Abstract
Nitrogen-fixing microbial populations in a Douglas fir forest on the western slope of the Oregon Cascade Mountain Range were analyzed. The complexity of the nifH gene pool (nifH is the marker gene which encodes nitrogenase reductase) was assessed by performing nested PCR with bulk DNA extracted from plant litter and soil. The restriction fragment length polymorphisms (RFLPs) of PCR products obtained from litter were reproducibly different than the RFLPs of PCR products obtained from the underlying soil. The characteristic differences were found during the entire sampling period between May and September. RFLP analyses of cloned nifH PCR products also revealed characteristic patterns for each sample type. Among 42 nifH clones obtained from a forest litter library nine different RFLP patterns were found, and among 64 nifH clones obtained from forest soil libraries 13 different patterns were found. Only two of the patterns were found in both the litter and the soil, indicating that there were major differences between the nitrogen-fixing microbial populations. A sequence analysis of clones representing the 20 distinct patterns revealed that 19 of the patterns had a proteobacterial origin. All of the nifH sequences obtained from the Douglas fir forest litter localized in a distinct phylogenetic cluster characterized by the nifH sequences of members of the genera Rhizobium, Sinorhizobium, and Azospirillum. The nifH sequences obtained from soil were found in two additional clusters, one characterized by sequences of members of the genera Bradyrhizobium, Azorhizobium, Herbaspirillum, and Thiobacillus and the other, represented by a single nifH clone, located between the gram-positive bacteria and the cyanobacteria. Our results revealed the distinctness of the nitrogen-fixing microbial populations in litter and soil in a Douglas fir forest; the differences may be related to special requirements for degradation and mineralization processes in the plant litter.
Nitrogen fixation is performed by phylogenetically diverse groups of prokaryotic organisms belonging to the domains Bacteria and Archaea (9, 30). Different growth requirements resulting from the different physiological properties of these prokaryotic microorganisms preclude their simultaneous cultivation (14, 27). In addition, the likelihood that some of these microorganisms are nonculturable (23) makes a general culture approach for evaluating nitrogen-fixing populations impractical, even though many important contributions to the characterization of nitrogen-fixing prokaryotes have been based on traditional culture techniques (14, 27).
Most forest soils are ecosystems in which nitrogen availability is limited (7). Nitrogen may become available through decomposition of organic material or fixation of molecular nitrogen. The low nitrogen contents of coniferous plant litter and the loss of nitrogen through mineralization contribute to its limited availability. Learning more about the organisms involved in nitrogen fixation in forest systems and about how anthropogenic activities may influence them is important in evaluating the long-term productivity of temperate coniferous forests. Perhaps not all input sources of fixed nitrogen in forest ecosystems have been described, and our understanding of the nitrogen fluxes in these systems is incomplete (5). Relating experimentally determined indigenous nitrogenase activities and nitrogen accumulation to the expected activities of known free-living and associative nitrogen fixers in forest systems leaves portions of the N input unaccounted for (5).
Molecular ecology has provided methods for analyzing environmental DNA extracts for specific gene pools (21). Since nitrogen fixation is exhibited by phylogenetically heterogeneous groups of prokaryotes, detection of a marker gene that is unique and is required for nitrogen fixation may provide a way to analyze the nitrogen fixation potential in an ecosystem. For evaluation of nitrogen-fixing populations in the environment, analysis of nifH, the gene encoding nitrogenase reductase, has been used with various PCR primers that amplify this gene from both microorganisms and environmental samples (13, 20, 25, 33). Due to the vast phylogenetic differences among nitrogen fixers, the sequences of nifH genes have diverged considerably (33), and even the DNA sequences encoding conserved protein regions may differ due to codon redundancy for most amino acids. The design of universal nifH primers requires a high degree of DNA sequence degeneracy and may result in reduced specificity during PCR amplification. However, the use of more sophisticated amplification protocols may make it relatively simple to study the complexity of nitrogen-fixing microorganisms in a given ecosystem.
Here we describe a protocol for general and selective PCR amplification of nifH gene fragments followed by a restriction fragment length polymorphism (RFLP) analysis. This PCR-RFLP protocol was used to analyze nifH gene pools in plant litter and soil samples obtained from a U.S. Environmental Protection Agency (EPA) experimental Douglas fir forest site on the western slope of the Oregon Cascade Mountain Range (18). Our results revealed novel insights into the genetic heterogeneity of the nitrogen fixation potential in forest ecosystems and demonstrated the power of molecular ecology analyses to detect and distinguish groups of nitrogen-fixing taxa in related habitats.
MATERIALS AND METHODS
Sampling and sample preparation.
Plant litter and soil cores (2.5 by 20 cm) were obtained on 3 days between May 1996 and September 1996. Samples were obtained from eight field plots (1 m2 each) at a U.S. EPA experimental Douglas fir forest site that was 40 by 50 m (18). At each plot, three soil cores and the overlying litter were collected. Each core was split into a topsoil portion (0 to 10 cm) and a deeper soil portion (10 to 20 cm). Corresponding samples from the three cores from each plot were pooled and mixed in a clean plastic bag. The samples were stored on ice, and DNA extraction was initiated within 24 h. Dry weights were determined from subsamples following drying at 110°C for 24 h.
DNA extraction.
Bulk DNA was extracted from 0.5 g (dry weight equivalent) of soil and 1 g (dry weight equivalent) of litter and was purified as described previously (22, 29). Briefly, crude DNA was obtained by using a hot extraction procedure performed with 2% sodium dodecyl sulfate, 250 mM NaCl, 100 mM EDTA, and 350 mM guanidine isothiocyanate at 68°C. The extracts were dissolved in 1 ml of TE (10 mM Tris, 1 mM EDTA; pH 8) per g (dry weight equivalent) of extracted sample. Crude DNA extracts were partially cleaned by chloroform extraction, polyethylene glycol 8000 precipitation, and filtration with Microcon 100 microconcentrators (Amicon, Beverly, Mass.). For each sample day, three composite DNA samples (litter, topsoil, and deeper soil) were prepared by mixing equal volumes of the eight purified bulk litter or soil DNA samples to produce a pooled sample that was representative of either the litter or soils obtained that day.
Nested PCR amplification.
Fragments of nifH genes were amplified by using nested PCR (4, 10). Three primers were used; these primers were originally developed by Zehr and McReynolds (33) and Ueda et al. (25). The lengths and degeneracies of the primers were adjusted for simultaneous use in a nested PCR, and the primers were designed to perfectly match all control nifH sequences shown in Fig. 1 with 14 nucleotides at their 3′ ends. The positions of the 20-mer primers were determined with reference to the Azotobacter vinelandii nifH coding sequence (873 bp; sequence positions 1240 to 2112 of the nif gene cluster [GenBank accession no. M20568]). DNA sequence degeneracies were indicated by using the International Union of Pure and Applied Chemistry conventions, as follows (15): R, A/G; Y, C/T; W, A/T; V, A/C/G; B, C/G/T; and N, A/C/G/T. Inosine (I) was used to reduce the degeneracy of the primers by replacing fourfold-degenerate positions (N) in the 5′ portions (1, 6, 25). The first PCR was performed with the forward primer nifH(forA) (GCIWTITAYGGNAARGGNGG; positions 19 to 38; degeneracy, 128 times) and the reverse primer nifH(rev) (GCRTAIABNGCCATCATYTC; positions 463 to 482; degeneracy, 48 times). The second (nested) PCR was performed with the forward primer nifH(forB) (GGITGTGAYCCNAAVGCNGA; positions 112 to 131; degeneracy, 96 times) and the same reverse primer that was used in the first reaction. The first reaction amplified nucleotides 19 to 482 (464 bp), and the nested reaction amplified nucleotides 112 to 482 (371 bp). The final PCR cocktails contained 1× reaction buffer (Boehringer Mannheim, Indianapolis, Ind.), each deoxynucleoside (Boehringer Mannheim) at a concentration of 200 nM, and each degenerated oligonucleotide primer mixture at a concentration of 1 mM. For the first PCR, 5 mg of bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) per ml was added (29), and the reaction volume was adjusted to 20 μl. These low-volume reactions were performed with an oil overlay. For the nested PCR, 0.3 mg of bovine serum albumin per ml was added, the reaction volume was adjusted to 95 μl, and no oil overlay was used. Amplification was performed by using a low-stringency PCR protocol and a model MJR PT-100 thermal cycler with Hot Bonnet (MJ Research, Inc., Watertown, Mass.) operated with block temperature control. After the initial denaturation consisting of 5 min at 95°C, samples were kept at 80°C to have hot start conditions when 5 μl of the enzyme solution (1 U of Taq DNA polymerase in 1× reaction buffer [Boehringer Mannheim]) was added. The cycling conditions used for both reactions were as follows: denaturation for 11 s at 94°C and for 15 s at 92°C, annealing for 8 s at 48°C and for 30 s 50°C, and extension for 10 s at 74°C and for 10 s at 72°C. A final 10-min extension step at 72°C was performed after the cycling steps and before the samples were maintained at 4°C. The first PCR was performed for 40 cycles with a 25-μl reaction mixture containing 2 μl of purified bulk DNA. The nested reaction was performed for 35 cycles with a 100-μl reaction mixture containing 2 μl of the first PCR product as the template. The quality and quantity of the amplification products were analyzed on 2% UltraPure agarose gels (Gibco/BRL, Life Technologies Inc., Gaithersburg, Md.). The nested PCR approach was tested and was found to amplify nifH gene fragments from pure cultures of 23 reference strains representing 14 proteobacterial genera and two gram-positive genera.
FIG. 1.
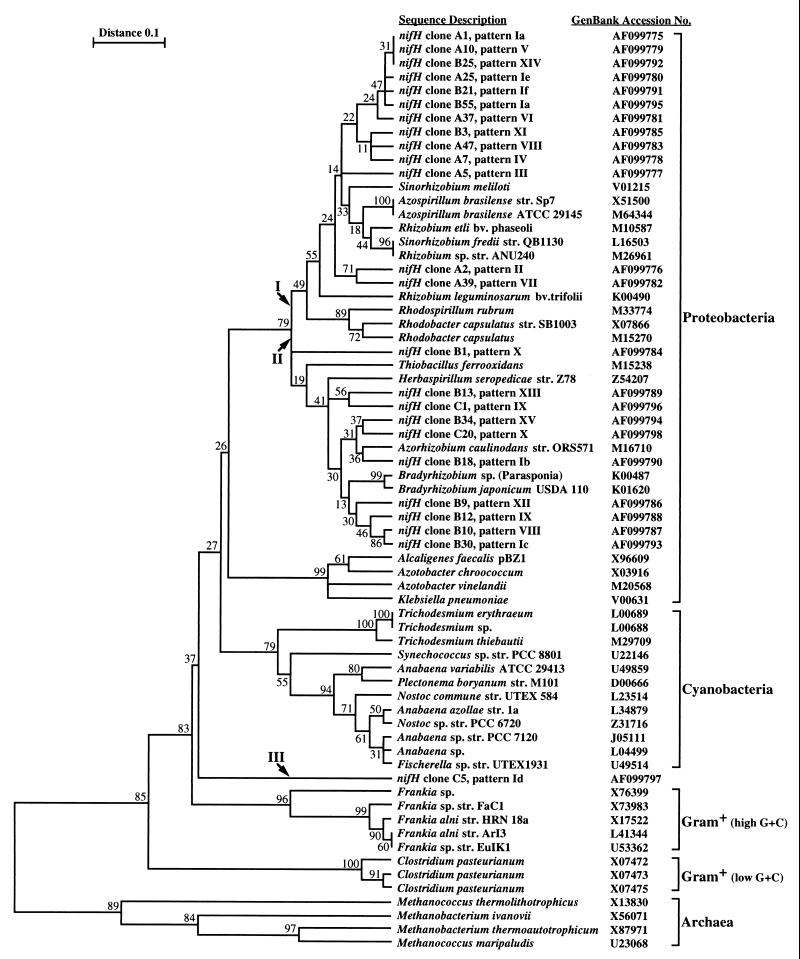
Phylogenetic inference cluster analysis based on the NifH amino acid sequences. An alignment of a 110-amino-acid portion of all 67 sequences was used. The percentage of 100 bootstrap samples that supported each branch is shown. The GenBank accession number of each sequence is also shown. Each of the nifH sequences determined in this study is identified by the clone designation (e.g., clone A1) and the corresponding HaeIII RFLP pattern (e.g., pattern Ia). Arrows I, II, and III indicate the three branches where the nifH clones clustered.
RFLP analyses.
Ninety microliters of each PCR product was mixed with 90 μl of precipitation solution (20% polyethylene glycol 8000, 2.5 M NaCl), incubated for 30 min at 37°C, and microcentrifuged at 8,000 × g for 15 min. The pellets were washed with 70% ethanol and air dried. A restriction analysis was performed by resuspending each air-dried pellet in 40 μl of a restriction enzyme mixture containing 1× restriction enzyme buffer and 2 U of restriction endonuclease. Either HaeIII with buffer M at 37°C or TaqI with buffer B at 65°C (Boehringer Mannheim) was used, and digestion was performed overnight to ensure that complete fragmentation occurred. RFLPs were analyzed in 4% MetaPhor gels (FMC Bioproducts, Rockland, Maine) by using the manufacturer’s recommendations.
DNA cloning and sequencing.
PCR products obtained from selected litter, topsoil, and deeper soil samples were cloned without prior purification by using a TA cloning kit (Invitrogen Co., San Diego, Calif.) according to the manufacturer’s recommendations. Libraries were screened by adding very small amounts of white bacterial colonies to 50-μl PCR amplification mixtures prepared as described above for the nested PCR with primers nifH(forB) and nifH(rev). Amplification was performed by using 30 amplification cycles and the conditions described above. The PCR products were analyzed on 2% UltraPure agarose gels, and the products of nifH-positive clones were subjected to RFLP analyses as described above. The clones were classified on the basis of their HaeIII restriction patterns, which were designated patterns I, II, III, etc. Clones with sequences that were not cleaved by HaeIII (pattern I clones) were subjected to TaqI digestion as described above. The different patterns resulting from these analyses were used to further differentiate the clones and were designated patterns Ia, Ib, Ic, etc. Plasmid DNA of selected clones were purified by using Qiagen 100 plasmid midiprep columns as recommended by the manufacturer (Qiagen, Inc., Chatsworth, Calif.). The sequences of both strands of the cloned nifH PCR products, which were approximately 370 bp long, were determined by using the T7 and M13 reverse sequencing primer sites of vector pCR 2.1 (Invitrogen Co.). Sequences were determined at the Center for Gene Research and Biotechnology, Oregon State University, Corvallis.
DNA sequence analyses.
DNA sequence information was used to perform various analyses. The RFLP patterns of cloned nifH fragments were confirmed by using sequence-based theoretical restriction fragmentation with MacDNASIS, version 3.6 (Hitachi Software Engineering America, Ltd., San Bruno, Calif.). Theoretical fragmentation patterns were calculated and arranged as fragment sizes on a logarithmic scale. DNA sequences were checked for the correct open reading frame, translated, and aligned by using MacDNASIS, version 3.6. The 110-amino-acid aligned sequences (excluding the primer sites) were used for phylogenetic analysis with TREECON (26). Pairwise protein sequence distances (12) and unweighted pair group with mathematical average cluster analysis of 100 bootstrap samplings were used to determine the phylogenetic relationships of the 24 new nifH sequences and 43 known sequences retrieved from GenBank (GenBank accession numbers are shown Fig. 1).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 24 new nifH clones have been deposited in the GenBank database under the following accession numbers: A1, AF099775; A2, AF099776; A5, AF099777; A7, AF099778; A10, AF099779; A25, AF099780; A37, AF099781; A39, AF099782; A47, AF099783; B1, AF099784; B3, AF099785; B9, AF099786; B10, AF099787; B12, AF099788; B13, AF099789; B18, AF099790; B21, AF099791; B25, AF099792; B30, AF099793; B34, AF099794; B55, AF099795; C1, AF099796; C5, AF099797; and C20, AF099798.
RESULTS
Amplification of nifH gene fragments by using a nested PCR approach.
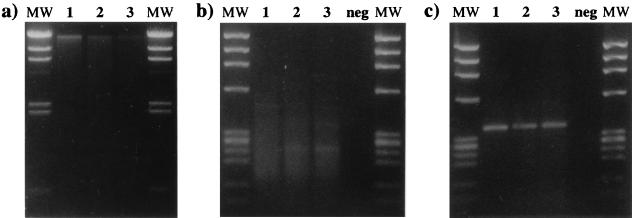
Samples were obtained from the experimental field plot on the west side of the Oregon Cascade Mountain Range three times in 1996. A gel analysis of pooled extracted bulk DNA obtained on the first sampling day (15 May 1996) is shown in Figure 2a. A nested PCR approach performed with degenerate primers was used to amplify nifH gene fragments from the bulk DNA. The first amplification was performed with nifH PCR primers nifH(forA) and nifH(rev) and usually yielded multiple amplification product bands and some less-well-resolved background smearing (Fig. 2b). The nested PCR amplification was performed with nifH PCR primers nifH(forB) and nifH(rev); for all three layers (i.e., litter, topsoil, and deeper soil) this amplification yielded single product bands at the expected nifH gene fragment size, about 370 bp (Fig. 2c). This specificity permitted us to perform RFLP analyses directly with the nifH PCR products and thus facilitated evaluation of the amplified gene pools.
FIG. 2.
DNA extraction and nested amplification of the nifH gene from forest litter and soil samples. (a) Agarose (1%) gel containing DNA extracted from 10 mg (dry weight equivalent) of litter (lane 1), topsoil (lane 2), and deeper soil (lane 3). The molecular weight marker (lanes MW) was bacteriophage λ DNA cleaved with HindIII. (b) Agarose (2%) gel containing 10-μl portions of the first amplification products obtained from litter (lane 1), topsoil (lane 2), and deeper soil (lane 3). Negative control amplification (lane neg) was performed without DNA. (c) Agarose (2%) gel containing 5-μl portions of the nested amplification products obtained from litter (lane 1), topsoil (lane 2), and deeper soil (lane 3). Negative control amplification (lane neg) was performed with the negative control product from gel b. The molecular weight marker (lanes MW) in gels b and c was bacteriophage φX174 DNA cleaved with HaeIII.
RFLP analysis of nifH PCR products from bulk DNA extracts from forest litter and soil.
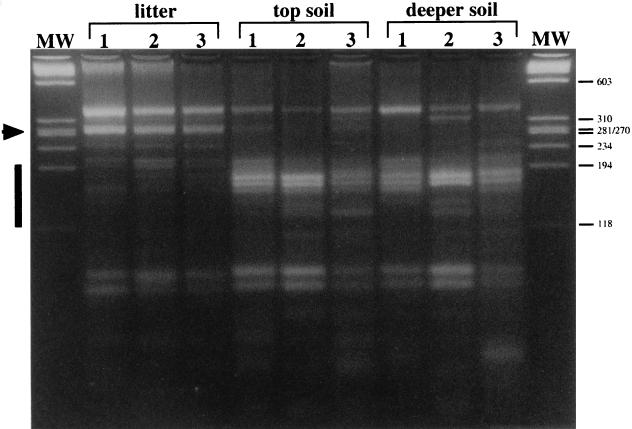
As shown in Fig. 2 for the first sampling day, nifH gene pools were amplified from the three sample types obtained on each of the three sampling dates (15 May 1996, 8 July 1996, and 3 September 1996). RFLP analyses of the amplification products with restriction endonuclease HaeIII revealed that the litter samples reproducibly yielded very similar patterns that were different from the patterns obtained with the samples derived from the underlying soil (Fig. 3). The main differences between the litter and soil patterns were an intense band at about 280 bp (Fig. 3, arrowhead) that was present in the litter sample fingerprints but not in the soil sample fingerprints and a series of bands between the 118- and 194-bp marker bands (Fig. 3, bar) that appeared to be more abundant in the soil fingerprints (Fig. 3). In addition, the litter samples yielded a highly reproducible pattern during this study (Fig. 3, litter lanes). The RFLP patterns produced by forest soil DNA (Fig. 3, topsoil and deeper soil lanes) were more variable over time but consistently exhibited the characteristic differences compared to the patterns produced by litter DNA. No clear difference between the patterns produced by the two soil layers was observed. These analyses indicated that there were detectable and stable differences between the nifH gene pools in the forest litter and soil samples.
FIG. 3.
RFLPs of nested nifH PCR products obtained from forest litter and soil on three sampling dates. The RFLPs obtained from litter, topsoil, and deeper soil are shown for all three sampling days, 15 May 1996 (lanes 1), 8 July 1996 (lanes 2), and 3 September 1996 (lanes 3). The arrowhead indicates the position of the 280-bp fragment characteristic of the litter fingerprints. The bar indicates the 118- to 194-bp size range where fragments characteristic of the soil samples migrated. This analysis was performed on a 4% MetaPhor gel, and the molecular weight marker (lanes MW) was bacteriophage φX174 DNA cleaved with HaeIII.
Characterization of nifH gene fragments amplified from bulk forest litter and soil DNA.
To identify nifH genes that were responsible for the differences between the litter and soil RFLP patterns, the PCR products of selected subsamples of the three layers collected on the first sampling date were cloned. The resulting libraries were screened for nifH clones by using the second step of the nested PCR procedure followed by agarose gel analysis. A total of 50 colonies from the litter library were screened, and 42 (84%) of these were nifH positive. A total of 60 topsoil and 20 deeper soil colonies were screened, and 47 (78%) and 17 (85%), respectively, of these colonies were nifH positive. A total of 130 colonies were screened, and 106 (82%) of them contained a cloned nifH fragment. RFLP analyses of the 106 nifH clones revealed 20 different RFLP patterns of different abundance in the three libraries (Table 1). Only two nifH gene fragments (characterized by patterns Ia and VIII) were obtained from both litter and soil samples. This finding supported the nifH RFLP results obtained with bulk DNA, which indicated that there were major differences in the nifH gene pools in forest litter and soil (Fig. 3).
TABLE 1.
RFLP patterns observed among cloned nifH PCR fragments
| RFLP patterna | No. (%) of clones
|
nifH clone(s) sequencedb | ||
|---|---|---|---|---|
| Litter | Topsoil | Deeper soil | ||
| Ia | 7 (17) | 2 (4) | A1, B55 | |
| Ib | 4 (9) | B18 | ||
| Ic | 3 (6) | B30 | ||
| Id | 1 (6) | C5 | ||
| Ie | 1 (2) | A25 | ||
| If | 1 (2) | B21 | ||
| II | 10 (24) | A2 | ||
| III | 3 (7) | A5 | ||
| IV | 8 (19) | A7 | ||
| V | 5 (12) | A10 | ||
| VI | 6 (15) | A37 | ||
| VII | 1 (2) | A39 | ||
| VIII | 1 (2) | 6 (13) | A47, B10 | |
| IX | 8 (17) | 15 (88) | B12, C1 | |
| X | 7 (15) | 1 (6) | B1, C20 | |
| XI | 6 (13) | B3 | ||
| XII | 7 (15) | B9 | ||
| XIII | 1 (2) | B13 | ||
| XIV | 1 (2) | B25 | ||
| XV | 1 (2) | B34 | ||
| Total | 42 (100) | 47 (100) | 17 (100) | |
RFLP patterns were designated as explained in the text.
The nifH clones were designated on the basis of their sources, as follows: A, litter; B, topsoil; and C, deeper soil.
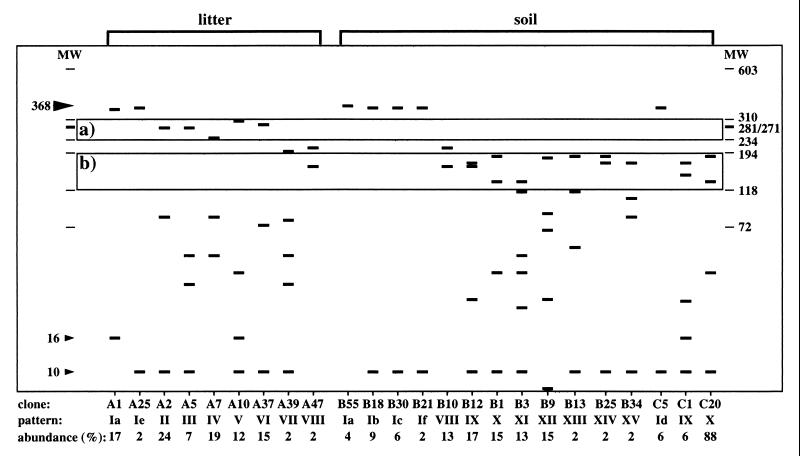
The DNA sequences of 24 clones representing the 20 patterns (Table 1) obtained for the three different sample types were determined. The sequences obtained from the nested PCR products were all 368 bp long and represented a portion of the NifH open reading frame. The expected HaeIII RFLP fragmentation patterns of the 24 DNA sequences are shown on a logarithmic scale in Fig. 4. These calculated patterns matched the patterns determined experimentally for each of the 24 clones. The position of the uncleaved PCR product (pattern I) is indicated by a large arrowhead in Figure 4. Pattern I clones were clones that either were not cleaved by HaeIII or were cleaved only in the primer region. Such cleavage sites were not considered because they may have resulted from using degenerate amplification primers. The resulting small size differences (i.e., 10 and 16 bp) (Fig. 4, small arrowheads) were not resolved on the agarose gels used. Consistent with the fingerprints obtained for the bulk DNA samples (Fig. 3), only clones isolated from the litter library (clones A2, A5, A7, A10, and A37) yielded fragments whose sizes were in the size range between the 234- and 310-bp markers. Similarly, the characteristic soil patterns with fragments that were between 194 and 118 bp long (Fig. 3) are reflected by the data for the 24 nifH clones (Fig. 4). All of the clones obtained from soil and that did not produce RFLP pattern I produced patterns in this size range (i.e., clones B1, B3, B9, B10, B12, B13, B25, B34, C1, and C20). Only clone A47 from the litter library produced a fragment in this size range; however, this clone produced pattern VIII, one of the two patterns obtained from both litter and soil samples.
FIG. 4.
Calculated HaeIII RFLP patterns of the 24 nifH clone sequences. One representative clone was sequenced for each of the patterns observed in the three libraries (A, litter; B, topsoil; C, deeper soil). The calculated fragment sizes for the 20 theoretical RFLP patterns (patterns Ia to If and II to XV) identified by the HaeIII and TaqI RFLP analyses were plotted on a logarithmic scale. Box a indicates the region between 234 and 310 bp where litter-specific RFLP fragments were observed. Box b indicates the region where prominent fragments of the soil clones migrated. The large arrowhead at 368 bp indicates the position of the uncleaved nested nifH PCR product. The small arrowheads at 10 and 16 bp indicate the positions of fragments resulting from cleavage in the primer regions. The abundance values indicate abundance within each library (A, litter; B, topsoil; C, deeper soil). Marker lanes MW show the positions of 72- to 603-bp fragments of bacteriophage φX174 DNA cleaved with HaeIII.
For phylogenetic analysis, the sequences of the 24 clones were compared to 43 nifH sequences retrieved from GenBank. These sequences were derived from members of the Archaea, as well as gram-positive bacteria (high G+C content and low G+C content) and gram-negative bacteria (cyanobacteria and members of the class Proteobacteria). All 67 amino acid sequences encoded by the nested PCR product were aligned. The 110-amino-acid-long alignment of these sequences (excluding the primer sites) was used for phylogenetic inference. In the initial analysis, a phylogenetic tree containing the 43 control sequences was constructed. The results obtained with this 43-×-110 data matrix revealed a clustering of the sequences which was consistent with the sequence descriptions and previously published NifH phylogenetic tree topologies (data not shown). In a second analysis step, the 24 cloned NifH sequences were included, which yielded a 67-×-110 data matrix. After the analytical steps described above were performed, we obtained a phylogenetic tree that perfectly maintained the topology of the tree obtained for the 43 control sequences. The nifH clones isolated clustered on three branches that were specified by the proximity of control sequences (Fig. 1). One group of sequences clustered with NifH sequences from members of the genera Rhizobium, Sinorhizobium, and Azospirillum (Fig. 1, group I). All nine clones obtained from the litter library clustered with this group together with 4 of the 12 topsoil clones (B3, B25, B21, and B55). The second cluster was represented by a more diverse group of NifH sequences which were derived from members of the genera Bradyrhizobium, Azorhizobium, Herbaspirillum, and Thiobacillus (Fig. 1, group II). Only clones that were isolated from the soil libraries (eight clones from topsoil and two clones from deeper soil) were present in this group. The third group was represented by nifH clone C5, which clustered between the gram-positive bacteria and the cyanobacteria (Fig. 1, group III). These results clearly confirmed that the nitrogen-fixing microbial taxa found in closely associated layers of forest litter and soil are distinct and were in strict agreement with the results of the RFLP analyses.
DISCUSSION
This molecular ecology study was undertaken in order to evaluate nifH diversity among nitrogen-fixing microorganisms in a Douglas fir forest. In order to increase the specificity and sensitivity of nifH detection, we developed a nested PCR approach based on previously described conserved primer target sites (13, 25). As shown in Fig. 2b and c, this approach improved both the sensitivity and the specificity of nifH amplification. The improvements resulted in highly specific detection of nifH genes in bulk environmental DNA samples and allowed RFLP analyses and cloning to be performed without prior purification of the PCR products.
PCR amplification of gene sequences has proven to be a powerful tool, but if applied to mixtures of related genes, it may be biased by two known factors. One factor is the potential for preferential amplification of certain sequences, which prevents quantitative correlation of sequence abundance in the DNA sample and sequence abundance in the PCR product (24). The results of our study clearly show, however, that RFLP fingerprints are reproducible and can have high diagnostic value (Fig. 3). It is also important to note that the abundance of clones in libraries derived from PCR products does not necessarily reflect the abundance of the sequences in the samples analyzed. A second limitation is the possibility of chimera formation, which has been observed in PCR products (16, 19, 28). Distinguishing these artifacts from natural variations in mixed PCR products with unknown but closely related sequences can be difficult. In this study the sequence NifH clone C5 clustered between the sequences of the gram-positive bacteria and the cyanobacteria and not close to known sequences (Fig. 1). This may indicate that it represented a PCR artifact. However, close inspection of this sequence revealed no chimera when it was compared to other NifH sequences used or evaluated in this study. We believe that the NifH clone C5 sequence is a unique sequence with no previously described close phylogenetic affinities.
RFLP analysis of nifH PCR products from environmental samples is a powerful tool for assessing the presence and diversity of nitrogen-fixing microorganisms in ecosystems. Although this approach does not directly allow evaluation of functional aspects of the nitrogen-fixing populations in a sample, structural information on the gene pool and the potential for nitrogen fixation may be assessed. The number and positions of the RFLP fragments reflect the diversity and heterogeneity of the nitrogen-fixing populations in a sample. The RFLP analyses performed in this study yielded highly reproducible patterns (Fig. 3) over time and revealed clear differences between the nifH gene pools present in the litter and soil from the Douglas fir forest site. The differences are consistent with reports that the nitrogen fixation activities associated with forest litter and logs are greater than the nitrogen fixation activities associated with soil (11). Nitrogen availability in litter may limit or regulate degradation processes (2, 3, 7), and controlled nitrogen fixation may provide nutrient levels that allow optimal mineralization activities.
The differences between the nifH gene pools in the Douglas fir forest litter and soil reported here were consistent for the PCR-RFLP analyses of bulk DNA (Fig. 3) and cloned PCR products (Table 2 and Fig. 4), as well as for the sequence data (Fig. 1). This suggested that stable differences between the litter and soil nifH gene pools persisted over the course of this experiment. RFLP analysis of the cloned PCR products revealed patterns that contributed to the characteristic differences between the total litter and soil nifH fingerprints. Patterns II, III, and VI, which were produced only by members of the litter library, had fragments at 276 and 285 bp, and these fragments might have contributed to the bright band at about 280 bp observed in the litter sample fingerprints (Fig. 3 and 4). Fragments in patterns IV (237 bp) and V (303 bp) may correspond to the weak bands that migrated above the 234-bp marker and below the 310-bp marker in the litter fingerprints. None of the patterns obtained from soil produced fragments between 234 and 281 bp. All of the soil clones produced either pattern I (5 clones not cleaved by HaeIII) or patterns IX to XV (10 clones), which produced fragments between 194 and 118 bp.
Cloning and sequencing of the nested amplification product were performed to obtain phylogenetic information on the nifH genes isolated from the samples. The approach that includes inferring phylogenetic information from gene sequences has been used particularly for the small-subunit rRNA sequence (17). It has been shown that phylogenetic inferences based on NifH amino acid sequence information agree with rRNA data (25, 30–32). The cluster analysis tree obtained with the 67-×-110 data set was consistent with previous reports based on NifH phylogenies (20, 25) and clustered the sequences derived from members of distinct genera, such as the genera Clostridium, Frankia, Klebsiella, Azotobacter, and Rhizobium (Fig. 1). Only a few sequences exhibited an irregular behavior; these sequences included the sequences of Anabaena sp., Nostoc sp., and Fischerella sp. in the cyanobacterial cluster and nifH clone C5, which clustered between the gram-positive bacteria and the cyanobacteria. Many of the bootstrap values obtained for the tree shown in Fig. 1 are rather low (<50%). This may be attributed to the large number (67) of relatively short (110-amino-acid) homologous sequences. However, very accurate and stable clustering of the sequences into the larger groups (i.e., the Proteobacteria, cyanobacteria, gram-positive bacteria, and Archaea) and at the genus level (e.g., the genera Clostridium, Frankia, Trichodesmium, Azotobacter, Klebsiella, Rhodobacter, and Azospirillum) was taken as an indication that this phylogenetic inference was valid (Fig. 1). Most interestingly, the sequences that produced patterns II, III, and VI, which contained fragments (276 and 285 bp) in the size range characteristic of litter, clustered with sequences of members of the genera Rhizobium and Sinorhizobium, which are known plant root symbionts. The association of specific nitrogen-fixing microorganisms with Douglas fir litter may be related to the specific requirements for mineralization of plant litter. The correct taxonomic classification of organisms in the environment may be determined by isolation and polyphasic characterization (8). The RFLP patterns identified in this study, together with the presumptive taxonomic identities, may facilitate screening for and subsequent isolation and detailed characterization of nitrogen-fixing organisms from forest litter and soil. The PCR-RFLP protocol used in the present study may also provide a rapid tool for detecting differences or changes in the nitrogen fixation potential in other environmental systems.
ACKNOWLEDGMENTS
This study was funded in part by the U. S. EPA. F.W. was supported by a postdoctoral research fellowship from the National Research Council, Washington, D.C., and acknowledges technical support received from the U.S. EPA. F.W. received additional support from the Ciba Geigy Jubiläumsstiftung, Basel, Switzerland.
We are grateful to K. Donegan, Dynamac Inc., for helping with sampling and to T. Townsend and D. Hahn, ETH Zürich, for critical comments on the manuscript.
REFERENCES
- 1.Bartl S, Weissman I L. PCR primers containing an inosine triplet to complement a variable codon within a conserved protein-coding region. BioTechniques. 1994;16:246–250. [PubMed] [Google Scholar]
- 2.Berg B. Nutrient release from litter and humus in coniferous forest soils—a mini review. Can J For Res. 1986;1:359–369. [Google Scholar]
- 3.Berg B, Tamm C O. Decomposition and nutrient dynamics of litter in long-term optimum nutrition experiments. II. Nutrient concentrations in decomposing Picea abies needle litter. Scan J For Res. 1994;9:99–105. [Google Scholar]
- 4.Beyer W, Glockner P, Otto J, Boehm R. A nested PCR method for the detection of Bacillus anthracis in environmental samples collected from former tannery sites. Microbiol Res. 1995;150:179–186. doi: 10.1016/S0944-5013(11)80054-6. [DOI] [PubMed] [Google Scholar]
- 5.Bormann B T, Bormann F H, Bowden W B, Pierce R S, Hamburg S P, Wang D, Snyder M C, Li C Y, Ingersoll R C. Rapid N2 fixation in pines, alder, and locust: evidence from the sandbox ecosystem study. Ecology. 1993;74:583–598. [Google Scholar]
- 6.Candrian U, Furrer B, Hofelein C, Luthy J. Use of inosine-containing oligonucleotide primers for enzymatic amplification of different alleles of the gene coding for heat-stable toxin type I of enterotoxigenic Escherichia coli. Appl Environ Microbiol. 1991;57:955–961. doi: 10.1128/aem.57.4.955-961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson J O. Nitrogen fixation in forests and agroforestry. In: Metting F B Jr, editor. Soil microbial ecology. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 227–253. [Google Scholar]
- 8.De Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, Collins M D, Dreyfus B, Kersters K, Gillis M. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol. 1994;44:715–733. [Google Scholar]
- 9.Eady R R. The dinitrogen-fixing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer Verlag; 1991. pp. 534–553. [Google Scholar]
- 10.El Fantroussi S, Mahillon J, Naveau H, Agathos S N. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested PCR monitoring. Appl Environ Microbiol. 1997;63:806–811. doi: 10.1128/aem.63.2.806-811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hope S M, Li C-Y. Respiration, nitrogen fixation, and mineralizable nitrogen spatial and temporal patterns within two Oregon Douglas-fir stands. Can J For Res. 1997;27:501–509. [Google Scholar]
- 12.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 13.Kirshtein J D, Paerl H W, Zehr J P. Amplification, cloning, and sequencing of a nifH segment from aquatic microorganisms and natural communities. Appl Environ Microbiol. 1991;57:2645–2650. doi: 10.1128/aem.57.9.2645-2650.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles R, Laserna Barraqio W. Free-living dinitrogen-fixing bacteria. In: Weaver R W, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A, editors. Methods of soil analysis. Part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America, Inc.; 1994. pp. 179–197. [Google Scholar]
- 15.Liébecq C, editor. International Union of Pure and Applied Chemistry and International Union of Biochemistry and Molecular Biology biochemical nomenclature and related documents. 2nd ed. London, United Kingdom: Portland Press; 1992. [Google Scholar]
- 16.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 17.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks D, Kimball J, Tingey D, Link T. The sensitivity of snowmelt processes to climate conditions and forest cover during rain-on-snow: a case study of the 1996 Pacific Northwest flood. Hydrolog Processes. 1998;12:1569–1587. [Google Scholar]
- 19.Odelberg S J, Weiss R B, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1995;23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pace N R. New perspective on the natural microbial world: molecular microbial ecology. ASM News. 1996;62:463–470. [Google Scholar]
- 22.Porteous L A, Watrud L S, Seidler R J. An improved method for purifying DNA from soil for polymerase chain reaction amplification and molecular ecology applications. Mol Ecol. 1997;6:787–791. [Google Scholar]
- 23.Roszak D B, Colwell R R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 27.Weaver R W, Graham P H. Legume nodule symbionts. In: Weaver R W, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A, editors. Methods of soil analysis. Part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America, Inc.; 1994. pp. 199–222. [Google Scholar]
- 28.Widmer F, Seidler R J, Gillevet P M, Watrud L S, Di Giovanni G D. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol. 1998;64:2545–2553. doi: 10.1128/aem.64.7.2545-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer F, Seidler R J, Watrud L S. Sensitive detection of transgenic plant marker gene persistence in soil microcosms. Mol Ecol. 1996;5:603–613. [Google Scholar]
- 30.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacy G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 43–86. [Google Scholar]
- 31.Young J P W. Molecular phylogeny of rhizobia and their relatives. In: Palacios R, Mora J, Newton W E, editors. New horizons in nitrogen fixation. London, United Kingdom: Kluwer Academic Publications; 1993. pp. 587–592. [Google Scholar]
- 32.Zehr J P, Capone D G. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb Ecol. 1996;32:263–281. doi: 10.1007/BF00183062. [DOI] [PubMed] [Google Scholar]
- 33.Zehr J P, McReynolds L A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]