Abstract
Phytases (myo-inositol hexakisphosphate phosphohydrolases) are found naturally in plants and microorganisms, particularly fungi. Interest in these enzymes has been stimulated by the fact that phytase supplements increase the availability of phosphorus in pig and poultry feed and thereby reduce environmental pollution due to excess phosphate excretion in areas where there is intensive livestock production. The wild-type phytases from six different fungi, Aspergillus niger, Aspergillus terreus, Aspergillus fumigatus, Emericella nidulans, Myceliophthora thermophila, and Talaromyces thermophilus, were overexpressed in either filamentous fungi or yeasts and purified, and their biophysical properties were compared with those of a phytase from Escherichia coli. All of the phytases examined are monomeric proteins. While E. coli phytase is a nonglycosylated enzyme, the glycosylation patterns of the fungal phytases proved to be highly variable, differing for individual phytases, for a given phytase produced in different expression systems, and for individual batches of a given phytase produced in a particular expression system. Whereas the extents of glycosylation were moderate when the fungal phytases were expressed in filamentous fungi, they were excessive when the phytases were expressed in yeasts. However, the different extents of glycosylation had no effect on the specific activity, the thermostability, or the refolding properties of individual phytases. When expressed in A. niger, several fungal phytases were susceptible to limited proteolysis by proteases present in the culture supernatant. N-terminal sequencing of the fragments revealed that cleavage invariably occurred at exposed loops on the surface of the molecule. Site-directed mutagenesis of A. fumigatus and E. nidulans phytases at the cleavage sites yielded mutants that were considerably more resistant to proteolytic attack. Therefore, engineering of exposed surface loops may be a strategy for improving phytase stability during feed processing and in the digestive tract.
Phytic acid (myo-inositol hexakisphosphate) is the major storage form of phosphorus in plants. In the context of human and animal nutrition, the following two aspects of phytic acid are critically important (18, 29): (i) monogastric animals have only low levels of phytate-degrading enzymes in their digestive tracts, and since phytic acid itself is not resorbed, feed for pigs and poultry commonly is supplemented with inorganic phosphate in order to meet the phosphorus requirements of these animals; and (ii) phytic acid is an antinutrient factor, since it forms complexes with proteins and a variety of metal ions and therefore decreases the dietary availability of these nutrients.
Because of these problems, there is considerable interest in phytate-degrading enzymes. The phytases (myo-inositol hexakisphosphate 3- and 6-phosphohydrolases; EC 3.1.3.8 and 3.1.3.26) are a subfamily of the histidine acid phosphatases (14) and are found naturally in plants and microorganisms, particularly fungi. As a class, the phytases have been rather poorly characterized biochemically (for a recent review see reference 29). In order to more clearly define this class of enzymes, several phytases of fungal origin have been cloned and overexpressed by workers in our company (14–16). In this paper we describe the purification of these fungal phytases, the structural and biophysical properties of these enzymes, and the results of a comparison with a prokaryotic phytase from Escherichia coli.
MATERIALS AND METHODS
Expression in Aspergillus niger.
The DNA fragments encoding the A. niger CB (30), Aspergillus terreus 9A1 (GenBank accession no. U59805), Aspergillus fumigatus (GenBank accession no. U59804), Emericella nidulans (GenBank accession no. U59803), and Myceliophthora thermophila (GenBank accession no. U59806) phytases were ligated as 5′ NcoI or BspHI (introduced sites at the ATG start codon) 3′ blunt-ended fragments downstream of the glaA promoter into the NcoI-EcoRV site of the expression vector, as described by Mitchell et al. (14) and Pasamontes et al. (15). Transformation of A. niger NW205 (ura− arg− nic−; kindly provided by F. Buxton) and screening for phytase-producing transformants were also done as described previously (15).
The phytase of A. niger (Natuphos; GenBank accession no. Z16414) was a commercial preparation obtained from BASF (Ludwigshafen, Germany) and was purified to homogeneity by anion-exchange chromatography.
Expression in Saccharomyces cerevisiae.
The phytase genes were cloned into pScer-Ro11, a 2μ-based vector harboring a shortened version of the gap(FL) promoter and the pho5 terminator (12), as well as the ura3 gene as a selection marker. The intronless phytase genes of A. fumigatus, A. niger CB, and A. terreus CBS were cloned as EcoRI-EcoRV fragments downstream of the gap(FL) promoter in the EcoRI-BamHI blunt-ended expression cassette. The gene for E. nidulans phytase was cloned as an EcoRI fragment into the corresponding site of pScer-Ro11. S. cerevisiae YMR4 (ura− his− leu− pho3− pho5−; kindly provided by M. Riederer) was used for transformation. Individual transformants were grown initially for 1 to 2 days in minimal medium. Phytase production was tested after subsequent culture for 2 to 3 days in YPD medium.
Expression in Hansenula polymorpha.
The phytase genes (intronless) of A. terreus CBS (GenBank accession no. U60412), A. fumigatus, and Talaromyces thermophilus (GenBank accession no. U59802) were cloned as EcoRI fragments into the corresponding site of the H. polymorpha expression vector pFP (4) downstream of the formate dehydrogenase (FMD) promoter (9). The resulting plasmids were transformed into H. polymorpha RB11 (ura−). About 300 to 400 transformants of each construct were inoculated into minimal medium (YNB containing 2% glucose). After several passages under selective pressure to force multiple integration of the expression plasmids into the genome of H. polymorpha, single stable clones were tested for phytase activity.
In order to obtain acceptable expression levels, the first 35 N-terminal amino acids of the T. thermophilus phytase were replaced by the amino acid sequence MGVFVVLLSIATLFGSTSGTALGPRGNHSKSCDTA35 (the underlined amino acids originate from A. terreus CBS phytase and contain the signal sequence). Amino acids K30SCDTA35 were unrelated residues resulting from the cloning strategy used to replace the N terminus. Computer modelling of the chimeric T. thermophilus phytase suggested that modification of the first 16 amino acids of the mature protein was not likely to have an effect on the biochemical properties of the enzyme.
Protein purification.
Independent of the expression system used, the culture broths (typically 500 to 1,000 ml) were centrifuged to remove the cells and were concentrated by ultrafiltration with Amicon 8400 cells (PM30 membranes; Grace AG, Wallisellen, Switzerland) and ultrafree-15 centrifugal filter devices (Biomax-30K; Millipore, Bedford, Mass.). The concentrates (typically 1.5 to 5 ml) were desalted with either Fast Desalting HR 10/10 or Sephadex G-25 Superfine columns (Pharmacia Biotech, Dübendorf, Switzerland); 10 mM sodium acetate (pH 5.0) was used as the elution buffer. The desalted A. fumigatus samples were directly loaded onto a 1.7-ml Poros HS/M cation-exchange chromatography column (PerSeptive Biosystems, Framingham, Mass.). When the other phytases were expressed in A. niger or H. polymorpha, they were loaded onto a 1.7-ml Poros HQ/M anion-exchange chromatography column. During both anion-exchange and cation-exchange chromatography, phytase was eluted in pure form by using an optimized sodium chloride gradient.
All of the phytases expressed in S. cerevisiae (except A. fumigatus phytase) were brought to 2 M (NH4)2SO4 after desalting and were loaded onto a 1-ml Butyl Sepharose 4 Fast Flow hydrophobic interaction chromatography column (Pharmacia Biotech). The enzymes were eluted with a linear 2 to 0 M (NH4)2SO4 gradient in 10 mM sodium acetate (pH 5.0). The phytases eluted in the breakthrough and were concentrated and loaded onto a 120-ml Sephacryl S-300 gel permeation chromatography column (Pharmacia Biotech). They eluted as symmetrical peaks and were determined to be pure by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
E. coli phytase.
In order to clone the appA gene of E. coli, total DNA from E. coli M15 (25) was prepared as described by Davis et al. (3). A 500-ng portion of this DNA was used in a PCR performed with the following primers designed by using the appA sequence published by Dassa et al. (2) (GenBank accession no. M58708): appA I (5′-AAACATATTCATGAAAGCGATCTTAATCCCA-3′), including an RcaI site (sequence in italics); and appA II (5′-ATATAGGATCCCAAACTGACAGCCGGTTATGCG-3′), including a BamHI site (sequence in italics). The PCR was performed as described by the manufacturer (Expand High Fidelity PCR kit; Boehringer, Mannheim, Germany) by using a hybridization temperature of 55°C. The resulting PCR product was separated from the primers by using a PCR purification kit obtained from Qiagen (Hilden, Germany), was digested with BamHI and RcaI, and was purified by agarose gel electrophoresis and a subsequent gel elution step (Qiaex II; Qiagen). The appA gene was ligated into the BamHI and NcoI sites of the pBluescript II SK vector (Stratagene, La Jolla, Calif.). A DNA sequence analysis of the gene revealed two differences when it was compared to the sequence deposited by Dassa et al. (2). One of these differences was a change from A to G at position 620. This difference also affected the amino acid sequence of the appA gene product; the glycine at position 207 was replaced by aspartic acid. The second sequence difference was at position 984 (change from G to A) and was a silent mutation. The appA gene was transferred into the pQE60 expression vector (Qiagen) containing a C-terminal 6xHis tag and a short linker sequence (Gly-Ser-Arg-Ser-His-His-His-His-His-His) and was transformed into E. coli BL21 (Stratagene).
A 500-ml portion of Luria-Bertani medium containing 200 μg of ampicillin per ml and 30 μg of kanamycin per ml was inoculated with 8 ml of an overnight culture of strain BL21 harboring the appA expression plasmid. When the culture reached an optical density at 600 nm of 1.0, the cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated for an additional 5 h at 37°C with vigorous shaking. The cells were harvested by centrifugation at 4000 × g for 20 min, resuspended in 30 ml of sonication buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 20 mM imidazole, 1 mg of lysozyme per ml). The suspension was incubated on ice for 15 min before the cells were disrupted by sonication (Vibra Cell 72408; Bioblock Scientific, Illkirch, France) at 60% power by using 2.5-s bursts, 2.5-s cooling periods, and a net sonication time of 3 min. The supernatant was clarified by centrifugation at 10,000 × g for 15 min, mixed with 8 ml of Ni2+-agarose (50% resin suspension), equilibrated with sonication buffer, and incubated for 1 h on ice. Then the mixture was poured into a column and washed with 85 ml of sonication buffer. Phytase was eluted with 40 ml of 20 mM sodium acetate buffer (pH 4.5) containing 300 mM NaCl.
SDS-PAGE and IEF.
SDS-PAGE was performed on 8 to 16% Tris–glycine gradient gels, and isoelectric focusing (IEF) was performed on IEF pH 3 to 7 or pH 3 to 10 gels (Novex, San Diego, Calif.). The gels were stained with colloidal Coomassie blue (Novex) or were semi-dry-blotted onto Immobilon PSQ (polyvinylidene difluoride) membranes (Millipore) and then stained with amido black (naphthol blue black).
Gel permeation chromatography.
The molecular sizes of the purified proteins were determined at room temperature by gel filtration performed with a calibrated Superdex 200 column (fast-protein liquid chromatography; Pharmacia Biotech). The elution buffer normally contained 50 mM sodium phosphate, 150 mM NaCl, 0.2 mM Na2EDTA, 2 mM 2-mercaptoethanol, and 1 mM sodium azide (pH 7.2). In order to determine the effect of phosphate on the molecular size of A. fumigatus phytase, gel permeation chromatography was also performed with an elution buffer containing 100 mM sodium acetate (pH 5.0). The gel filtration column was calibrated for both elution buffers with high- and low-molecular-weight kits obtained from Pharmacia Biotech, which contained thyroglobulin (Mr, 749,000; Stokes radius, 85.0 Å), ferritin (Mr, 421,000; Stokes radius, 61.0 Å), catalase (Mr, 211,000; Stokes radius, 52.2 Å), aldolase (Mr, 163,000; Stokes radius, 48.1 Å), bovine serum albumin (Mr, 71,700; Stokes radius, 35.5 Å), ovalbumin (Mr, 45,700; Stokes radius, 30.5 Å), chymotrypsinogen A (Mr, 20,200; Stokes radius, 20.9 Å), and RNase A (Mr, 15,700; Stokes radius, 16.4 Å).
Analytical ultracentrifugation.
Analytical ultracentrifugation was performed in 10 mM sodium acetate (pH 5.0) by using a model Optima XL-A ultracentrifuge (Beckman, Palo Alto, Calif.) equipped with a type An-60 Ti rotor and cells having standard double-sector Epon aluminum-filled centerpieces. The data were analyzed with the program DISCREEQ of Schuck (20), which is based on an iterative Marquardt procedure. The specific volume of a protein depends not only on the amino acid sequence but also on the extent and type of glycosylation. Since glycosylation of fungal phytases most likely is the high-mannose type of glycosylation, all sugars were assumed to be mannose. This assumption was a simplification but introduced only a small error into the Mr calculation.
Mass spectrometry.
The peptides of interest obtained from trypsin digestion of A. niger phytase (Fig. 1) were analyzed by electrospray mass spectrometry. All analyses were performed in the positive ion mode with a triple quadrupole instrument (model API III; SCIEX, Concord, Ontario, Canada). Scans between m/z 400 and m/z 1600 were recorded with 0.2-average-mass-unit steps. A. fumigatus phytase was analyzed with a PerSeptive Biosystems Voyager Elite mass spectrometer equipped with a reflectron and delayed extraction. The ion acceleration voltage was 20 kV, and the results of 100 to 200 scans were averaged.
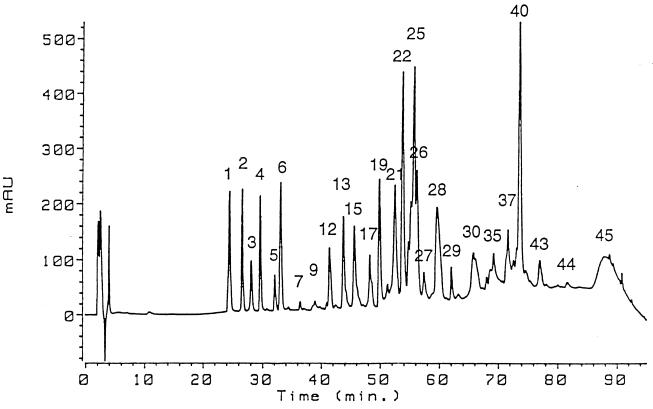
FIG. 1.
Separation of A. niger phytase fragments by reversed-phase HPLC after carboxymethylation and trypsin digestion of the protein. For experimental details see the text. AU, arbitrary units.
N-terminal sequencing and calculation of theoretical Mr and pI values.
Automated Edman degradation of purified phytases was performed with a model ABD 494HT sequencer (Perkin-Elmer, Foster City, Calif.) with on-line microbore phenylthiohydantoin-amino acid detection. Theoretical Mr and pI values were calculated from the amino acid sequences of the native proteins (without a signal sequence) with the programs EditSeq and Protean from the Lasergene software package of DNASTAR Inc. (Madison, Wis.).
Proteolytic susceptibility of A. fumigatus phytase mutants.
Purified A. fumigatus wild-type phytase and the A. fumigatus phytase mutants S126N and R125L/S126N were pretreated for 20 min at 90°C in order to inactivate potentially interfering traces of protease and then were renatured in the cold. Subsequently, they were incubated at 50°C at a concentration of 175 μg/ml in 10 mM sodium acetate (pH 5.0) with a 1:400-diluted 3-day culture supernatant of A. niger NW205 containing proteolytic activity. After 0, 20, 40, 60, 90, and 120 min of incubation, aliquots were subjected to SDS-PAGE or phytase activity was measured at 37°C. In control experiments, A. fumigatus wild-type phytase was incubated under the same conditions without NW205 culture supernatant or with NW205 culture supernatant that had been pretreated for 20 min at 90°C in order to inactivate the protease(s).
Identification of the glycosylation sites of A. niger phytase.
A. niger phytase was S-carboxymethylated and digested with trypsin in 100 mM ammonium bicarbonate buffer (pH 8.0) at a phytase-to-protease ratio of 50:1 (wt/wt) for 18 h at 37°C. The reaction was stopped by adding 10% trifluoroacetic acid. The proteolytic fragments were separated at room temperature by reversed-phase high-performance liquid chromatography (HPLC) by using a C18 column (250 by 2.1 mm; Vydac, Hesperia, Calif.) and a linear 0 to 95% acetonitrile gradient in 0.1% trifluoroacetic acid (Fig. 1). All of the peptides were collected and analyzed by electrospray mass spectrometry. In addition, fractions 9, 17, 21, 27 to 30, 35, 37, and 43 to 45 (Fig. 1) were analyzed by Edman degradation. Assignments of glycosylated Asn residues were made on the basis of the absence of the phenylthiohydantoin derivative of Asn in the corresponding Edman cycles and by comparison of the peptide sequences with the predicted sequence of A. niger phytase.
Phytase activity measurements.
Phytase activity was measured in an assay mixture containing 0.5% (∼5 mM) phytic acid and 200 mM sodium acetate (pH 5.0). After 15 min of incubation at 37°C (or at temperatures between 37 and 90°C), the reaction was stopped by adding an equal volume of 15% trichloroacetic acid. The liberated phosphate ions were quantified by mixing 100 μl of the assay mixture with 900 μl of H2O and 1 ml of 0.6 M H2SO4–2% ascorbic acid–0.5% ammonium molybdate. After 20 min of incubation at 50°C, absorbance at 820 nm was measured. Standard solutions of potassium phosphate were used as a reference. One unit of phytase activity was defined as the amount of activity that liberates 1 μmol of phosphate per min at 37°C.
RESULTS
Purification of the phytases.
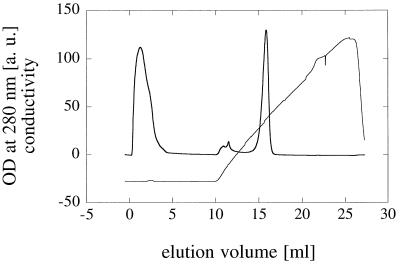
Phytase genes from different fungal species were expressed in three different host organisms. A. fumigatus phytase is the only fungal phytase having an isoelectric point greater than 7.0 (Table 1). Since the pI of this phytase is considerably higher than the pIs of all of the extracellular proteins produced by the different expression strains, purification of A. fumigatus phytase was essentially a one-step procedure. As Fig. 2 shows, A. fumigatus phytase eluted as a symmetrical peak from a cation-exchange chromatography column. Probably because of acidic glycosylation, the pI of A. fumigatus phytase expressed in H. polymorpha was less than 4.0 (Fig. 3A). Nevertheless, cation-exchange chromatography could still be used for purification of this protein, although the capacity was much lower (data not shown).
TABLE 1.
Isoelectric points of fungal and E. coli phytases
| Source of phytase | pI calculated by using amino acid sequence of mature protein | pI determined experimentally by IEF | n |
|---|---|---|---|
| A. niger (Natuphos) | 4.78 | 5.06 ± 0.01 (m)b | 4 |
| 5.24 ± 0.01 | 4 | ||
| 4.92 ± 0.01 | 4 | ||
| 4.80 ± 0.01 | 4 | ||
| A. niger CB | 4.79 (27)a, 4.85 (24, 25) | 5.21 ± 0.01 | 4 |
| A. terreus 9A1 | 5.08 | 4.97 ± 0.02 (double band) | 3 |
| 4.93 ± 0.01 | 3 | ||
| A. terreus CBS | 5.50 (20), 5.38 (26) | —c | |
| A. fumigatus | 7.28 | 8.56 ± 0.02 (m) | 6 |
| 8.12 ± 0.02 | 6 | ||
| E. nidulans | 5.27 (23, 26) | 5.29 ± 0.01 (m) | 3 |
| 5.39 ± 0.01 | 3 | ||
| 5.14 ± 0.01 | 3 | ||
| M. thermophila | 4.95 | 4.83 ± 0.01 (quadruplet) | 6 |
| 4.73 ± 0.01 | 6 | ||
| 4.63 ± 0.01 | 6 | ||
| 4.55 ± 0.01 | 6 | ||
| T. thermophilus | 5.23 | — | |
| E. coli | 7.01d | 7.52 ± 0.01 | 3 |
| 7.37 ± 0.02 (m) | 3 |
N-terminal residue of the mature protein species (see Table 2) having the respective calculated pI value.
(m), major band.
—, There was a broad smear below pI 4.0 due to acidic glycosylation when the phytase was expressed in H. polymorpha; all of the other fungal phytases were expressed in A. niger.
The rather high pI of E. coli phytase was probably due to the His6 tag at the C terminus of the protein. Without this His6 tag, the predicted pI was 6.35.
FIG. 2.
Purification of A. fumigatus phytase by cation-exchange chromatography. An aliquot of a concentrated A. niger culture supernatant containing A. fumigatus phytase was loaded onto a 1.7-ml Poros HS/M cation-exchange chromatography column and eluted with a linear sodium chloride gradient. A. fumigatus phytase eluted as a symmetrical peak at an elution volume of approximately 15.5 ml. OD, optical density; a.u., arbitrary units.
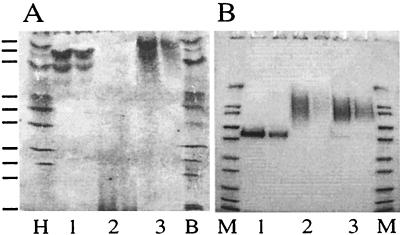
FIG. 3.
Extent of glycosylation and its effect on the isoelectric point of A. fumigatus phytase in different expression systems. (A) IEF pH 3 to 10 gel. (B) SDS-PAGE gel. Purified A. fumigatus phytase was expressed in A. niger (lanes 1), H. polymorpha (lanes 2), or S. cerevisiae (lanes 3) (two lanes with different protein concentrations were used for each expression system). Lane H, high-pI kit (Pharmacia Biotech); lane B, broad-pI kit (Pharmacia Biotech) (from top to bottom, pI 8.65, 8.45, 8.15, 7.35, 6.85, 6.55, 5.85, 5.20, 4.55, and 3.50); lanes M, Mark 12 molecular weight standard (Novex) containing myosin (Mr, 200,000), β-galactosidase (116,300), phosphorylase b (97,400), albumin (66,300), glutamic dehydrogenase (55,400), lactate dehydrogenase (36,500), carbonic anhydrase (31,000), trypsin inhibitor (21,500), lysozyme (14,400), and aprotinin (6,000).
The other fungal phytases expressed in A. niger or H. polymorpha had pI values that were less than 5.5 and, accordingly, could be purified by anion-exchange chromatography (data not shown). Although the high level of glycosylation associated with expression in S. cerevisiae had no effect on the pI of A. niger CB, A. terreus CBS, or E. nidulans phytase (data not shown), all attempts to bind these proteins to anion-exchange chromatography columns failed. Consequently, we used an alternative approach, in which these proteins were subjected to hydrophobic interaction chromatography followed by preparative gel permeation chromatography, which yielded homogeneous products (data not shown).
N-terminal sequences.
In no instance did the N terminus of the mature fungal phytase that was determined experimentally (Table 2) correspond to the N terminus that was predicted on the basis of the von Heijne rules (26, 27). This finding is surprising and contrasts with observations made with other acid phosphatases (data not shown). Whether this deviation from the theoretical results is an inherent property of phytases or whether the initial processing conformed to the von Heijne rules but was followed by additional proteolytic cleavage could not be determined in this study. However, the latter possibility was supported by the finding that the A. niger NRRL 3135 (24), A. niger CB, A. terreus CBS, and E. nidulans phytases had multiple N-terminal sequences (Table 2). In the case of E. coli phytase, our sequence data agree with previously published N-terminal sequence data for the protein (2, 6). However, the amount of amino acid recovered in the Edman cycles was small compared with the amount of purified protein applied, suggesting that a large proportion of E. coli phytase is N-terminally blocked.
TABLE 2.
N-terminal sequences of fungal and E. coli phytases
| Source of phytase | Sequenceb | Position of first residue |
|---|---|---|
| A. niger (Natuphos) | XQSSXDTVDQGYQXF | 27 (19)c |
| A. niger CB | XQSTXDTV | 27 (19) |
| SRXQSTX | 25 | |
| ASRXQST | 24 | |
| A. terreus 9A1 | SDXNSVDHGY | 29 (19) |
| A. terreus CBSa | TALGPXGX | 20 (19) |
| GXHSDXTSVD | 26 | |
| A. fumigatus | SKSXDTVDLGY | 27 (21) |
| E. nidulans | VVQNHSX | 23 (20) |
| NHSXNTA | 26 | |
| M. thermophila | SESRP | 21 (18) |
| T. thermophilusa | TALGPRGXHSK(S)XDTAX(G)(G)XQ | 20 (19) |
| E. coli | QSE(P)ELKL(E) | 23 (23) |
These proteins were expressed in H. polymorpha; all of the other fungal phytases were expressed in A. niger.
Some of the Xs represent Asn residues that were probably glycosylated.
The number in parentheses indicates the first residue of the mature protein predicted by using the von Heijne rules.
Biophysical properties of fungal phytases are not affected by different extents of glycosylation.
A. fumigatus phytase was expressed in three different expression systems, A. niger, H. polymorpha, and S. cerevisiae. In all of these expression systems, the protein was glycosylated, although to different extents (Fig. 3B). While glycosylation was moderate in A. niger, it was excessive and highly variable in H. polymorpha and S. cerevisiae, as indicated by the broad bands spanning Mrs ranging from 75,000 to 160,000. Different extents of glycosylation were observed even with different batches of A. fumigatus phytase expressed in the same expression system (Table 3).
TABLE 3.
Molecular sizes of fungal and E. coli phytases as determined experimentally or by using the amino acid sequences of the mature proteins
| Source of phytase |
Mr as determined by:
|
Gel filtration analysis
|
Mr as determined by mass spectrometry | ||||
|---|---|---|---|---|---|---|---|
| Sequence analysis | SDS-PAGE | Analytical ultracentrifugation | Mr | Stokes radius (Å) | nc | ||
| A. niger (Natuphos) | 48, 423 | 66,360 ± 2,440 (5)b | 64,890 | 82,360 ± 1,710 | 39.1 ± 0.3 | 4 | NDc |
| A. niger CB | 48,658-48,973 | 74,530 ± 2,800 (5) | 71,020 | 103,030 ± 1,840 | 42.3 ± 0.3 | 4 | ND |
| A. terreus 9A1 | 48,189 | 60,550 ± 1,340 (8) | 70,380 | 79,900 ± 910 | 38.7 ± 0.2 | 3 | ND |
| A. terreus CBSa | 48,570-49,166 | 82,110 ± 720 (3) | 78,030 | 115,100 ± 300 | 43.8 ± 0.1 | 3 | ND |
| A. fumigatus batch 1 | 48,276 | 72,360 ± 520 (5) | 70,740 | 90,500 ± 1,630 | 40.5 ± 0.3 | 4 | |
| A. fumigatus batch 2 | 60,770 ± 2,150 (9) | 62,500-62,800 | |||||
| E. nidulans batch 1 | 49,034-49,360 | 66,430 ± 2,300 (5) | 67,400 | 77,850 ± 600 | 38.4 ± 0.1 | 3 | ND |
| E. nidulans batch 2 | 86,930 ± 1330 | 39.9 ± 0.2 | 3 | ||||
| M. thermophila | 50,524 | 62,890 ± 1,210 (4) | 66,150 | 73,800 ± 200 | 37.6 ± 0.1 | 3 | ND |
| T. thermophilusa | 49,775 | 128,400 ± 1,700 (3) | 137,140 | ND | ND | ND | |
| E. coli | 45,846 | 47,270 ± 60 (3) | 47,060 | 39,190 ± 200 | 28.7 ± 0.1 | 3 | 45,833 |
These proteins were expressed in H. polymorpha; all of the other fungal phytases were expressed in A. niger.
The numbers in parentheses are the numbers of preparations examined.
ND, not determined.
When the protein was expressed in A. niger and S. cerevisiae, the different extents of glycosylation either had no or only a minor effect on the pI of the protein, suggesting that glycosylation was neutral (Fig. 3A). On the other hand, the pI of A. fumigatus phytase expressed in H. polymorpha was less than 4.0. Since deglycosylation with endoglycosidase F1 resulted in a pI of >8.0 (data not shown), the latter finding must have been due to acidic glycosylation. Remarkably, the different extents and patterns of glycosylation had no significant effect on the specific activity of the enzyme (31).
Virtually identical observations were made with A. niger CB, A. terreus CBS, and E. nidulans phytases which were also produced in different expression systems (data not shown).
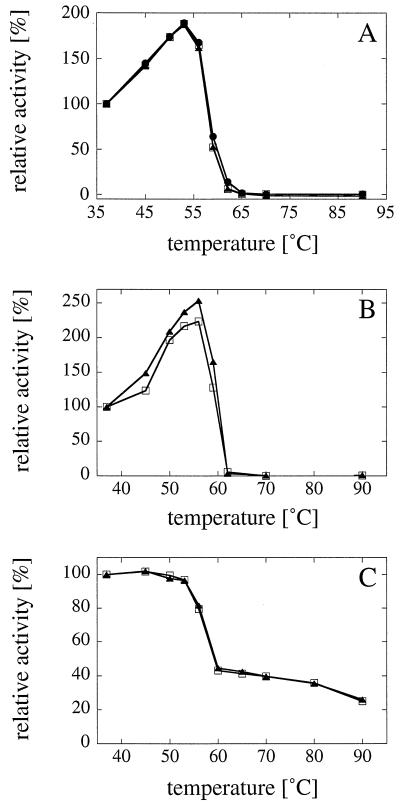
Different extents of glycosylation may have an impact on the structure, stability, and function of proteins (11, 19, 28). In order to determine whether the extent of glycosylation had an effect on phytase stability, the activities of A. fumigatus phytase expressed in A. niger, H. polymorpha, or S. cerevisiae (Fig. 4A) and the activities of A. niger CB phytase expressed in A. niger or S. cerevisiae (Fig. 4B) were measured at a range of temperatures between 37 and 90°C. Evidently, the different extents of glycosylation had no effect on the thermostabilities of these two phytases. In addition, A. niger CB phytase expressed in either A. niger or S. cerevisiae was incubated for 20 min at temperatures between 37 and 90°C and then was incubated for 1 h at 4°C in order to allow (partial) refolding of the heat-denatured protein. Subsequent activity measurements at 37°C (Fig. 4C) revealed that the different extents of glycosylation had no effect on the refolding properties.
FIG. 4.
Different extents of glycosylation do not affect the thermostability and refolding of phytase. The enzymatic activity of A. fumigatus phytase (A) expressed in A. niger (□), H. polymorpha (•), or S. cerevisiae (▴) and the enzymatic activity of A. niger CB phytase (B) expressed in A. niger (□) or S. cerevisiae (▴) were measured at a series of temperatures between 37 and 90°C. (C) A. niger CB phytase expressed in A. niger (□) or S. cerevisiae (▴) was incubated for 20 min at 37, 45, 50, 53, 56, 60, 65, 70, 80, or 90°C and then for 1 h at 4°C. Subsequently, phytase activity was measured at 37°C. It is evident that the different extents of glycosylation had no or only minor effects on the thermostability and refolding properties of the phytases.
Fungal phytases are monomeric proteins.
The molecular sizes of the phytases were determined by SDS-PAGE, analytical ultracentrifugation, gel permeation chromatography, and mass spectrometry and were calculated theoretically by using the amino acid sequences of the mature proteins (Table 3). While SDS-PAGE, mass spectrometry, and amino acid sequence determinations provided the Mr of the protomers, analytical ultracentrifugation and gel permeation chromatography provided values for the native proteins. The results obtained by the different methods agreed closely, and the data showed that all of the phytases investigated are monomeric proteins. There was no indication of higher oligomeric forms of phytase in any experiment. This interpretation is consistent with the results of an analysis of the three-dimensional structure, which also showed that the A. niger phytase is a monomeric protein (13).
The difference between the Mr determined by SDS-PAGE and the Mr calculated by using the amino acid sequence is an indication of the extent of glycosylation of a protein. In the examples listed above, the extent of glycosylation ranged from 20 to 65% of the total Mr, but it may be even higher. While mass spectrometry of E. coli phytase was straightforward, a broad, diffuse peak was obtained for A. fumigatus phytase. This finding is consistent with the difficulties encountered with mass spectrometry of highly glycosylated proteins.
For all of the glycosylated phytases, gel permeation chromatography consistently gave a higher Mr than expected. The difference between the values determined by gel filtration and SDS-PAGE (or analytical ultracentrifugation) increased with the extent of glycosylation, which was calculated from the difference between the Mr obtained by SDS-PAGE (or analytical ultracentrifugation) and the Mr obtained by amino acid sequence analysis (Fig. 5). Therefore, the higher Mrs obtained by gel filtration are most likely artifacts due to glycosylation. Similar observations have previously been made for Sephadex columns, in which glycoproteins also eluted earlier than expected (1).
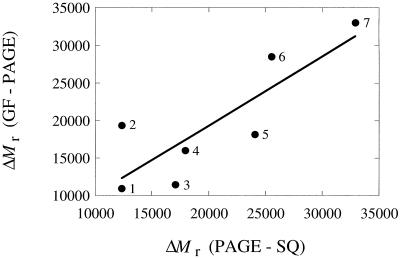
FIG. 5.
Gel filtration artifacts due to protein glycosylation. Gel permeation chromatography of the glycosylated phytases resulted in higher Mrs than expected on the basis of SDS-PAGE, mass spectrometry, or analytical ultracentrifugation data. The overestimation, as expressed by the difference between the Mrs obtained by gel filtration and SDS-PAGE (y axis), increased with the extent of protein glycosylation, as expressed by the difference between the Mrs obtained by SDS-PAGE and amino acid sequence analysis (x axis). The regression line has the equation y = 904.1 + 0.9203x and an R value of 0.837. The data were obtained from Table 3. Data point 1, M. thermophila phytase; data point 2, A. terreus 9A1 phytase; data point 3, E. nidulans phytase; data point 4, A. niger phytase (Natuphos); data point 5, A. fumigatus phytase; data point 6, A. niger CB phytase; data point 7, A. terreus CBS phytase. GF, gel filtration; SQ, sequence analysis.
Isoelectric points.
The isoelectric points of the different phytases were calculated by using the amino acid sequences of the mature proteins (without signal sequences) and were determined experimentally by IEF (Table 1). In general, there was a good correlation between the calculated and observed pI values, and the maximum difference was 1.28 pH units. This suggests that glycosylation in most instances either had no effect or had only a minor effect on the pI of the protein. The only exceptions were phytases expressed in H. polymorpha, in which a pronounced shift to acidic pI values and considerable pI heterogeneity were observed (Fig. 3A). As Table 1 shows, A. fumigatus phytase is peculiar in that it has a much higher pI than all of the other fungal phytases. Since the pI of this phytase also is higher than the pI values of most extracellular proteins produced by the expression strains used, A. fumigatus phytase is particularly suited to easy and efficient purification on an industrial scale. Because of the differences in pI values between individual phytases or for a single phytase expressed in different production hosts, different strategies had to be used for protein purification (see above).
Site-directed mutagenesis of surface-exposed cleavage sites decreases the proteolytic susceptibility of fungal phytases.
When expressed in A. niger and stored as concentrated culture supernatants at 4°C, the phytases from A. fumigatus, E. nidulans, A. terreus 9A1, and M. thermophila had a tendency to undergo proteolytic degradation (data not shown). The results of N-terminal sequencing of the fragments (V153VPFIRASGS for A. fumigatus phytase, R187ATPVVNV and A188TPVVNV for E. nidulans phytase, XP192SPRVDVAI for A. terreus 9A1 phytase, and G282RPLSPFXR for M. thermophila phytase) suggested that cleavage occurred between amino acids S-152 and V-153, K-186 and R-187 or R-187 and A-188, H-190 and Q-191, and N-281 and G-282, respectively. A comparison with the three-dimensional structure of A. niger phytase (13) and homology modelling of the other phytases (23a) revealed that all of the cleavage sites occur within surface-exposed loop structures or turns and are therefore accessible to proteases. While limited proteolysis of the A. fumigatus phytase between amino acids 152 and 153 was associated with pronounced or even complete inactivation of the enzyme (Fig. 6B), cleavage of the E. nidulans phytase between amino acids 186 and 187 or between amino acids 187 and 188 seemingly had no effect on the specific activity (data not shown). The two proteolytic fragments of both phytases remained associated with each other, as shown by gel permeation chromatography, as well as by copurification (data not shown). Site-directed mutagenesis at the protease-sensitive sites of A. fumigatus phytase (S152N and R151L/S152N) (Fig. 6B) and E. nidulans phytase (K186G/R187Q) (data not shown) yielded mutant proteins whose susceptibility to proteolysis was reduced considerably. On the other hand, the R151L and S152N mutations had no effect on the specific activity of A. fumigatus phytase, which was 20.3, 20.5, and 21.2 U/mg after heat pretreatment for the wild-type, the R151L/S152N, and the S152N enzymes, respectively.
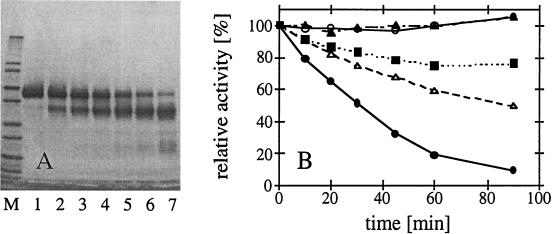
FIG. 6.
Proteolytic susceptibility of A. fumigatus phytase expressed in A. niger. (A) Purified A. fumigatus phytase was incubated at 50°C with a diluted A. niger NW205 culture supernatant containing proteolytic activity. After 0, 10, 20, 30, 45, 60, and 90 min of incubation, aliquots were subjected to SDS-PAGE (lanes 1 to 7, respectively). Lane M contained the Mark 12 molecular weight standard (see the legend to Fig. 3). (B) A. fumigatus wild-type phytase (•), A. fumigatus S126N (■), and A. fumigatus R125L/S126N (▵) were incubated with diluted NW205 culture supernatant at 50°C for 0, 10, 20, 30, 45, 60, and 90 min, and then phytase activity was measured at 37°C. As controls, A. fumigatus wild-type phytase was incubated under the same conditions without NW205 culture supernatant (○) or with NW205 culture supernatant that had been pretreated for 20 min at 90°C in order to inactivate the protease(s) (▴). The decreased rate of inactivation of the mutants was paralleled by much slower accumulation of degradation products on SDS-PAGE gels (data not shown).
In contrast to expression in A. niger, problems of proteolytic degradation were not encountered when the phytases were expressed in H. polymorpha (data not shown).
Identification of the glycosylation sites of A. niger phytase.
A. niger phytase has 10 potential sites for N-linked glycosylation of the Asn-X-Ser/Thr type; these sites occur at residues 27, 59, 105, 120, 207, 230, 339, 352, 376, and 388, all of which are exposed on the surface of the molecule, as determined by an examination of the three-dimensional structure (13). In order to identify which of these residues are actually glycosylated in the mature protein, A. niger phytase was reduced, carboxy-methylated, and digested with trypsin. The proteolytic fragments were separated by reversed-phase HPLC (Fig. 1) and analyzed by electrospray mass spectrometry and Edman sequencing. Asn residues 27 (four peptides analyzed), 105 (one peptide analyzed), 207 (two peptides analyzed), 230 (two peptides analyzed), 339 (one peptide analyzed), and 376 (three peptides analyzed) were glycosylated in all of the peptides analyzed. In contrast, Asn-59 and Asn-120 were both glycosylated in only one of the two peptides examined. No glycosylation was observed for Asn-352 and Asn-388 in the one and three peptides analyzed, respectively. The incomplete glycosylation of Asn residues 59 and 120 may be one of the reasons for the Mr heterogeneity of A. niger phytase.
DISCUSSION
Depending on the application, an enzyme in which there is commercial interest should fulfill a series of predefined quality criteria. In the case of phytase, which is added to animal feed to increase the availability of phosphorus, these criteria include high specific activity, broad substrate specificity, a broad pH optimum, and good stability during storage, feed pelleting, and passage through the digestive tract. Thermostability is a particularly important issue since feed pelleting is commonly performed at temperatures between 65 and 95°C. In order to obtain information which can be used for engineering improved phytases, the range of phytase properties that occur in nature was investigated in this study by biochemically and biophysically characterizing different wild-type phytases, most of which were fungal phytases.
A comparison of the molecular sizes obtained by analytical ultracentrifugation, gel permeation chromatography, mass spectrometry, SDS-PAGE, and amino acid sequence analyses (Table 3), as well as by X-ray crystallography (13), clearly showed that all of the fungal phytases considered as well as E. coli phytase are monomeric proteins. This conclusion is consistent with the findings of Ullah and coworkers for fungal and soybean seed phytases (5; for a review see reference 29), of Greiner et al. (6, 7) for E. coli and Klebsiella terrigena phytases, and of Shimizu (22) for Bacillus subtilis phytase. On the other hand, the phytases of A. terreus, Aspergillus oryzae, Schwanniomyces castellii, and maize roots were reported to be homohexameric, heterotetrameric or dimeric proteins (10, 21, 23, 32). The proposed hexameric structure of A. terreus phytase is particularly surprising, since in our study there was no indication of oligomeric forms of A. terreus phytases isolated from two different strains. Since inorganic phosphate might act as an effector molecule for phytases and the phosphate normally present in the gel filtration buffer may therefore have caused dissociation of phytase oligomers into monomers, gel permeation chromatography was also performed in the absence of inorganic phosphate. Again, there was no indication of higher oligomeric forms (data not shown).
In the four conflicting examples mentioned above, the molecular sizes of the phytases were determined by gel filtration on Sephadex G-200, Sepharose S-200, and Sephacryl HR S300 (10, 21, 32) or by native PAGE (23). As shown previously (1) and in this study (Fig. 5), gel filtration of glycosylated proteins results in overestimates of the molecular sizes, and the error increases with the extent of glycosylation. Calculation of the Mr from gel filtration experiments is based on the assumption that the proteins have a compact globular shape. As indicated by Andrews (1), glycoproteins may “have more expanded structures [… ], which may well be due to a greater hydration in solution of carbohydrate chains as compared with polypeptide chains.” In conclusion, gel permeation chromatography and possibly also native PAGE should not be considered reliable techniques for determining the molecular sizes of fungal phytases and of (highly) glycosylated proteins in general. Most, if not all, phytases of fungi, bacteria, and plants are monomeric proteins, and conflicting conclusions should be regarded with caution at least until there is more convincing evidence that oligomeric forms occur.
We found that glycosylation of the phytases was highly variable. It differed not only among the three expression systems used (Fig. 2) but also among different batches of a phytase produced in the same expression system (Table 3). As shown by the analysis of the glycosylation pattern of A. niger phytase (Fig. 1), some of the heterogeneity is due to the fact that 2 of the 10 potential N-glycosylation sites, Asn-59 and Asn-120, are nonstoichiometrically modified. Remarkably, different extents of glycosylation, such as those obtained when a phytase was produced in different expression systems (Fig. 3), had no effect on phytase thermostability and refolding properties (Fig. 4). The importance of glycosylation for phytase structure and function is further questioned by the fact that only two potential N-glycosylation sites, Asn-207 and Asn-339, are conserved in all of the fungal phytases whose primary structures are known (16).
Glycosylation may have a number of effects on the properties of an enzyme. First, it may have an impact on the stability of a protein, or it may influence the catalytic properties. Second, in case of acidic carbohydrate modification, it may influence the pI of a protein (Fig. 3) and thereby change the behavior of the protein during purification. And third, by diverting metabolic energy, it may lower the level of expression of a phytase. With regard to basic research, it is noteworthy that heterogeneous glycosylation may impede crystallization of a protein and therefore determination of its three-dimensional structure. Strategies for circumventing this problem by deglycosylating glycoproteins, particularly phytase, with recombinant fusion protein glycosidases have recently been described by Grueninger-Leitch et al. (8).
All of the fungal, bacterial, and plant phytases investigated so far have acidic pI values (5–7, 10, 22, 23; for a review see reference 29). The difference between the pI values for E. coli phytase determined previously (about 6) (6) and in the present investigation (7.4 to 7.5) is most probably due to the His6 tag attached to the C terminus in our preparation. In any case, A. fumigatus phytase, which has a pI of 8.1 to 8.6, is by far the most basic wild-type phytase. Since the great majority of contaminating proteins secreted by the expression systems into the culture supernatants have acidic pI values, this property makes A. fumigatus phytase a particularly well-suited enzyme for straightforward purification on an industrial scale (Fig. 2).
Finally, some of the phytases in some of the expression systems which we used showed a tendency to slowly undergo limited proteolysis. Identification of the cleavage sites by N-terminal sequencing of the fragments revealed that cleavage invariably occurred at exposed loops on the surface of the molecule. Site-directed mutagenesis of A. fumigatus phytase (Fig. 6) and E. nidulans phytase (data not shown) at these cleavage sites in fact reduced the susceptibility to proteases. Engineering of surface-exposed loops may therefore be a promising strategy for improving the resistance of phytases to proteases present in the culture supernatant or in the digestive tract.
ACKNOWLEDGMENTS
Ulrike Dahlems is gratefully acknowledged for constructing the phytase-expressing strains of H. polymorpha; Andrea Tomschy, Dirk Kostrewa, and Clemens Broger are acknowledged for stimulating discussions; and Hanno Langen is acknowledged for mass spectrometry of A. fumigatus phytase.
REFERENCES
- 1.Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965;96:595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dassa J, Marck C, Boquet P L. The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphatase and glucose-1-phosphatase. J Bacteriol. 1990;172:5497–5500. doi: 10.1128/jb.172.9.5497-5500.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. pp. 120–123. [Google Scholar]
- 4.Gellissen G, Janowicz Z A, Merckelbach A, Piontek M, Keup P, Weydemann U, Hollenberg C P, Strasser A W M. Heterologous gene expression in Hansenula polymorpha: efficient secretion of glucoamylase. Bio/Technology. 1991;9:291–295. doi: 10.1038/nbt0391-291. [DOI] [PubMed] [Google Scholar]
- 5.Gibson D M, Ullah A H J. Purification and characterization of phytase from cotyledons of germinating soybean seeds. Arch Biochem Biophys. 1988;260:503–513. doi: 10.1016/0003-9861(88)90475-4. [DOI] [PubMed] [Google Scholar]
- 6.Greiner R, Konietzny U, Jany K-D. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;303:107–113. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- 7.Greiner R, Haller E, Konietzny U, Jany K-D. Purification and characterization of a phytase from Klebsiella terrigena. Arch Biochem Biophys. 1997;341:201–206. doi: 10.1006/abbi.1997.9942. [DOI] [PubMed] [Google Scholar]
- 8.Grueninger-Leitch F, D’Arcy A, D’Arcy B, Chène C. Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci. 1996;5:2617–2622. doi: 10.1002/pro.5560051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollenberg C P, Janowicz Z A. DNA molecules coding for FMDH control region. European Patent Application 0299108; 1987. [Google Scholar]
- 10.Hübel F, Beck E. Maize root phytase. Plant Physiol. 1996;112:1429–1436. doi: 10.1104/pp.112.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham K C, Brew S A, Novokhatny V V. Influence of carbohydrate on structure, stability, and function of gelatin-binding fragments of fibronectin. Arch Biochem Biophys. 1995;316:235–240. doi: 10.1006/abbi.1995.1033. [DOI] [PubMed] [Google Scholar]
- 12.Janes M, Meyhack B, Zimmermann W, Hinnen A. The influence of GAP promoter variants on hirudin production, average plasmid copy number and cell growth in Saccharomyces cerevisiae. Curr Genet. 1990;18:97–103. doi: 10.1007/BF00312596. [DOI] [PubMed] [Google Scholar]
- 13.Kostrewa D, Grüninger-Leitch F, D’Arcy A, Broger C, Mitchell D, van Loon A P G M. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat Struct Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell D B, Vogel K, Weimann B, Pasamontes L, van Loon A P G M. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology. 1997;143:245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- 15.Pasamontes L, Haiker M, Wyss M, Tessier M, van Loon A P G M. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol. 1997;63:1696–1700. doi: 10.1128/aem.63.5.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasamontes L, Haiker M, Henriquez-Huecas M, Mitchell D B, van Loon A P G M. Cloning of the phytases from Emericella nidulans and the thermophilic fungus Talaromyces thermophilus. Biochim Biophys Acta. 1997;1353:217–223. doi: 10.1016/s0167-4781(97)00107-3. [DOI] [PubMed] [Google Scholar]
- 17.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A M J J. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 18.Reddy N R, Sathe S K, Salunkhe D K. Phytates in legumes and cereals. Adv Food Res. 1982;28:1–92. doi: 10.1016/s0065-2628(08)60110-x. [DOI] [PubMed] [Google Scholar]
- 19.Rudd P M, Woods R J, Wormald M R, Opdenakker G, Downing A K, Campbell I D, Dwek R A. The effects of variable glycosylation on the functional activities of ribonuclease, plasminogen and tissue plasminogen activator. Biochim Biophys Acta. 1995;1248:1–10. doi: 10.1016/0167-4838(94)00230-e. [DOI] [PubMed] [Google Scholar]
- 20.Schuck P. Simultaneous radial and wavelength analysis with the Optima XL-A analytical centrifuge. Prog Colloid Polym Sci. 1994;94:1–13. [Google Scholar]
- 21.Segueilha L, Lambrechts C, Boze H, Moulin G, Galzy P. Purification and properties of the phytase from Schwanniomyces castellii. J Ferment Bioeng. 1992;74:7–11. [Google Scholar]
- 22.Shimizu M. Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci Biotechnol Biochem. 1992;56:1266–1269. [Google Scholar]
- 23.Shimizu M. Purification and characterization of phytase and acid phosphatase produced by Aspergillus oryzae K1. Biosci Biotechnol Biochem. 1993;57:1364–1365. [Google Scholar]
- 23a.Tomschy, A. Unpublished data.
- 24.van Hartingsveldt W, van Zeijl C M J, Harteveld G M, Gouka R J, Suykerbuyk M E G, Luiten R G M, van Paridon P A, Selten G C M, Veenstra A E, van Gorcom R F M, van den Hondel C A M J J. Cloning, characterization and overexpression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene. 1993;127:87–94. doi: 10.1016/0378-1119(93)90620-i. [DOI] [PubMed] [Google Scholar]
- 25.Villarejo M R, Zabin J. Beta-galactosidase from termination and deletion mutant strains. J Bacteriol. 1974;120:466–476. doi: 10.1128/jb.120.1.466-474.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 29.Wodzinski R J, Ullah A H J. Phytase. Adv Appl Microbiol. 1996;42:263–302. doi: 10.1016/s0065-2164(08)70375-7. [DOI] [PubMed] [Google Scholar]
- 30.Wyss M, Pasamontes L, Rémy R, Kohler J, Kusznir E, Gadient M, Müller F, van Loon A P G M. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl Environ Microbiol. 1998;64:4446–4451. doi: 10.1128/aem.64.11.4446-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyss M, Brugger R, Kronenberger A, Rémy R, Fimbel R, Oesterhelt G, Lehmann M, van Loon A P G M. Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl Environ Microbiol. 1999;65:367–373. doi: 10.1128/aem.65.2.367-373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto S, Minoda Y, Yamada K. Chemical and physicochemical properties of phytase from Aspergillus terreus. Agric Biol Chem. 1972;36:2097–2103. [Google Scholar]