Abstract
Endothelial cells have a crucial role in nervous system function, and mounting evidence points to endothelial impairment as a major contributor to a wide range of neurological diseases. However, tools to genetically interrogate these cells in vivo remain limited. Here, we describe AAV-BI30, a capsid that specifically and efficiently transduces endothelial cells throughout the central nervous system. At relatively low systemic doses, this vector transduces the majority of arterial, capillary, and venous endothelial cells in the brain, retina, and spinal cord vasculature of adult C57BL/6 mice. Furthermore, we show that AAV-BI30 robustly transduces endothelial cells in multiple mouse strains and rats in vivo and human brain microvascular endothelial cells in vitro. Finally, we demonstrate AAV-BI30’s capacity to achieve efficient and endothelial-specific Cre-mediated gene manipulation in the central nervous system. This combination of attributes makes AAV-BI30 uniquely well-suited to address outstanding research questions in neurovascular biology and aid the development of therapeutics to remediate endothelial dysfunction in disease.
INTRODUCTION
Recent strides in AAV development have produced engineered capsids capable of transducing well-defined cellular populations in the central nervous system (CNS) far more efficiently than their naturally occurring counterparts 1–7. Leveraged as a rapid and flexible in vivo gene transfer platform, these vectors are poised to act as transformative catalysts for research when used in conjunction with – or as a substitute for – existing mouse genetics tools. However, capsid development has predominantly focused on vectors designed to transduce neurons or astrocytes. By comparison, relatively few vectors have been described that specifically target other cellular populations within the CNS, despite emerging appreciation that a vast repertoire of non-neuronal cell types is critical for nervous system function.
Among these, CNS endothelial cells – specialized cells that line the luminal face of blood vessels – have been shown to orchestrate a number of key physiological processes. Moreover, their dysfunction is increasingly appreciated to contribute to a wide range of neurodegenerative and neurological diseases 8,9. While endothelial cells are often regarded as a relatively homogenous entity, recent work has highlighted a striking degree of molecular and functional specialization across the cerebrovascular arterio-venous axis 10. For example, arterial endothelial cells play a critical role in dynamically coupling blood flow with neural activity to meet local energetic demand 11–13, capillary endothelial cells actively suppress transcytotic trafficking to maintain blood-brain barrier integrity 14–16, and venous endothelial cells appear to act as essential intermediaries in neuroimmune crosstalk 9,17,18. However, a mismatch between the expanding functions ascribed to endothelial cells and the relatively limited tools available to study them in vivo is a major obstacle to research progress. A highly efficient, endothelial-specific vector with broad tropism encompassing arterial, capillary, and venous endothelial cells would be ideally suited to accelerate neurovascular research. Similarly, the ability to effectively transduce spinal cord and retinal vasculature – widely studied systems in the field of neurovascular biology – in addition to brain vasculature would dramatically expand the applications of a CNS-directed, endothelial-specific vector.
To date, only two viral vectors with the ability to efficiently transduce cerebrovascular endothelial cells have been described – AAV-PHP.V1 6 and AAV-BR1 2 – both of which have caveats that restrict their use. Within the brain, AAV-PHP.V1 does not selectively transduce endothelial cells; it transduces astrocytes with similarly high efficiency. Because astrocytes are intimately associated with the brain vasculature, this lack of specificity limits the vector’s utility. AAV-BR1, by contrast, transduces brain microvascular endothelial cells with high specificity and has been successfully leveraged by a number of groups since its initial discovery 19–26. However, while it is well-established that AAV-BR1 efficiently transduces capillary endothelial cells, it is unclear whether the vector robustly targets the endothelium of arteries and veins. A recent study found that AAV-BR1’s efficacy is diminished in these larger vessel segments 20, suggesting that it is not well-suited to address rapidly emerging interest in the specialized functions of arterial and venous endothelial cells. Moreover, the vector’s ability to effectively target spinal cord and retinal vasculature is ambiguous. Initial characterization of AAV-BR1 revealed that its transduction of the spinal cord endothelium was significantly less efficient than that observed in the brain 2, and while there is some evidence that the vector transduces endothelial cells of the retinal vasculature, its performance in this context remains poorly characterized 23
Here, we describe a viral capsid that meets the need for a specific, high-efficiency vector to target endothelial cells throughout the entire CNS: AAV-BI30, an engineered variant of AAV9. At relatively low systemic doses in adult mice, this capsid transduces the majority of endothelial cells across the arterio-venous axis in brain, retina, and spinal cord vasculature. Furthermore, we demonstrate that the capsid’s transduction profile extends across species: we observed robust endothelial transduction in C57BL/6 and BALB/cJ mouse strains, in rats, and in human brain microvascular endothelial cells. Finally, we show that the capsid can be utilized for efficient endothelial cell-specific Cre recombination and gene knockout in the brain vasculature. Taken together, these attributes make AAV-BI30 exceptionally well-suited to accelerate our understanding of neurovascular interactions in normal physiology and pioneer therapies to address their dysfunction in disease.
RESULTS
AAV-BI30 transduces brain endothelial cells in mice and rats in vivo.
To develop capsids with improved transduction of CNS endothelial cells, we generated an AAV9 capsid library and selected for capsids that more efficiently transduced human and mouse endothelial cells. The library comprised AAV9 variants modified with a randomized 7-mer insertion between amino acids 588 and 589 (AAV9 VP1 position). The library was built within a recombinant AAV backbone, AAV9-CMV-Express, which expresses the capsid gene in transduced cells. By sequencing capsid mRNA this approach allows for the selective recovery of functional capsids, eliminating AAV variants that traffic to the tissue or organ of interest but fail to achieve transgene expression. Similar RNA-based selection methods have recently been used to identify capsids with enhanced blood-brain barrier penetrance 7 and muscle transduction 27.
Using AAV9-CMV-Express, we selected for capsids expressed in human and mouse cells in vitro and in the brains of mice in vivo. After two rounds of selection, we identified a variant with the 7-mer amino acid sequence NNSTRGG that was enriched in the expressed capsid pool across five assays: in immortalized human cerebral microvascular endothelial cell (hCMEC/D3) transduction, in human and mouse brain microvascular endothelial cell (BMVEC) transduction, and in C57BL/6J and BALB/cJ mouse brain transduction in vivo (Supplementary Figure 1). In contrast, AAV-PHP.eB – a previously described capsid with enhanced CNS transduction 4 selective to a subset of mice including C57BL/6J’s 28,29 – was only enriched in C57BL/6J brain and BMVECs derived from this strain.
To individually assess the transduction characteristics of AAV-BI30, we used the capsid to package a single-stranded recombinant AAV2 reporter genome. AAV-BI30 transduced BMVECs from mouse (282- to 2261-fold) and human (72- to 96-fold) more efficiently than AAV9 (Figure 1A). In addition, AAV-BI30 transduced hCMEC/D3 (22.7 ± 1.4-fold; mean ± SD) more efficiently than AAV9, an increase that was observed across a wide range of doses (Extended Data Figure 1 & Supplementary Figure 2). Fitting with our library enrichment data, this cross-species transduction enhancement differentiates AAV-BI30 from AAV-PHP.eB, which exhibited an enhanced transduction phenotype restricted to mouse (Figure 1A), and AAV-BR1, which was not found to transduce hCMEC/D3 cells more efficiently than its parental vector AAV2 2.
Figure 1. AAV-BI30 enables efficient transduction of brain endothelial cells across species.

(a) Quantification of transduction by AAV-BI30 and AAV-PHP.eB relative to AAV9 in several independent batches of mouse & human BMVECs and hCMEC/D3s assessed by luciferase activity (relative light units). (b) Representative image of AAV-BI30 transduction in a sagittal section of adult C57BL/6 brain cropped to show cortex and hippocampus. Samples were collected 10 days after administration of 1x1011 vg/animal of AAV-BI30 carrying a CAG-NLS-GFP-WPRE genome (c) High-magnification images of mouse liver harvested 10 days following intravenous injection of 3x1011 vg/animal of AAV-BI30. Note abnormal nuclear morphology in hepatocytes expressing the highest levels of GFP. (d) AAV-BI30 carrying a CAG-NLS-GFP-WPRE or CAG-NLS-GFP-miR122-WPRE genome was intravenously injected into adult C57BL/6 mice at 1x1011 vg/animal. Left: low-magnification images of liver collected 10 days after injection, demonstrating effective miR122-mediated suppression of transgene expression in hepatocytes. Right: Starting immediately after viral injection, mice were weighed every 24 hours for 20 consecutive days (mean ± s.e.m.; n = 3 animals per group). (e) AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered at 1x1011 vg/animal (BALB/cJ) or 1.42x1013 vg/kg (rat). Transduction was assessed after three (BALB/cJ) or four (rat) weeks. Representative images show AAV-BI30 transduction in the cortical microvasculature of each animal. (f) AAV-BI30 or AAV-BR1 carrying a CAG-NLS-GFP-miR122-WPRE construct were intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal. Transduction was assessed after three weeks. Representative confocal images of viral transduction in cerebral cortex; note the high cell-type specificity of ERG immunostaining. (g) Quantification of endothelial transduction measured as the fraction of ERG+ cells expressing GFP in whole sagittal sections of brain (t4 = 3.631; P = 0.0221). For quantification: n = 3 animals per group, mean ± s.e.m. ; unpaired, two-tailed t-test (* P < 0.05). Scale bars are as follows: 500μm in (d); 250μm in (b); 100μm in (e) and (f); 25μm in (c).
Next, to evaluate AAV-BI30’s performance in vivo we used the capsid to package a genome expressing a nuclear localization signal (NLS) tagged GFP from the ubiquitous CAG promoter (AAV-BI30:CAG-NLS-GFP). We then intravenously administered the AAV at 1x1011 vg/mouse in C57BL/6 mice and assessed transduction after 10 days. Encouragingly, AAV-BI30 transduced endothelial cells throughout the brain with remarkable efficiency and specificity at this dose (Figure 1B, Extended Data Figure 2 & Supplementary Figure 3).
However, approximately one week post-administration we observed unexpected dose-dependent toxicity at doses as low as 1x1011 vg/mouse. This adverse response manifested in weight loss, lethargic behavior, and ultimately mortality at the highest dose tested, 1x1012 vg/mouse. Necropsy revealed strong transduction of liver hepatocytes accompanied by abnormal nuclear morphology (Figure 1C). To determine whether toxic overexpression of the NLS-transgene in hepatocytes contributed to systemic toxicity, we incorporated three repeats of the hepatocyte-specific miR-122 target sequence 30 into the 3’UTR of our viral construct - a strategy successfully employed by a number of groups to selectively degrade transgene mRNA in hepatocytes 31–33
This modification efficiently suppressed hepatocyte expression of the NLS-GFP transgene and prevented transient weight loss (Figure 1D) without compromising AAV-BI30’s transduction of the CNS vasculature, results consistent with virtually undetectable expression of Mir122a in brain endothelial cells 10. A higher 5x1011 vg dose of AAV-BI30 carrying the modified NLS-GFP-miR122-WPRE construct similarly produced no discernable weight loss (Supplementary Figure 4) illustrating the microRNA binding element’s ability to effectively detarget transgene expression from the liver across the experimental working range of the vector. Further, measuring less than 80 bp, the element did not constrain AAV-BI30’s functional packaging capacity. A survey of peripheral tissues following incorporation of the miR-122 repeat element revealed transduction of several non-CNS endothelial populations in addition to hepatocytes, including endothelial cells in the lung microvasculature, aorta, and interlobular vessels of the kidney (Extended Data Figure 3). That said, the vector’s transduction profile was strongly biased towards the CNS.
C57BL/6-restricted tropism has frustrated past efforts to deploy engineered AAV vectors in genetically intractable organisms 34,35. To test whether AAV-BI30’s applications were similarly constrained, we evaluated the capsid’s performance in a second mouse strain (BALB/cJ) and a distinct mammalian species (rat). Consistent with AAV-BI30’s cross-species transduction observed in vitro, the capsid achieved robust endothelial transduction in the BALB/cJ and rat brain following systemic administration (Figure 1E & Extended Data Figure 4).
AAV-BI30 transduces endothelial cells across the entire arterio-venous axis.
We subsequently sought to assess AAV-BI30’s transduction efficiency in brain endothelial cells. Previous studies have quantified the efficiency of endothelial-targeted vectors by measuring the co-localization of a viral transgene with endothelial-specific markers such as CD31 or GLUT1 2,6. This strategy relies on an implicit assumption that the size and morphology of individual endothelial cells is roughly consistent throughout the CNS – a generalization that does not hold true across the arterio-venous axis 36. To improve upon existing approaches, we developed an automated workflow to estimate endothelial transduction efficiency by measuring co-localization of NLS-GFP with ERG, an endothelial-specific transcription factor 10,37. Because ERG expression is sharply restricted to the nucleus, this strategy enabled fast and reliable identification of individual endothelial cells throughout the brain microvasculature (Figure 1F).
Observing that AAV-BI30’s tropism resembled AAV-BR1’s despite its highly divergent sequence, we used this approach to directly compare the vectors and identify capsid-specific properties. Quantifying transduction across entire sagittal brain sections, we found that AAV-BI30 transduced 84 ± 4% (mean ± s.e.m.) of brain endothelial cells at 1x1011 vg/mouse. By comparison, AAV-BR1 transduced 66 ± 2% of this population at the same dose, consistent with previous reports 2,19,20,24 (Figure 1G). AAV-BI30’s efficacy showed no appreciable region-to-region variation throughout the brain; cortex, hippocampus, thalamus, and cerebellum all exhibited > 80% endothelial cell transduction (Extended Data Figure 5). Further, the vector was highly endothelial-specific in this dose regime – isolated instances of neuronal or astrocytic transduction were rare (Extended Data Figure 6). Notably, while both AAV-BI30 and AAV-BR1 were predominantly endothelial-directed, AAV-BI30 transduced significantly fewer non-endothelial cells than AAV-BR1 (which is known to sporadically transduce neurons and astrocytes 2,38).
Next, we measured AAV-BI30’s transduction efficiency in endothelial cells of larger arteries and veins. While surveying sagittal or coronal sections is well-suited to the gauge the overall efficiency of AAV transduction across the brain microvasculature, it provides limited information about a vector’s ability to transduce different segments of the arterio-venous axis. Capillaries, the brain’s smallest blood vessels, constitute the vast bulk of the cerebrovascular network. Furthermore, arteries and veins are disproportionately confined to the pia surface and poorly sampled by sectioning approaches. As a result, the overwhelming majority of microvessels surveyed in a given sagittal or coronal plane are capillaries. Therefore, to assess AAV-BI30 and AAV-BR1’s ability to target the endothelium of arteries and veins we examined whole-mount preparations of the intact pia vasculature (Figure 2A). Because ERG is ubiquitously expressed across arteries, capillaries, and veins our analysis workflow could be rapidly adapted to calculate segment-specific transduction efficiency. We manually identified arteries and veins based on (i) the presence of α-Smooth Muscle Actin (high in arteries, low-to-absent in veins) 10,39 and (ii) the nuclear morphology of endothelial cells (ellipsoidal in arteries, circular in veins) 36 (Figure 2B). Strikingly, we found that AAV-BI30 captured these vessel segments efficiently, transducing 62 ± 4% of arterial ECs and 71 ± 3% of venous ECs. By contrast, AAV-BR1’s tropism appeared strongly biased against large-vessel transduction: the vector only transduced 23 ± 3% of arterial ECs and 35 ± 3% of venous ECs in the pia vasculature (Figure 2C). In line with these findings, we were able to visualize robust AAV-BI30-mediated endothelial GFP expression in the arteries, capillaries, and veins of live mice via two-photon imaging through surgically implanted cranial windows (Figure 2D). Interestingly, AAV-BI30’s tropism extended to the largest arteries of the brain; at a higher dose (5x1011 vg/mouse) we observed efficient endothelial transduction throughout the Circle of Willis and associated cerebral arteries (Extended Data Figure 7). Collectively, these results demonstrate that AAV-BI30 can be leveraged to genetically interrogate the majority of brain endothelial cells across the entire arterio-venous axis at relatively low systemic doses.
Figure 2. AAV-BI30 efficiently transduces endothelial cells across the arterio-venous axis.

AAV-BI30 or AAV-BR1 carrying a CAG-NLS-GFP-miR122-WPRE construct was intravenously administered to adult C57BL/6 mice at 1 x 1011 vg/animal. Transduction was assessed after three weeks. (a) Representative images of AAV-BI30 and AAV-BR1 transduction in whole-mount preparations of the pia vasculature. Note the strong GFP signal present in arteries, veins, and capillaries following AAV-BI30 transduction; by contrast, GFP+ endothelial cells transduced by AAV-BR1 are predominantly restricted to capillary microvessels. (b) Illustration of semi-automated image-processing workflow used to calculate arterial and venous transduction efficiency. Left: input image – note that arteries, veins, and capillaries are clearly separable based on nuclear morphology of endothelial cells and SMA expression. Middle: manual annotation of arteries and veins. Artery-vein overlap regions were intentionally omitted from analysis. Right: arterial and venous EC nuclei identified by automated Cell Profiler pipeline superimposed on ERG channel of input image. (d) Quantification of endothelial transduction measured as the fraction of ERG+ cells expressing GFP within manually annotated arterial (t4 = 7.172; P = 0.0020) and venous (t4 = 9.488; P = 0.0007) vessel segments. (e) Representative two-photon z-stacks of brain vasculature imaged in live, awake mice demonstrate AAV-BI30’s robust transduction of cerebrovascular arteries, veins, and capillaries. For quantification: n = 3 animals per group, mean ± s.e.m. ; unpaired, two-tailed t-test (** P < 0.01, *** P < 0.001). Scale bars are as follows: 100μm in (a) and (b); 25μm in (d).
AAV-BI30 transduces endothelial cells in retina and spinal cord.
While the majority of endothelial-targeted AAV research to-date has focused on the brain vasculature, the retina and spinal cord vasculature are highly tractable systems crucial to the study of angiogenesis, blood-brain barrier dynamics, neurovascular pathology, and a host of other key processes 16,40–42. Accordingly, we investigated AAV-BI30’s capacity to transduce the endothelial cells of these CNS tissues. Across all segments of the retina’s stereotyped vasculature, AAV-BI30 dramatically outperformed AAV-BR1, transducing 73 ± 3% versus 14 ± 3% superficial plexus arterial ECs; 69 ± 4% versus 18 ± 1% intermediate plexus ECs; 75 ± 3% versus 30 ± 5% deep plexus ECs; and 81 ± 4% versus 23 ± 2% superficial plexus venous ECs (Figure 3A–C). The difference between the vectors was similarly apparent in the spinal cord, where AAV-BI30 transduced 76 ± 4% of ECs compared to AAV-BR1’s 46 ± 5% – an estimate consistent with the capsid’s initial characterization 2 (Figure 3D,E). Thus, AAV-BI30’s highly efficient, endothelial cell-specific tropism is not limited to the brain; instead, it extends the entirety of the CNS. In addition, AAV-BI30-mediated transgene expression persists across long timescales. We observed robust endothelial transduction in brain, retina, and spinal cord 152 days after administration of a single 1x1011 vg dose of the vector (Extended Data Figure 8) – a result consistent with the relatively slow turnover of CNS endothelial cells 43 and longitudinally stable endothelial transduction previously demonstrated with AAV-BR1 2.
Figure 3. AAV-BI30 targets endothelial cells throughout the retina and spinal cord vasculature.

AAV-BI30 or AAV-BR1 carrying a CAG-NLS-GFP-miR122-WPRE construct was intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal. Transduction was assessed after three weeks. Representative low (a) and high (b) magnification images of AAV-BI30 and AAV-BR1 transduction in retina are shown. (c) Quantification of retinal endothelial transduction measured as the fraction of ERG+ cells expressing GFP in superficial plexus arteries (SP aECs: t4 = 13.05; P = 0.0002), intermediate plexus vessels (IP ECs: t4 = 11.44; P = 0.0003), deep plexus vessels (DP ECs: t4 = 8.107; P = 0.0013), and superficial plexus veins (SP vECs: t4 = 12.45; P = 0.0002). (d) Representative images of AAV-BI30 and AAV-BR1 transduction in spinal cord; high-zoom co-localization of GFP with endothelial markers is shown in bottom row. (e) Quantification of endothelial transduction measured as the fraction of ERG+ cells expressing GFP in whole transverse sections of spinal cord (t4 = 4.815; P = 0.0086). For quantification: n = 3 animals per group, mean ± s.e.m. ; unpaired, two-tailed t-test (** P < 0.01, *** P < 0.001). Scale bars are as follows: 100μm in (a) and middle column of (d); 50μm in (b); 25μm in rightmost column of (d).
AAV-BI30 can be leveraged for brain endothelial-specific gene manipulation.
To evaluate AAV-BI30’s capacity to genetically manipulate CNS endothelial cells in vivo, we used the capsid to package Cre recombinase (AAV-BI30:CAG-Cre-miR122-WPRE) and delivered a 1x1011 vg dose of the vector to Rosa26:CAG-LSL-tdTomato (Ai9) Cre-dependent reporter mice 44. Consistent with our prior results, we observed remarkably efficient, endothelial-specific tdTomato expression throughout the brain (Figure 4A,B & Supplementary Figure 5). By quantifying ERG co-localization with tdTomato (Figure 4C), we found that AAV-BI30-mediated Cre delivery drove recombination in 94 ± 1% of endothelial cells within the cortical microvasculature.
Figure 4. AAV-BI30 can be leveraged to achieve efficient endothelial-specific genetic manipulation.
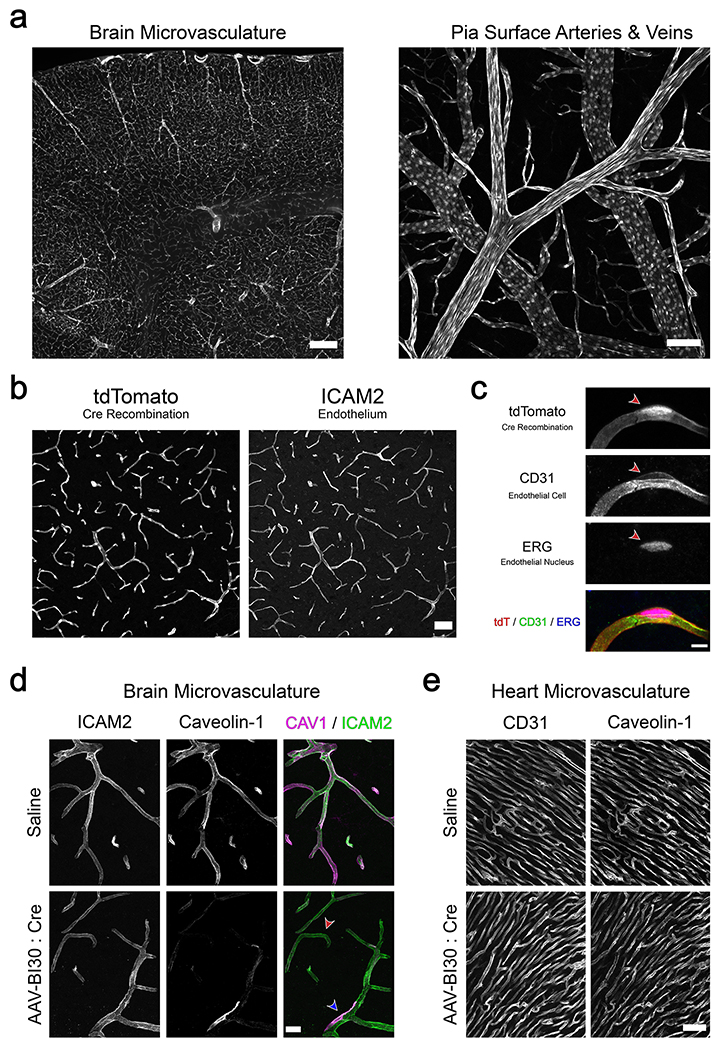
(a) AAV-BI30:CAG-Cre-miR122-WPRE was intravenously administered to adult Ai9 Cre-dependent reporter mice at 1x1011 vg/animal and recombination was assessed after 12 days. Robust tdTomato expression was observed throughout the brain microvasculature (left) as well as arteries and veins situated at the pia surface (right). Representative low (b) and high (c) magnification images of BI30-mediated Ai9 recombination in the cortical microvasculature; co-localization with endothelial-specific markers demonstrates cell-type-specificity. Recombination efficiency, measured as the fraction of ERG+ cells expressing tdTomato, was 94 ± 1% (mean ± s.e.m.; n = 3 animals) in this brain region. (d, e) A 1x1011 vg/animal dose of AAV-BI30:CAG-Cre-miR122-WPRE or saline was intravenously administered to adult Cav1 fl/fl mice and Caveolin-1 protein levels were assessed after four weeks by immunohistochemistry. (d) Representative images of brain microvasculature demonstrate reduction of endothelial Caveolin-1 in AAV-BI30-injected animals. The majority of endothelial cells in these animals exhibited a spectrum of moderate to near-complete protein loss (red arrow), likely reflecting variable viral genome count and consequent stage of protein turnover. A small fraction of endothelial cells showed no evidence of Caveolin-1 loss (blue arrow). (e) Representative images of Caveolin-1 in heart microvasculature in AAV-BI30:CAG-Cre-miR122-WPRE and saline-injected mice. All images are representative of n = 3 animals per group. Scale bars are as follows: 250μm in left panel of (a); 100μm in right panel of (a); 50μm in (b) and (e); 20μm in (d); 5μm in (c).
We next tested this strategy’s ability to achieve acute genetic loss-of-function in CNS endothelial cells. For a proof-of-concept we chose to target Caveolin-1 (encoded by the Cav1 gene), an essential component of caveolae 45. Caveolae are flask-shaped vesicular invaginations of the plasma membrane found in a number of cell types, including endothelial cells. Within CNS endothelial cells, these subcellular structures play a key role in blood-brain barrier dynamics 15,16 and neurovascular coupling 13 among other important functions.
To genetically ablate Caveolin-1 expression, we delivered a 1x1011 vg dose of AAV-BI30 carrying a CAG-Cre-miR122-WPRE genome to Cav1fl/fl mice 46. Four weeks post-administration we observed strong reduction of Caveolin-1 protein in the brain endothelial cells of AAV-BI30-injected animals relative to saline-injected controls (Figure 4D). Importantly, this effect was brain endothelial cell-specific; protein levels in arterial smooth muscle cells, which also express high levels of Caveolin-1 13,47, were unaltered following AAV-BI30 administration (Extended Data Figure 9). Consistent with our results in the Ai9 reporter line, we saw a small population of endothelial cells that escaped transduction in the 1x1011 vg dose regime with no apparent reduction in Caveolin-1 expression. By titrating dosage, experimenters could use AAV-BI30 to achieve mosaic recombination – an approach which would be particularly useful to investigate the cell-autonomous function of blood-brain-barrier genes whose widespread loss throughout CNS endothelial cells is lethal, such as Claudin-5 48 or β-Catenin 49–52.
In addition, we verified that AAV-BI30-mediated Cre delivery did not drive widespread loss of endothelial Caveolin-1 outside of the CNS. Protein levels in the heart microvasculature – an organ where endothelial Caveolin-1 is known to be robustly expressed 53 – were not appreciably altered in AAV-BI30-injected animals (Figure 4E). Given that no CNS endothelial cell-specific transgenic drivers currently exist, the ability to rapidly perform CNS-directed loss-of-function experiments may make AAV-BI30-mediated Cre delivery an attractive alternative to commonly used pan-endothelial driver lines such as CDH5:CreERT2 54 or TIE2:Cre 55.
DISCUSSION
Here we demonstrate that AAV-BI30 is ideally suited to accelerate neurovascular research, providing a rapid and easily adaptable means to access CNS endothelial cells with clear advantages over existing vectors (Extended Data Figure 10). Unlike AAV-PHP.V1, at a 1x1011 vg dose AAV-BI30’s tropism within the CNS is almost exclusively limited to endothelial cells, obviating the need for complex workarounds – such as intersectional Cre-dependent approaches – to restrict cell-type specificity. And compared to AAV-BR1, AAV-BI30 is more efficient and versatile, with particularly evident benefits for applications targeting retina, spinal cord, or cerebrovascular arteries and veins.
While AAV-BI30’s tropism appears largely biased towards the CNS vasculature, we observed transduction of liver hepatocytes and endothelial cells in the lung microvasculature, aorta, and interlobular vessels of the kidney. Importantly, peripheral endothelial transduction was sharply restricted to these populations – most organs had little-to-no endothelial cell transduction. For most research applications, AAV-BI30’s advantageous properties likely outweigh this caveat. However, in cases where CNS-specificity is critical, a Cre-dependent viral genome could be used in tandem with a CNS-specific transgenic driver – such as MFSD2A:CreERT2 56 or SLCO1C1:CreERT2 57 – to minimize AAV-BI30’s peripheral transduction.
During our characterization of AAV-BI30 we encountered unexpected dose-dependent hepatoxicity. By incorporating a hepatocyte-specific microRNA binding site into our viral genome, we were able to effectively rescue toxicity without compromising the vector’s on-target transduction efficiency. This microRNA-based strategy functions by suppressing translation of the AAV transgene in hepatocytes without affecting stages of transduction involving the capsid protein; as a result, the strategy’s efficacy argues against the possibility that the AAV-BI30 capsid’s modifications have resulted in de novo immunogenicity. Instead, the hepatotoxicity observed in our study most likely stems from cytotoxic transgene overexpression 58,59. Future engineering efforts could modify additional features of the AAV-BI30 capsid to limit hepatocyte transduction, similar to a recent study which generated a variant of the AAV-PHP.eB capsid de-targeted from peripheral organs 60. Alternatively, the development of robust endothelial-specific promoters would likely obviate the need to include microRNA binding sites in the viral genome. This latter approach could even be used to restrict transgene expression to one or more of the endothelial subpopulations transduced by AAV-BI30 in a manner similar to vessel segment-specific transgenic drivers like BMX:CreERT2 61.
In contrast with a number of recently discovered AAV vectors – including AAV-PHP.B 34 and AAV-PHP.V1 6 – AAV-BI30’s transduction profile was broadly similar between C57BL/6 and BALB/cJ mouse strains. The BALB/cJ strain’s hypomorphic Ly6a allele has been directly linked to impaired CNS transduction by capsids of the AAV-PHP.B family following systemic administration 28,29,62. As a result, our findings suggest that AAV-BI30’s mechanism of transduction is LY6A-independent. We speculate that the virus may instead enter cells using a novel surface receptor present on endothelial cells throughout the body but enriched in CNS and lung microvasculature. Moving forward, recent efforts to profile the transcriptomes of endothelial cells isolated from a wide variety of murine organs 53 could be leveraged to identify candidate receptors.
AAV-BI30 appears well-suited for a wide range of applications such as gene editing 63,64, live-imaging sensors, loss- and gain-of-function studies, or two-component tunable expression systems 4. The vector’s high efficacy is likely understated by the binary quantification metric used throughout our analyses; throughout the brain, retina, and spinal cord we observed a wide range of NLS-GFP intensities, suggesting that at a relatively low 1x1011 vg dose we delivered multiple viral genomes to a large number of CNS endothelial cells. Moreover, given that structural modifications to AAV capsids can dramatically reduce their manufacturability, it is important to note that when used to package a variety of genomes for this study – fluorescent reporters, bioluminescence producing enzymes, and gene editors – post-purification AAV-BI30 yields were in line with other engineered and natural AAV vectors (Supplementary Figure 6). This versatility – combined with the capsid’s conserved tropism across mouse strains and mammalian species – highlight AAV-BI30’s potential as a powerful tool to access CNS endothelial cells in vivo and catalyze our growing understanding of neurovascular processes in health and disease.
MATERIALS AND METHODS
Animals
All procedures were approved by the Harvard University and Broad Institute of MIT and Harvard Institutional Animal Care and Use Committees (IACUC). The following commercially available mouse strains were used: C57BL/6NCrl (Charles River 027), C57BL/6J (Jackson Laboratory 000664), BALB/cJ (Jackson Laboratory 000651), and Ai9 (Jackson Laboratory 007909; originally generated by Madisen et al. 44). CD® (Sprague Dawley) IGS rat was obtained from Charles River (Strain Code 001).
Cav1flox mice were originally generated by Asterholm et al. 46 and generously shared by Philipp Scherer. The line was genotyped using Phire Green Hot Start II DNA Polymerase (Thermo Fischer F124L) and the following primers: 5’-GTGCATCAGCCGCGTCTACTCC-3’ and 5’-GGCCGTAACCTGAATCTCTTCCCTTTG-3’. PCR reaction with genomic DNA samples produces ~490bp wild-type and ~445bp floxed products.
Recombinant AAV vectors were administered intravenously via the tail vein or retro-orbital sinus in young adult male or female animals.
Virus production
Recombinant AAVs were generated following a previously published protocol 66 with minor modifications as described below. HEK293T/17 cells (ATCC, CRL-11268) were seeded at 22 million cells per 15 cm plate the day before transfection and grown in DMEM with GlutaMAX (Gibco, 10569010) supplemented with 5% FBS and 1X non-essential amino acid solution (NEAA) (Gibco, 11140050). The next day, the cells were triple transfected with 39.93 μg of total plasmid DNA encoding Rep-Cap, pHelper, and an ITR-flanked transgene at a plasmid ratio of 4:2:1, respectively, using polyethyleimine (PEI) MAX (Polyscience, 24765-1) at a DNA:PEI ratio of 1:3.5. Twenty hours post-transfection, the media was changed to fresh DMEM with GlutaMAX supplemented with 5% FBS and 1X NEAA. Seventy two hours post-transfection cells were scraped and pelleted at 2000 RCF x 10 minutes. The pellets were resuspended in 7 mL of Salt Active Nuclease (SAN) digestion buffer (500 mM NaCl, 40 mM Tris-base, 10 mM MgCl2, SAN enzyme (ArcticZymes, #70920-202) at 100 U/mL) for every 10 plates and incubated at 37°C for 1.5 hours. Afterwards, the lysate was clarified at 2000 RCF x 10 min and loaded onto a density step gradient containing OptiPrep (Cosmo Bio, AXS-1114542) at 60%, 40%, 25%, and 15% at a volume of 5, 5, 6, and 6 mL respectively in OptiSeal tubes (Beckman, 361625). The step gradients were spun in a Beckman Type 70ti rotor (Beckman, 337922) in a Sorvall WX+ ultracentrifuge (Thermo Scientific, 75000090) at 69,000 RPM for 1 hour at 18°C. Afterwards, 4-4.5 mL of the 40-60% interface was extracted using a 16-gauge needle, filtered through a 0.22 μm PES filter, and then buffer exchanged with 100K MWCO protein concentrators (Thermo Scientific, 88532) into PBS containing 0.001% Pluronic F-68 and concentrated down to a volume of 500 μL. The concentrated virus was then filtered through a 0.22 μm PES filter and stored at 4°C or −80°C.
Virus titering
Purified virus was incubated with 1000U/mL Turbonuclease (Sigma T4330-50KU) with 1X DNase I reaction buffer (NEB B0303S) at 37°C for one hour. The endonuclease solution was inactivated with 0.5M, pH 8.0 EDTA at room temperature for 5 minutes and then at 70°C for 10 minutes. AAV genomes were released by incubation with 100μg/mL Proteinase K (Qiagen, 19131) in 1M NaCl, 1% N-lauroylsarcosine, and in UltraPure DNase/RNase-Free water at 56°C for 2 to 16 hours before heat inactivation at 95°C for 10 minutes. The nuclease-resistant AAV genomes were diluted between 460-460,000X and 2μL of the diluted samples were used as input in a ddPCR supermix for probes (Bio-Rad, 1863023) with 900nM ITR2_Forward (5’-GGAACCCCTAGTGATGGAGTT-3’), 900nM ITR2_Reverse (5’-CGGCCTCAGTGAGCGA-3’), and 250nM ITR2_Probe (5’-HEX-CACTCCCTC-ZEN-TCTGCGCGCTCG-IABkFQ-3’). The ITR2_Probe contained the following modifications - 5’ HEX dye, ZEN internal quencher, and 3’ Iowa Black fluorescent quencher (IDT, PrimeTime qPCR Probes). Droplets were generated using a QX100 Droplet Generator, transferred to thermocycler, and cycled according to the manufacturer’s protocol with an annealing/extension of 58°C for 1 minute. Finally, droplets were read on a QX100 Droplet Digital System to determine titers.
In vitro transduction assays
Multiple lots of mBMVEC and hBMVEC cells were obtained from CellBiologics (H-6023 & C57-6023) and maintained in endothelial cell media (H1168 & M1168). hCMEC/D3 cells were obtained from Millipore (SCC0066) and maintained in EndoGRO™-MV Complete Media (SCME004). All cells were handled according to the manufacturer’s instructions. For the Luciferase assays, 5000 cells/well were seeded in 96 well plates (PerkinElmer, 6005680). One day later, AAV9, AAV-PHP.eB or AAV-BI30 carrying pAAV-CAG-eGFP-p2A-luciferase was added at 20,000 vg/cell. 24 hours after transduction, a luciferase reporter assay was performed according to the manufacturer’s instructions (PerkinElmer, 6066761) on an EnSpire plate reader (PerkinElmer). For flow cytometry, hCMEC/D3 cells were plated at 434,000 cells/well in 24 well plates and exposed to the indicated dose of AAV-BI30 or AAV9. The media was exchanged for fresh media after 24 hours and transduction was assessed at 4 days post-administration on a Beckman CytoFLEX S Flow Cytometer using FlowJo 10.8.1 (BD Biosciences).
Capsid library generation
The mRNA selection vector (AAV9-CMV-Express) was designed to enrich for functional AAV capsid sequences by recovering capsid mRNA from transduced cells. AAV9-CMV-Express uses a ubiquitous CMV enhancer and AAV5 p41 gene regulatory elements to drive AAV9 Cap expression. The AAV9-CMV-Express plasmid was constructed by cloning the following elements into an AAV genome plasmid in the listed order: a cytomegalovirus (CMV) enhancer-promoter, a synthetic intron, and the AAV5 P41 promoter along with the 3’ end of the AAV2 Rep gene, which includes the splice donor sequences for the capsid RNA. The capsid gene splice donor sequence in AAV2 Rep was modified from CAGGTACCA to a consensus donor sequence CAGGTAAGT. The AAV9 capsid gene sequence was synthesized with nucleotide changes at 1344, 1346, and 1347 (which introduces a K449R mutation) and at 1782 (which is a silent mutation) to introduce restriction enzyme recognition sites for NNK library PCR insert fragment cloning. The AAV2 polyadenylation sequence was replaced with a simian virus 40 (SV40) late polyadenylation signal. A previously described cap-deficient Rep-AAP AAV helper plasmid 1 was supplied in trans to generate virus with the AAV9-CMV-Express vector.
The initial random 7-mer library was produced using 5’-CGGACTCAGACTATCAGCTCCC-3’ and 5’-GTATTCCTTGGTTTTGAACCCAACCGGTCTGCGCCTGTGCMNNMNNMNNMNNMNNMNNMNNTTGGGCACTCTGGTGGTTTGTG-3’ primers (IDT) to PCR amplify a modified AAV9 template (K449R) using Q5 Hot Start High-Fidelity 2X Master Mix (NEB, M0494S) following the manufacturer’s protocol. The PCR products were cleaned up using AMPure XP beads (Beckman, A63881) following the manufacturer’s protocol. The NNK PCR insert was assembled into a linearized mRNA selection vector (AAV9-CMV-Express) with NEBuilder HiFi DNA Assembly Master Mix (NEB, E2621L). Afterwards, Quick CIP (NEB, M0508S) was spiked into the reaction and incubated at 37°C for 30 minutes to dephosphorylate unincorporated dNTPs. Finally, T5 Exonuclease (NEB M0663S) was added to the reaction mixture and incubated at 37°C for 30 minutes to remove unassembled products. The final assembled product was cleaned up using AMPure XP beads (Beckman, A63881) following the manufacturer’s protocol quantified using the Qubit dsDNA HS Assay Kit (ThermoFisher Scientific, Q32851). Capsid variants chosen for secondary screening were synthesized as an oligopool (Agilent). Each capsid 7-mer was represented by two unique nucleotide sequences and cloned into the AAV9-CMV-Express backbone as described above.
Capsid library screening
For in vivo screening, the AAV capsid library was injected at 1x1011 vg/animal in either C57BL/6J or BALB/cJ mice. 21 days post-injection animals were euthanized, and tissue was extracted and flash-frozen for RNA isolation. For in vitro screening, 1x1011 vg of the AAV capsid library was added to confluent BMVECs or hCMEC/D3 cells and cellular mRNA was collected 3 days post-administration. mRNA from in vivo and in vitro assays were recovered using TRIzol (Invitrogen, 15596026) followed by RNA cleanup with RNeasy Mini Kit (Qiagen, 74104). The recovered mRNA was next converted to cDNA using an oligo dT primer using Maxima H Minus Reverse Transcriptase (ThermoFisher, EP0751). To prepare AAV libraries for sequencing, qPCR was performed on the converted cDNA from each sample type to identify the minimum number of cycles necessary for amplification. Once cycle thresholds were determined SeqF1 (5’-CTTTCCCTACACGACGCTCTTCCGATCTNCCAACGAAGAAGAA-ATTAAAACTACTAACCCG-3’) and SeqR1 (5’-GGAGTTCAGACGTGTGCTCTTCCGATCTCATCTCTGTCCTGCCA-AACCATACC-3’) primers were used to attach Illumina Read 1 and Read 2 sequences using Q5 Hot Start High-Fidelity 2X Master Mix with an annealing temperature of 65°C for 20 seconds and an extension time of 1 minute with a cycle number determined from the qPCR. These PCR products were purified using AMPure XP beads following the manufacturer’s protocol and eluted in 25 μL UltraPure Water (ThermoScientific) and then 2 μL was used as input in a secondary PCR to attach Illumina adaptors and dual index primers (NEB, E7600S) for 5 cycles using Q5 HotStart-High-Fidelity 2X Master Mix with an annealing temperature of 65°C for 20 seconds and an extension time of 1 minute. The second PCR products were purified using AMPure XP beads following the manufacturer’s protocol and eluted in 25 μL UltraPure DNase/RNase-Free distilled water (ThermoScientific, 10977015). To quantify the PCR product for NGS, an Agilent High Sensitivity DNA Kit (Agilent, 5067-4626) was used with an Agilent 2100 Bioanalyzer system. The secondary PCR products were then pooled and diluted to 2-4 nM in 10 mM Tris-HCl, pH 8.5 and sequenced on an Illumina NextSeq 550 following the manufacturer’s instructions using a NextSeq 500/550 Mid or High Output Kit (Illumina, 20024904 or 20024907). Reads were allocated as follows: I1: 8, I2: 8, R1: 150, R2: 0.
Cloning
The AAV-BI30 rep-cap plasmid was generated by assembling an oligo (IDT) containing the AAV-BI30 DNA sequence (5’-AACAACTCAACCCGCGGCGGC-3’) into a synthesized kanamycin resistant rep-tTA-Cap helper (iCap-Kan-BI30; GenScript) containing a K449R point mutation in AAV9. The AAV-BR1 cap gene was synthesized and cloned into the same iCap-Kan backbone. To generate pAAV-CAG-NLS-GFP-miR122-WPRE gene fragments were synthesized (GenScript) and cloned into pAAV-CAG-mTurquoise2, a gift from Viviana Gradinaru (Addgene #99122). pAAV-CAG-Cre-miR122-WPRE was cloned by amplifying Cre from pAAV-EF1a-Cre (Addgene, 55636), a gift from Karl Deisseroth, with (5’-GAATTGCGGCCCAACGGTACCATGCCCAAGAAGAAGAGGAAGGT-3’) and (5’-TGACAATGGTGTTTGACCGGTGAATTCTCAGTCACCATCTTCGAGCAGTC-3’) and then cloned into the pAAV-CAG-NLS-GFP-miR122-WPRE backbone using the KpnI and EcoRI restriction sites.
Immunohistochemistry
Mice were deeply anesthetized with an intraperitoneal injection of a ketamine / xylazine solution and transcardially perfused with ~15 mL of room-temperature PBS followed by ~20 mL of ice-cold 4% PFA using a peristaltic pump set to a flow rate of ~9 mL/min. Tissues were subsequently harvested and processed as follows: Pia Vasculature Whole Mounts. Brain was dissected out of the skull and partially immersed in PBS in a glass dish. A razor blade was used to make a cut along the sagittal midline followed by a cut along the horizontal axis to separate each hemisphere into dorsal and ventral pieces. Dorsal-facing brain samples were then transferred into a 48-well plate and post-fixed in 4% PFA on ice for 30 minutes. All subsequent wash steps were carried out in the plate, and care was taken to ensure the ventral surface of the brain sample always remained in contact with the bottom of the dish. Samples were washed 3x with PBS, blocked with a 10% Donkey Serum / 0.5% PBST (Triton X-100) solution for 2 hours at room temperature, and then incubated with primary antibodies made up in blocking solution for 48 hours at 4°C with agitation. Next, samples were washed 3x with 0.5% PBST and 1x with PBS. To capture images, brain sample was placed dorsal side down in a 2-well glass bottom slide (Ibidi 80287) partially filled with PBS such that the pia vasculature faced the objective on an inverted microscope. Retina Whole Mounts. Eyes were removed from the eye sockets and briefly post-fixed in room-temperature 4% PFA for 5 minutes. Next, retinas were isolated via fine dissection in PBS and further post-fixed in room-temperature 4% PFA for 30 minutes. Retinas were washed 3x with PBS, blocked with a 10% Donkey Serum / 0.5% PBST solution for 1 hour at room temperature, and then incubated with primary antibodies made up in blocking solution overnight at 4°C with agitation. Next, retinas were washed 3x with 0.5% PBST, 1x with PBS, and flat-mounted on glass coverslips. Aorta Whole Mounts. Thoracic aorta was grossly dissected and immersed in PBS. Fat, connective tissue, and arterial branches were subsequently removed via fine dissection. A post-fixation step was omitted. Aortas were washed 3x with PBS, blocked with a 10% Donkey Serum / 0.5% PBST solution for 1 hour at room temperature, and then incubated with primary antibodies made up in blocking solution overnight at 4°C with agitation. Next, aortas were washed 3x with 0.5% PBST and immersed in PBS. A single cut was made lengthwise along the vessels to expose the lumen and the aortas were flat-mounted en face on glass coverslips. Tissue Sections. Organs were post-fixed in 4% PFA at 4°C overnight. Alternatively, brain was post-fixed 4 hours in 4% PFA on ice for Caveolin-1 immunostaining due to fixation-sensitivity of the antibody. Samples were then washed 3x with PBS to remove residual PFA. For cryosections, samples were cryopreserved in a 30% sucrose solution at 4°C overnight, and then frozen in NEG-50 (Richard-Allan Scientific 6502). 18μm (Fig 1F; Fig 3D; Fig 4D; Ext. Data Fig 5, 9) or 30μm (Fig 1C & E [BALB/cJ]; Fig 4B, C & E; Ext. Data Fig 2, 3, 4 [BALB/cJ], 6, 8) sections were cut using a cryostat. For vibratome sections, 60μm (Fig 1B & E [rat]; Ext. Data Fig 4 [rat]; Supp. Fig 3) or 100μm (Fig 1D; Fig 4A; Supp. Fig 5) sections were cut using a LeicaVT1200S vibratome. Sections were washed 3x in PBS, permeabilized in 0.5% PBST for 10 minutes, and then blocked with a 5% Donkey Serum / 0.1% PBST solution for 1 hour at room temperature. Sections were subsequently incubated with primary antibodies made up in blocking solution overnight at 4°C, washed 3x with 0.1% PBST, and incubated with secondary antibodies and DAPI made up in blocking solution for 1 hour at room temperature. Finally, sections were washed 3x with 0.1% PBST, 1x with PBS, and coverslipped for imaging.
The following primary antibodies were used at the indicated concentrations: Rabbit anti-ERG ◦ Alexa Fluor 647 (1:100; Abcam ab196149), Rabbit anti-GFP ◦ Alexa Fluor 488 (1:200; Thermo Fisher Scientific A-21311), Mouse anti-α Smooth Muscle Actin ◦ FITC (1:500; Sigma-Aldrich F3777), Mouse anti-α Smooth Muscle Actin ◦ Cy3 (1:500; Sigma-Aldrich C6198), Mouse anti-α Smooth Muscle Actin ◦ Alexa Fluor 647 (1:100; Santa Cruz Biotechnology sc-32251), Goat anti-CD31 (1:100; R&D Systems AF3628), Rat anti-ICAM2 (1:100; BD Biosciences 553326), Rabbit anti-Caveolin-1 (1:200; Cell Signaling Technology 3267), Mouse anti-RECA-1 (1:200; Abcam ab9774), and Chicken anti-GFP (1:1000; Aves Labs GFP-1020). In addition, Isolectin GS-IB4 ◦ Alexa Fluor 568 (1:100; Invitrogen I21412) was used to stain vasculature in retina whole mounts. Specificity of the monoclonal anti-Cav1 antibody used in this study has been previously demonstrated by our group in Caveolin-1 knockout mice 13.
Donkey anti-Goat IgG ◦ AF488 (1:250; Jackson ImmunoResearch 705-545-003), Donkey anti-Goat IgG ◦ Cy3 (1:250; Jackson ImmunoResearch 705-165-147), Donkey anti-Rat IgG ◦ AF488 pre-adsorbed against Mouse IgG (1:250; Jackson ImmunoResearch 712-546-153), Donkey anti-Rat IgG ◦ Cy3 pre-adsorbed against Mouse IgG (1:250; Jackson ImmunoResearch 712-165-153), Donkey anti-Rabbit IgG ◦ AF647 (1:250; Jackson ImmunoResearch 711-605-152), Donkey anti-Mouse IgG ◦ AF546 highly cross-adsorbed (1:1000; Invitrogen A10036), and Goat anti-Chicken ◦ AF488 (1:1000 Invitrogen, A-11039) were used in conjunction with unconjugated primary antibodies.
Representative images were acquired with a Leica TCS SP8 confocal microscope, a Keyence BZ-X810, or a Nikon Ti-E inverted microscope / Andor CSU-X1 spinning disc confocal with an Andor DU-888 EMCCD camera.
Quantification of viral transduction efficiency
NLS-GFP Overexpression.
Cell Profiler 67 was used to construct unbiased, semi-automated or fully automated analysis pipelines for all tissues examined. Viral transduction was assessed using a binary metric: GFP+ ERG+ cells taken as a fraction of total ERG+ cells. Staining and imaging of AAV-BI30 and AAV-BR1 samples was performed on the same day to ensure identical imaging conditions. All quantification was performed in C57BL/6NCrl mice. Retina, spinal cord, and brain data were collected from the same set of animals. Tissue-specific workflow was as follows: Brain Microvasculature. A single sagittal brain section was acquired from each animal and imaged in its entirety using an Olympus VS120 whole slide-scanning microscope with a UPLSAPO 20x/0.75 objective lens. An average of 15,310 ± 3,929 (mean ± s.d.) endothelial cells were identified by the analysis pipeline in each replicate. Pia Vasculature. Three ~1164μm x 1164μm FOVs of the pia vasculature were acquired for each animal using a Leica TCS SP8 confocal microscope with a HC PL APO 10x/0.40 objective lens. Relatively flat regions of the brain surface were sampled with shallow z-stacks to clearly separate pia vasculature from the underlying capillary plexus. Arteries and veins were manually identified using (i) the presence of α-Smooth Muscle Actin (high in arteries, low-to-absent in veins) and (ii) the nuclear morphology of endothelial cells (ellipsoidal in arteries, circular in veins). Artery-vein overlap regions were omitted from analysis. Average endothelial cell counts identified by analysis pipeline per animal were as follows: 1,197 ± 182 (aECs), 1,908 ± 197 (vECs). Retina Vasculature. Organization of the retinal vasculature is highly stereotyped; vessels form three distinct vascular plexuses. Blood enters via the retinal artery which emerges alongside the optic nerve, branching into spoke-like radial arteries that spread across the superficial plexus. These arteries elaborate to form a dense capillary network that constitute the majority of the intermediate and deep plexuses. Capillaries in the deep plexus finally merge into draining venules which ascend to the superficial plexus and form veins interleaved among the retina’s arteries. To quantify viral transduction efficiency across the entirety of the retinal arterio-venous axis, four sub-regions of each retina whole-mount were sampled: radial arteries in the superficial plexus; capillaries in the intermediate plexus; capillaries in the deep plexus; and radial veins in the superficial plexus. Three ~290μm x 290μm FOVs of each of these vascular sub-regions were acquired for each animal using a Leica TCS SP8 confocal microscope with a HC PL APO CS2 20x/0.75 objective lens. Special care was taken when setting the z-axis bounds on each image in order to isolate signal from a single vascular plexus. Superficial plexus arteries and veins within a given FOV were manually identified using SMA expression and nuclear morphology as described for pia vasculature. All ERG+ nuclei within intermediate and deep plexus FOVs were counted – the vast majority of these endothelial cells belong to capillary microvessels. Average endothelial cell counts identified by analysis pipeline per animal were as follows: 155 ± 12 (superficial plexus aECs), 157 ± 13 (intermediate plexus ECs), 229 ± 19 (deep plexus ECs), 102 ± 11 (superficial plexus vECs). Spinal Cord Microvasculature. Imaging was performed using a whole-slide scanning microscope as described for the brain microvasculature. Images of two transverse sections were acquired for each animal and used to identify an average of 1,061 ± 200 endothelial cells in each replicate. Non-Endothelial Transduction. (Fiji is Just) ImageJ’s Cell Counter plugin was used to manually quantify non-endothelial transduction in cortex. These counts were collected from the aforementioned sagittal sections of brain used to quantify endothelial transduction efficiency in brain microvasculature (18μm thick; imaged with an Olympus VS120 whole slide-scanning microscope). DAPI+ GFP+ ERG−nuclei were identified as instances of non-endothelial transduction. Ai9 Recombination Efficiency. Three ~582μm x 582μm FOVs of the cortical microvasculature per replicate were acquired from 30 μm sagittal sections using a Leica TCS SP8 confocal microscope with a HC PL APO CS2 20x/0.75 objective lens. A modified Cell Profiler pipeline similar to those used to quantify AAV-mediated NLS-GFP overexpression was employed to count tdT+ ERG+ cells taken as a fraction of total ERG+ cells. An average of 481 ± 73 endothelial cells were identified by the analysis pipeline per replicate.
Cranial window implantation and two-photon imaging
The cranial window implantation workflow was based on the protocol described by Goldey et al 68. The craniotomy was centered over somatosensory cortex at approximately 2mm posterior and 2.5mm lateral to bregma. The perimeter of the craniotomy was traced using a 4mm circular biopsy punch (VWR 21909-140) marked with a surgical marker (Aspen Surgical 1000-00-PDG). A custom titanium headplate was then centered on this trace and bonded to the skull with dental cement (C&B Metabond; Parkell S396, S398, S371). Next, a micro-motor drill (Foredom, MH-170) outfitted with a 0.2mm miniature carbide burr bit (Stoelting, 51451) was used to carefully remove the bone along the circumference of the craniotomy trace. The resulting circular bone flap was subsequently removed with fine forceps while continuously irrigating with saline so as to avoid damage to the pia vasculature. A cranial window – composed of a 4mm round cover glass (Warner Instruments CS-4, 64-0724) glued to a 5mm round cover glass (Warner Instruments CS-5R, 64-0700) with a UV-curable adhesive (Norland Products NOA68) – was carefully lowered onto the exposed brain and bonded to surrounding regions of the skull with dental cement. Finally, a well – composed of O-Rings (USA Sealing, ZUSAH1X10 & ZUSAH1X10.5 ) adhered to one another with cyanoacrylate (3M Vetbond) – was constructed around the window to facilitate use of a water-immersion objective. Immediately prior to imaging, 1mg of 70kDa Dextran ◦ Texas Red (Thermo Fisher D1830) was dissolved in 75μL sterile saline and injected intravenously via the tail vein to visualize the vasculature. In vivo imaging was performed with a previously described custom-built two photon microscope 13 using a Mai Tai Ti:Sapphire (Spectra-Physics) laser tuned to 900nm and ScanImage 5.1 (Vidrio Technologies).
Image processing and data analysis
(Fiji is Just) ImageJ 2.0.0-rc-69 and Adobe Illustrator 24.2 were used to process images displayed in figures throughout the manuscript. In cases where AAVs were directly compared, matched images were treated identically. GraphPad Prism 9.1.1 was used for statistical analysis and graph generation.
Statistics & Reproducibility
No statistical method was used to predetermine sample size. Sample size was instead designed to approximately match similar studies characterizing engineered AAVs 1,4,6. All attempts at replication were successful - no data were excluded from the analyzes. All replicates were assigned to treatment groups at random. Investigators were not blinded to allocation during experiments and outcome assessment.
Extended Data
Extended Data Figure 1. AAV-BI30 is more efficient at transducing hCMEC/D3 cells than AAV9 across a wide range of doses.

hCMEC/D3 cells were grown to confluence in a 24-well plate format. AAV9 or AAV-BI30 carrying a CAG-NLS-GFP-miR122-WPRE genome was applied to cells at 0, 500, 1,000, 5,000, 10,000, or 50,000 vg/cell. 4 days post-treatment the cells were analyzed for GFP expression via flow cytometry. The profiles show data from a single sample from each condition but are representative of n = 2 replicates.
Extended Data Figure 2. Comparison of the tropism of AAV-BI30 and AAV9.

AAV-BI30 or AAV9 vectors carrying a CAG-NLS-GFP-WPRE genome were intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal and transduction was assessed after 7 days. Panels show endogenous NLS-GFP transgene expression in organs throughout the body. Scale bar shown is 200μm. Images are representative of n = 3 animals per group.
Extended Data Figure 3. Characterization of the peripheral tropism of AAV-BI30.

AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal and transduction was assessed after three weeks. Representative images of AAV-BI30 transduction throughout the periphery; high-zoom co-localization of GFP with endothelial markers is shown in the rightmost column. With the notable exception of lung, AAV-BI30 rarely transduced endothelial cells in the microvasculature of peripheral organs – a striking contrast to efficient transduction seen throughout the CNS vasculature. Note residual NLS-GFP expression in hepatocytes of the liver persisting in the presence of miR122 repeats in the viral genome, illustrating AAV-BI30’s potent transduction of this cell type. Transduction observed in kidney glomeruli was non-endothelial; GFP+ cells are most likely mesangial cells. Relatively strong endothelial transduction in the interlobular vessels of the renal medulla and the aorta suggest that AAV-BI30 may achieve transduction of large-diameter arteries and veins throughout the systemic circulation. Scale bars are as follows: 100μm in fourth column from left, 15μm in rightmost column, and 25μm in aorta panel. Images are representative of n = 3 animals.
Extended Data Figure 4. AAV-BI30 transduces endothelial cells throughout the BALB/cJ and rat brain.

AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered at 1x1011 vg/animal (BALB/cJ; top) or 1.42x1013 vg/kg (rat; bottom). Transduction was assessed after three (BALB/cJ) or four (rat) weeks. Images demonstrate endothelial expression of NLS-GFP transgene throughout brains of each species. Scale bars shown are 200μm (BALB/cJ third row from left) or 100μm (BALB/cJ rightmost row & rat). Images are representative of n = 3 BALB/cJ mice, n = 1 rat.
Extended Data Figure 5. Endothelial transduction by AAV-BI30 is consistent across brain regions.

AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal and transduction was assessed after three weeks. Images demonstrate high endothelial expression of NLS-GFP transgene throughout the brain. Region-specific endothelial transduction efficiency was as follows: 86 ± 6% in cortex, 81 ± 5% in hippocampus, 85 ± 3% in thalamus, 83 ± 2% in cerebellum (mean ± s.e.m.; n = 3 animals). Compare to 84 ± 4% efficiency measured across entire brain. Scale bars are as follows: third row from left 100μm; rightmost row 50μm.
Extended Data Figure 6. The transduction profile of AAV-BI30 within the brain is highly endothelial-specific.

AAV-BI30 or AAV-BR1 vectors carrying a CAG-NLS-GFP-miR122-WPRE genome were intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal and transduction was assessed after three weeks. Left: Representative images of rare instances of neuronal and astrocytic transduction observed following AAV-BI30 administration at the 1x1011 vg/animal dose; cell types of interest are demarcated with red arrowheads. Scale bar shown is 50μm. Right: Quantification of AAV transduction in non-endothelial (GFP+ ERG−) cells per mm2 of cortex using 18μm sagittal sections of brain. An average of 0.6 ± 0.3 and 10.8 ± 3.0 cells/mm2 (mean ± s.e.m.; n = 3 animals per group) were identified in AAV-BI30 and AAV-BR1 administered cohorts, respectively. Consistent with previous reports, neurons constituted the majority of non-endothelial cells transduced by AAV-BR1. The data presented were compared using an unpaired, two-tailed t-test (t4 = 3.37; P = 0.0281).
Extended Data Figure 7. AAV-BI30 efficiently transduces the brain’s largest arteries.

AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered to an adult C57BL/6 mouse at 5x1011 vg/animal and transduction was assessed after 3 weeks. Robust, endothelial-specific transduction was observed throughout the cerebral arteries, Circle of Willis, and the head of the basilar artery. Scale bar shown is 100μm. Images are representative of n = 1 mouse.
Extended Data Figure 8. AAV-BI30-mediated gene transfer enables long-term transgene expression in CNS endothelial cells.

AAV-BI30:CAG-NLS-GFP-miR122-WPRE was intravenously administered to adult C57BL/6 mice at 1x1011 vg/animal and transduction was assessed after approximately 5 months (152 days). Vascular counterstains displayed are as follows: ICAM2 in brain parenchyma and spinal cord; isolectin in retina; and ERG in pia vasculature. High-zoom co-localization with GFP shown in rightmost column. Scale bars are as follows: middle row 200μm; rightmost row 50μm. Images are representative of n = 3 mice.
Extended Data Figure 9. AAV-BI30-mediated Cre delivery achieves efficient endothelial-specific gene deletion.

A 1x1011 vg/animal dose of AAV-BI30:CAG-Cre-miR122-WPRE or saline was intravenously administered to adult Cav1fl/fl mice, and Caveolin-1 protein levels were assessed after four weeks. Representative images of the brain microvasculature show strongly reduced endothelial Caveolin-1 protein levels in AAV-BI30 injected animals. Note lack of Caveolin-1 loss in the smooth muscle cell layer of arterioles, demarcated with red arrowheads. High-zoom co-localization of Caveolin-1 and ICAM2 shown in rightmost column. Scale bars are as follows: second column from right 100μm; rightmost column 50μm. Images are representative of n = 3 mice per group.
Extended Data Figure 10. Comparison of engineered AAV capsids with enhanced CNS endothelial cell transduction.

The heptameric peptides used to generate the listed capsids were uniformly inserted within the surface exposed hypervariable region VIII of the AAV sequence 65. Specifically, heptamers are inserted between amino acids 587 & 588 in AAV-PPS and 588 & 589 in AAVs BI30, BR1, PHP.B, PHP.V1, and PHP.V2. AAV-PHP.eB shares AAV-PHP.B’s TLAVPFK insert and additionally contains an AQ to DG mutation at amino acids 587-588 (bracketed sequence).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Gu and Deverman laboratories for their insightful feedback throughout the study and their comments on the manuscript during drafting. This research was supported by a National Science Foundation Graduate Research Fellowship (Grant No. DGE1745303) (T.K.), an HMS Mahoney Postdoctoral Fellowship (L.K.), an award from the NIH Common Fund and the National Institute of Neurological Disorders and Stroke through the Somatic Cell Genome Engineering (SCGE) Consortium (UG3NS111689) (B.D.), a Brain Initiative award funded through the National Institute of Mental Health (UG3MH120096) (B.D.), the Stanley Center for Psychiatric Research (B.D.), a Fidelity Biosciences Research Initiative (C.G.), an Allen Distinguished Investigator Award (C.G.), an AHA-Allen Initiative in Brain Health and Cognitive Impairment Award (C.G.), NIH R35NS116820 (C.G), NIH RF1DA048786 (C.G.), and NIH R01 HL153261 (C.G.). The research of C.G. was supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute. C.G. is an investigator of the Howard Hughes Medical Institute. Imaging was in part performed in the Neurobiology Imaging Facility at Harvard Medical School. This facility is supported in part by the HMS/BCH Center for Neuroscience Research as part of an NINDS P30 Core Center grant (NS072030).
COMPETING INTERESTS
K.C., Q.H., and B.D. are inventors on a provisional patent application filed by the Broad Institute (applicant). The specific aspects of the manuscript covered include modified AAV vectors and methods of making and using the same. B.D. is a scientific founder and scientific advisor of Apertura Gene Therapy. B.D. receives research funding from Apertura Gene Therapy, which was used to generate some of the data in this manuscript. B.D. is on the scientific advisory board of Tevard Biosciences. The remaining authors declare no competing interests.
Data Availability Statement
Statistical data used to generate the graphs presented throughout the manuscript is provided in the form of Source Data and Supplementary Statistical Data files. The AAV-BI30 rep-cap, pAAV-CAG-NLS-GFP-miR122-WPRE-pA, and pAAV-CAG-Cre-miR122-WPRE-pA plasmids generated in this study will be made available on Addgene shortly following publication; in the meantime, the reagents will be available from the corresponding authors upon request.
REFERENCES
- 1.Deverman BE et al. Cre-Dependent Selection Yields AAV Variants for Widespread Gene Transfer to the Adult Brain. Nat Biotechnol 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Körbelin J et al. A Brain Microvasculature Endothelial Cell-Specific Viral Vector With the Potential to Treat Neurovascular and Neurological Diseases. EMBO Mol Med 8, 609–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tervo DGR et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan KY et al. Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems. Nat Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanlon KS et al. Selection of an Efficient AAV Vector for Robust CNS Transgene Expression. Mol Ther Methods Clin Dev 15, 320–332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar SR et al. Multiplexed Cre-Dependent Selection Yields Systemic AAVs for Targeting Distinct Brain Cell Yypes. Nat Methods 17, 541–550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nonnenmacher M et al. Rapid Evolution of Blood-Brain-Barrier-Penetrating AAV Capsids by RNA-Driven Biopanning. Mol Ther Methods Clin Dev 20, 366–378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney MD, Kisler K, Montagne A, Toga AW & Zlokovic BV The Role of Brain Vasculature in Neurodegenerative Disorders. Nat Neurosci 21, 1318–1331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mastorakos P & McGavern D The Anatomy and Immunology of Vasculature in the Central Nervous System. Sci Immunol 4, eaav0492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanlandewijck M et al. A Molecular Atlas of Cell Types and Zonation in the Brain Vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Chen BR, Kozberg MG, Bouchard MB, Shaik MA & Hillman EM A Critical Role for the Vascular Endothelium in Functional Neurovascular Coupling in the Brain. J Am Hear. Assoc 3, e000787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longden TA et al. Capillary K+ - Sensing Initiates Retrograde Hyperpolarization to Increase Local Cerebral Blood Flow. Nat Neurosci 20, 717–726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow BW et al. Caveolae in CNS Arterioles Mediate Neurovascular Coupling. Nature 579, 106–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Zvi A et al. Mfsd2a is Critical for the Formation and Function of the Blood-Brain Barrier. Nature 509, 507–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreone BJ et al. Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron 94, 581–594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow BW & Gu C Gradual Suppression of Transcytosis Governs Functional Blood-Retinal Barrier Formation. Neuron 93, 1325–1333.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerfoot SM & Kubes P Overlapping Roles of P-Selectin and Alpha 4 Integrin to Recruit Leukocytes to the Central Nervous System in Experimental Autoimmune Encephalomyelitis. J Immunol 169, 1000–1006 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Piccio L et al. Molecular Mechanisms Involved in Lymphocyte Recruitment in Inflamed Brain Microvessels: Critical Roles for P-Selectin Glycoprotein Ligand-1 and Heterotrimeric G(i)-Linked Receptors. J Immunol 168, 1940–1949 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Tan C et al. Endothelium-Derived Semaphorin 3G Regulates Hippocampal Synaptic Structure and Plasticity via Neuropilin-2/PlexinA4. Neuron 101, 920–937.e13 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Santisteban MM et al. Endothelium-Macrophage Crosstalk Mediates Blood-Brain Barrier Dysfunction in Hypertension. Hypertension 76, 795–807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X xiu et al. Endothelial Cdk5 Deficit Leads to the Development of Spontaneous Epilepsy Through CXCL1/CXCR2-Mediated Reactive Astrogliosis. J Exp Med 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dogbevia G, Grasshoff H, Othman A, Penno A & Schwaninger M Brain Endothelial Specific Gene Therapy Improves Experimental Sandhoff Disease. J Cereb Blood Flow Metab 40, 1338–1350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen DY et al. Endothelium-Derived Semaphorin 3G Attenuates Ischemic Retinopathy by Coordinating β-Catenin-Dependent Vascular Remodeling. J Clin Invest 131, e135296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolakopoulou AM et al. Endothelial LRP1 Protects Against Neurodegeneration By Blocking Cyclophilin A. J Exp Med 218, e20202207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Y et al. Brain Endothelial PTEN / AKT / NEDD4-2 / MFSD2A Axis Regulates Blood-Brain Barrier Permeability. Cell Rep. 36, 109327 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Song X et al. Genome Editing with AAV-BR1-CRISPR in Postnatal Mouse Brain Endothelial Cells. Int J Biol Sci 18, 652–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabebordbar M et al. Directed Evolution of a Family of AAV Capsid Variants Enabling Potent Muscle-Directed Gene Delivery Across Species. Cell 184, 4919–4938.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Q et al. Delivering Genes Across the Blood-Brain Barrier: LY6A, A Novel Cellular Receptor for AAV-PHP.B Capsids. PLoS One 14, e0225206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hordeaux J et al. The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier. Mol Ther 27, 912–921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagos-Quintana M et al. Identification of Tissue-Specific microRNAs from Mouse. Curr Biol 12, 735–739 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T et al. miR-122a-Regulated Expression of a Suicide Gene Prevents Hepatotoxicity Without Altering Antitumor Effects in Suicide Gene Therapy. Mol Ther 16, 1719–1726 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Xie J et al. MicroRNA-Regulated , Systemically Delivered rAAV9: A Step Closer to CNS-Restricted Transgene Expression. Mol Ther 19, 526–535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geisler A et al. microRNA122-Regulated Transgene Expression Increases Specificity of Cardiac Gene Transfer Upon Intravenous Delivery of AAV9 Vectors. Gene Ther 18, 199–209 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Hordeaux J et al. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol Ther 26, 664–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuzaki Y et al. Intravenous Administration of the Adeno-Associated Virus-PHP.B Capsid Fails to Upregulate Transduction Efficiency in the Marmoset Brain. Neurosci Lett 665, 182–188 (2018). [DOI] [PubMed] [Google Scholar]
- 36.dela Paz NG & D’Amore PA Arterial Versus Venous Endothelial Cells. Cell Tissue Res 335, 5–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah AV, Birdsey GM & Randi AM Regulation of Endothelial Homeostasis, Vascular Development and Angiogenesis by the Transcription Factor ERG. Vasc. Pharmacol 86, 3–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graßhoff H et al. Short Regulatory DNA Sequences to Target Brain Endothelial Cells for Gene Therapy. J Cereb Blood Flow Metab (2021). doi: 10.1177/0271678X211039617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill RA et al. Regional Blood Flow in the Normal and Ischemic Brain Is Controlled by Arteriolar Smooth Muscle Cell Contractility and Not by Capillary Pericytes. Neuron 87, 95–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartanusz V, Jezova D, Alajajian B & Digicaylioglu M The Blood-Spinal Cord Barrier: Morphology and Clinical Implications. Ann Neurol 70, 194–206 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Stahl A et al. The Mouse Retina as an Angiogenesis Model. Invest Ophthalmol Vis Sci 51, 2813–2826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman EA Functional Hyperemia and Mechanisms of Neurovascular Coupling in the Retinal Vasculature. J Cereb Blood Flow Metab 33, 1685–1695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobson B & Denekamp J Endothelial Proliferation in Tumours and Normal Tissues: Continuous Labelling Studies. Br J Cancer 49, 405–413 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madisen L et al. A Robust and High-Throughput Cre Reporting and Characterization System for the Whole Mouse Brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Razani B et al. Caveolin-1 Null Mice are Viable but Show Evidence of Hyperproliferative and Vascular Abnormalities. J Biol Chem 276, 38121–38138 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Asterholm IW, Mundy DI, Weng J, Anderson RGW & Scherer PE Altered Mitochondrial Function and Metabolic Inflexibility Associated with Loss of Caveolin-1. Cell Metab 15, 171–185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanlandewijck M et al. A Molecular Atlas of Cell Types and Zonation in the Brain Vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Nitta T et al. Size-Selective Loosening of the Blood-Brain Barrier in Claudin-5-Deficient Mice. J Cell Biol 161, 653–660 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenman JM et al. Canonical Wnt Signaling Regulates Organ-Specific Assembly and Differentiation of CNS Vasculature. Science (80-.). 322, 1247–1250 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Liebner S et al. Wnt/Beta-Catenin Signaling Controls Development of the Blood-Brain Barrier. J Cell Biol 183, 409–417 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daneman R et al. Wnt/β-Catenin Signaling is Required for CNS, but not Non-CNS, Angiogenesis. Proc Natl Acad Sci U S A 106, 641–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran KA et al. Endothelial β-Catenin Signaling Is Required for Maintaining Adult Blood-Brain Barrier Integrity and Central Nervous System Homeostasis. Circulation 133, 177–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalucka J et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Wang Y et al. Ephrin-B2 Controls VEGF-Induced Angiogenesis and Lymphangiogenesis. Nature 465, 483–6 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Kisanuki YY et al. Tie2-Cre Transgenic Mice: A New Model for Endothelial Cell-Lineage Analysis In Vivo. Dev Biol 230, 230–242 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Pu W et al. Mfsd2a+ Hepatocytes Repopulate the Liver During Injury and Regeneration. Nat Commun 7, 13369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ridder DA et al. TAK1 in Brain Endothelial Cells Mediates Fever and Lethargy. J Exp Med 208, 2615–2623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawada Y et al. High Transgene Expression by Lentiviral Vectors Causes Maldevelopment of Purkinje Cells In Vivo. Cerebellum 9, 291–302 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Khabou H, Cordeau C, Pacot L, Fisson S & Dalkara D Dosage Thresholds and Influence of Transgene Cassette in Adeno-Associated Virus-Related Toxicity. Hum Gene Ther 29, 1235–1241 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Goertsen D et al. AAV Capsid Variants With Brain-Wide Transgene Expression and Decreased Liver Targeting After Intravenous Delivery in Mouse and Marmoset. Nat Neurosci (2021). doi: 10.1038/s41593-021-00969-4 [DOI] [PubMed] [Google Scholar]
- 61.Ehling M, Adams S, Benedito R & Adams RH Notch Controls Retinal Blood Vessel Maturation and Quiescence. Development 140, 3051–61 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Batista AR et al. Ly6a Differential Expression in Blood-Brain Barrier Is Responsible for Strain Specific Central Nervous System Transduction Profile of AAV-PHP.B. Hum Gene Ther 31, 90–102 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Tabebordbar M et al. In Vivo Gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells. Science (80-.). 351, 407–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levy JM et al. Cytosine and Adenine Base Editing of the Brain, Liver, Retina, Heart and Skeletal Muscle of Mice Via Adeno-Associated Viruses. Nat Biomed Eng 4, 97–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DiMattia MA et al. Structural Insight into the Unique Properties of Adeno-Associated Virus Serotype 9. J Virol 86, 6947–6958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES - METHODS
- 66.Challis RC et al. Systemic AAV Vectors for Widespread and Targeted Gene Delivery in Rodents. Nat Protoc 14, 379–414 (2019). [DOI] [PubMed] [Google Scholar]
- 67.McQuin C et al. CellProfiler 3.0: Next-Generation Image Processing for Biology. PLoS Biol 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldey GJ et al. Removable Cranial Windows for Long-Term Imaging in Awake Mice. Nat Protoc 9, 2515–2538 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistical data used to generate the graphs presented throughout the manuscript is provided in the form of Source Data and Supplementary Statistical Data files. The AAV-BI30 rep-cap, pAAV-CAG-NLS-GFP-miR122-WPRE-pA, and pAAV-CAG-Cre-miR122-WPRE-pA plasmids generated in this study will be made available on Addgene shortly following publication; in the meantime, the reagents will be available from the corresponding authors upon request.


