target miRNA of hsa_circ_0018289, participate in hsa_circ_0018289‐mediated CC cell growth, metastasis, and metabolism remain to be explored.
Isoprenylcysteine carboxyl methyltransferase (ICMT) is a necessary enzyme that catalyzes the carboxymethylation process during prenylation. 18 ICMT was verified as an oncogene in many human cancers, such as pancreatic cancer, prostate cancer, and hepatoma. 19 , 20 , 21 In CC, ICMT inhibition facilitated the sensitization of tumor cells to doxorubicin or paclitaxel treatment. 22 TargetScan estimated that ICMT was a target gene of miR‐1294. The association of ICMT and miR‐1294 in the malignant properties of CC cells needs to be validated.
In this work, the expression enrichment of hsa_circ_0018289 was determined. Moreover, functional effects and molecular action modes of hsa_circ_0018289 on the CC cell malignancy phenotype were assessed.
2. MATERIALS AND METHODS
2.1. Patients and samples
Fifty‐three CC tissues and paired normal tissues were collected from 53 patients who underwent surgery at the Linyi Central Hospital, then all preserved at −80°C. The present study was performed under permission from the Ethics Committee of the Linyi Central Hospital and written informed consent was obtained from each patient.
2.2. Cell culture and transfection
Human cervical epithelial cells HcerEpic (#7060; ScienCell Research Laboratories), CC cells HeLa (ATCC® CCL‐2; American Type Culture Collection), and SiHa (ATCC® HTB‐35; American Type Culture Collection) were maintained in Roswell Park Memorial Institute (RPMI)‐1640 medium (Sigma‐Aldrich) added with 10% fetal bovine serum (Gibco) and 1% penicillin‐streptomycin (Sigma‐Aldrich). Cell flasks were kept in an incubator at 37°C with 5% CO2.
Hsa_circ_0018289small interference RNA: si‐hsa_circ_0018289‐1 (5′‐CAG CGG CGG CCU UGA GCA GCU‐3′) and si‐hsa_circ_0018289‐2 (5′‐CGG CCU UGA GCA GCU GGC CCU‐3′), as well as their negative control si‐control (5′‐AAC AGU CGC GUU UGC GAC UGG‐3′), mimic of miR‐1294 (miR‐1294, 5′‐AGA CAA CAA UGC CAA CCU CAC A‐3′) and its negative control miR‐NC (5′‐UUC UCC GAA CGU GUC ACG U‐3′), inhibitor of miR‐1294 (anti‐miR‐1294, 5′‐UCU GUU GUU ACG GUU GGA GUG U‐3′) and its negative control anti‐miR‐NC (5′‐UGAGCUGCAUAGAGUAGUGAUUA‐3′), ICMT (OE‐ICMT) over‐expressing plasmid and negative control OE‐NC (pcDNA 3.1) were all supplied by GenePharma. Cell transfection was executed with the help of Lipofectamine 3000 (Invitrogen), then cells were collected at 48 h after transfection.
2.3. Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
Total RNA was extracted from tissues or cells exploiting TRIzol Reagent (Invitrogen), followed by quality detection using a spectrophotometer (Thermo Fisher Scientific). For evaluating the relative expression of circ_0018289, Synaptotagmin 15 (SYT15) mRNA and ICMT, M‐MLV reverse transcriptase (Beyotime), and SYBR Green PCR Kit (Qiagen) were used for complementary DNA (cDNA) synthesis and qRT‐PCR assay. For miR‐1294 expression analysis, qRT‐PCR assay was implemented with qRT‐PCR miRNA Detection Kit (Thermo Fisher Scientific) after reverse transcription experiment using MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). QRT‐PCR primers are exhibited in Table 1. In addition, relative expression was analyzed via 2−ΔΔCt method, normalized to GAPDH or U6.
TABLE 1.
Primers used for qRT‐PCR assay
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| circ_0018289 | CCTGCACAGTGGAGAGTGG | CTTCTCCACAGACAGCAGCTT |
| SYT15 mRNA | TCAGCCTGACTGTGGTGCAGAA | TCCTTGGGCTTGCTGAGCATCT |
| miR‐1294 | GTGAGGTTGGCATTGTTG | GAACATGTCTGCGTATCTC |
| ICMT | CGCTTGGTTTCGGCATCCTTCT | CGGAAGAATCGCCACACTGTCA |
| GAPDH | TGCACCACCAACTGCTTAGC | AGCTCAGGGATGACCTTGCC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
2.4. RNase R digestion
To identify the character of hsa_circ_0018289, 5 μg total RNA isolated from HeLa and SiHa CC cells was subjected for RNase R (Epicentre Biotechnologies) at 37°C. 30 min later, the obtained total RNA was assessed via qRT‐PCR assay.
2.5. Nuclear/cytoplasmic fractionation assay
Hsa_circ_0018289 content in the nucleus and cytoplasm of CC cells was determined using a PARIS Kit (Thermo Fisher Scientific), with positive controls (U6 for nuclear) and (GAPDH for cytoplasmic RNA).
2.6. 5‐Ethynyl‐2’‐deoxyuridine (EdU) incorporation assay
In short, after being cultured 24 h, 5 × 104 CC cells in 6‐well plates were reacted using EdU (50 μM, Ribobio) for 2 h. After being fixed in 4% formaldehyde (Sigma‐Aldrich), the cells were reacted with Apollo reaction cocktail for 30 min, and then DAPI was used to identify the nuclei. At length, the stained cells were visualized under laser scanning confocal microscopy.
2.7. Colony formation assay
This experiment was executed to evaluate CC cell clonogenicity. After transfection, 600 HeLa and SiHa CC cells were seeded in 6‐well plates and continually cultured for 2 weeks. Generated colonies were subjected for immobilization with 4% paraformaldehyde (Sigma‐Aldrich), staining by crystal violet (Beyotime), and counting by a feat of Image J software.
2.8. Transwell assay
Transwell chambers (BD Biosciences) pre‐enveloped with Matrigel (BD Biosciences) or not were utilized to measure invasion and migration. Cells suspended in upper chambers with non‐serum medium, and lower ones were filled with the complete medium. The next day, cells moved through the lower membrane were dyed with crystal violet (Beyotime), photographed in 5 stochastically selected fields under a microscope (magnification: 100×) (Olympus).
2.9. Assessment of glucose consumption and lactate production
Glucose Colorimetric Assay Kit II (K686; Biovision) and Lactate Colorimetric Assay Kit II (K627; Biovision) were applied to determine glucose consumption and lactate production, respectively, as per the supplier's instructions.
2.10. Western blot assay
The loading protein samples were prepared as previously described. 23 After 10% separation gel treatment, the protein was electro‐transferred on polyvinylidene difluoride membranes (Roche). After being incubated with primary anti‐proliferating cell nuclear antigen (anti‐PCNA) antibody (ab29; Abcam, 1:1000), anti‐MMP9 antibody (ab76003; Abcam, 1:1500), anti‐hexokinase‐2 (anti‐HK2) antibody (ab209847; Abcam, 1:1500), anti‐M2 isoform of pyruvate kinase (anti‐PKM2) antibody (ab137852; Abcam, 1:1000), anti‐ICMT (NBP1‐69579; Bio‐Techne, 1:1500 dilution), or loading control anti‐β‐actin (ab8226; Abcam, 1:3000), the membranes were mixed with secondary antibody (ab205718; Abcam, 1:5000). Protein signals were activated and detected ECL kit.
2.11. Target gene prediction and confirmation
Wild‐ and mutant‐type luciferase reporters of hsa_circ_0018289 (hsa_circ_0018289‐WT and hsa_circ_0018289‐MUT) and ICMT (ICMT‐3’UTR‐WT and ICMT‐3’UTR‐MUT) containing the predicted binding sites with miR‐1294 were designed with Hanbio Biotechnology. HeLa and SiHa cells were co‐introduced with a luciferase reporter and miR‐NC or miR‐1294. Luciferase Reporter Gene Assay Kit (Beyotime) was used for measuring luciferase density according to the producer's guidance, with Renilla luciferase activity as control.
For RNA pull‐down assay, CC cells were transfected with biotinylated (Bio)‐miR‐1294 (GenePharma) or Bio‐miR‐NC (GenePharma). 48 h later, cells were disintegrated, and lysate was mixed with magnetic beads (M‐280; Invitrogen) coated with RNase‐free bovine serum albumin (Sigma‐Aldrich) and yeast tRNA (Sigma‐Aldrich) at 4°C for 3 h. After being purified, bound RNA was subjected to the analysis of qRT‐PCR.
2.12. Xenograft experiment
SiHa cells were stably transfected with short hairpin hsa_circ_0018289 (sh‐ hsa_circ_0018289; GenePharma) or sh‐NC (GenePharma). 3 × 106 transfected SiHa cells were subcutaneously inoculated into BALB/c nude mice (5 weeks old; Beijing Vital River Laboratory Animal Technology Co., Ltd., n = 5 per group), called sh‐hsa_circ_0018289 group and sh‐NC group. Then, the generated xenograft tumors volume (Volume (mm3) = 0.5 × length × width2) and the weight of mice were recorded every 5 days for 6 times. After 30 days, all mice were euthanatized, and tumors were cut off for further analyses. Procedures of this assay were ethically approved by the Ethics Committee of the Linyi Central Hospital.
2.13. Immunofluorescence (IHC)
Xenograft tumors derived from nude mice were immobilized using formalin then enveloped with paraffin. Later, a paraffin section (5‐μm thick) was prepared 24 and immunostained with anti‐Ki‐67 (ab231172; Abcam) at 4°C overnight, then probed with secondary antibody (ab205718; Abcam) at indoor temperature for 1 h. Subsequently, sections were dyed with diaminobenzidine and hematoxylin in succession, followed by observation using a light microscope (magnification: 200×) (Olympus).
2.14. Statistical analysis
Data were all from three biological replicates. Statistical analysis was implemented with SPSS software (version 20.0; SPSS). After processing, data were expressed as mean ± standard deviation. Student's t test and analysis of variance were applied for comparison. p < 0.05 indicated significantly different.
3. RESULTS
3.1. Hsa_circ_0018289 enrichment in CC tissues and cells
According to analysis from circBase, hsa_circ_0018289 (circSYT15_003) is derived from exon 2–3 of SYT15 gene located on chr10: 46968580–46969453 by back‐splicing and is 348 bp in length (Figure 1A). To explore the role of hsa_circ_0018289 in CC progression, its expression level was firstly evaluated in CC tissues. Besides, hsa_circ_0018289 in CC tissues was higher than neighboring normal tissues (N = 53) (Figure 1B). Analogously, enrichment of hsa_circ_0018289 was upregulated in CC cells versus HcerEpic cells (Figure 1C). To determine the structure of hsa_circ_0018289, RNase R digestion assay was performed. We found that SYT15 mRNA expression was apparently declined after RNase R treatment, while hsa_circ_0018289 was changeless (Figure 1D and E). Subsequently, the majority of hsa_circ_0018289 was distributed in the cytoplasm of HeLa and SiHa cells (Figure 1F and G). The above results confirmed that hsa_circ_0018289 was upregulated in CC.
FIGURE 1.
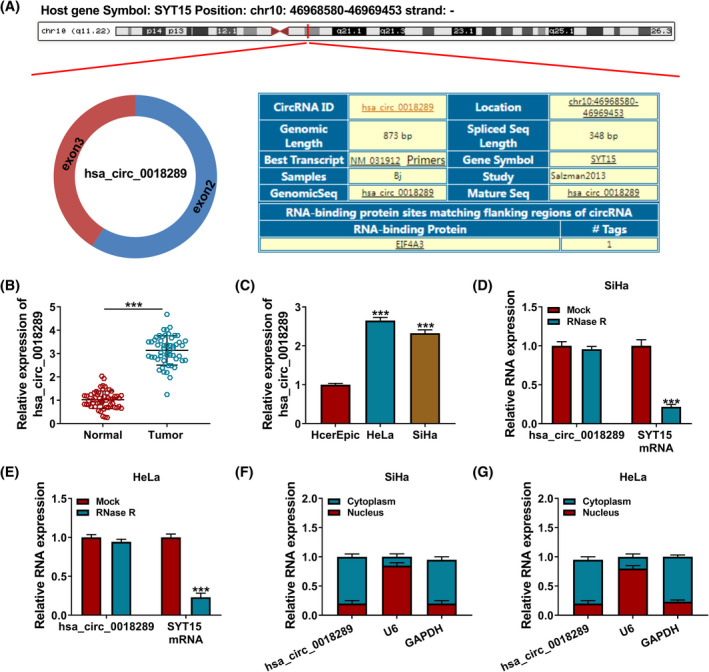
Enrichment of hsa_circ_0018289 in CC. (A) Schematic illustration indicates the formation of circ_0018289. (B and C) QRT‐PCR assay for hsa_circ_0018289 level in CC tissues, normal tissues, HcerEpic, HeLa, and SiHa cells. (D and E) The effects of RNase R on hsa_circ_0018289 and SYT15 mRNA were detected using qRT‐PCR assay. (F and G) Subcellular location of hsa_circ_0018289 using Nuclear/cytoplasmic fractionation assay in CC cells. ***p < 0.001
3.2. Depletion of hsa_circ_0018289 inhibited proliferation, metastasis, and glycolysis in CC cells
SiRNA against hsa_circ_0018289 was constructed to induce hsa_circ_0018289 knockdown in CC cells (Figure 2A). Knockdown efficiency was validated according to qRT‐PCR assay (Figure 2B). Hsa_circ_0018289 knockdown could drastically reduce cell proliferative ability of CC cells (Figure 2C and D). Following transwell assay suggested that silencing of hsa_circ_0018289 could suppress migration and invasion of HeLa and SiHa cells (Figure 2E and F). Western blot assay further confirmed hsa_circ_0018289 inhibition might repress proliferation, migration, and invasion (Figure 2G). Commercial kits revealed that hsa_circ_0018289 deficiency decreased glucose consumption and lactate production in CC cells (Figure 2H and I). By detecting the levels of glycolysis‐associated proteins utilizing Western blot assay, we discovered that levels of HK2 and PKM2 were blocked in si‐ hsa_circ_0018289‐1 and si‐hsa_circ_0018289‐2 groups compared with si‐control group (Figure 2J). Taken together, hsa_circ_0018289 depletion decreased CC cell proliferation, metastasis, and glycolysis.
FIGURE 2.
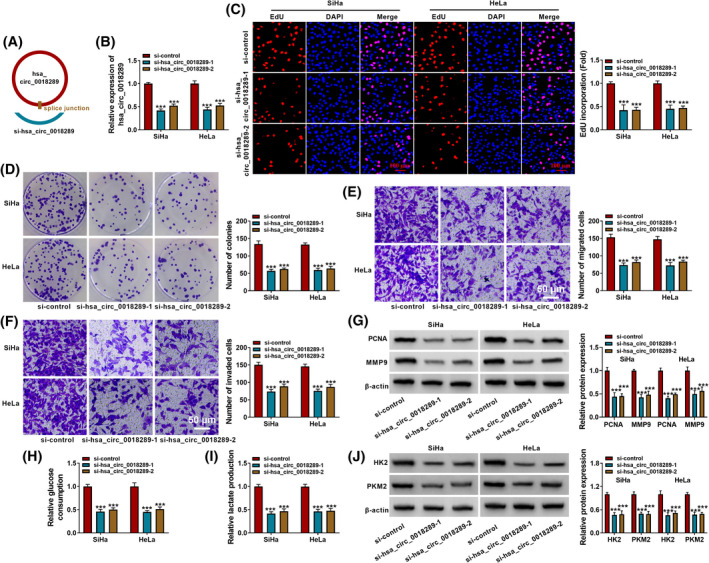
Depletion of hsa_circ_0018289 inhibited proliferation, metastasis, and glycolysis in CC cells. (A) Schematic illustration of siRNA of hsa_circ_0018289. (B‐H) HeLa and SiHa cells were transfected with si‐control, si‐hsa_circ_0018289‐1, or si‐hsa_circ_0018289‐2. (B) Hsa_circ_0018289 level was determined using qRT‐PCR assay. (C and D) EdU incorporation and colony formation assay for cell proliferation and colony formation capacity. (E and F) Transwell assay for the cell migration and invasion. (G) PCNA and MMP9 protein levels were determined. (H and I) Glucose consumption and lactate production were assessed using kits. (J) HK2 and PKM2 were measured. ***p < 0.001
3.3. Hsa_circ_0018289 was capable of binding to miR‐1294
The subcellular location of hsa_circ_0018289 in CC cells uncovered its potential to sponge miRNAs. Then, using by Circular RNA Interactome, seven potential target miRNAs of hsa_circ_0018289 were predicted. Among them, miR‐1294 was significantly upregulated in response to hsa_circ_0018289 knockdown in HeLa and SiHa cells (Figure 3A and B). Hence, we selected miR‐1294 for in‐depth investigations. Subsequently, qRT‐PCR assay demonstrated that miR‐1294 expression was downregulated in CC tissues (Figure 3C and D). Also, their binding sites were exhibited in Figure 3E. Subsequently, the miR‐1294‐induced the obvious reduction (exceeding 50%) in the luciferase density of CC cells co‐transfected hsa_circ_0018289‐WT, while there was little change in hsa_circ_0018289‐MUT (Figure 3F and G). Also, Bio‐miR‐1294‐enriched the hsa_circ_0018289 content was strikingly higher than Bio‐miR‐NC‐enriched one in RNA pull‐down assay (Figure 3H). Therefore, hsa_circ_0018289 could sponge miR‐1294.
FIGURE 3.
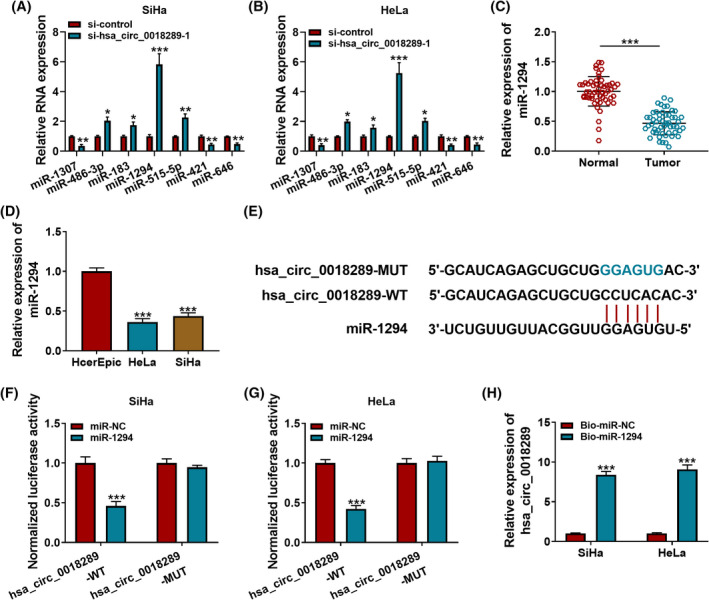
Hsa_circ_0018289 was capable of binding to miR‐1294. (A and B) Using Circular RNA Interactome, seven potential target miRNAs of hsa_circ_0018289 were predicted, and relative levels of seven miRNAs were detected in si‐hsa_circ_0018289‐1 or si‐control‐transfected HeLa and SiHa cells using qRT‐PCR assay. (C and D) QRT‐PCR analysis of miR‐1294 in CC tissues, normal tissues, HcerEpic, HeLa, and SiHa cells. (E) Bioinformatics analysis of the binding sites between hsa_circ_0018289 and miR‐1294. (F and G) Dual‐luciferase reporter assay was used to verify the binding. (H) Their interaction was also confirmed using RNA pull‐down assay. ***p < 0.001
3.4. Hsa_circ_0018289 functioned as an oncogene by sponging miR‐1294
As illustrated in Figure 4A, the introduction with miR‐1294 inhibitor evidently decreased miR‐1294 expression in HeLa and SiHa cells. To further explore the functional significance of hsa_circ_0018289 combined with miR‐1294, we carried out rescue assays. Data revealed that hsa_circ_0018289 knockdown‐induced decline in cell proliferative ability (Figure 4B–F), migration and invasion (Figure 4E–H), and inhibited glycolysis (Figure 4I–P) were all mitigated by miR‐1294 inhibitor. To conclude, interference of miR‐1294 eliminated hsa_circ_0018289 deficiency‐caused CC cell proliferation, metastasis, and glycolysis inhibitions.
FIGURE 4.
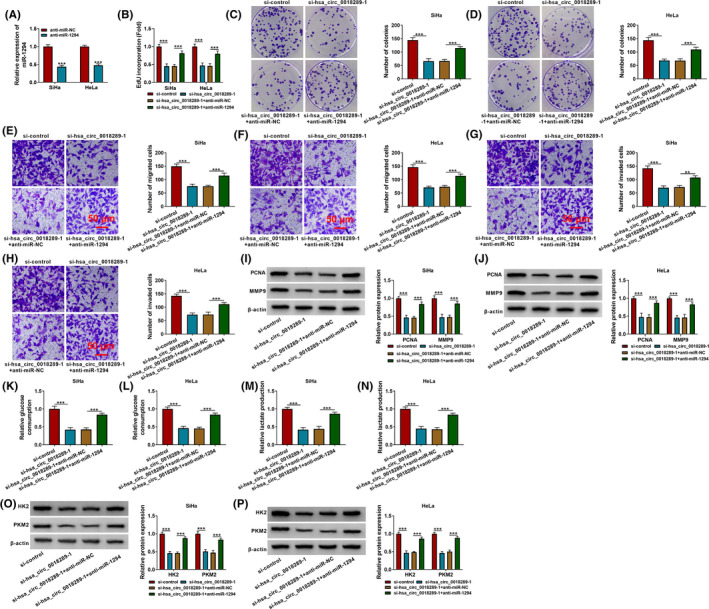
Hsa_circ_0018289 functioned as an oncogene by sponging miR‐1294. (A) QRT‐PCR analysis of miR‐1294 expression in anti‐miR‐NC or anti‐miR‐1294‐transfected HeLa and SiHa cells. (B–M) HeLa and SiHa cells were transfected with si‐control, si‐hsa_circ_0018289‐1, si‐hsa_circ_0018289‐1+anti‐miR‐NC or si‐hsa_circ_0018289‐1+anti‐miR‐1294. (B–D) EdU incorporation assay and colony formation assay for cell proliferation. (E–H) Transwell assay for the cell migration and invasion. (I–J) Western blot assay was used for detecting the protein expression of PCNA and MMP9. (K–N) Assay for glucose consumption and lactate production using kits. (O and P) HK2 and PKM2 protein levels were detected using Western blot assay. ***p < 0.001
3.5. ICMT was targeted by miR‐1294
To clarify the potential mechanism by which the hsa_circ_0018289/miR‐1294 axis affects CC cell growth, metastasis, and glycolysis, we searched TargetScan and Gene Expression Profiling Interactive Analysis (GEPIA) to seek the downstream genes of miR‐1294. As a result, we found eight target mRNAs of miR‐1294, which were subjected to qRT‐PCR analysis responding to miR‐1294 overexpression. Among these, ICMT exhibited the highest fold change (Figure 5A–B), Therefore, ICMT was chosen for further study. Besides, ICMT was obviously upregulated in CC tissues and cells (Figure 5C–F). The complementary sequence between miR‐1294 and ICMT‐3’UTR was shown in Figure 5G. Accumulation of miR‐1294 distinctly decreased the luciferase activity of CC cells in the ICMT‐3’UTR‐WT group, while the luciferase activity of cells in the ICMT‐3’UTR‐MUT group remained unchanged (Figure 5H and I). HeLa and SiHa cells with miR‐1294 overexpression were constructed via transfecting with miR‐1294 mimic (Figure 5J). In addition, the overexpression of miR‐1294 suppressed ICMT protein level in CC cells (Figure 5K). We also found that hsa_circ_0018289 depletion could decrease ICMT protein level in HeLa and SiHa cells, which was rescued via miR‐1294 inhibitor (Figure 5L and M). Collectively, miR‐1294 directly targeted ICMT.
FIGURE 5.
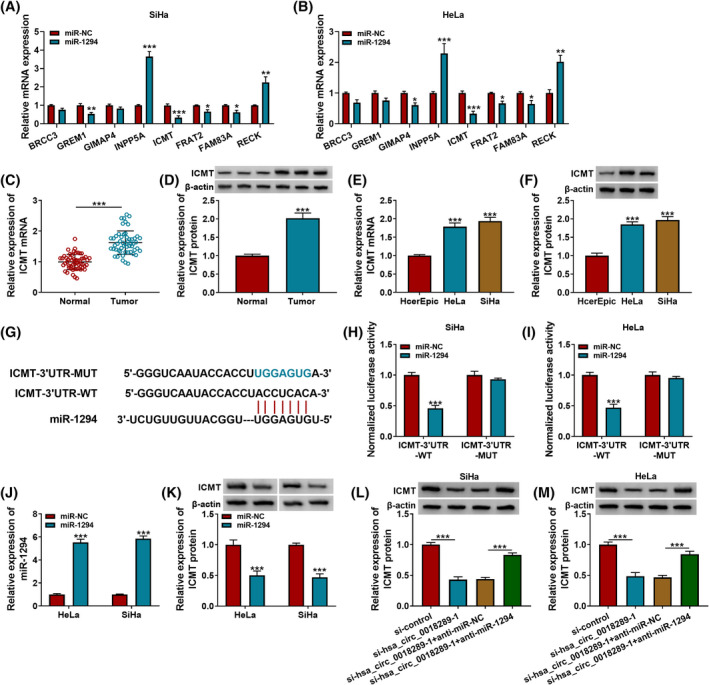
ICMT was targeted by miR‐1294. (A and B) TargetScan and GEPIA were used to predict the underlying target mRNAs of miR‐1294, followed by measurement using qRT‐PCR assay. (C and D) QRT‐PCR and Western blot assay the relative mRNA and protein expression of ICMT in CC tissues and normal tissues. (E and F) QRT‐PCR assay and Western blot assay the relative mRNA and protein expression of ICMT in HcerEpic, HeLa, and SiHa cells. (G) Bioinformatics analysis for the complementary sequence between miR‐1294 and ICMT‐3’UTR. (H and I) Dual‐luciferase reporter assay for the luciferase activity of HeLa and SiHa cells co‐transfected with miR‐NC or miR‐1294 and ICMT‐3’UTR‐WT or ICMT‐3’UTR‐MUT. (J and K) miR‐1294 and ICMT were determined in miR‐NC or miR‐1294‐transfected HeLa and SiHa cells using qRT‐PCR and Western blot assay. (L and M) ICMT protein level was analyzed in CC cells transfected with si‐control, si‐hsa_circ_0018289‐1, si‐hsa_circ_0018289‐1+anti‐miR‐NC, or si‐hsa_circ_0018289‐1+anti‐miR‐1294. ***p < 0.001
3.6. ICMT overexpression rescued the role of hsa_circ_0018289 knockdown in CC cell growth, metastasis, and glycolysis
The protein expression of ICMT in HeLa and SiHa cells was evidently enhanced due to transfection with OE‐ICMT, OE‐NC serving as negative control (Figure 6A). Furthermore, ICMT overexpression could attenuate the hsa_circ_0018289 knockdown mediated the repressed cell proliferative ability (Figure 6B–E), migration, and invasion (Figure 6F–K), as well as glycolysis (Figure 6L–Q) in CC cells. Hence, hsa_circ_0018289 silencing hampered CC progression via decreasing ICMT expression.
FIGURE 6.
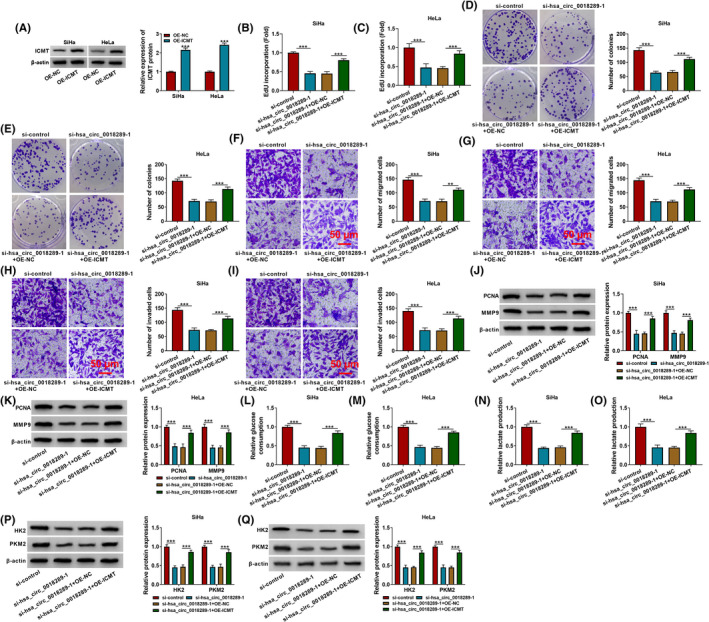
ICMT overexpression could rescue hsa_circ_0018289 knockdown‐mediated CC cell development. (A) ICMT protein level in OE‐NC‐ or OE‐ICMT‐transfected CC cells. (B‐Q) HeLa and SiHa cells were transfected with si‐control, si‐hsa_circ_0018289‐1, si‐hsa_circ_0018289‐1+OE‐NC, or si‐hsa_circ_0018289‐1+OE‐ICMT. (B–E) EdU incorporation assay and colony formation assay for the cell proliferative capacity of transfected cells. (F–I) Transwell assay for the cell migration and invasion in transfected cells. (J and K) Western blot assay was used for detecting the protein expression of PCNA and MMP9 in transfected cells. (L‐O) Assay for glucose consumption and lactate production using kits. (P and Q) Western blot assay for the protein levels of HK2 and PKM2 in transfected cells. **p < 0.01. ***p < 0.001
3.7. Hsa_circ_0018289 depletion impeded CC tumor growth in vivo
Xenograft model was established to evaluate the effect of hsa_circ_0018289 on tumor growth in vivo. SiHa cells stably expressing sh‐NC or sh‐hsa_circ_0018289 were hypodermically injected into nude mice. After injection, the tumor volume in the sh‐hsa_circ_0018289 group was smaller (Figure 7A). Although the whole body weight of mice was changeless (Figure 7B), hsa_circ_0018289 depletion indeed decreased the weight of tumors (Figure 7C). IHC experiment revealed that cell proliferation marker Ki‐67 was reduced by hsa_circ_0018289 knockdown (Figure 7D). In the sh‐hsa_circ_0018289 group, hsa_circ_0018289and ICMT were downregulated, while miR‐1294 was upregulated (Figure 7E and F). Silencing of hsa_circ_0018289 hindered tumor growth in vivo, which might be achieved through the miR‐1294/ICMT axis.
FIGURE 7.
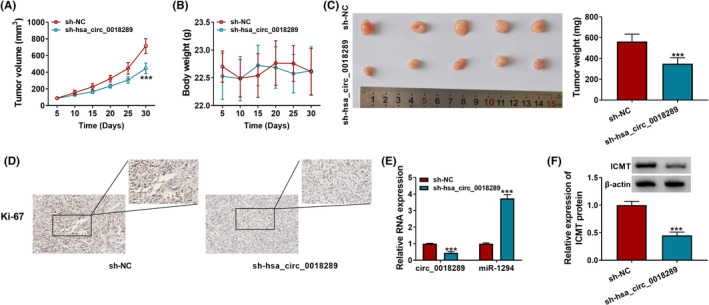
Hsa_circ_0018289 depletion impeded CC tumor growth in vivo. SiHa cells stably expressing sh‐NC or sh‐hsa_circ_0018289were hypodermically injected into nude mice (n = 5). (A) The volume of tumors was measured every 5 days. (B) The body weight of nude mice was measured every 5 days. (C) Weight of xenografts measured at 30 days after injection. (D) IHC analysis for Ki‐67 level in tumors. (E and F) hsa_circ_0018289, miR‐1294, and ICMT in tumors were detected using qRT‐PCR and Western blot assay. ***p < 0.001
4. DISCUSSION
In recent years, circRNAs have emerged as research hotspots due to their close relation to gynecological diseases, including CC. 25 In addition, circRNAs could regulate CC progression through influencing cell proliferation, metastasis, and angiogenesis. 26 In‐depth functional experiments revealed that silencing of hsa_circ_0018289 effectively repressed the growth, metastasis, glycolysis, and tumorigenesis of CC cells, which was attributed to regulation on miR‐1294/ICMT axis (Figure 8).
FIGURE 8.
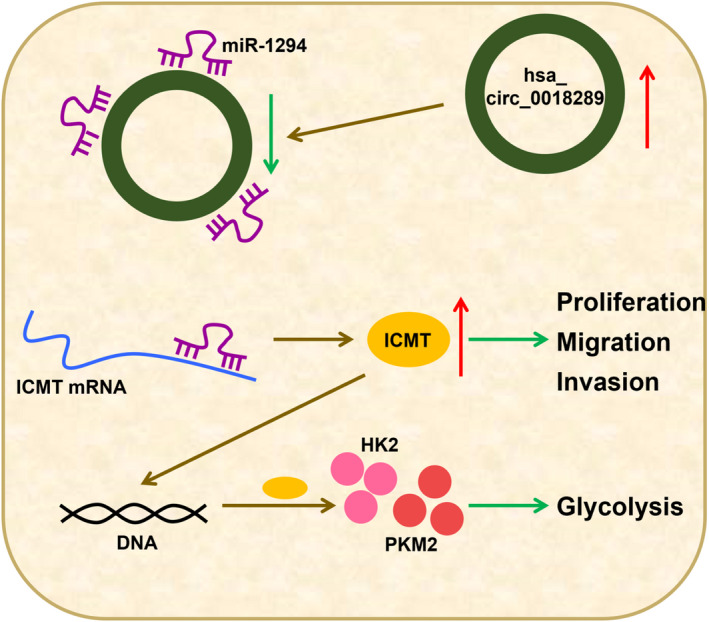
Schematic diagram of hsa_circ_0018289 regulating CC cell malignancy behavior
CircRNAs could perform as biomarkers for CC diagnosis, due to their cell or tissue‐specific expression. 27 Moreover, certain circRNAs were circRNAs in CC development. CircRNA circGSE1 accelerated CC cell migration and invasion by acting as a sponge of miR‐138‐5p and increasing Vimentin expression level. 28 Circ‐CCDC66 could facilitate the malignant properties of CC cells by sponging miR‐452‐5p to enhance REXO1 expression. 29 Enforced expression of circHIPK3 promoted growth, migration, and invasion of CC cells by modulating the miR‐485‐3p/FGF2 axis. 30 He et al. reported that hsa_circ_0018289 was positively correlated with the tumor size, lymph node metastasis, and stage of CC patients, suggesting that it was a promising biomarker for CC prognosis and disease monitoring. 31 Moreover, hsa_circ_0018289 was manifested to positively regulate CC cell proliferation and mobility via absorbing miR‐497. 11 In keeping with former reports, hsa_circ_0018289was freakishly enhanced in CC tissues and cells, and hsa_circ_0018289 deficiency hampered growth, metastasis, and glycolysis of CC cells.
Since circRNAs are able to serve as miRNAs sponges, thereby altering target genes expression, 32 we intended to search the possible action mechanism of hsa_circ_0018289 in CC development using bioinformatics software. Among these predicted candidates, miR‐1294 was of particular interest. Moreover, miR‐1294 expression was decreased in GC and epithelial ovarian cancer tissues, whose low expression could indicate poor prognosis of patients. 33 , 34 Furthermore, miR‐1294 was uncovered to exert a cancer‐suppressing role in clear cell renal cell carcinoma, 35 glioma, 36 osteosarcoma, 37 osteosarcoma, 38 and CC. 17 Similar to the report of Kan et al., our data also revealed the downregulation of miR‐1294 in CC, and its overexpression repressed CC cell metastasis and glycolysis. 17 In addition, miR‐1294 inhibition mitigated the role of hsa_circ_0018289 depletion in CC cells, implying hsa_circ_0018289 played a cancerogenic role in CC via sponging miR‐1294.
Notably, ICMT was a target gene of miR‐1294. According to previous research, interference of ICMT‐hindered growth and facilitated autophagy in several cell lines, suggesting its oncogenic role in human malignancies. 19 ICMT was closely associated with the sensitivity of CC cells to chemotherapy. 22 Gain of ICMT reversed miR‐1294 induced the repressed impact on CC cell malignant behavior, establishing the hsa_circ_0018289/miR‐1294/ICMT axis.
In summary, hsa_circ_0018289 knockdown had an inhibitory impact on CC cell growth, metastasis, glycolysis, and tumorigenesis, indicating that hsa_circ_0018289 was a CC‐promoting agent, at least in part by modulating miR‐1294/ICMT axis. This study highlighted a therapeutic target for CC prevention.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Li Y, Gao X, Yang C, Yan H, Li C. CircRNA hsa_circ_0018289 exerts an oncogenic role in cervical cancer progression through miR‐1294/ICMT axis. J Clin Lab Anal. 2022;36:e24348. doi: 10.1002/jcla.24348
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37(3):2779‐2788. [DOI] [PubMed] [Google Scholar]
- 2. Zheng RS, Sun KX, Zhang SW, et al. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41(1):19‐28. [DOI] [PubMed] [Google Scholar]
- 3. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169‐182. [DOI] [PubMed] [Google Scholar]
- 4. Johnson CA, James D, Marzan A, Armaos M. Cervical cancer: an overview of pathophysiology and management. Semin Oncol Nurs. 2019;35(2):166‐174. [DOI] [PubMed] [Google Scholar]
- 5. Li Z, Ruan Y, Zhang H, et al. Tumor‐suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao W, Zhang Y, Zhu Y. Circular RNA circbeta‐catenin aggravates the malignant phenotype of non‐small‐cell lung cancer via encoding a peptide. J Clin Lab Anal. 2021;35(9):e23900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015‐5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134‐142. [DOI] [PubMed] [Google Scholar]
- 11. Gao YL, Zhang MY, Xu B, et al. Circular RNA expression profiles reveal that hsa_circ_0018289 is up‐regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8(49):86625‐86633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lynam‐Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55‐71. [DOI] [PubMed] [Google Scholar]
- 13. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857‐866. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Q, Lv R, Guo W, Li X. microRNA‐802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine‐rich splicing factor 9. J Cell Biochem. 2019;120(6):10370‐10379. [DOI] [PubMed] [Google Scholar]
- 15. Xiao H, Yu L, Li F, et al. MiR‐340 suppresses the metastasis by targeting EphA3 in cervical cancer. Cell Biol Int. 2018;42(9):1115‐1123. [DOI] [PubMed] [Google Scholar]
- 16. Xu J, Wang H, Wang H, et al. The inhibition of miR‐126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas. 2019;156:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kan XQ, Li YB, He B, et al. MiR‐1294 acts as a tumor inhibitor in cervical cancer by regulating FLOT1 expression. J Biol Regul Homeost Agents. 2020;34(2). doi: 10.23812/20-10A. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 18. Winter‐Vann AM, Casey PJ. Post‐prenylation‐processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5(5):405‐412. [DOI] [PubMed] [Google Scholar]
- 19. Wang M, Hossain MS, Tan W, et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic‐dependent apoptosis and impairs tumor growth. Oncogene. 2010;29(35):4959‐4970. [DOI] [PubMed] [Google Scholar]
- 20. Manu KA, Chai TF, Teh JT, et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces cell‐cycle arrest and apoptosis through p21 and p21‐regulated BNIP3 induction in pancreatic cancer. Mol Cancer Ther. 2017;16(5):914‐923. [DOI] [PubMed] [Google Scholar]
- 21. Lau HY, Ramanujulu PM, Guo D, et al. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther. 2014;15(9):1280‐1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan Q, Liu R, Banu H, Ma L, Li H. Inhibition of isoprenylcysteine carboxylmethyltransferase sensitizes common chemotherapies in cervical cancer via Ras‐dependent pathway. Biomed Pharmacother. 2018;99:169‐175. [DOI] [PubMed] [Google Scholar]
- 23. Tang Q, Chen Z, Zhao L, Xu H. Circular RNA hsa_circ_0000515 acts as a miR‐326 sponge to promote cervical cancer progression through up‐regulation of ELK1. Aging (Albany NY). 2019;11(22):9982‐9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR‐138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50(5):1801‐1809. [DOI] [PubMed] [Google Scholar]
- 25. Huang J, Zhou Q, Li Y. Circular RNAs in gynecological disease: promising biomarkers and diagnostic targets. Biosci Rep. 2019;39(5):BSR20181641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tornesello ML, Faraonio R, Buonaguro L, et al. The Role of microRNAs, long non‐coding RNAs, and circular RNAs in cervical cancer. Front Oncol. 2020;10:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718‐724. [DOI] [PubMed] [Google Scholar]
- 28. Fan S, Zhao S, Gao X, et al. Circular RNA circGSE1 promotes cervical cancer progression through miR‐138‐5p/Vimentin. Onco Targets Ther. 2020;13:13371‐13386. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Zeng L, Liu YM, Yang N, Zhang T, Xie H. Hsa_circRNA_100146 promotes prostate cancer progression by upregulating TRIP13 via sponging miR‐615‐5p. Front Mol Biosci. 2021;8: 693477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu S, Liu S, Song H, Xia J. Circular RNA HIPK3 plays a carcinogenic role in cervical cancer progression via regulating miR‐485‐3p/FGF2 axis. J Investig Med. 2021;69(3):768‐774. [DOI] [PubMed] [Google Scholar]
- 31. He J, Lv X, Zeng Z. A potential disease monitoring and prognostic biomarker in cervical cancer patients: The clinical application of circular RNA_0018289. J Clin Lab Anal. 2020;34(8):e23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838‐1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi YX, Ye BL, Hu BR, Ruan XJ. Expression of miR‐1294 is downregulated and predicts a poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22(17):5525‐5530. [DOI] [PubMed] [Google Scholar]
- 34. Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. Downregulation of miR‐1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(22):7646‐7652. [DOI] [PubMed] [Google Scholar]
- 35. Pan W, Pang LJ, Cai HL, et al. MiR‐1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. Eur Rev Med Pharmacol Sci. 2019;23(9):3719‐3725. [DOI] [PubMed] [Google Scholar]
- 36. Wang J, Li J, Wang H, Lv L, Sun J. Overexpression of circ_0005198 sponges miR‐1294 to regulate cell proliferation, apoptosis, migration, and invasion in glioma. J Cell Biochem. 2019;120(9):15538‐15545. [DOI] [PubMed] [Google Scholar]
- 37. Zhang ZF, Li GR, Cao CN, et al. MicroRNA‐1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(24):8582‐8588. [DOI] [PubMed] [Google Scholar]
- 38. Wang Z, Yan J, Zou T, Gao H. MicroRNA‐1294 inhibited oral squamous cell carcinoma growth by targeting c‐Myc. Oncol Lett. 2018;16(2):2243‐2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


