Abstract
Background
Chemoresistance is one of the major obstacles for tumor treatment. Circular RNAs (circRNAs) have been confirmed to play vital roles in chemoresistance of cancer, including esophageal squamous cell carcinoma (ESCC). We investigated the roles and mechanisms of circ_0007142 in cisplatin (DDP) resistance of ESCC.
Methods
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was conducted to determine the levels of circ_0007142, DOCK1 mRNA, microRNA‐494‐3p (miR‐494‐3p) and LIM And SH3 Protein 1 (LASP1) mRNA. RNase R assay was conducted to analyze the characteristic of circ_0007142. Cell Counting Kit‐8 (CCK‐8) assay was performed to evaluate IC50 of DDP. Flow cytometry analysis, 5‐ethynyl‐2’‐deoxyuridine (EdU) assay and transwell assay were carried out to examine cell apoptosis, proliferation and invasion, respectively. Dual‐luciferase reporter assay was employed to verify the association between miR‐494‐3p and circ_0007142 or LASP1. Murine xenograft assay was conducted to investigate the role of circ_0007142 in DDP resistant in vivo. The protein level of LASP1 in tumors was measured by Immunohistochemistry (IHC) analysis.
Results
Circ_0007142 was upregulated in DDP‐resistant ESCC tissues and cells. Circ_0007142 knockdown improved DDP sensitivity, induced cell apoptosis and hampered cell proliferation and invasion in DDP‐resistant ESCC cells. Circ_0007142 functioned as the sponge for miR‐494‐3p and miR‐494‐3p inhibition reversed the impacts of circ_0007142 knockdown on DDP resistance, cell apoptosis, proliferation, and invasion. LASP1 was a target of miR‐494‐3p, and the effects on DDP resistance, cell apoptosis, growth, and invasion mediated by LASP1 downregulation were rescued by miR‐494‐3p inhibition. Moreover, circ_0007142 knockdown enhanced DDP sensitivity in vivo.
Conclusion
Circ_0007142 improved DDP resistance of ESCC by upregulating LASP1 via sponging miR‐494‐3p.
Keywords: circ_0007142, DDP, ESCC, LASP1, miR‐494‐3p, resistance
Circ_0007142 promoted DDP resistance, cell proliferation, and invasion in DDP‐resistant ESCC cells by regulating miR‐494‐3p and LASP1.
1. INTRODUCTION
Esophageal squamous cell carcinoma (ESCC) is a common malignant gastrointestinal tumor with high mortality and morbidity. 1 , 2 The overall prognosis is still not optimistic though great efforts have been made in the treatment of ESCC. 3 , 4 Chemotherapy is one of the methods for the treatment of tumors, but multidrug resistance (MDR) is the main cause of tumor chemotherapy failure. 5 Cisplatin (DDP) is a first‐line drug for the treatment of diverse human tumors in the clinic, including ESCC. 6 However, the acquisition of DDP resistance in the process of therapy seriously affects the treatment effect of ESCC. 7 Thus, it is of great significance to understand the mechanism of DDP resistance in ESCC.
Circular RNAs (circRNAs) are a family of covalently closed RNA molecules, which are not affected by RNA exonuclease and the expression is more stable and difficult to be degraded. 8 , 9 , 10 , 11 In recent years, more and more studies have shown that circRNAs are closely related to drug resistance of diverse human cancers. 12 , 13 For instance, Zhu et al. reported that circPVT1 was evidently elevated in drug‐resistant osteosarcoma (OS) cells and associated with the resistance of OS cells to doxorubicin and DDP. 14 Huang et al. implicated that circAKT3 was increased in DDP‐resistant gastric cancer (GC) cells and improved DDP resistant of GC. 15 Zhao et al. revealed that Cdr1as was conspicuously reduced in DDP‐resistant ovarian cancer and its upregulation hampered cell growth and facilitated cell apoptosis mediated by DDP in ovarian cancer. 16 Moreover, circ_0007142 has been identified as an oncogene in colorectal cancer 17 and lung adenocarcinoma. 18 But up to now, there has been no report on the function of circ_0007142 in the carcinogenesis and drug resistance of ESCC.
MicroRNAs (miRNAs), a set of ncRNAs with about 22 nucleotides, modulate genes expression at the post‐transcriptional level via recognizing the 3’‐untranslated region (3’UTR) of target messenger RNAs (mRNAs). 19 Mounting evidence verified that miRNAs were associated with drug resistance of ESCC. For example, Imanaka et al. suggested that there was an elevation in miR‐141 in DDP‐resistant ESCC cells and the ectopic expression of miR‐141 improved DDP resistance via promoting cell growth. 20 Liu et al. manifested that downregulation of miR‐455‐3p repressed chemoresistance and tumorigenesis in ESCC. 21 MiR‐494‐3p has been confirmed to repress ESCC malignancy by targeting CLPTM1L 22 and PFN2. 23 Nevertheless, the function of miR‐494‐3p in the resistance of ESCC to DDP remains unclear.
LIM and SH3 protein 1 (LASP1), an actin‐binding protein, was aberrantly expressed in some human cancers, including ESCC. 24 , 25 A previous report implicated that LASP1 contributed to ESCC progression via promoting cell growth and metastasis. 25 Moreover, several studies claimed that LASP1 was associated with chemoresistance in some human cancers. 26 , 27 However, it has not been reported the precise role of LASP1 in drug resistance of ESCC.
Here, we investigated the expression patterns of circ_0007142, miR‐494‐3p and LASP1 in DDP‐resistant ESCC. Moreover, their functions and underlying mechanisms in DDP resistance of ESCC were further explored.
2. MATERIALS AND METHODS
2.1. Tissues collection
After the study was permitted by the Ethics Committee of The First Affiliated Hospital of USTC, and written informed consents were signed by all participants, 30 DDP‐resistant ESCC tissues and 30 DDP‐sensitive ESCC tissues were collected from ESCC patients who received DDP‐based treatment at the First Affiliated Hospital of USTC. The collected tissues were preserved at −80°C. The clinicopathologic features of these patients were presented in Table 1.
TABLE 1.
The clinicopathologic features of esophageal squamous cell carcinoma patients
|
Characteristics n = 60 |
|
|---|---|
| Age (years) | |
| ≤60 | 22 |
| >60 | 38 |
| TNM grade | |
| I+II | 27 |
| III/IV | 33 |
| Lymph node metastasis | |
| Positive | 35 |
| Negative | 25 |
| Tumor size | |
| ≤5 cm | 29 |
| >5 cm | 31 |
Abbreviation: TNM, tumor‐node‐metastasis.
2.2. Cell culture
ESCC cells (KYSE150, ECA109, TE1, and KYSE410) and normal human esophageal epithelial cells (HEEC) were bought from BeNa Culture Collection (Beijing, China). DDP‐resistant ESCC cells (TE1/DDP and KYSE410/DDP) were constructed by treating TE1 cells and KYSE410 cells with gradually increased doses of DDP (10%–20% increase per time, Solarbio, Beijing, China) at an initial dose of 2 mmol/L until the concentration reached 35 mmol/L over a 2‐month period. All cells grew in Dulbecco's modified Eagle's medium (DMEM; Gibco) including 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco) in an incubator containing 5% CO2 at 37°C. To maintain DDP resistance of TE1/DDP and KYSE410/DDP cells, 2 µM DDP was added into the medium.
2.3. Cell transfection
Short hairpin RNA (shRNA) targeting circ_0007142 (sh‐circ_0007142), shRNA targeting LASP1 (sh‐LASP1), and their control (sh‐NC); circ_0007142 overexpression vector (circ_0007142) and its control (pcDNA); mimics of miR‐494‐3p (miR‐494‐3p) and its control (miR‐NC); inhibitors of miR‐494‐3p (anti‐miR‐494‐3p); and its control (anti‐miR‐NC) were bought from GenePharma. The transfection of TE1/DDP and KYSE410/DDP cells was conduced using Lipofectamine 2000 (Invitrogen).
2.4. Real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR)
Total RNA in ESCC tissues and cells was isolated via RNAiso Plus (Takara). For RNase R treatment, total RNA was treated with or without RNase R (Epicentre) for 20 min at 37°C. Reverse transcription experiment was conducted through HiScript® II Reverse Transcriptase Kit (Takara) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme). Then, RT‐qPCR was performed with BeyoFast™ SYBR Green qPCR Mix (Beyotime) on ABI 7900 Real‐Time PCR system (Applied Biosystems). The relative expression was measured with glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) or small nuclear RNA U6 as internal control via the 2−ΔΔCt method. The primers were as follows: circ_0007142: (F: 5’‐CTGGAACTCTGCCTCAGGAT‐3’ and R: 5’‐CCTCGGTACCACCCTTCATA‐3’); DOCK1: (F: 5’‐ACCGAGGTTACACGTTACGAA‐3’ and R: 5’‐TCGGAGTGTCGTGGTGACTT‐3’); LASP1: (F: 5’‐GGTGCGGCAAGATCGTGTA‐3’ and R: 5’‐TGCAGGTCTCGCAATGGAA‐3’); miR‐494‐3p: (F: 5’‐GGGTGAAACACACACGGGAA‐3’ and R: 5’‐GGCAGGTCCGAGGT‐3’); GAPDH: (F: 5’‐AGAAGGCTGGGGCTCATTTG‐3’ and R: 5’‐AGGGGCCATCCACAGTCTTC‐3’); and U6: (F: 5’‐GCTCGCTTCGGCAGCACATA‐3’ and R: 5’‐CCTCGCTTCGGCAGCACATA‐3’).
2.5. Subcellular fraction assay
The nuclear and cytoplasm were separated using PARIS Kit (Invitrogen) in line with the manufacturers’ instructions. The levels of circ_0007142, U6 (control for nuclear transcript) and GAPDH (control for cytoplasm transcript) were examined by RT‐qPCR assay.
2.6. Cell Counting Kit‐8 (CCK‐8) assay
CCK‐8 assay was conducted to analyze DDP resistance. In brief, TE1, KYSE410, TE1/DDP, and KYSE410/DDP cells were treated with different doses (0, 10, 20, 30, 40, and 50 μm) of DDP (Solarbio) and then seeded into 96‐well plates. Afterward, 10 μL CCK‐8 (Beyotime) was added and incubated for an additional 2 h. The optical density value at 450 nm was examined on a microplate reader (Bio‐Rad). The 50% inhibitory concentration of DDP (IC50) was analyzed by the relative survival curve.
2.7. Flow cytometry analysis
After relevant transfection, the apoptosis of TE1/DDP and KYSE410/DDP cells was evaluated through the Annexin V‐fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection Kit (Beyotime). Briefly, transfected cells were harvested, washed, and resuspended. Then, 5 μl Annexin V‐FITC and 5 μl PI were added and maintained for 15 min in the dark to stain cells. The apoptotic cells were analyzed with a flow cytometry (BD Biosciences).
2.8. 5‐Ethynyl‐2’‐deoxyuridine (EdU) assay
Cell proliferation was assessed by using EdU assay kit (Solarbio). In brief, the transfected TE1/DDP and KYSE410/DDP cells (1 × 104 cells/well) were plated into 24‐well plates and incubated for 2 h with EdU. After washing with PBS (Solarbio), the cells were fixed in 4% paraformaldehyde (Sigma‐Aldrich), and then, 2 mg/ml glycine (Sigma‐Aldrich) was added to neutralize paraformaldehyde (Sigma‐Aldrich). Next, the cells added with penetrant (0.5% Triton X‐100 in PBS) for 10 min and washed with PBS (Solarbio). The cells were then dyed with EdU according to the manufacturers’ instructions. The results were detected under a fluorescence microscope (Olympus).
2.9. Transwell assay
A Transwell chamber (Corning Incorporated) coated with Matrigel (Solarbio) was utilized to examine the invasion of TE1/DDP and KYSE410/DDP cells. Transfected TE1/DDP and KYSE410/DDP cells were suspended in serum‐free DMEM (Gibco) and plated in the upper chamber. DMEM (Gibco) including 10% FBS (Gibco) was added to the bottom chamber. After 48 h, the invasive cells were treated with methanol and stained with crystal violet (Solarbio). The stained cells were observed and counted with a microscope (Olympus).
2.10. Western blot assay
Total protein was isolated from ESCC tissues and cells using RIPA buffer (Beyotime). The proteins were separated through sodium dodecyl sulfonate‐polyacrylamide gel (SDS‐PAGE; Solarbio) after being quantified via a BCA Protein Quantification Kit (Vazyme). Then, the samples were transferred onto polyvinylidene difluoride membranes (PVDF; Pall Corporation, New York, NYC, USA) and blocked in skim milk for 2 h. Next, the membranes were incubated with primary antibodies against P‐glycoprotein (P‐gp; ab3366; Abcam), glutathione S‐transferase π (GST‐π; ab53942; Abcam), LASP1 (ab156872; Abcam), or GAPDH (ab8245; Abcam) and corresponding secondary antibody (ab150077; Abcam). The protein bands were analyzed with an enhanced chemiluminescence assay kit (Beyotime).
2.11. Dual‐luciferase reporter assay
The sequences of circ_0007142 including wild‐type or mutant binding sequences of miR‐494‐3p were amplified and inserted into a psi‐CHECK2 vector (Promega) to generate luciferase reporter vectors WT‐circ_0007142 and MUT‐circ_0007142, respectively. The sequences of LASP1 3’UTR including wild‐type or mutant binding sequences of miR‐494‐3p were cloned into a pmirGLO vector (Promega) to form LASP1 3’UTR‐WT and LASP1 3’UTR‐MUT, respectively. MiR‐494‐3p or miR‐NC was transfected into TE1/DDP or KYSE410/DDP cells together with relevant vector. The luciferase activity was examined through dual‐luciferase reporter assay kit (Promega) 48 h post‐co‐transfection.
2.12. In vivo experiment
Six‐week‐old nude mice were bought from Shanghai SLAC Laboratory Animals Co., Ltd and divided into four groups (n = 7). TE1/DDP cells transfected with sh‐NC or sh‐circ_0007142 were injected into the nude mice. 8 days later, the mice were intraperitoneally administrated with phosphate buffer saline (PBS; Solarbio) or 6 mg/kg of DDP (Solarbio) every 3 days. Tumor volume was examined every 3 days and calculated using the formula: (length × width2)/2. On Day 26, the mice were euthanized and tumors were harvested, weighed and preserved at −80°C for RT‐qPCR assay. The research was permitted by the Ethics Committee of Animal Research of The First Affiliated Hospital of USTC.
2.13. Immunohistochemistry (IHC) assay
The tumor tissues were fixed in formalin (Sigma‐Aldrich) overnight, dehydrated with ethanol, embedded with paraffin (Sigma‐Aldrich), and then sliced to 5 mm. Next, the samples were treated with xylene (Sigma‐Aldrich) and ethanol to remove paraffin (Sigma‐Aldrich). Next, the sections were blocked with normal nonimmune animal serum for 30 min, incubated with antibody against LASP1 (ab156872; Abcam), Ki67 (ab15580; Abcam), MMP9 (ab38898; Abcam) overnight and corresponding secondary antibody (ab150077; Abcam) for 1 h. Thereafter, IHC reactions were examined with DAB kit (Sigma‐Aldrich). Finally, the results were observed under a microscope (Olympus).
2.14. Statistical analysis
The data collected from three independent experiments were exhibited as mean ± standard deviation (SD), processed by using GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA). The difference was analyzed via Student's t‐test or one‐way analysis of variance (ANOVA). Spearman's correlation analysis was utilized to analyze the linear relationship between miR‐494‐3p and circ_0007142 or LASP1. p‐value less than 0.05 was considered to be significantly different.
3. RESULTS
3.1. Circ_0007142 was upregulated in DDP‐resistant ESCC tissues and cells
Through analyzing GEO dataset GSE131969 (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE131969), hsa_circ_0007142 was found to be elevated in ESCC patients compared with normal patients (Figure 1A–C). To determine the role of circ_0007142 in DDP resistance of ESCC, RT‐qPCR was firstly performed to examine the expression of circ_0007142 in DDP‐resistant and DDP‐sensitive ESCC tissues. The data exhibited that circ_0007142 was highly expressed in DDP‐resistant ESCC tissues in reference to that in DDP‐sensitive ESCC tissues (Figure 1D). Next, the expression of circ_0007142 in HEEC cells, ESCC cells (KYSE150, ECA109, TE1, and KYSE410 cells) and DDP‐resistant ESCC cells (TE1/DDP and KYSE410/DDP cells) was measured. The results implied that circ_0007142 was conspicuously elevated in ESCC cells compared with that in HEEC cells; moreover, the expression of circ_0007142 was higher in TE1/DDP and KYSE410/DDP cells compared with that in TE1 and KYSE410 cells (Figure 1E,F). RNase R assay indicated that circ_0007142 was resistant to RNase R treatment, while linear DOCK1 was digested by RNase R (Figure 1G,H). Moreover, we found that circ_0007142 was mainly enriched in the cytoplasm of TE1/DDP and KYSE410/DDP cells (Figure 1I,J). All these data implied that the dysregulation of circ_0007142 might be associated with DDP resistance in ESCC.
FIGURE 1.
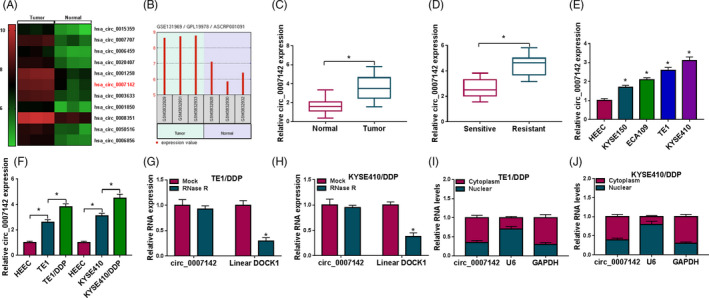
High expression of circ_0007142 in DDP‐resistant ESCC tissues and cells. (A–C) According to GEO dataset GSE131969, circ_0007142 was upregulated in ESCC tissues. (D) The level of circ_0007142 in DDP‐resistant and DDP‐sensitive ESCC tissues was detected via RT‐qPCR. (E) The level of circ_0007142 in ESCC cells (KYSE150, ECA109, TE1, and KYSE410 cells) and HEEC cells was determined through RT‐qPCR. (F) The expression of circ_0007142 in HEEC, TE1, TE1/DDP, KYSE410, and KYSE410/DDP cells was examined by RT‐qPCR. (G and H) The expression levels of circ_0007142 and linear DOCK1 mRNA in TE1/DDP and KYSE410/DDP cells treated with or without RNase R were examined by RT‐qPCR assay. (I and J) The expression of circ_0007142 in the nuclear and cytoplasm of TE1/DDP and KYSE410/DDP was determined by RT‐qPCR assay. *p < 0.05
3.2. Knockdown of circ_0007142 enhanced DDP sensitivity in DDP‐resistant ESCC cells
To investigate the functional role of circ_0007142 in DDP resistance in ESCC cells, DDP‐resistant cells (TE1/DDP and KYSE410/DDP cells) were established by incubating TE1 and KYSE410 cells with different concentrations of DDP. As illustrated by CCK‐8 assay, IC50 of DDP in TE1/DDP and KYSE410/DDP cells was drastically increased compared with that in TE1 and KYSE410 cells, indicating the production of DDP resistance in TE1/DDP and KYSE410/DDP cells (Figure 2A). Then, we transfected sh‐circ_0007142 or sh‐NC into TE1/DDP and KYSE410/DDP cells to knock down the expression of circ_0007142 and the knockdown efficiency was evaluated by RT‐qPCR. As shown in Figure 2B, sh‐circ_0007142 transfection resulted in a distinct decrease in circ_0007142 expression in TE1/DDP and KYSE410/DDP cells compared with that in sh‐NC transfected cells. CCK‐8 assay displayed that IC50 of DDP in TE1/DDP and KYSE410/DDP cells transfected with sh‐circ_0007142 was reduced, suggesting that the sensitivity of TE1/DDP and KYSE410/DDP cells to DDP was enhanced after circ_0007142 knockdown (Figure 2C). The results of flow cytometry analysis implicated that depletion of circ_0007142 caused an obvious increase in the apoptosis of TE1/DDP and KYSE410/DDP cells in reference to control group (Figure 2D). EdU assay indicated that circ_0007142 silencing repressed the capacity of TE1/DDP and KYSE410/DDP cells to proliferate (Figure 2E). Transwell assay manifested that cell invasion was markedly inhibited in TE1/DDP and KYSE410/DDP cells transfected with sh‐circ_0007142 compared with sh‐NC group (Figure 2F). Furthermore, we determined the protein levels of drug‐resistant markers (P‐gp and GST‐π) using Western blot assay. We found that P‐gp and GST‐π were all decreased in TE1/DDP and KYSE410/DDP cells following transfection with sh‐circ_0007142 when compared to those in TE1/DDP and KYSE410/DDP cells transfected with sh‐NC (Figure 2F,G). Collectively, circ_0007142 downregulation led to an enhancement in DDP sensitivity in DDP‐resistant ESCC cells.
FIGURE 2.
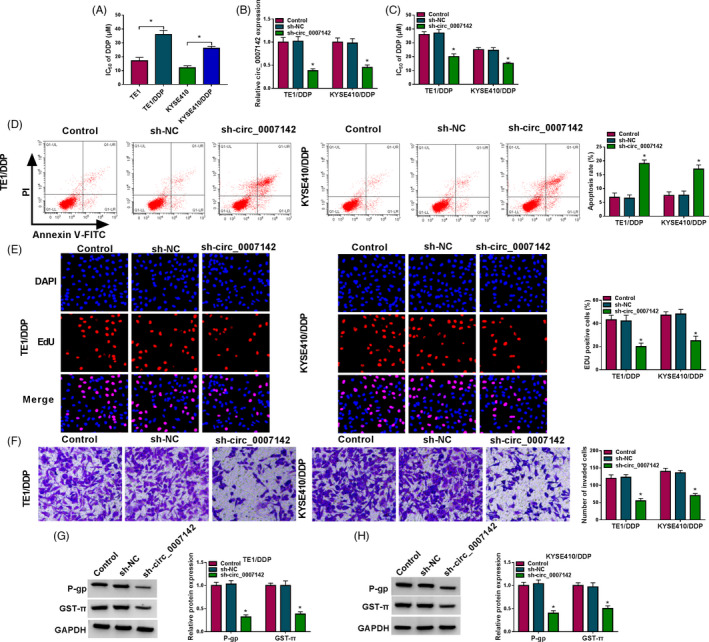
Silencing of circ_0007142 evidently reduced DDP resistance in DDP‐resistant ESCC cells. (A) IC50 of DDP in TE1/DDP and KYSE410/DDP cells was assessed by CCK‐8 assay. (B–G) TE1/DDP and KYSE410/DDP cells were transfected with sh‐NC or sh‐circ_0007142. (B) The expression of circ_0007142 in TE1/DDP and KYSE410/DDP cells was measured by RT‐qPCR. (C) IC50 value of DDP in TE1/DDP and KYSE410/DDP cells was analyzed by CCK‐8 assay. (D) The apoptosis of TE1/DDP and KYSE410/DDP cells was determined via flow cytometry analysis. (E) The proliferation of TE1/DDP and KYSE410/DDP cells was analyzed by EdU assay. (F) The invasion of TE1/DDP and KYSE410/DDP cells was evaluated through Transwell assay. (G and H) The protein levels of P‐gp and GST‐π in TE1/DDP and KYSE410/DDP cells were examined through Western blot assay. *p < 0.05
3.3. Silencing of circ_0007142 enhanced DDP sensitivity by modulating miR‐494‐3p in DDP‐resistant ESCC cells
In order to investigate the underlying mechanism of circ_0007142 in regulating DDP resistance in DDP‐resistant ESCC cells, online software packages starBase v2.0 and circinteractome were utilized to search the potential target of circ_0007142. As presented in Figure 3A, miR‐1277, miR‐186, and miR‐494‐3p were predicted to be the targets of circ_0007142 and circ_0007142 knockdown led to a distinct elevation in miR‐494‐3p expression in TE1/DDP cells (Figure 3A). The binding sites between circ_0007142 and miR‐494‐3p were exhibited in Figure 3B. Then, dual‐luciferase reporter assay was performed to verify the relationship between circ_0007142 and miR‐494‐3p. The results indicated that the luciferase activity in TE1/DDP and KYSE410/DDP cells co‐transfected with WT‐circ_0007142 and miR‐494‐3p was remarkably suppressed compared with that in WT‐circ_0007142 and miR‐NC co‐transfected group; however, the luciferase activity was not affected in MUT‐circ_0007142 group (Figure 3C,D). As exhibited in Figure 3E, miR‐494‐3p level was reduced in DDP‐resistant ESCC tissues compared with DDP‐sensitive ESCC tissues (Figure 3E). Moreover, miR‐494‐3p was weakly expressed in ESCC cells compared with that in HEEC cells; moreover, there was a lower expression of miR‐494‐3p in DDP‐resistant ESCC cells compared with that in DDP‐sensitive ESCC cells (Figure 3F,G). As suggested by Spearman's correlation analysis, there was an inverse correlation between circ_0007142 expression and miR‐494‐3p expression in ESCC tissues (Figure 3H). Next, the expression levels of circ_0007142 and miR‐494‐3p in TE1/DDP and KYSE410/DDP cells transfected with sh‐NC, sh‐circ_0007142, pcDNA, or circ_0007142 were determined via RT‐qPCR. The data presented that the expression of circ_0007142 was drastically reduced and the expression of miR‐494‐3p was dramatically improved in TE1/DDP and KYSE410/DDP cells transfected with sh‐circ_0007142, while circ_0007142 transfection led to opposite results (Figure 3I,J). As presented in Figure 3K, the elevated expression of miR‐494‐3p caused by sh‐circ_0007142 was effectively overturned following anti‐miR‐494‐3p transfection in TE1/DDP and KYSE410/DDP cells. The above data suggested that circ_0007142 negatively modulated miR‐494‐3p expression by direct interaction.
FIGURE 3.
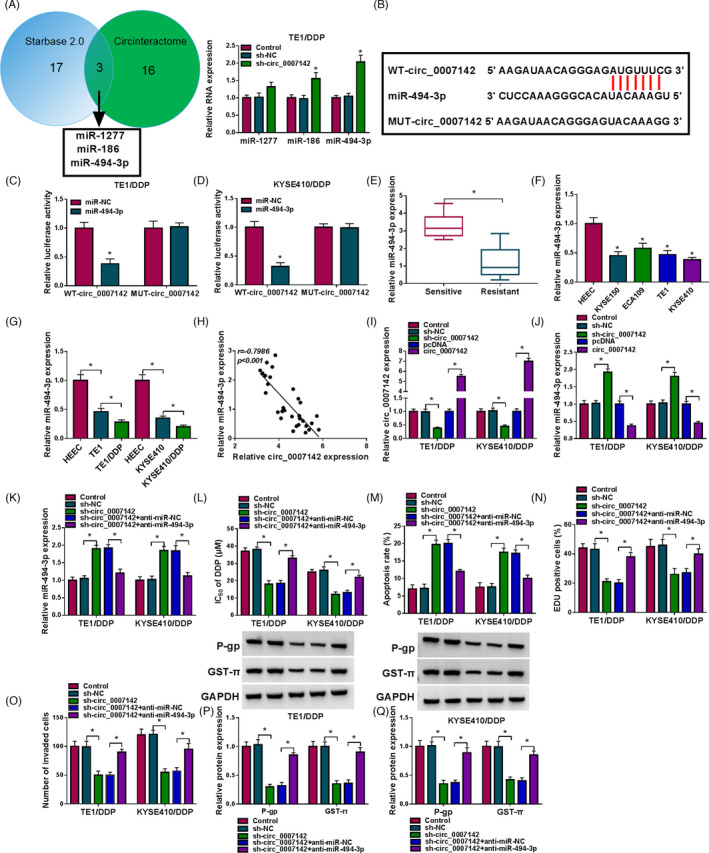
Circ_0007142 downregulation repressed DDP resistance by negatively regulating miR‐494‐3p expression in DDP‐resistant ESCC cells. (A) The potential targets of circ_0007142 were predicted by starBase v2.0 and circinteractome, and their expression in TE1/DDP cells transfected with sh‐NC or sh‐circ_0007142 was detected by RT‐qPCR assay. (B) The potential binding sites between circ_0007142 and miR‐494‐3p were shown. (C and D) The luciferase activity in WT‐circ_0007142 or MUT‐circ_0007142 and miR‐NC and miR‐494‐3p co‐transfected TE1/DDP and KYSE410/DDP cells was determined by dual‐luciferase reporter assay. (E) The expression of miR‐494‐3p in DDP‐resistant and DDP‐sensitive ESCC tissues was detected by RT‐qPCR assay. (F and G) The expression of miR‐494‐3p in HEEC, KYSE150, ECA109, TE1, KYSE410, TE1/DDP, and KYSE410/DDP cells was determined by RT‐qPCR assay. (H) The linear correlation between circ_0007142 and miR‐494‐3p in ESCC tissues was analyzed. (I and J) The levels of circ_0007142 and miR‐494‐3p in TE1/DDP and KYSE410/DDP cells transfected with sh‐NC, sh‐circ_0007142, pcDNA, or circ_0007142 were examined using RT‐qPCR. (K‐Q) Sh‐NC, sh‐circ_0007142, sh‐circ_0007142+anti‐miR‐NC, or sh‐circ_0007142+anti‐miR‐494‐3p was transfected into TE1/DDP and KYSE410/DDP cells. (K) The expression of miR‐494‐3p in TE1/DDP and KYSE410/DDP cells was detected via RT‐qPCR. (L–O) IC50 of DDP, cell apoptosis, proliferation, and invasion of TE1/DDP and KYSE410/DDP cells were assessed through CCK‐8 assay, flow cytometry analysis, EdU assay, and Transwell assay, respectively. (P and Q) The levels of P‐gp and GST‐π in TE1/DDP and KYSE410/DDP cells were measured through Western blot assay. *p < 0.05
Subsequently, the functional roles of circ_0007142 and miR‐494‐3p in altering DDP resistance in DDP‐resistant ESCC cells were investigated. The decreased IC50 value of DDP in TE1/DDP and KYSE410/DDP cells mediated by sh‐circ_0007142 was notably restored following miR‐494‐3p inhibition, as revealed by CCK‐8 assay (Figure 3L). Flow cytometry analysis showed that the promotion in the apoptosis of TE1/DDP and KYSE410/DDP cells caused by circ_0007142 knockdown was conspicuously repressed following miR‐494‐3p depletion (Figure 3M and Figure S1A). As revealed by EdU assay and Transwell assay, downregulation of circ_0007142 resulted in remarkable reduction in cell proliferation and invasion in TE1/DDP and KYSE410/DDP cells, whereas the effects were weakened after the knockdown of miR‐494‐3p expression (Figure 3N,O and Figure S1B,C). Furthermore, the decreased levels of P‐gp and GST‐π mediated by circ_0007142 deficiency were also reversed by miR‐494‐3p inhibition in TE1/DDP and KYSE410/DDP cells (Figure 3P,Q). To sum up, circ_0007142 knockdown inhibited DDP resistance in DDP‐resistant ESCC cells via sponging miR‐494‐3p.
3.4. Inhibition of miR‐494‐3p attenuated the enhancement of DDP sensitivity mediated by LASP1 knockdown in DDP‐resistant ESCC cells
Through searching starBase v2.0, LASP1 was predicted to be a target gene of miR‐494‐3p and their potential binding sites were displayed in Figure 4A. The data of dual‐luciferase reporter assay revealed that the co‐transfection of LASP1 3’UTR‐WT and miR‐494‐3p resulted in a suppression in the luciferase activity in TE1/DDP and KYSE410/DDP cells in comparison with LASP1 3’UTR‐WT and miR‐NC co‐transfected group, while the co‐transfection of LASP1 3’UTR‐MUT and miR‐494‐3p or miR‐NC did not affect the luciferase activity (Figure 4B,C). The mRNA and protein levels of LASP1 were all obviously elevated in DDP‐resistant ESCC tissues compared with that in DDP‐sensitive ESCC tissues (Figure 4D,E). The protein level of LASP1 was distinctly increased in TE1 and KYSE410 cells in reference to that in HEEC cells; moreover, the protein level of LASP1 was higher in TE1/DDP and KYSE410/DDP cells than that in TE1 and KYSE410 cells (Figure 4F). As determined by Spearman's correlation analysis, there was a negative correlation between the expression of LASP1 and miR‐494‐3p in ESCC tissues (Figure 4G). Subsequently, to explore the roles of miR‐494‐3p and LASP1 in regulating DDP resistance in DDP‐resistant ESCC tissues, we transfected sh‐NC, sh‐LASP1, sh‐LASP1+anti‐miR‐NC, or sh‐LASP1+anti‐miR‐494‐3p into TE1/DDP and KYSE410/DDP cells. The results of Western blot assay implicated that the expression of LASP1 was strikingly reduced in TE1/DDP and KYSE410/DDP cells transfected with sh‐LASP1, while anti‐miR‐494‐3p transfection partially recovered the reduction (Figure 4H,I). CCK‐8 assay proved that the resistance of TE1/DDP and KYSE410/DDP cells to DDP was reduced following the downregulation of LASP1, as demonstrated by decreased IC50 value; however, the effect was abolished after miR‐494‐3p inhibition (Figure 4J). The data of flow cytometry analysis manifested that LASP1 knockdown markedly facilitated cell apoptosis in TE1/DDP and KYSE410/DDP cells, whereas miR‐494‐3p inhibition partly overturned this impact (Figure 4K and Figure S1A). The proliferation of TE1/DDP and KYSE410/DDP cells was repressed by LASP1 knockdown, while miR‐494‐3p inhibition reversed the impact (Figure 4L and Figure S1B). Transwell assay indicated that the invasion of TE1/DDP and KYSE410/DDP cells was greatly hampered by LASP1 knockdown, while miR‐494‐3p downregulation effectively abrogated this influence (Figure 4M and Figure S1C). Silencing of LASP1 led to marked downregulation in the levels of P‐gp and GST‐π in TE1/DDP and KYSE410/DDP cells, while deficiency of miR‐494‐3p restored the effects, as illustrated by Western blot assay (Figure 4N,O). All these data demonstrated that miR‐494‐3p modulated DDP resistance in DDP‐resistant ESCC cells via targeting LASP1.
FIGURE 4.
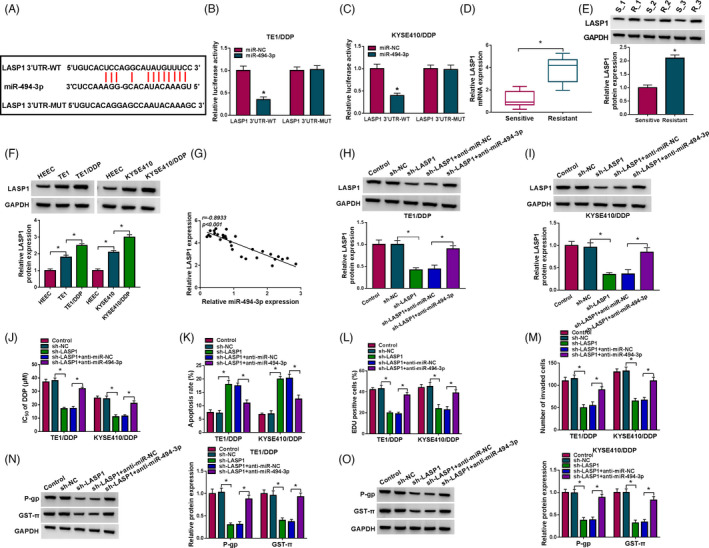
MiR‐494‐3p inhibition ameliorated the influence of LASP1 knockdown on DDP resistance in DDP‐resistant ESCC cells. (A) The potential binding sequences between miR‐494‐3p and LASP1 were predicted by starBase v2.0. (B and C) The interaction between miR‐494‐3p and LASP1 was verified through dual‐luciferase reporter assay. (D and E) The mRNA and protein levels of LASP1 in DDP‐resistant and DDP‐sensitive ESCC tissues were determined through RT‐qPCR assay and Western blot assay, respectively. (F) The protein expression of LASP1 in HEEC, TE1, TE1/DDP, KYSE410, and KYSE410/DDP cells was examined via Western blot assay. (G) The correlation between miR‐494‐3p and LASP1 was analyzed via Spearman's correlation analysis. (H–O) TE1/DDP and KYSE410/DDP cells were transfected with sh‐NC, sh‐LASP1, sh‐LASP1+anti‐miR‐NC, or sh‐LASP1+anti‐miR‐494‐3p. (H and I) The protein level of LASP1 in TE1/DDP and KYSE410/DDP cells was examined through Western blot assay. (J‐M) IC50 of DDP, cell apoptosis, proliferation, and invasion in TE1/DDP and KYSE410/DDP cells was assessed by CCK‐8 assay, flow cytometry analysis, EdU assay, and Transwell assay, respectively. (N and O) The protein levels of P‐gp and GST‐π in TE1/DDP and KYSE410/DDP cells were determined via Western blot assay. *p < 0.05
3.5. Circ_0007142 knockdown suppressed LASP1 expression via sponging miR‐494‐3p in DDP‐resistant ESCC cells
In order to further confirm the relationship of circ_0007142, miR‐494‐3p, and LASP1, sh‐NC, sh‐circ_0007142, sh‐circ_0007142+anti‐miR‐NC, or sh‐circ_0007142+anti‐miR‐494‐3p was transfected into TE1/DDP and KYSE410/DDP cells, and then, the protein level of LASP1 was examined by western blot assay. The results implicated that there was an obvious reduction in LASP1 expression in TE1/DDP and KYSE410/DDP cells transfected with sh‐circ_0007142, whereas this reduction was abolished by anti‐miR‐494‐3p transfection (Figure 5A,B). Taken together, circ_0007142 could positively regulate the expression of LASP1 by sponging miR‐494‐3p in DDP‐resistant ESCC cells.
FIGURE 5.
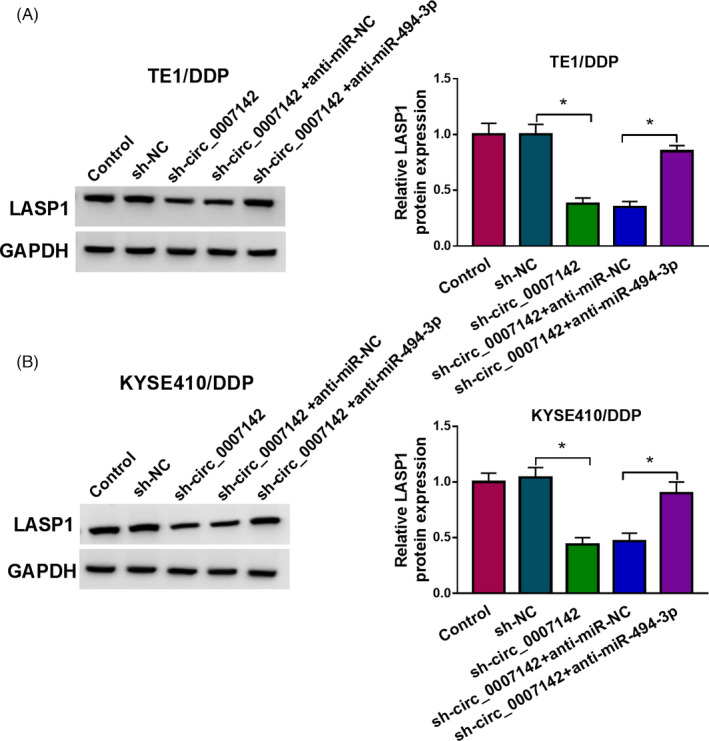
Downregulation of circ_0007142 downregulated LASP1 expression through sponging miR‐494‐3p in DDP‐resistant ESCC cells. (A and B) TE1/DDP and KYSE410/DDP cells were transfected with sh‐NC, sh‐circ_0007142, sh‐circ_0007142+anti‐miR‐NC, or sh‐circ_0007142+anti‐miR‐494‐3p, and the level of LASP1 in TE1/DDP and KYSE410/DDP cells was measured by Western blot assay. *p < 0.05
3.6. Knockdown of circ_0007142 reduced DDP resistance of ESCC in vivo
To further verify the functional role of circ_0007142 in vivo, we established a xenograft tumor mouse model by injecting sh‐NC or sh‐circ_0007142 transfected TE1/DDP cells into the mice. After 8 days, the mice were treated with PBS or 6 mg/kg of DDP every 3 days. As shown in Figure 6A,B, tumor volume and weight were notably decreased by circ_0007142 knockdown or DDP treatment. The data of RT‐qPCR revealed that circ_0007142 was evidently reduced and miR‐494‐3p was evidently elevated in the tumor tissues harvested from sh‐circ_0007142 group compared with that in tumor tissues harvested from sh‐NC group (Figure 6C,D). Moreover, Western blot assay and IHC assay showed that LASP1, Ki67, and MMP9 levels were reduced in the tumor tissues harvested from sh‐circ_0007142 group compared with sh‐NC group (Figure 6E,F). These data suggested that circ_0007142 knockdown enhanced DDP sensitivity in vivo.
FIGURE 6.
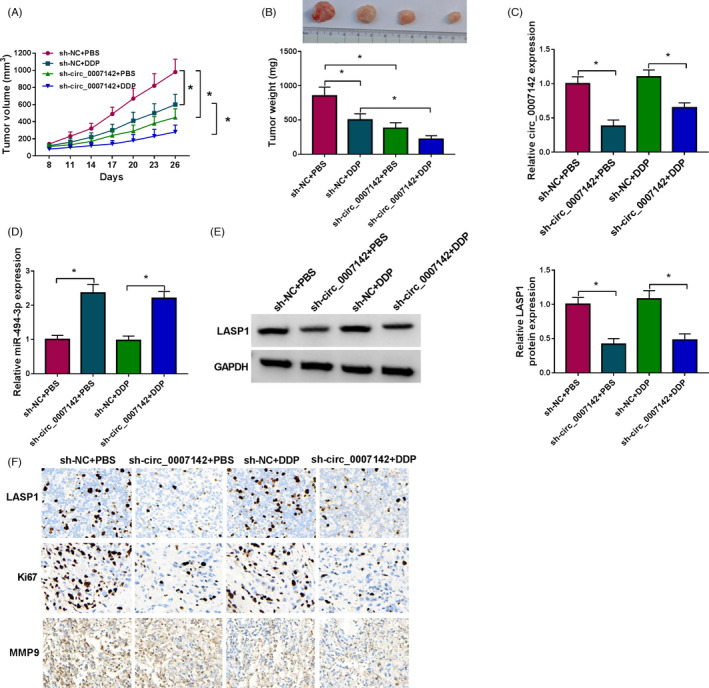
Downregulation of circ_0007142 improved DDP sensitivity of ESCC in vivo. sh‐NC or sh‐circ_0007142 transfected TE1/DDP cells were introduced into the mice, and the mice were treated with PBS or 6 mg/kg of DDP every 3 days after 8 days. (A) Tumor volume was examined every 3 days. (B) Tumor weight was measured after 26 days. (C and D) The levels of circ_0007142 and miR‐494‐3p in the tissues collected from the mice were detected through RT‐qPCR. (E and F) The levels of LASP1, Ki67, and MMP9 in the tissues collected from the mice were measured by Western blot assay and IHC assay. *p < 0.05
4. DISCUSSION
As a new type of ncRNAs, circRNAs have been proved to play vital roles in many pathological processes, including drug resistance. 28 , 29 In the study, we explored the functional role of circ_0007142 in DDP resistance of ESCC. We found that circ_0007142 was increased in DDP‐resistant ESCC and silencing of circ_0007142 enhanced DDP sensitivity in ESCC via miR‐494‐3p/LASP1 axis.
It has been reported that circ_0007142 promoted colorectal cancer cell growth and metastasis and hampered apoptosis by altering miR‐455‐5p/SGK1 axis and miR‐122‐5p/CDC25A axis. 30 , 31 Multiple circRNAs could serve as oncogenes and participate in drug resistance in human cancers. 32 , 33 In this study, we observed that circ_0007142 was markedly improved in DDP‐resistant ESCC tissues and cells. Thus, we speculated that circ_0007142 participated in the development of DDP resistance in ESCC. Functionally, depletion of circ_0007142 enhanced DDP sensitivity, induced cell apoptosis, and repressed cell proliferation and migration in DDP‐resistant ESCC cells. GST‐π is secreted by multidrug resistance‐associated proteins and can reduce the cytotoxicity of chemotherapeutic agents. 34 P‐gp is an energy‐dependent transporter and can bind to drugs and ATP to provide energy for the efflux of chemotherapy drugs and reduce the concentration of drugs. 35 Interestingly, hsa_circ_0007142 silencing decreased the levels of P‐gp and GST‐π in DDP‐resistant ESCC cells. Moreover, hsa_circ_0007142 knockdown markedly suppressed the resistance of ESCC to DDP in vivo. These data suggested that circ_0007142 was closely related to DDP resistance of ESCC.
It has been determined that circRNAs can act as miRNAs sponges to alter genes expression in cancers. 36 In this study, circ_0007142 was identified as a sponge for miR‐494‐3p. MiR‐494‐3p expression was drastically reduced in ESCC cells. Zhang et al. determined that miR‐494‐3p was weakly expressed in ESCC, and the elevated expression of miR‐494‐3p hampered cell growth and metastasis and facilitated cell apoptosis in ESCC by targeting CLPTM1L. 22 Herein, we explored the function of miR‐494‐3p in the modulation of DDP resistance in ESCC for the first time. We observed that miR‐494‐3p was reduced in DDP‐resistant ESCC. Inhibition of miR‐494‐3p effectively restored the impacts of circ_0007142 deficiency on DDP resistance, cell apoptosis, and metastasis in DDP‐resistant ESCC. These findings suggested that miR‐494‐3p improved DDP sensitivity in ESCC.
In addition, we identified that LASP1 was a target gene of miR‐494‐3p. Takeshita et al. reported that LASP1 knockdown repressed ESCC cell progression via acting as a target of miR‐203. 37 , 38 Zhong et al. disclosed that LASP1 was increased in glioblastoma and its downregulation enhanced temozolomide sensitivity in vitro and in vivo. 27 LASP1 was drastically elevated in DDP‐resistant ESCC tissues and cells. Knockdown of LASP1 reduced DDP resistance and cell metastasis and promoted cell apoptosis in DDP‐resistant ESCC cells, whereas these influences were abolished by miR‐494‐3p inhibition.
In summary, we explored the role of circ_0007142 in DDP resistance for the first time. These results uncovered that circ_0007142 knockdown reduced the resistance of DDP‐resistant ESCC cells to DDP via miR‐494‐3p/LASP1 axis. This study is of great significance for the comprehensive understanding of drug resistance mechanism in tumor cells.
CONFLICT OF INTEREST
The authors declare that they have no financial conflicts of interest.
Supporting information
Fig S1
Fig S2
ACKNOWLEDGMENT
None.
Chang N, Ge N, Zhao Y, Yang L, Qin W, Cui Y. Hsa_circ_0007142 contributes to cisplatin resistance in esophageal squamous cell carcinoma via miR‐494‐3p/LASP1 axis. J Clin Lab Anal. 2022;36:e24304. doi: 10.1002/jcla.24304
Funding information
This study was supported by The Fundamental Research Funds for the Central Universities [No. WK9110000071]; Youth Fund of Anhui Cancer Hospital [No. 2018YJQN019、 2020YJQN004、 and 2020YJQN022]
Contributor Information
Wei Qin, Email: qinwei20212021@126.com.
Yayun Cui, Email: fangcui0711@163.com.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Abnet CC, Arnold M, Wei W‐Q. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360‐373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rustgi AK, El‐Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371(26):2499‐2509. [DOI] [PubMed] [Google Scholar]
- 3. Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149(7):1700‐1715. [DOI] [PubMed] [Google Scholar]
- 4. Yuequan J, Shifeng C, Bing Z. Prognostic factors and family history for survival of esophageal squamous cell carcinoma patients after surgery. Ann Thor Surg. 2010;90(3):908‐913. [DOI] [PubMed] [Google Scholar]
- 5. Gillet J‐P, Gottesman MM. Mechanisms of multidrug resistance in cancer. Methods Mol Biol. 2010;596:47‐76. doi: 10.1007/978-1-60761-416-6_4 [DOI] [PubMed] [Google Scholar]
- 6. Darnton S, Archer V, Stocken D, Mulholland P, Casson A, Ferry DR. Preoperative mitomycin, ifosfamide, and cisplatin followed by esophagectomy in squamous cell carcinoma of the esophagus: pathologic complete response induced by chemotherapy leads to long‐term survival. J Clin Oncol. 2003;21(21):4009‐4015. [DOI] [PubMed] [Google Scholar]
- 7. Phatak P, Byrnes KA, Mansour D, et al. Overexpression of miR‐214‐3p in esophageal squamous cancer cells enhances sensitivity to cisplatin by targeting survivin directly and indirectly through CUG‐BP1. Oncogene. 2016;35(16):2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. [DOI] [PubMed] [Google Scholar]
- 9. Chen L‐L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greene J, Baird A‐M, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H‐D, Jiang L‐H, Sun D‐W, Hou J‐C, Ji Z‐L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1‐7. [DOI] [PubMed] [Google Scholar]
- 14. Kun‐Peng Z, Xiao‐Long M, Chun‐Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in Ovarian Cancer via miR‐1270 suppression. Mol Ther Nucleic Acids. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Zhu CL, Sha X, Wang Y, et al. Circular RNA hsa_circ_0007142 Is upregulated and targets miR‐103a‐2‐5p in colorectal cancer. J Oncol. 2019;2019:9836819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma D, Liu H, Qin Y, et al. Circ_0007142/miR‐186/FOXK1 axis promoted lung adenocarcinoma progression. Am J Transl Res. 2020;12(8):4728‐4738. [PMC free article] [PubMed] [Google Scholar]
- 19. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281‐297. [DOI] [PubMed] [Google Scholar]
- 20. Imanaka Y, Tsuchiya S, Sato F, Shimada Y, Shimizu K, Tsujimoto G. MicroRNA‐141 confers resistance to cisplatin‐induced apoptosis by targeting YAP1 in human esophageal squamous cell carcinoma. J Hum Genet. 2011;56(4):270. [DOI] [PubMed] [Google Scholar]
- 21. Liu A, Zhu J, Wu G, et al. Antagonizing miR‐455‐3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang R, Chen X, Zhang S, et al. Upregulation of miR‐494 inhibits cell growth and invasion and induces cell apoptosis by targeting cleft lip and palate transmembrane 1‐like in esophageal squamous cell carcinoma. Dig Dis Sci. 2015;60(5):1247‐1255. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Q, Pan X, You D. Overexpression of long non‐coding RNA SBF2‐AS1 promotes cell progression in esophageal squamous cell carcinoma (ESCC) by repressing miR‐494 to up‐regulate PFN2 expression. Biol Open. 2020. doi: 10.1242/bio.048793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chew CS, Chen X, Parente JA, Tarrer S, Okamoto C, Qin H‐Y. Lasp‐1 binds to non‐muscle F‐actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J Cell Sci. 2002;115(24):4787‐4799. [DOI] [PubMed] [Google Scholar]
- 25. He B, Yin B, Wang B, et al. Overexpression of LASP1 is associated with proliferation, migration and invasion in esophageal squamous cell carcinoma. Oncol Rep. 2013;29(3):1115‐1123. [DOI] [PubMed] [Google Scholar]
- 26. Chen K, Quan J, Yang J, Chen Z. The potential markers of endocrine resistance among HR+/HER2+ breast cancer patients. Clin Transl Oncol. 2020;22(4):576‐584. [DOI] [PubMed] [Google Scholar]
- 27. Zhong C, Chen Y, Tao B, et al. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer. 2018;18(1):722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829‐1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472. [PMC free article] [PubMed] [Google Scholar]
- 30. Wen T, Wu H, Zhang L, et al. Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR‐455‐5p/SGK1 axis in colorectal cancer. Anticancer Drugs. 2021;32(1):22‐33. [DOI] [PubMed] [Google Scholar]
- 31. Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR‐122‐5p. OncoTargets Ther. 2020;13:3689‐3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shang J, Chen W‐M, Wang Z‐H, Wei T‐N, Chen Z‐Z, Wu W‐B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR‐153‐5p/miR‐183‐5p–XIAP axis. Exp Hematol. 2019;70:42‐54.e3. [DOI] [PubMed] [Google Scholar]
- 33. Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 sponges hsa‐miR‐335 to promote the gemcitabine‐resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Comm. 2019;519(1):172‐178. [DOI] [PubMed] [Google Scholar]
- 34. Kulaksiz‐Erkmen G, Dalmizrak O, Dincsoy‐Tuna G, Dogan A, Ogus IH, Ozer N. Amitriptyline may have a supportive role in cancer treatment by inhibiting glutathione S‐transferase pi (GST‐pi) and alpha (GST‐alpha). J Enzyme Inhib Med Chem. 2013;28(1):131‐136. [DOI] [PubMed] [Google Scholar]
- 35. Chen YL, Yang TY, Chen KC, Wu CL, Hsu SL, Hsueh CM. Hypoxia can impair doxorubicin resistance of non‐small cell lung cancer cells by inhibiting MRP1 and P‐gp expression and boosting the chemosensitizing effects of MRP1 and P‐gp blockers. Cell Oncol (Dordr). 2016;39(5):411‐433. [DOI] [PubMed] [Google Scholar]
- 36. Hou L‐D, Zhang J. Circular RNAs: An Emerging Type of RNA in Cancer. SAGE Publications Sage UK; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li P‐D, Hu J‐L, Ma C, et al. Upregulation of the long non‐coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR‐203 and LASP1. Oncotarget. 2017;8(21):34164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeshita N, Mori M, Kano M, et al. miR‐203 inhibits the migration and invasion of esophageal squamous cell carcinoma by regulating LASP1. Int J Oncol. 2012;41(5):1653‐1661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Data Availability Statement
Not applicable.


