Abstract
Supplementation with phytase is an effective way to increase the availability of phosphorus in seed-based animal feed. The biochemical characteristics of an ideal phytase for this application are still largely unknown. To extend the biochemical characterization of wild-type phytases, the catalytic properties of a series of fungal phytases, as well as Escherichia coli phytase, were determined. The specific activities of the fungal phytases at 37°C ranged from 23 to 196 U · (mg of protein)−1, and the pH optima ranged from 2.5 to 7.0. When excess phytase was used, all of the phytases were able to release five phosphate groups of phytic acid (myo-inositol hexakisphosphate), which left myo-inositol 2-monophosphate as the end product. A combination consisting of a phytase and Aspergillus niger pH 2.5 acid phosphatase was able to liberate all six phosphate groups. When substrate specificity was examined, the A. niger, Aspergillus terreus, and E. coli phytases were rather specific for phytic acid. On the other hand, the Aspergillus fumigatus, Emericella nidulans, and Myceliophthora thermophila phytases exhibited considerable activity with a broad range of phosphate compounds, including phenyl phosphate, p-nitrophenyl phosphate, sugar phosphates, α- and β-glycerophosphates, phosphoenolpyruvate, 3-phosphoglycerate, ADP, and ATP. Both phosphate liberation kinetics and a time course experiment in which high-performance liquid chromatography separation of the degradation intermediates was used showed that all of the myo-inositol phosphates from the hexakisphosphate to the bisphosphate were efficiently cleaved by A. fumigatus phytase. In contrast, phosphate liberation by A. niger or A. terreus phytase decreased with incubation time, and the myo-inositol tris- and bisphosphates accumulated, suggesting that these compounds are worse substrates than phytic acid is. To test whether broad substrate specificity may be advantageous for feed application, phosphate liberation kinetics were studied in vitro by using feed suspensions supplemented with 250 or 500 U of either A. fumigatus phytase or A. niger phytase (Natuphos) per kg of feed. Initially, phosphate liberation was linear and identical for the two phytases, but considerably more phosphate was liberated by the A. fumigatus phytase than by the A. niger phytase at later stages of incubation.
The phosphatases are a diverse class of enzymes. According to one classification, alkaline phosphatases, purple acid phosphatases, high-molecular-weight acid phosphatases, low-molecular-weight acid phosphatases, and protein phosphatases can be distinguished (13). These classes differ in their pH optima, metal ion requirements, substrate specificities, and possibly even reaction mechanisms. The phytases (myo-inositol hexakisphosphate phosphohydrolases; EC 3.1.3.8 and 3.1.3.26) are a subfamily of the high-molecular-weight histidine acid phosphatases. The phytase reaction mechanism is a two-step mechanism which includes a covalent phosphohistidine adduct as an obligatory reaction intermediate (6).
Phytases are found naturally in plants and microorganisms, particularly fungi (for a review see reference 15). They catalyze phosphate monoester hydrolysis of phytic acid (myo-inositol hexakisphosphate), which results in the stepwise formation of myo-inositol pentakis-, tetrakis-, tris-, bis-, and monophosphates, as well as the liberation of inorganic phosphate. Phytic acid is the major storage form of phosphorus in plant seeds and, thus, in seed-based animal feed (for reviews see references 1 and 8). Monogastric animals, such as pigs and poultry, are not able to utilize phytic acid phosphorus, since they have only low levels of phytase activity in their digestive tracts and since phytic acid cannot be resorbed. Therefore, pig and poultry feed commonly is supplemented with either inorganic phosphate or a phytase of fungal origin.
Despite considerable economic interest, only limited data on the catalytic properties of fungal phytases are available. In order to get an impression of the natural diversity of phytases, the enzymatic properties of six fungal phytases (phytases from Aspergillus niger, two strains of Aspergillus terreus, Aspergillus fumigatus, Emericella nidulans, and Myceliophthora thermophila) and of Escherichia coli phytase were characterized in more detail by addressing the following questions. (i) What are the specific activities and pH optima of wild-type phytases? (ii) What are the kinetics of phytic acid degradation, and what are the end products? (iii) Does the substrate specificity profile correlate with the results of in vitro experiments performed to determine phosphate liberation from feed samples? And (iv) what is the potential influence of modulators of enzymatic activity?
MATERIALS AND METHODS
Sources of purified proteins.
The A. niger phytase (Natuphos) and the phytases from A. terreus 9A1, A. fumigatus ATCC 13073 (erroneously designated ATCC 34625 in previous papers by members of our group), E. nidulans, and M. thermophila were overexpressed in filamentous fungi and were purified to homogeneity by using the methods of Pasamontes et al. (7) and Wyss et al. (16). A. terreus CBS phytase was overexpressed in Hansenula polymorpha, E. coli phytase was overexpressed in E. coli, and both proteins were purified as described previously (16). E. coli phytase with the amino acid sequence originally described by Dassa et al. (2) was used in this study. However, the G207D mutation in the E. coli phytase used in the study described in the accompanying paper (16) had no effect on the specific activity (data not shown). A. niger pH 2.5 acid phosphatase was overexpressed in A. niger and was purified by anion-exchange chromatography.
Measurements of enzymatic activity.
Standard phytase activity measurements were obtained at 37°C by using the method described in the accompanying paper (16). To determine the pH optimum curves, purified enzymes were diluted in 10 mM sodium acetate (pH 5.0). Incubations were started by mixing aliquots of the diluted protein with equal volumes of 1% (∼10 mM) phytic acid in the following buffers: 0.4 M glycine, pH 2.5 (HCl); 0.4 M acetate, pH 3.0, 3.5, 4.0, 4.5, 5.0, or 5.5 (NaOH); 0.4 M imidazole, pH 6.0, 6.5, or 7.0 (HCl); and 0.4 M Tris, pH 7.5, 8.0, 8.5, or 9.0 (HCl). Control experiments showed that the pH was only slightly affected by the mixing step. Preparations were incubated for 15 min at 37°C as described above.
To determine the substrate specificities, the phytic acid in the assay mixture was replaced with different phosphate compounds at a concentration of 5 mM. Besides, the activity test was performed exactly as described above. One unit of phytase (or acid phosphatase, glucose-6-phosphatase, etc.) activity catalyzed the liberation of 1 μmol of inorganic phosphate per min.
For end-point determinations, excess enzyme (0.5 U of phytase per ml and/or 0.5 U of pH 2.5 acid phosphatase per ml) was incubated with a limiting substrate concentration (0.2 mM phytic acid, 0.75 mM myo-inositol 1-monophosphate, or 0.75 mM myo-inositol 2-monophosphate) for 5 to 120 min at 37°C. The inorganic phosphate liberated was subsequently quantified as described above.
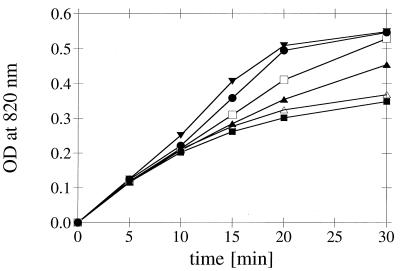
In order to compare the kinetics of phosphate release by the different wild-type phytases (see Fig. 7), reaction mixtures were adjusted so that they had identical initial reaction rates. After 5, 10, 15, 20, and 30 min of incubation with 0.2 mM phytic acid at 37°C, each reaction was stopped by adding trichloroacetic acid, and the inorganic phosphate liberated was quantified as described above.
FIG. 7.
Kinetics of phosphate liberation from phytic acid catalyzed by different wild-type phytases. The phytase preparations were adjusted so that they had identical initial reaction rates. For the phytic acid concentration used (0.2 mM) an optical density at 820 nm of ∼0.55 corresponded to hydrolysis of five of the six phosphomonoester bonds. Symbols: •, A. fumigatus phytase; ▾, E. nidulans phytase; □, M. thermophila phytase; ▴, A. niger phytase; ▵, A. terreus CBS phytase; ■, A. terreus 9A1 phytase. OD, optical density.
Some adaptations were necessary in order to determine the dependence of the initial reaction rates on the phytic acid concentration, which was the basis for calculating Km values. The assay mixtures contained either 1.0, 0.5, 0.2, 0.1, 0.05, 0.02, or 0.01 mM phytic acid in 200 mM sodium acetate (pH 5.0). Each reaction was stopped after 5, 10, 15, 20, or 30 min of incubation by adding an equal volume of 15% trichloroacetic acid. The inorganic phosphate liberated was quantified by mixing 1 ml of the assay mixture with 1 ml of 0.6 M H2SO4–2% ascorbic acid–0.5% ammonium molybdate, incubating the preparation for 20 min at 50°C, and measuring the absorbance at 820 nm.
Effects of potential modulators on the enzymatic activities of phytases and pH 2.5 acid phosphatase.
The effects of cations, sulfhydryl compounds, and sulfhydryl reagents on the enzymatic activities were determined under standard assay conditions (5 mM phytic acid, 200 mM sodium acetate; pH 5.0). Experiments to determine the effects of anions on the enzymatic activities of phytases and pH 2.5 acid phosphatase were performed in 50 mM MES (morpholinoethanesulfonic acid) buffer (pH 5.0) containing either 5 mM phytic acid or 5 mM p-nitrophenyl phosphate as the substrate.
Gas chromatography-mass spectrometry.
Each solution investigated (0.5 ml) was lyophilized, and the residue was dissolved and incubated in 500 μl of silylating reagent [250 μl of pyridine, 250 μl of bis(trimethylsilyl)trifluoroacetamide] at room temperature for 1 day. The silylated products were injected at 270°C into a model 5890A gas chromatograph (Hewlett-Packard) which was coupled to a model 5989B mass spectrometer (Hewlett-Packard). The stationary phase was methylsilicon (type DB-1; film thickness, 0.25 μm) in a fused-silica column (15 m by 0.25 mm). Helium was used as the carrier gas at a flow rate of 0.5 m · s−1. A heating program was used for the column (100 to 340°C; rate of increase, 4°C · min−1); ionization was performed by electron impact at 70 eV and 250°C.
Phosphate liberation from feed samples.
Mash feed (containing 50% maize, 30% rapeseed meal, 17% barley, and 3% minerals, vitamins, and amino acids) was autoclaved for 20 min at 120°C in order to inactivate any phytases present in the feed. Aliquots (5 g) of this autoclaved mash feed were resuspended in 40 ml of 200 mM sodium acetate (pH 5.0). The suspensions were incubated as such or after supplementation with 250 or 500 U of either A. fumigatus phytase or A. niger phytase per kg of feed for various periods of time at 37°C on a shaker. Aliquots were then mixed with an equal volume of 15% trichloroacetic acid, and the inorganic phosphate liberated was quantified as described above.
HPLC analysis of the time course of accumulation of phytic acid degradation intermediates.
Phytase was incubated at 37°C in 200 μl of an assay mixture containing 200 mM sodium acetate (pH 5.0) and 0.2 mM (final concentration) phytic acid which was radioactively labelled with 1 μCi of myo-[2-3H(N)]inositol hexakisphosphate (NEN) per ml. The reaction was stopped after 2.5, 5, 10, 15, 20, 25, 30, 40, 50, 60, or 90 min by adding an equal volume of acetonitrile and then heating the preparation at 95°C for 2 min. After centrifugation for 10 min at 10,000 × g, a 150-μl aliquot of the supernatant was diluted with an equal volume of H2O. A 200-μl portion of this sample was assayed by anion-exchange high-performance liquid chromatography (HPLC) by using a Zorbax SAX (5 μm) column (4.6 by 250 mm) and a flow rate of 1 ml/min as described by van der Kaay and van Haastert (11). An HPLC system consisting of two pumps (Kontron model 422), a sample injector (Hitachi model AS-4000 autosampler), and a flowthrough radioactivity monitor (Packard Radiomatic Flo-one) was used in combination with Ultima-Flo AP scintillation cocktail (Packard) which was delivered at a flow rate of 3 ml/min.
Other methods.
Protein concentrations were calculated from optical densities at 280 nm by using theoretical absorption values calculated from the known protein sequences with the DNA* software (DNASTAR, Inc.). One unit of absorption at 280 nm corresponded to 1.03 mg of A. niger phytase per ml, 0.81 mg of A. terreus 9A1 phytase per ml, 0.82 mg of A. terreus CBS phytase per ml, 0.94 mg of A. fumigatus phytase per ml, 0.81 mg of E. nidulans phytase per ml, 0.91 mg of M. thermophila phytase per ml, 0.89 mg of E. coli phytase per ml, and 0.63 mg of A. niger pH 2.5 acid phosphatase per ml.
RESULTS
Specific activities, Km values, and pH optima.
Under the standard assay conditions (i.e., 5 mM phytic acid), only the rate of the reaction from myo-inositol hexakisphosphate to pentakisphosphate is measured. The specific activities of the fungal phytases for this reaction varied almost 10-fold, from 23 to 196 U · (mg of protein)−1 (Table 1). The difference between the specific activities of the phytases of the two A. terreus wild-type strains, whose amino acid sequences differed at 58 positions, was modest, and no attempt was made to identify the amino acids responsible for this difference.
TABLE 1.
Specific activities and Km values for phytic acid of fungal phytasesa
| Phytaseb | Sp act (U/mg) | Km (μM) |
|---|---|---|
| A. niger phytase | 102.5 ± 19.9 (8) | <5 |
| A. terreus 9A1 phytase | 141.6 ± 7.1 (3) | 10.6 ± 0.9 (5) |
| A. terreus CBS phytase | 195.8 ± 17.8 (7) | 23.2 ± 2.2 (6) |
| A. fumigatus phytase from A. niger | 26.5 ± 5.2 (22) | ≤10 |
| A. fumigatus phytase from H. polymorpha | 28.1 ± 3.7 (6) | NDc |
| A. fumigatus phytase from S. cerevisiae | 23.4 ± 3.6 (3) | ND |
| E. nidulans phytase from A. niger | 28.6 ± 4.1 (8) | —d |
| E. nidulans phytase from H. polymorpha | 33.1 ± 2.7 (4) | ND |
| M. thermophila phytase | 41.8 ± 4.4 (3) | — |
| E. coli phytase | 811.2 ± 215.5 (5) | ND |
Means ± standard deviations. The values in parentheses are the numbers of preparations examined.
A. niger phytase (Natuphos) was obtained from a commercial source (BASF); A. terreus 9A1 and M. thermophila phytases were overexpressed in A. niger; A. terreus CBS phytase was overexpressed in H. polymorpha; and E. coli phytase was overexpressed in E. coli.
ND, not determined.
—, non-Michaelis-Menten behavior.
While glycosylation of the fungal phytases was moderate when they were overexpressed in A. niger, glycosylation was excessive and highly variable when the phytases were expressed in H. polymorpha or Saccharomyces cerevisiae (16). However, the different extents and patterns of glycosylation did not have a significant effect on the specific activities (Table 1) or on other catalytic properties of the fungal phytases (data not shown). Therefore, with the exception of the A. terreus CBS phytase which was expressed in H. polymorpha, only data for fungal phytases expressed in A. niger are presented below.
In experiments performed to determine the dependence of the initial reaction velocity on the substrate concentration, the A. terreus 9A1 and CBS phytases followed Michaelis-Menten kinetics, with Km values of 10.6 ± 0.9 and 23.2 ± 2.2 μM, respectively (Table 1). On the other hand, the Km values of the A. fumigatus and A. niger phytases could not be determined reliably due to the limited sensitivity of the assay, while the M. thermophila and E. nidulans phytases displayed non-Michaelis-Menten behavior.
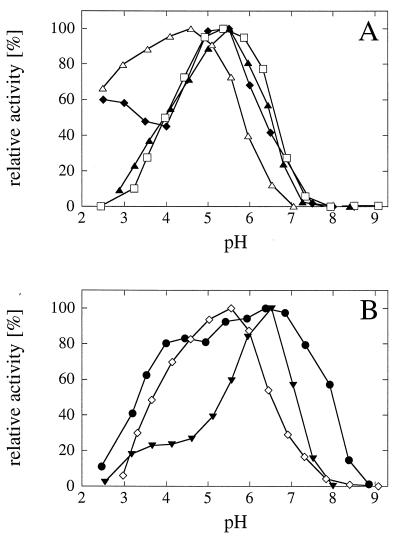
The pH optima of the fungal and E. coli phytases ranged from 2.5 to 7.0 (Fig. 1). A. fumigatus phytase had a broad pH optimum, and at least 80% of the maximal activity was observed at pH values between 4.0 and 7.3; in contrast, E. nidulans phytase had a rather narrow pH optimum around pH 6.5.
FIG. 1.
pH optimum curves for wild-type phytases. The data are expressed as percentages of the maximal activity. (A) Symbols: ⧫, A. niger phytase; □, A. terreus CBS phytase; ▴, A. terreus 9A1 phytase; ▵, E. coli phytase; (B) Symbols: •, A. fumigatus phytase; ▾, E. nidulans phytase; (◊) M. thermophila phytase.
Substrate specificity.
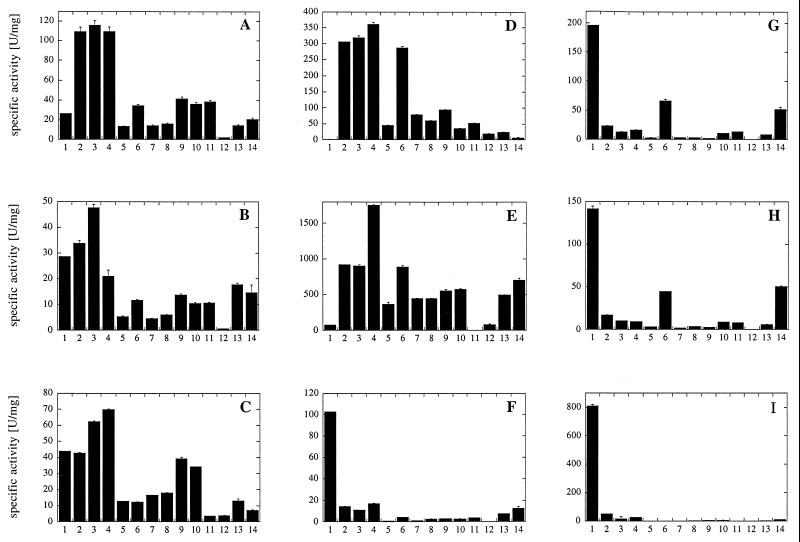
To determine the substrate specificities of fungal phytases, E. coli phytase, and A. niger pH 2.5 acid phosphatase, we investigated the specific activities of the purified proteins with a series of phosphate compounds (Fig. 2). A. niger pH 2.5 acid phosphatase displayed rather broad substrate specificity at both pH 2.5 and pH 5.0 but only low activity with phytic acid. The A. fumigatus, E. nidulans, and M. thermophila phytases also displayed broad substrate specificities. On the other hand, A. niger, A. terreus CBS, A. terreus 9A1, and, in particular, E. coli phytases were rather specific for phytic acid.
FIG. 2.
Substrate specificities of wild-type phytases and A. niger pH 2.5 acid phosphatase. All substrates were used at a concentration of 5 mM. 1, phytic acid; 2, p-nitrophenyl phosphate; 3, phenyl phosphate; 4, fructose 1,6-bisphosphate; 5, fructose 6-phosphate; 6, glucose 6-phosphate; 7, ribose 5-phosphate; 8, α-glycerophosphate; 9, β-glycerophosphate; 10, 3-phosphoglycerate; 11, phosphoenolpyruvate; 12, AMP; 13, ADP; 14, ATP. (A) A. fumigatus phytase. (B) E. nidulans phytase. (C) M. thermophila phytase. (D) A. niger pH 2.5 acid phosphatase. (E) A. niger pH 2.5 acid phosphatase (measurements obtained at pH 2.5 instead of pH 5.0). (F) A. niger phytase. (G) A. terreus CBS phytase. (H) A. terreus 9A1 phytase. (I) E. coli phytase.
End product of phytic acid degradation.
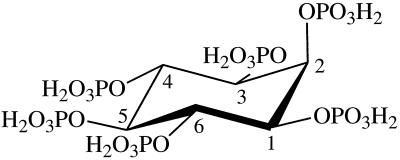
Phytic acid has six phosphate groups that may be released by the fungal phytases at different rates and in different orders. In the energetically most favorable conformation, five of the six phosphate groups of phytic acid are in an equatorial position, while the 2-phosphate group is in an axial position (Fig. 3).
FIG. 3.
Energetically most favorable conformation of phytic acid (myo-inositol hexakisphosphate). The numbering of the carbon atoms is the numbering for the d-configuration.
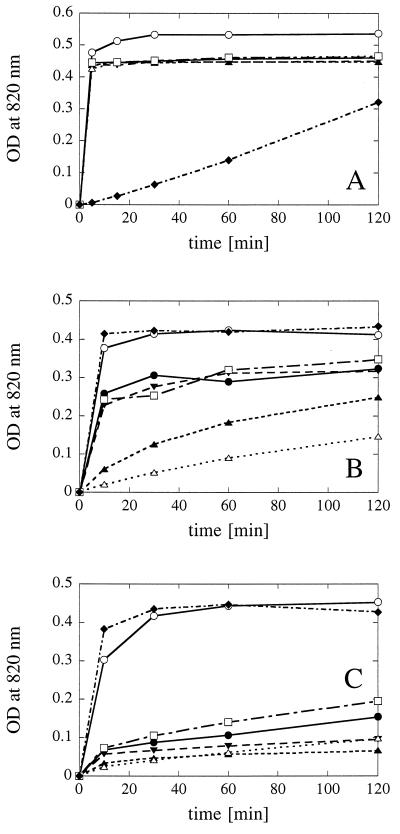
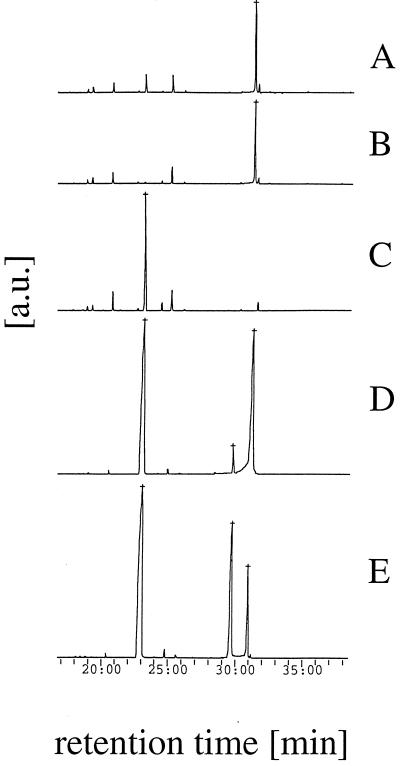
While detailed characterization of all of the degradation intermediates was beyond the scope of this study, we attempted to investigate the kinetics of phosphate release and the kinetics of accumulation of intermediates, as well as the end products of phytic acid degradation. The end products of phytic acid degradation were determined by incubating excess protein (0.5 U of phytase activity per ml and/or 5 U of acid phosphatase activity per ml) with a limiting substrate concentration (0.2 mM phytic acid). Both the time course of phosphate release (Fig. 4A) and the results of a gas chromatography-mass spectrometry analysis of the 2-h samples (Fig. 5) revealed that a combination of A. fumigatus phytase and A. niger pH 2.5 acid phosphatase liberated all six phosphate groups. All of the fungal phytases and E. coli phytase released five of the six phosphate groups, and the end product was identified by gas chromatography-mass spectrometry as myo-inositol 2-monophosphate. Only in rare cases were traces of free myo-inositol or myo-inositol 1-monophosphate detected.
FIG. 4.
Phytic acid (A), myo-inositol 1-monophosphate (B), and myo-inositol 2-monophosphate (C) degradation kinetics observed with a limiting concentration of substrate (0.2 or 0.75 mM) and excess enzyme (approximately 0.5 U/ml). Symbols: ○, A. fumigatus phytase plus A. niger pH 2.5 acid phosphatase; •, A. fumigatus phytase; ▾, E. nidulans phytase; ▴, A. niger phytase; ▵, A. terreus CBS phytase; □, M. thermophila phytase; ⧫, A. niger pH 2.5 acid phosphatase. OD, optical density.
FIG. 5.
Gas chromatography analysis of the end products of phytic acid degradation. Two-hour samples from the incubations shown in Fig. 4 (A through C) or commercially available reference compounds (D and E) were analyzed. (A) A. fumigatus phytase. (B) A. niger phytase. (C) A. fumigatus phytase plus A. niger pH 2.5 acid phosphatase. (D) myo-Inositol plus myo-inositol 2-monophosphate. (E) myo-Inositol plus myo-inositol 1-monophosphate. a.u., arbitrary units.
The preference of the fungal phytases for equatorial phosphate groups instead of the axial 2-phosphate group was corroborated by phosphate liberation experiments performed with myo-inositol 1-monophosphate (Fig. 4B) and myo-inositol 2-monophosphate (Fig. 4C). Interpretation of the data in Fig. 4B is facilitated when it is taken into account that the preparation of myo-inositol 1-monophosphate used in this experiment (which was obtained from Sigma Chemical Co., St. Louis, Mo.) contained 27% myo-inositol 2-monophosphate as an impurity (Fig. 5E). No preference for the 1- or 2-phosphate group was observed with A. niger pH 2.5 acid phosphatase or with a combination of A. niger pH 2.5 acid phosphatase and A. fumigatus phytase. In contrast, all of the fungal phytases, particularly the A. fumigatus, E. nidulans, and M. thermophila phytases, exhibited much higher activity with myo-inositol 1-monophosphate than with myo-inositol 2-monophosphate.
In conclusion, all of the phytases investigated were able to release all five equatorial phosphate groups of phytic acid. Invariably, myo-inositol 2-monophosphate was identified as the end product.
Kinetics of phosphate release and accumulation of intermediates.
Using a previously described HPLC procedure (11), we separated myo-inositols with different numbers of phosphate groups (one to six phosphate groups) in a single run (data not shown). However, no attempt was made to separate the different pentakis-, tetrakis-, tris-, or bisphosphate isomers or to characterize in detail the stereoisomers formed.
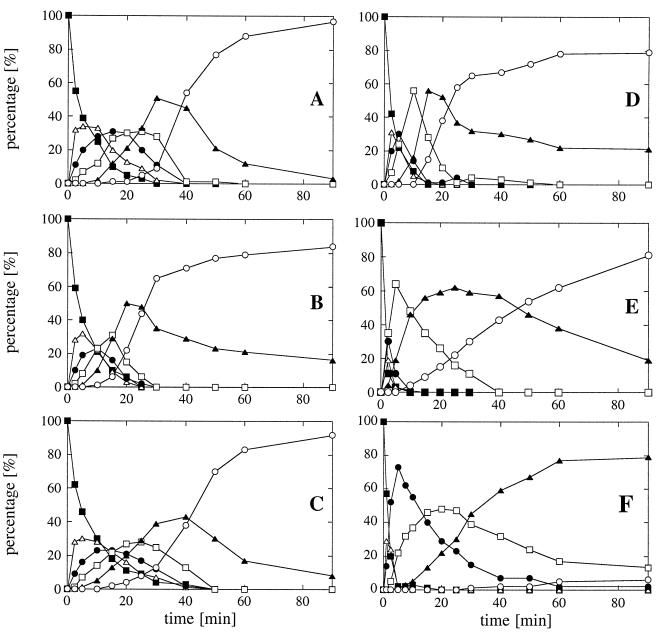
HPLC analysis of the degradation intermediates obtained in a time course experiment (Fig. 6), as well as phosphate liberation kinetics (Fig. 7), showed that A. fumigatus phytase readily degraded phytic acid to myo-inositol 2-monophosphate, and that only the bisphosphate accumulated to some extent. However, the pentakis-, tetrakis-, and trisphosphates seemed to be cleaved by A. fumigatus phytase as efficiently as phytic acid was. In contrast, A. niger and A. terreus phytases had to be used at much higher initial activities in order to obtain degradation down to myo-inositol 2-monophosphate, and considerable amounts of the tris- and bisphosphates accumulated during degradation (Fig. 6C and D). This fact was also reflected by the decrease in the rate of phosphate liberation with time (Fig. 7). When E. coli phytase was used at an even higher initial activity, there was a pronounced accumulation of the tetrakisphosphate and almost no monophosphate was detected at the end of the 90-min incubation period (Fig. 6F). Therefore, myo-inositols with lower numbers of phosphate groups seem to be markedly less efficient substrates for A. niger, A. terreus, and E. coli phytases than phytic acid is. In identical experiments, E. nidulans phytase behaved like A. fumigatus phytase, while the behavior of M. thermophila phytase was intermediate (Fig. 6 and 7).
FIG. 6.
HPLC analysis of phytic acid degradation kinetics. (A) A. fumigatus phytase (0.015 U/ml). (B) E. nidulans phytase (0.025 U/ml). (C) M. thermophila phytase (0.05 U/ml). (D) A. niger phytase (0.075 U/ml). (E) A. terreus CBS phytase (0.2 U/ml). (F) E. coli phytase (0.5 U/ml). Symbols: ■, myo-inositol hexakisphosphate; ▵, myo-inositol pentakisphosphate; •, myo-inositol tetrakisphosphate; □, myo-inositol trisphosphate; ▴, myo-inositol bisphosphate; ○, myo-inositol monophosphate. Note that the initial activities used for the different phytases were different in order to get degradation of phytic acid down to the monophosphate stage within 90 min.
In vitro phosphate liberation from feed samples.
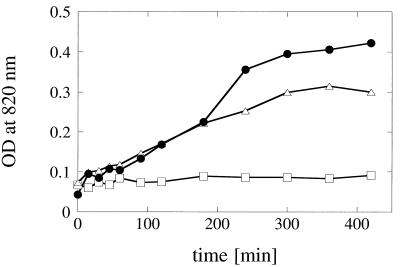
The in vitro system used to mimic the conditions in the digestive tract consisted of aliquots of autoclaved feed samples that were resuspended in buffer solution (pH 5.0) and incubated with agitation for different periods of time at 37°C. Supplementation of the feed samples with 250 or 500 U of either A. fumigatus or A. niger phytase per kg of feed resulted in identical initial rates of phosphate liberation (Fig. 8). However, at later stages of incubation, significantly more inorganic phosphate was liberated by A. fumigatus phytase than by A. niger phytase. Therefore, the differences in kinetic properties observed with the purified enzymes were also reflected in the results of this in vitro feed experiment, as well as in the results of animal trials (12).
FIG. 8.
In vitro phosphate liberation from animal feed. Autoclaved feed samples were resuspended in 200 mM sodium acetate (pH 5.0) and incubated for various periods of time at 37°C; the samples were either not supplemented (□) or supplemented with 250 U of A. fumigatus phytase per kg of feed (•) or 250 U of A. niger phytase per kg of feed (▵). OD, optical density.
Modulators of enzymatic activity.
Several authors have shown previously that metal ions (3, 4, 9, 10, 17) and sulfhydryl reagents (9) can modulate phytase activity. When activity assays were performed with A. fumigatus, E. nidulans, A. niger, and A. terreus CBS phytases in the absence or presence of several metal ions at a concentration of 0.1 or 1 mM or in the presence or absence of 0.1, 1, or 10 mM EDTA, no major effects on the enzymatic activity of A. niger phytase were observed (data not shown). On the other hand, 1 mM Cu2+ depressed considerably the enzymatic activities of E. nidulans and A. terreus CBS phytases.
It is unlikely that the inhibitory effects of Cu2+ (and to some extent the inhibitory effects of Fe3+, Fe2+, Al3+, and Mn2+) are due to direct binding of the metal ions to the phytases. Rather, metal ions were found to form poorly soluble complexes with phytic acid and may therefore decrease the active concentration of phytic acid in an activity assay. At pH 7.4, the stability of metal ion-phytic acid complexes was found to decrease in the following order: Cu2+ > Zn2+ > Ni2+ > Co2+ > Mn2+ > Fe3+ > Ca2+ (14). Evidently, A. terreus CBS phytase with a Km of 23 μM should be more susceptible to a Cu2+-induced decrease in the phytic acid concentration than A. niger phytase with a Km of <5 μM, which is actually the case.
The results obtained for A. fumigatus phytase were somewhat different. Several metal ions inhibited this phytase, whereas 1 and 10 mM EDTA stimulated the enzymatic activity by up to 50%. Whether this protein in fact has a nonspecific cation binding site should be further determined.
The effects of metal ions mentioned above and the fact that EDTA either has no effect or even stimulates phytase activity shows that the fungal phytases clearly differ from the metal ion-dependent phytase of Bacillus subtilis that is readily inhibited by EDTA (4a). This conclusion is supported by the lack of a metal ion in the crystal structure of A. niger phytase (5) and by the lack of protein sequence similarity between fungal phytases and B. subtilis phytase (4a).
When A. fumigatus, E. nidulans, A. niger, and A. terreus CBS phytases, as well as A. niger pH 2.5 acid phosphatase, were incubated with 1 or 10 mM 2-mercaptoethanol, 1 or 10 mM dithiothreitol, 1 or 10 mM reduced glutathione, 1 or 10 mM iodoacetamide, or 100 μM p-chloromercuribenzoate before the activity assay was started by adding phytic acid (or p-nitrophenyl phosphate in the case of A. niger pH 2.5 acid phosphatase), the enzymatic activity was not affected by more than 11% (data not shown). This suggests that either these proteins have no free and accessible sulfhydryl groups or the free sulfhydryl groups play a negligible role in enzyme structure and activity. This interpretation is supported by the fact that all of the fungal acid phosphatases which we considered except the A. terreus phytases (which have 11 Cys residues) have an even number of cysteine residues (i.e., 10 Cys residues) and by the fact that all of the cysteines of the mature A. niger phytase are implicated in disulfide bridges (5).
In MES buffer (pH 5.0), 100 mM sodium acetate, 100 mM KNO3, and 10 mM sulfate had only minor effects (<23%) on the activities of A. fumigatus, E. nidulans, A. niger, and A. terreus CBS phytases (data not shown). Since inorganic phosphate interferes with the standard phytase activity assay, the effects of anions were also determined with 5 mM p-nitrophenyl phosphate as substrate. While 10 mM sulfate in the assay mixture inhibited only E. nidulans and A. terreus CBS phytases (by 32 and 55%, respectively), 10 mM inorganic phosphate inhibited all of the phytases by 61 to 85% (data not shown).
DISCUSSION
Despite the high degree of amino acid sequence homology among the fungal phytases which we studied, these enzymes had substantially different catalytic properties. The specific activities with phytic acid as substrate ranged from 23 to 196 U · (mg of protein)−1; the pH optima obtained with the same substrate varied between 2.5 and 7.0 and were either broad (e.g., pH 4.0 to 7.3 for A. fumigatus phytase) or narrow (e.g., pH 6.5 for E. nidulans phytase). Marked differences were also noted in substrate specificity, not only with different myo-inositol phosphates but also with classic acid phosphatase substrates (phenyl phosphate and p-nitrophenyl phosphate [pNPP]), sugar phosphates, and other phosphate compounds.
On the other hand, when excess fungal phytases or excess E. coli phytase was used, the end product of phytic acid degradation was the same, namely, myo-inositol 2-monophosphate. This finding demonstrates that all of these phytases have pronounced stereospecificity and a strong preference for equatorial phosphate groups, while they are virtually unable to cleave axial phosphate groups. However, the fact that the end products were identical does not necessarily mean that the degradation pathways for phytic acid are identical for all phytases. For instance, plant and E. coli phytases are 6-phytases; i.e., they preferentially yield l-myo-inositol 1,2,3,4,5-pentakisphosphate as the first intermediate (1, 3), while fungal and most bacterial phytases are 3-phytases, which give rise to d-myo-inositol 1,2,4,5,6-pentakisphosphate (1, 4).
On the basis of substrate specificity, two classes of phytases could be discriminated, phytases with broad substrate specificity (A. fumigatus, E. nidulans, and M. thermophila phytases) and phytases which are rather specific for phytic acid (A. niger, A. terreus 9A1, A. terreus CBS, and E. coli phytases). The phytases with broad substrate specificity (i) exhibited significant levels of activity with a range of phosphate compounds that were not necessarily structurally similar to phytic acid (e.g., phenyl phosphate, pNPP, and phosphoenolpyruvate) (Fig. 2); (ii) readily degraded phytic acid to myo-inositol 2-monophosphate with no major accumulation of intermediates (Fig. 6); and (iii) readily cleaved myo-inositol 1-monophosphate (Fig. 4). In contrast, the phytases with narrow substrate specificity (i) were rather specific for myo-inositol phosphates; (ii) resulted in myo-inositol tris- and bisphosphate accumulation during phytic acid degradation, coupled with a progressive decrease in the rate of phosphate release, which suggested that lower inositol phosphates are worse substrates than phytic acid is; and (iii) exhibited much lower levels of activity with myo-inositol 1-monophosphate. However, the phytases with broad substrate specificity inherently had rather low specific activities when phytic acid was the substrate (23 to 42 U · [mg of protein]−1), while the phytases with narrow substrate specificity had specific activities of 103 to 811 U · (mg of protein)−1. The difference in the substrate specificities of the two classes of phytases, therefore, reflects to a considerable extent a selective difference in the specific activities with phytic acid.
The results of time course experiments in which the intermediates were analyzed by HPLC (Fig. 6) and the phosphate liberation kinetics observed both with purified enzymes (Fig. 7) and in in vitro feed experiments (Fig. 8) suggest that phytases with broad substrate specificity are better suited for animal nutrition purposes than phytases with narrow substrate specificity are, since when the initial phytase activities are the same, the phytases with broad substrate specificity more readily liberate all five equatorial phosphate groups of phytic acid. Unfortunately, broad substrate specificity currently must be paid for dearly in that it is coupled with low specific phytase activity. In the future it will be a challenge to combine high specific phytase activities with broad substrate specificities.
REFERENCES
- 1.Cosgrove D J. Inositol phosphates. Their chemistry, biochemistry and physiology. Studies in organic chemistry. Vol. 4. Amsterdam, The Netherlands: Elsevier Scientific Publishing Co.; 1980. [Google Scholar]
- 2.Dassa J, Marck C, Boquet P L. The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphatase and glucose-1-phosphatase. J Bacteriol. 1990;172:5497–5500. doi: 10.1128/jb.172.9.5497-5500.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greiner R, Konietzny U, Jany K-D. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;303:107–113. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- 4.Greiner R, Haller E, Konietzny U, Jany K-D. Purification and characterization of a phytase from Klebsiella terrigena. Arch Biochem Biophys. 1997;341:201–206. doi: 10.1006/abbi.1997.9942. [DOI] [PubMed] [Google Scholar]
- 4a.Kerovuo J, Lauraeus M, Nurminen P, Kalkkinen N, Apajalahti J. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:2079–2085. doi: 10.1128/aem.64.6.2079-2085.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostrewa D, Grüninger-Leitch F, D’Arcy A, Broger C, Mitchell D, van Loon A P G M. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat Struct Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- 6.Ostanin K, Harms E H, Stevis P E, Kuciel R, Zhou M-M, van Etten R L. Overexpression, site-directed mutagenesis, and mechanism of Escherichia coli acid phosphatase. J Biol Chem. 1992;267:22830–22836. [PubMed] [Google Scholar]
- 7.Pasamontes L, Haiker M, Wyss M, Tessier M, van Loon A P G M. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl Environ Microbiol. 1997;63:1696–1700. doi: 10.1128/aem.63.5.1696-1700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy N R, Sathe S K, Salunkhe D K. Phytates in legumes and cereals. Adv Food Res. 1982;28:1–92. doi: 10.1016/s0065-2628(08)60110-x. [DOI] [PubMed] [Google Scholar]
- 9.Segueilha L, Lambrechts C, Boze H, Moulin G, Galzy P. Purification and properties of the phytase from Schwanniomyces castellii. J Ferment Bioeng. 1992;74:7–11. [Google Scholar]
- 10.Shimizu M. Purification and characterization of phytase from Bacillus subtilis (natto) N-77. Biosci Biotechnol Biochem. 1992;56:1266–1269. [Google Scholar]
- 11.van der Kaay J, van Haastert P J M. Stereospecificity of inositol hexakisphosphate dephosphorylation by Paramecium phytase. Biochem J. 1995;312:907–910. [PMC free article] [PubMed] [Google Scholar]
- 12.van Loon A P G M, Simoes-Nunes C, Wyss M, Tomschy A, Hug D, Vogel K, Pasamontes L. A heat-resistant phytase of Aspergillus fumigatus with superior performance in animal experiments. Phytase optimization and natural variability. In: Rasmussen S K, Raboy V, Dalbøge H, Loewus F, editors. The biochemistry of phytate and phytases. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 13.Vincent J B, Crowder M W, Averill B A. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci. 1992;17:105–110. doi: 10.1016/0968-0004(92)90246-6. [DOI] [PubMed] [Google Scholar]
- 14.Vohra P, Gray G A, Kratzer F H. Phytic acid-metal complexes. Proc Soc Exp Biol Med. 1965;120:447–449. doi: 10.3181/00379727-120-30559. [DOI] [PubMed] [Google Scholar]
- 15.Wodzinski R J, Ullah A H J. Phytase. Adv Appl Microbiol. 1996;42:263–302. doi: 10.1016/s0065-2164(08)70375-7. [DOI] [PubMed] [Google Scholar]
- 16.Wyss M, Pasamontes L, Friedlein A, Rémy R, Tessier M, Kronenberger A, Middendorf A, Lehmann M, Schnoebelen L, Röthlisberger U, Kusznir E, Wahl G, Müller F, Lahm H-W, Vogel K, van Loon A P G M. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular sizes, glycosylation patterns, and engineering of proteolytic resistance. Appl Environ Microbiol. 1999;65:359–366. doi: 10.1128/aem.65.2.359-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon S J, Choi Y J, Min H K, Cho K K, Kim J W, Lee S C, Jung Y H. Isolation and identification of phytase-producing bacterium, Enterobacter sp. 4, and enzymatic properties of phytase enzyme. Enzyme Microb Technol. 1996;18:449–454. [Google Scholar]