Abstract
Lactobacillus rhamnosus GG is one of the most thoroughly studied probiotic strains. Its advantages in the treatment of gastrointestinal disorders are well documented. The aim of the present study was to demonstrate with colonic biopsies the attachment of strain GG to human intestinal mucosae and the persistence of the attachment after discontinuation of GG administration. A whey drink fermented with strain GG was fed to human volunteers for 12 days. Fecal samples were collected before, during, and after consumption. L. rhamnosus GG-like colonies were detected in both fecal and colonic biopsy samples. Strain GG was identified by its characteristic colony morphology, a lactose fermentation test, and PCR. This study showed that strain GG was able to attach in vivo to colonic mucosae and, although the attachment was temporary, to remain for more than a week after discontinuation of GG administration. The results demonstrate that the study of fecal samples alone is not sufficient in evaluating colonization by a probiotic strain.
Oral consumption of health-promoting lactic acid bacteria or probiotics has been associated with the prevention, alleviation, or cure of diverse intestinal disorders such as lactose intolerance, viral and bacterial diarrhea, adverse effects of abdominal radiotherapy, constipation, inflammatory bowel disease, and food allergy (3, 5, 11). Much of the early evidence on the actual health effects of probiotics was anecdotal, but during the last few years data based on rigorous clinical studies indicating real health-promoting properties of certain well-characterized strains have started to accumulate (8).
Adhesion to the intestinal epithelium is one of the selection criteria for new probiotic strains (6). The adhesion properties have generally been deduced from in vitro experiments with intestinal cell lines, although, for example, rectal mucosal samples have been successfully used to demonstrate intestinal colonization by lactobacillar strains (7).
Lactobacillus rhamnosus GG (ATCC 53103) (previously known as Lactobacillus casei GG) is one of the most thoroughly studied probiotics (11). The reviewed beneficial effects (9, 12, 13) include prevention of antibiotic-associated diarrhea, treatment and prevention of rotavirus diarrhea, treatment of relapsing Clostridium difficile diarrhea, prevention of acute diarrhea, and enhancement of intestinal immunity. The ability of strain GG to survive passage through the gastrointestinal tract has been demonstrated in both adults and children by the use of fecal samples (4, 10, 14). Recently, adhesion of the strain to human colonic mucosae has been demonstrated with colonic biopsy samples (1). The aims of the present study were to confirm with colonic biopsy samples the attachment of L. rhamnosus GG to human intestinal mucosae and to evaluate the persistence of this attachment after discontinuation of strain GG administration.
Volunteers and L. rhamnosus GG administration.
The three experimental groups in this study each consisted of six to eight adults undergoing routine diagnostic colonoscopy. The experimental protocol was designed to fit within the normal diagnostic schedule of the volunteers. Informed consent from all subjects was obtained before the experiment. With the exception of various gastric symptoms, all subjects considered themselves healthy. No antibiotic therapy was applied either during the trial or during the month immediately preceding the administration period. The volunteers had no immediate past history of consuming L. rhamnosus GG-containing products. For this study they took 100 ml of a commercial drink based on lactose-hydrolyzed whey fermented with strain GG and flavored with a peach-apricot concentrate (Gefilus; Valio Ltd., Kouvola Dairy, Kouvola, Finland) twice daily for 12 days. The daily dose of strain GG was approximately 6 × 1010 CFU. After administration of strain GG, the volunteers were divided into three groups (see Fig. 1): those having undergone colonoscopy immediately after the 12-day GG administration period (one male, five females, 34 to 78 years old), those having undergone colonoscopy 1 week after stopping GG administration (five males, three females, 42 to 68 years old), and those having undergone colonoscopy 2 weeks after stopping GG administration (four males, three females, 27 to 73 years old).
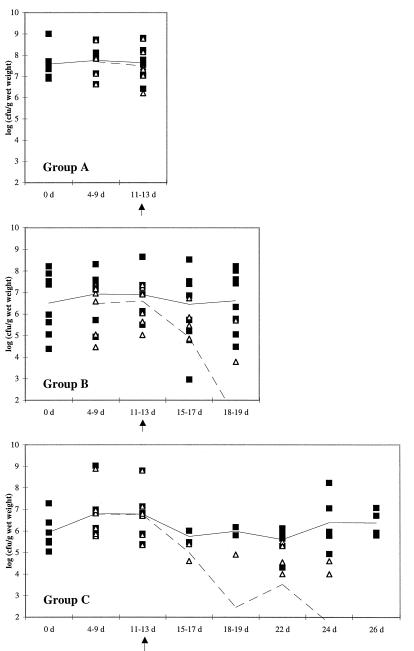
FIG. 1.
Fecal counts of lactic acid bacteria (■) and L. rhamnosus GG-like colonies (▵). The solid line shows the mean counts of lactic acid bacteria, and the dashed line shows the mean counts of strain GG. In this context, lactic acid bacteria are defined as colonies growing on MRS agar without further taxonomic characterization, with the exception of GG-like colonies. The end of L. rhamnosus GG administration is marked by a vertical arrow below the horizontal axis.
Colonoscopy and biopsies.
In preparation for colonoscopy, evacuation of the colon was induced by three doses of a laxative (Pico-salax; Malmö, Sweden) consumed within 36 h. The instrument used for colonoscopy and sampling of biopsies was a Pentax ES-3801L (Tokyo, Japan). The diameter of the biopsies was approximately 3 mm. Three parallel biopsies were taken from the descending colon. This location was selected on the basis of previous results (1) showing preferential adhesion of L. rhamnosus GG to this part of the large intestine.
Cultivation of L. rhamnosus GG from fecal and biopsy samples.
Fecal samples were collected as indicated in Fig. 1. The samples were immediately stored at about −20°C in the home freezers of the patients (for up to 3 weeks) and afterwards at −20°C in the laboratory until analysis (within 9 weeks after collection). Biopsy samples from the descending colon were immediately transferred into a thioglycolate medium (Difco, Detroit, Mich.) and stored at 4°C until analysis (within a day). The samples were homogenized for 30 s in a stomacher (Stomacher 400; Seward, London, United Kingdom) before dilution and cultivation on MRS agar (Merck, Darmstadt, Germany). The plates were incubated under anaerobic conditions (Anaerocult A; Merck) for 3 days at 37°C.
L. rhamnosus GG forms large, creamy, white colonies on MRS agar that are generally distinct from other lactic acid bacterial colonies. Strain GG is further distinguished from most other lactic acid bacteria by its inability to efficiently ferment lactose (4), which was tested by selecting one to four typical GG-like colonies from each fecal and biopsy sample and further cultivating them for 48 h in lactose MRS broth with indicator dye (bromocresol purple, 0.04 g/liter). One or two lactose-negative isolates per sample were further confirmed as L. rhamnosus by species-specific PCR.
PCR confirmation of L. rhamnosus isolates.
Bacterial cells were collected from 1 ml of an overnight culture by centrifugation, washed with 50 mM Tris buffer (pH 8.0), and suspended in 100 μl of 50 mM Tris-EDTA buffer (pH 8.0). Lysozyme (100 μl, 20 mg/ml) (Sigma, St. Louis, Mo.) and mutanolysin (8 μl, 0.5 mg/ml) (Sigma) were added, and the mixture was incubated at 37°C for 1 h. The cells were lysed by addition of 20 μl of 20% sodium dodecyl sulfate and 12 μl of proteinase K solution (14.6 mg/ml) (Boehringer, Mannheim, Germany) followed by a 10-min incubation at 65°C. The volume was adjusted to 500 μl with sterile ultrapure water. Deproteinization was done by extraction with 1 volume of Tris-saturated phenol (Amresco, Solon, Ohio). The water phase was extracted once more with phenol-chloroform (1:1). Finally, DNA was precipitated by adding 0.1 volume of 3 M sodium acetate to the water phase followed by 2 volumes of 94% ethanol and incubating the mixture in an ice bath for 30 min. The DNA was collected by centrifugation at 13,000 rpm for 15 min, and the pellet was washed with 70% ethanol and finally dissolved in 20 μl of sterile ultrapure water.
The universal 16S rRNA gene forward and reverse primers (5′ to 3′) were AGAGTTTGATCCTGGCTCAGG and ACGGCAACCTTGTTACGAGTT, respectively. The species-specific primers (CTTGCATCTTGATTTAATTTTG, forward; CCGTCAATTCCTTTGAGTTT, reverse) were designed on the basis of the L. rhamnosus (previously L. casei subsp. rhamnosus) 16S ribosomal DNA sequence (GenBank accession no. M58815) specifying the 863-bp fragment between positions 91 and 953 in the gene. The primers were made with a PCR Mate EP 391 DNA synthesizer, model 391 (Applied Biosystems, Foster City, Calif.), according to the manufacturer’s instructions.
Taq DNA polymerase and PCR buffer (final concentrations of 10 mM Tris-HCl, 1.5 mM MgCl2 and 50 mM KCl [pH 8.3]) were obtained from Boehringer, and the deoxynucleotides were purchased from Sigma. The primer concentrations were 0.5 μM with specific primers and 0.25 μM with universal primers, and those of the deoxynucleotides were 200 μM. The amount of template was 1 μl of the DNA extracted from fecal isolates or 1 μl of an appropriate dilution of the DNA extracted from pure cultures. The amount of Taq DNA polymerase used was 2.0 U in a total reaction volume of 100 μl. A Gene Amp PCR System 9600 apparatus (Perkin-Elmer Cetus, Norwalk, Conn.) was used for PCR cycling. Initial denaturation was carried out at 94°C for 5 min followed by a touch-down thermocycling program with 30 amplification cycles (annealing for 30 s at 62°C in cycles 1 to 10, at 60°C in cycles 11 to 20, and at 58°C in cycles 21 to 30, with extension for 1 min at 72°C and denaturation for 40 s at 94°C) and a final extension for 10 min at 72°C. Reaction mixtures were subsequently cooled to 4°C. In the PCR with universal primers, the annealing temperature was 55°C.
The specificity of the L. rhamnosus primers was confirmed with 8 different L. rhamnosus strains and 17 other lactobacillar species or strains as references (data not shown). To exclude the possibility of DNA extraction failure or the presence of inhibitors in samples, reference strains were subjected to PCR with universal primers prior to PCR with specific primers.
L. rhamnosus GG-like colonies in biopsy and fecal samples of different test groups.
The counts of total fecal lactic acid bacteria and strain GG-like colonies in the three experimental groups are presented in Fig. 1. The results of PCR (Fig. 2) were in good agreement (88%) with screening based on colony morphology and the lactose fermentation test, confirming the general reliability of identification of the strain. The counts of strain GG-like colonies decreased as a function of time after discontinuation of GG administration. Strain GG was detected in biopsy specimens and final fecal samples of all volunteers in group A (Table 1). The counts of lactic acid bacteria in biopsy samples were 3 × 102 to 4 × 104 CFU per biopsy (mean, 6 × 103 CFU per biopsy). The corresponding counts of strain GG-like colonies were 6 × 101 to 4 × 104 CFU per biopsy.
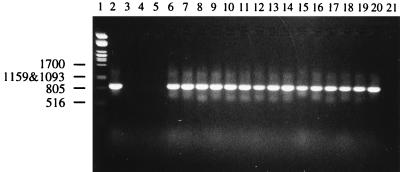
FIG. 2.
Detection of L. rhamnosus by PCR coupled with gel electrophoresis. Lanes: 1, molecular weight marker; 2 through 19, strain GG-like findings from fecal samples; 20, positive control (L. rhamnosus GG VTT E-96666); 21, control reaction with no template DNA.
TABLE 1.
Recovery of L. rhamnosus GG from colonic biopsy samples and final fecal samplesa
| Group | Day of colonoscopy | No. of subjects with the indicated characteristics/total no. of subjects
|
|||
|---|---|---|---|---|---|
| Both biopsy and feces positive | Biopsy positive, feces negative | Biopsy negative, feces positive | Both biopsy and feces negative | ||
| A | 14 | 6/6 | 0/6 | 0/6 | 0/6 |
| B | 21 | 2/8 | 5/8 | 0/8 | 1/8 |
| C | 28 | 0/7 | 2/7 | 0/7 | 5/7 |
Final fecal samples were obtained a day before evacuation and 2 days before colonoscopy.
In group B, L. rhamnosus GG-like colonies were detected in seven of eight biopsy samples (Tables 1 and 2), with counts varying between 2 × 103 and 1 × 106 CFU per biopsy. The total counts of lactic acid bacteria were 3 × 103 to 2 × 106 CFU per biopsy (mean, 1 × 105 CFU per biopsy). Only two of the eight subjects, however, had strain GG-like colonies at detectable levels in the final fecal samples; these counts were 6 × 103 and 5 × 105 CFU/g (wet weight). The individual counts of GG-like colonies in the biopsies and final fecal samples of the group B volunteers are presented in Table 2.
TABLE 2.
Counts of L. rhamnosus GG-like colonies in biopsy specimens and final fecal samples in group B
| Volunteer | Strain GG-like colony count in:
|
|
|---|---|---|
| Biopsy (CFU/biopsy)a | Final fecal sample (CFU/g [wet weight])b | |
| 1 | —c | — |
| 2 | 1.0 × 106 | — |
| 3 | 7.8 × 104 | — |
| 4 | 4.6 × 105 | 6.0 × 103 |
| 5 | 2.5 × 103 | — |
| 6 | 2.2 × 103 | — |
| 7 | 6.3 × 103 | — |
| 8 | 1.0 × 104 | 6.0 × 105 |
Detection level, 102 CFU per biopsy. Biopsies were performed on day 21.
Detection level, 103 CFU/g [wet weight]. Final fecal samples were obtained on day 18 or 19.
—, below detection level.
None of the seven subjects in group C had strain GG-like colonies in the final fecal samples (Table 1). However, GG-like colonies were detected in the biopsy samples of two of the seven volunteers at counts of 1 × 102 and 1 × 104 CFU per biopsy. The total counts of lactic acid bacteria in biopsies of group C were 6 × 102 to 2 × 105 CFU per biopsy (mean, 2 × 104 CFU per biopsy).
L. rhamnosus GG has been shown to adhere in vitro to the Caco-2 intestinal cell line (2) and in vivo to human colonic mucosae (1). The finding reported here that strain GG can persist in colonic mucosae even after its disappearance from fecal samples may have significance in the elucidation of the colonization mechanisms of probiotic strains. The fact that the strain GG counts observed in the biopsy samples from group B are rather similar to those obtained from group A is particularly interesting, since it indicates that GG can survive in high numbers in colonic mucosae despite its rapid turnover. This finding suggests that L. rhamnosus GG can multiply on the colonic surface at a rate that partially counterbalances its shedding. However, as can be seen from the results from group C, even an adherent strain can be gradually diluted out of the colon unless it is replenished with a fresh inoculum. The high counts of endogenous lactic acid bacteria associated with colonic biopsies mean that the probiotic strain faces strong competition when establishing itself. This may well be one of the reasons that permanent colonization by a probiotic strain seldom, if ever, occurs.
The present study confirms that L. rhamnosus GG is able to attach in vivo to colonic mucosae and to persist there for prolonged periods after discontinuation of administration of strain GG. In accounting for the findings reported here, the study of fecal samples alone may underestimate colonization by probiotic strains.
Acknowledgments
This work was conducted as a part of the FAIR PROBDEMO CT96-1028 project. Support from the Ministry of Agriculture and Forestry of Finland is gratefully acknowledged.
We thank Sherwood Gorbach for commenting on the manuscript and Helena Toivanen, Marja-Liisa Jalovaara, Anu Miettinen, Marja-Leena Kekäläinen, and Saara Tirkkonen for technical assistance.
REFERENCES
- 1.Alander M, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett Appl Microbiol. 1997;24:361–364. doi: 10.1046/j.1472-765x.1997.00140.x. [DOI] [PubMed] [Google Scholar]
- 2.Elo S, Saxelin M, Salminen S. Attachment of Lactobacillus casei strain GG to human colon carcinoma cell line Caco-2: comparison with other dairy strains. Lett Appl Microbiol. 1991;13:154–156. [Google Scholar]
- 3.Gilliland S E. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol Rev. 1990;87:175–188. doi: 10.1111/j.1574-6968.1990.tb04887.x. [DOI] [PubMed] [Google Scholar]
- 4.Goldin B R, Gorbach S L, Saxelin M, Barakat S, Gualtieri L, Salminen S. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci. 1992;37:121–128. doi: 10.1007/BF01308354. [DOI] [PubMed] [Google Scholar]
- 5.Hammes W P, Tichaczek P S. The potential of lactic acid bacteria for the production of safe and wholesome food. Z Lebensm-Unters -Forsch. 1994;198:193–201. doi: 10.1007/BF01192595. [DOI] [PubMed] [Google Scholar]
- 6.Huis In’t Veld J H J, Shortt C. Selection criteria for probiotic microorganisms. R Soc Med Int Congr Symp Ser. 1996;219:27–36. [Google Scholar]
- 7.Johansson M-L, Molin G, Jeppsson B, Nobaek S, Ahrné S, Bengmark S. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol. 1993;59:15–20. doi: 10.1128/aem.59.1.15-20.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y K, Salminen S. The coming age of probiotics. Trends Food Sci Technol. 1995;6:241–245. [Google Scholar]
- 9.Majamaa H, Isolauri E. Probiotics: a novel approach in the management of food allergy. J Allergy Clin Immunol. 1997;99:179–185. doi: 10.1016/s0091-6749(97)70093-9. [DOI] [PubMed] [Google Scholar]
- 10.Millar M R, Bacon C, Smith S L, Walker V, Hall M A. Enteral feeding of premature infants with Lactobacillus GG. Arch Dis Child. 1993;69:483–487. doi: 10.1136/adc.69.5_spec_no.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salminen S, Deighton M, Benno Y, Gorbach S. Lactic acid bacteria in health and disease. In: Salminen S, von Wright A, editors. Lactic acid bacteria. Microbiology and functional aspects. New York, N.Y: Marcel Dekker; 1998. pp. 211–253. [Google Scholar]
- 12.Salminen S, Isolauri E, Salminen E. Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie Leeuwenhoek J Microbiol. 1996;70:347–358. doi: 10.1007/BF00395941. [DOI] [PubMed] [Google Scholar]
- 13.Saxelin M. Lactobacillus GG—a human probiotic strain with thorough clinical documentation. Food Rev Int. 1997;13:293–313. [Google Scholar]
- 14.Saxelin M, Ahokas M, Salminen S. Dose response on the faecal colonisation of Lactobacillus strain GG administered in two different formulations. Microb Ecol Health Dis. 1993;6:119–122. [Google Scholar]