ABSTRACT
Colon cancer (CC) is a common malignant tumor of the digestive tract. Circular RNAs (circRNAs) play important roles in the progression of CC. This study aimed to explore the role and mechanism of circRNA_0085315 in CC. In this study, we used qRT-PCR and Western blot assays to analyze the expressions of circRNA, miRNA, and mRNA as well as the expression of the related proteins. Luciferase reporter, RNA pull-down, and qRT-PCR assays were used to prove the relationship among circRNA, miRNA, and mRNA. CCK-8, colony formation, and transwell assays were used to perform the analysis of cell proliferation, migration, and invasion. Our results showed that the higher circRNA_0085315 expression led to the poorer prognosis of CC patients. The function of circRNA_0085315 as a ceRNA in competing with MAP3K1 mRNA to sponge miR-1200. CircRNA_0085315 sponged miR-1200 to promote cell proliferation, migration, and invasion and affected the expression of Ki67, MMP2, E-cadherin, and N-cadherin, but not circRNA_0085315-mut without the binding site of miR-1200. MAP3K1-overexpression or miR-1200 mimics prevented the suppression on the enhanced cell proliferation, migration, and invasion caused by circRNA_0085315-overexpression. circRNA_0085315 increased the phosphorylation levels of JNK, p38, and ERK1/2 by stimulating MAP3K1 up-regulation caused by miR-1200 inhibition. In conclusion, circRNA_0085315 serves as a ceRNA and promotes CC progression through the activation of the MAPK signaling pathway mediated via the miR-1200/MAP3K1 axis, suggesting that circRNA_0085315 may be a promising diagnostic and therapeutic target for CC.
KEYWORDS: Circ_0085315, colon cancer, MAPK
Introduction
Colon cancer (CC), one of the most frequently occurring malignant tumors in the world, especially in cities and regions with rapid economic development, exhibits an increasing trend year by year [1,2]. Since the early symptoms of CC are not obvious and the onset is hidden, most patients are already in the middle and late stages when they see a doctor. What is more worrying is that CC has the characteristics of high recurrence and distant metastasis, resulting in many patients with a 5-year survival rate of less than 50% [3–6]. This poses a major threat to patients’ life. The occurrence and metastasis of CC is a complex process involving multiple steps such as the activation of oncogenes and the inactivation of tumor suppressor genes. Therefore, it is particularly important to study the pathogenesis of colon cancer in-depth, to find markers for early diagnosis, prognostic judgment, thereby providing theoretical support for the effective control of the progression of CC and the improvement of patient prognosis.
Circular RNAs (circRNAs) are endogenous non-coding RNAs (ncRNAs), which form a covalently closed-continuous loop by using back-splice without 3ʹ-end or 5ʹ-end. This explains the multiple activities of circRNAs. In recent years, many circRNAs are identified to be involved in the occurrence and development of many diseases [7]. As circRNAs have a unique closed-loop structure that can resist the effect of degradation imposed by exonuclease, their stability is superior to most of the other linear RNAs. Accordingly, circRNAs can be considered as the biomarkers for the diagnosis and treatment of cancers [8,9]. Hsa_circ_0085315, with its encoding gene positioned at chromosome (Chr) 8:109,213,971–109,254,142, is a recently uncovered new circRNA with a length of 1390 bp. Hsa_circ_0085315 is derived from eukaryotic initiation factors 3E (EIF3E), and it has not been reported in any cancer, and its functions in regulating CC progression and the underlying mechanism should be further explored.
By interacting with RNA-binding proteins, circRNAs can also regulate the expression of target genes. What is more, circRNAs can be used as the competitive endogenous RNAs (ceRNAs) to adsorb miRNAs, so as to control the degradation or translation of target genes by miRNAs [10,11]. MiRNA is a class of non-coding single-stranded RNA molecules with a length of approximately 22 nucleotides encoded by endogenous genes, which are involved in post-transcriptional gene expression regulation. More and more studies have shown that miRNA expression and function could be inhibited by circRNAs that act like sponges, and ultimately promote gene transcription at the post-transcriptional level. This indicates that miRNA plays an indispensable role in circRNAs-involved cancers. Many studies have proved that miR-1200 is a key miRNA in tumor progression. It has been reported that circ_0001785 acts as a ceRNA to sponge miR-1200 and increase HOXB2 expression, thereby promoting the proliferative ability of osteosarcoma cells [12]. In addition, Pan B et al. reported that RGMB-AS1 promoted glioma progression by targeting the miR-1200/HOXB2 axis [13]. The circ_0026359-mediated miR-1200/POLD4 pathway functions in regulating the cisplatin resistance of GC [14]. The above reports show that the effect of miR-1200 on the development and progression of CC is still unclear. Based on the results of the circRNA interactome database, we found that miR-1200 was one of the target miRNAs of hsa_circ_0085315. Thus, we speculated that circ_0085315 and miR-1200 play an important role in the development of colon cancer.
Materials and methods
Patients and tumor tissues
We obtained 70 quick-frozen CC tissues and paired with non-tumorous tissues from nearby CC patients. Prior-informed consent from patients has been obtained for the study, as well as approval from the ethics committee of Ningbo First Hospital. The study was performed following the guidelines of the committee and the Declaration of Helsinki. The demographics of the patients and clinical findings are given in Table 1. All of the operations were carried out strictly following applicable guidelines, laws, and regulations.
Table 1.
Correlation Between circ_0085315 Expression and Clinicopathological Characteristics in Colon Cancer Patients
| Characteristics | Number | circ_0085315 Expression |
χ2 | P values | |
|---|---|---|---|---|---|
| Low(n = 32) | High(n = 38) | ||||
| Age (years) | 0.306 | 0.580 | |||
| ≤60 | 26 | 13 | 13 | ||
| >60 | 44 | 19 | 25 | ||
| Sex | 0.270 | 0.603 | |||
| Male | 24 | 12 | 12 | ||
| Female | 46 | 20 | 26 | ||
| Tumor size (cm) | 0.549 | 0.459 | |||
| ≤5 | 34 | 14 | 20 | ||
| >5 | 36 | 18 | 18 | ||
| Stage | 7.775 | 0.05 | |||
| I/II | 27 | 18 | 9 | ||
| III/IV | 43 | 14 | 29 | ||
| Differentiation | 0.320 | 0.572 | |||
| Well | 31 | 13 | 18 | ||
| Poor | 39 | 19 | 20 | ||
| Lymph nodes metastasis | 5.757 | 0.016* | |||
| Negative | 35 | 21 | 14 | ||
| Positive | 35 | 11 | 24 | ||
| Distant metastasis | 0.425 | 0.514 | |||
| Negative | 13 | 7 | 25 | ||
| Positive | 57 | 6 | 32 | ||
Note: *P < 0.05.
Cell cultures
The human colon cancer cell lines SW480, LOVO, and normal colon epithelial NCM460 cells (HCoEpic) were obtained from Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco). The cells were grown in a humidified chamber with CO2 and air at a volume ratio of 5:95 at 37°C
Quantitative real-time polymerase chain reaction (qRT-PCR)
By using the Trizol reagent (Invitrogen), we isolated total RNA from cells and tissues. The cDNA synthesis for miR-1200 was performed by virtue of TaqMan MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA), and the cDNA synthesis for mRNAs (MAP3K1, MAP3K2, MAP3K3, Ki67, MMP2, E-cadherin, and N-cadherin) and circRNA_0085315 by One Step PrimeScript cDNA kit (Beyotime, Shanghai, China). The qRT-PCR assay was used to determine the expression change of mRNAs and circRNA_0085315 in different groups via using the GeneAmp 7500 system (Applied Biosystems) in triplicate. The TaqMan MicroRNA assay was also adopted to evaluate miR-1200 expression. Respectively, GAPDH was taken as the endogenous reference gene for mRNA and circRNA_0085315. U6 was taken as the endogenous reference gene for the miR-1200. The expressions exposed by mRNA, miR-1200 and circRNA_0085315 were assessed by using the 2−ΔΔCt method. The primer sequences are shown in Table 2.
Table 2.
The sequences of the primers
| Genes | Forward (5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) |
|---|---|---|
| circRNA_0085315 | AGCGAAAAGAGGCACAGCTA | GGAAAAACTAAGCAATGTCC |
| miR-1200 | AGTGCTGACGTCAAGTACCCATAAA | CTGCAGATAAGTGCGGTAGGT |
| miR-548p | CTGCTGAACTGCAATGAAAT | TGCGGTAGCGTGAAAGTGA |
| miR-330-3p | GCAAAGCACACGGCCTG | TCCTCCTCTCCTCTCCTCTC |
| miR-516b | AGGCATCTGGAGGTAAGAAG | GTTGTGGTTGGTTGGTTTGT |
| miR-1276 | AGGGTAAAGAGCCCTGTG | GTTGTTGGTTGGTTGGTTGT |
| miR-557 | GTTTGCACGGGTGGGCC | GAGAGGAGAGGGAGAGGAGA |
| miR-604 | ATCGTGTAGTGCCATGTACTG | CCTGTAAGTCATGCAAAC |
| miR-217 | ATTGGCTGCAACCACACATGGGT | AATGTCTGAATGCCCCTGCAAA |
| miR-324-5p | CGCATCCCCTAGGGCAT | CACACTCTCACTCACGCATC |
| miR-659 | CTGAACTGCAATGAAATGGGTA | TGCGTGAAAAGTCAAAACC |
| miR-936 | TGCTGAATGTACAACTGGTAAA | GCGCGCATGTAGCGCTGGAA |
| miR-1257 | TGCTACATGATAGATACTGTTAA | CGACTCATGCATGACACATGTA |
| miR-451 | AGGGGAAACCGTTACCATTAC | ACCCAGACACAAACATAGCC |
| miR-570 | AGGGGAAAGGTAATTGCAGTT | CAGCAGTTCCAGTTCCAGTT |
| miR-644 | AGGCAGTGTGGCTTTCT | GTTGTGGTTGGTTGGTTTGT |
| miR-543 | GAAACATTCGCGGTGCA | GAGAGGAGAGGAAGAGGGA |
| miR-520 g | GGGTCTAGAGGAAGCACTTT | GTTGTTGGTTGGTTGGTTGT |
| miR-1305 | AGCGCTTTTCAACTCTAATGG | GAGAGGAGAGGAAGAGGGA |
| miR-1289 | GGTGGAGTCCAGGAATCTG | CAGGAAGGACAGGAAGACAG |
| miR-576-5p | AGCGCATTCTAATTTCTCCAC | TCCTCCTCTCCTTCCTTCTC |
| miR-215 | AGGGGCATGACCTATGAATTG | GCGTTGTGTTGTGTTGTGTT |
| miR-1258 | AGCGCAGTTAGGATTAGGTC | AACAACCAACACAACCCAAC |
| miR-584 | CCTCTTATGGTTTGCCTGGG | GTTGTGGTTGGTTGGTTTGT |
| miR-224 | GGGCAAGTCACTAGTGGT | GAGAGGAGAGGAAGAGGGAA |
| miR-568 | GGGGGGGGATGTATAAATGTAT | GTTGTTGGTTGGTTGGTTGT |
| miR-127-5p | GCTGAAGCTCAGAGGGC | AACCAACCAACCACAACAAC |
| miR-1299 | AGGGTTCTGGAATTCTGTGT | AACAACCAACACAACCCAAC |
| miR-153 | TGCGTTGCATAGTCACAAAA | GAGAGGAGAGGAAGAGGGAA |
| miR-1243 | AGGGGCAACTGGATCAATTATA | AGGAAGGAAGGAGAAGGGAT |
| miR-634 | ACTGGCTGACACACTGCCCCC | TGTGTCAGACACACTGAAGTGG |
| miR-656 | GGAGGTTGCCTGTGAGG | TTGTTGTCCCGTTCTGCTAT |
| miR-653 | AGGGGGTGTTGAAACAATCT | GAGAGGAGAGGAAGAGGGAA |
| miR-1225-3p | TGAGCCCCTGTGCCGC | AACAACCAACACAACCCAAC |
| miR-155 | AGGGGTTAATGCTAATCGTGAT | AACAACCAACACAACCCAAC |
| miR-431 | GTGTCTTGCAGGCCGT | GAGAGGAGAGGGAGAGGAGA |
| miR-660 | AGGGTACCCATTGCATATCG | GAGGAGGAAGAAGAGGAGGA |
| miR-149 | TCTGGCTCCGTGTCTTC | TCCTCTCCTCCTTCCTCTTC |
| miR-571 | GGGTGAGTTGGCCATCTG | TAGAGGAGGGAGAGGGAGAG |
| miR-633 | AGGCGCTAATAGTATCTACCAC | AACAACCAACACAACCCAAC |
| miR-548 m | AGGGGCAAAGGTATTTGTGG | GTGTGGTGTGGTATGGTGTG |
| miR-507 | AGGGTTTTGCACCTTTTGG | GAGAGGAGAGGAAGAGGGAA |
| miR-188-3p | GCTCCCACATGCAGGG | AAACCAACCAACCACTACCA |
| miR-1287 | GGTGCTGGATCAGTGGTT | GTTGTGGTTGGTTGGTTTGT |
| miR-647 | GGTGGCTGCACTCACT | GAGAGGAGAGGAAGAGGGAA |
| miR-495 | GGGGAAGTTGCCCATGTTA | GAGGAGGAAGAAGAGGAGGA |
| miR-1231 | GGTGTCTGGGCGGAC | ACAACAACCAACCAACCAAC |
| miR-421 | AGGGGCATCAACAGACATTAATT | TACCAACCAACCCACTCACT |
| miR-520 h | GGGACAAAGTGCTTCCCTT | CAGGTTCAGGTTCAGGTTCA |
| miR-1233 | AGTGGGAGGCCAGGGC | GCGTTGTGTTGTGTTGTGTT |
| miR-569 | TGCGCAGTTAATGAATCCTG | GTTGTGGTTGGTTGGTTTGT |
| miR-192 | AGGGGCTGACCTATGAATTG | GCGTTGTGTTGTGTTGTGTT |
| miR-942 | AGGGTCTTCTCTGTTTTGGC | TCCTCTCCTCCTTCCTCTTC |
| MAP3K1 | GGGAGCAGTTGTAGTAGGTA | CTCTCCGTCTGATTAGCCAT |
| MAP3K2 | TGTCCTTCATAAGGCCAGT | TCCATAGACTGTCCAAAGGC |
| MAP3K3 | CCGAGGAGGTGTCTCAGG | TGGGCTTTCCCTTCTACCAG |
| Ki67 | AGGTTCCTAAGAGAGGAGGGA | TCCGAAGCACCACTTCTTCT |
| MMP2 | TACGACCGCGACAAGAAGTA | TCATCGTAGTTGGCTGTGGT |
| E-cadherin | TGCCAACTGGCTGGAGATTA | AGTGTCCCTGTTCCAGTAGC |
| N-cadherin | ACACCAGGTTTGGAATGGGA | TTGGAGCCTGAGACACGATT |
| U6 | TTCGGCACACATAGGTACTCGC | TAACTCACGAAATGCTAAGG |
| GAPDH | AGCCTTAACCCAACTTATTGCC | ACAGGCACCATACACACGG |
RNase R treatment
Total RNAs were treated with the RNase R (Epicenter Technologies). The total RNAs from SW620 and LOVO cells were then divided into two parts in this experimenter: one for the RNase R digestion and the other with the digestion buffer as the control group. As designed, we had the total RNA (2 μg) mixed with 2 μl 10× RNase R Reaction Buffer plus 2 μl RNase R (20 U/μl) for the first part, and RNase R replaced by DEPC-treated water for the second part. Following that we had RNA samples incubated in water at 37°C for 30 min. We used the qRT-PCR assay to help with the analysis of the GAPDH mRNA and circRNA_0085315 detected. We further adopted RNA treated with RNase R to help with the detection of the resistance presented by circRNA_0085315 to the exonuclease digestion of RNase R.
Cell transfection
The CC cells (2 × 106) were seeded in a 6-well plate and incubated at 37℃ in humidified 5% CO2 atmosphere overnight. The fragment of circ_0085315 (TTGACTTGTAGTTCCCTGCAATAACAGTATCTCTTTTTTAGTTT) and the fragment of MAP3K1 (TGTTTCCAGCCTTGTCAACCCCTTCTTCTT) was introduced as a complete sequence to cloned into overexpression plasmids (pcDNA3.1 vector; BGI, Beijing, China), respectively. Mutant circ_0085315 without the binding site of miR-1200 was designed by VectorBuilder (Yunzhou Biological Technology Co., LTD, Guangzhou, China). Then, the circ_0085315-mut-overexpressing plasmid (CGAATGTGGGAATTACTTGAAGG) was synthesized from Yunzhou Biological Technology Co., LTD. The small interference RNA (siRNA)#1-circ_0085315 (ACTTGTAGTTCCCTGCAATAACAGTATCTCTTTTTTAGTTT) and siRNA#2-circ_0085315 (TTGACTTGTAGTTCCCTGCAATAACAGTATCTCTTTTTTAG) were purchased from Gene Pharmacy (Shanghai, China). miR-1200 mimics (sense: 5′-AGAUGCAGGGCUGCACUAUAG-3′; antisense: 3′-CUUGACCUUGCCAGCACUUU-5′), and miR-1200 inhibitors (5′-AGGCCUCUAUCUUGCCAGCAU-3′) were purchased from Qcbio Science&Technologies Co., Ltd (Shanghai, China). Then, pcDNA3.1 vector-circ_00851315, pcDNA3.1 vector-MAP3K1, siRNA#1/or#2-circ_0085315, miR-1200 mimics, miR-1200 inhibitors, and their corresponding negative control (NC) was transfected into CC cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, respectively. The above cells were harvested at 48 h for further experiments.
Colony formation assay
We put every group of the treated cells (1 × 103 per well) into a 10 cm culture dish and started a culturing for 2 weeks. Going through the culturing in RPMI-1640 consisting of 80 U/ml penicillin, 20% FBS and 100 μg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) with 5% CO2 at 37°C, the cells were stained with 1% crystal violet (Beyotime, Shanghai, China) for number counting.
Transwell migration & invasion assay
In the process of the migration assay, the SW620 and LOVO cells suspended in serum-free medium on the top chamber were seeded following the transfection, and the medium with 10% FBS was added to the lower chamber. In the process of the invasion assay, we applied a coating of Matrigel (BD, Franklin Lakes, NJ, USA) on the surface of transwell inserts (Fisher Scientific, Waltham, MA, USA). Following a 24-hour incubation, we used a cotton swab to gently remove the cells on the upper surface of the transwell member, methanol to fix the cells on the lower surface of the transwell member, and 0.5% crystal violet (Solarbio) to stain all the cells. Then, we counted the numbers in five microscopic fields selected randomly.
Western blot analysis
Protein’s isolation, quantification, and Western blotting were performed as previously mentioned. Cells were washed, and proteins were extracted with lysis buffer (100 μl/50 ml). Protein samples of equal amount were separated by SDS-PAGE and shifted to PVDF membranes (Millipore, Bedford, Mass). The membranes were then put into incubation overnight with primary antibodies anti-MAP3K1 (dilution = 1:500), Ki67 (dilution = 1:800), MMP2 (dilution = 1:1000), E-cadherin (dilution = 1:500), N-cadherin (dilution = 1:800), JNK (dilution = 1:1000), p38 (dilution = 1:800), ERK (dilution = 1:500), GAPDH (dilution = 1:500) (the above antibodies were purchased from Abcam company (Cambridge, MA, USA)), phosphorylation (phospho-) JNK (dilution = 1:500), phospho-p38 (dilution = 1:800), and phospho-ERK1/2 (dilution = 1:600) (the above antibodies were purchased from Cell Signaling Technology Company (USA)) at 4°C, and then washed thoroughly using Tris-buffered saline-Tween 20 (TBST) before an incubation for 1 h with horseradish peroxidase-labeled secondary antibody (Santa Cruz Technology, Santa Cruz, CA, USA) at room temperature. Following the membranes’ washing in TBST, an enhanced chemiluminescence (ECL) system was applied to observe the results. The mean values were drawn from three experiments.
Pull-down assay with biotinylated circ_0085315 probe
The biotinylated probe was specifically designed to bind to the junction area of circ_0082315, while the oligo probe was taken as a control. Then, 1 × 107 CC cells were harvested and lysed. The circ_0085315 probe (Tsingke, Wuhan, China) was incubated with streptavidin magnetic beads (Invitrogen) at room temperature for 2 h to generate probe-coated beads. The cell lysates were incubated with probe-coated beads at 4 ℃ overnight. The beads were washed, and the bound miRNAs in the pull-down materials were extracted using Trizol reagent and analyzed by qRT-PCR assay.
Pull-down assay with biotinylated miRNA
Colon cancer cells were transfected with 50 nM of biotinylated miRNA mimics or nonsense control (Tsingke, Wuhan, China) at 50% confluence using Lipofectamine 3000 (Invitrogen). The cells were harvested and lysed in lysis buffer 24 h after transfection. The cell lysates were incubated with washed streptavidin magnetic beads (Life Technologies) for 3 h. The beads were washed, and Trizol reagent was used to extract RNA interacting with miRNA. The abundance of circ_0085315 was evaluated by qRT-PCR analysis.
Luciferase reporter assay
A wild-type or mut-MAP3K1 fragment was constructed and inserted downstream of the luciferase reporter gene of the psiCHECK2 plasmid (HH-LUC-015, HedgehogBio Science and Technology Ltd, Shanghai, China). Lipofectamine® 3000 (Invitrogen) was used to transfect the reporter plasmid into cells. Then, then co-transfected the miR-1200 mimics, circ_0085315-siRNA#1, circ_0085315 overexpressing plasmid, or circ_0085315-mut overexpressing plasmid (10 μM) with the reporter gene into cells. After 48 h following the transfection, we performed an examination for luciferase activity using the luciferase report analysis kit (Promega, Beijing, China). We conducted 3 times of all the experiments for sake of the data reliability.
Cell viability assay
We inoculated the transfected cells 1 × 104 cells per well into 96-well plate. Following 0, 24, 48, and 72-hour incubation, we seeded 10 µl Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China) solution to each well, and cultured the 96-well plate for 4 h in the medium in darkness at 37°We then we measured the optical density of each well using microporous plate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA) at 450 nm.
CircRNA localization
To accomplish the localization of circRNA, we used Cytoplasmic & Nuclear RNA Purification Kit (Amyjet Scientific Inc, Wuhan, Hubei, China) to separate and draw the nuclear and cytoplasmic RNA of CC cells, and qRT-PCR to identify the circ_0085315 expression in the two types of RNA mentioned above. We took GAPDH and U1 as the cytoplasm control and the nuclear control, respectively.
Statistics analysis
We adopted SPSS 17.0 (SPSS Inc., IL, and USA) to analyze the results from abovementioned experiments expressing in the form of means ± SD. We conducted student’s t-tests as well as one-way ANOVA for the purpose of data comparison. With the help of the chi-squared test, we analyzed the way hsa_circ_0085315 expression impacts the patients’ clinical findings. By combining Kaplan-Meier curves with log-rank tests, we made a comparison among survival outcomes. We repeated all the experiments three times and considered it significant if P < 0.05.
Results
CircRNA_0085315 was highly expressed in tumor tissues and significantly associated with the poor prognosis of CC patients
The structure of hsa_circ_0085315 was obtained from the Cancer-Specific CircRNA Database (CSCD) (Figure 1a). Afterward, a qRT-PCR assay was conducted to detect the circRNA_0085315 mRNA levels in tumor tissues and adjacent normal tissues. As a result, a high expression of circ_0085315 was detected in tumor tissues (Figure 1b). Besides, circ_0085315 was also highly expressed in CC cell lines (Figure 1c). However, circ_0085315 expression in SW620 cells was higher than that in LOVO cells. circRNAs were able to resist the exonuclease-mediated degradation, while linear_RNA did not have such resistance. Therefore, we used the RNase R assay to prove the resistance of circ_0085315 in CC cell lines to RNase R; Compared with mock LOVO cells, circRNA_0085315 expression was down-regulated in RNase R-treated LOVO cells (Figure 1d and 1E). The expression of EIF3E mRNA was significantly down-regulated in RNase R-treated SW620 cells and LOVO cells. However, the degradation degree of circRNA_0085315 was much lower than that of EIF3E mRNA, indicating circRNA_0085315 exists in CC cells. Then, the intracellular localization of circ_0085315 was identified via qRT-PCR, detecting the nuclear and cytoplasmic RNA. U1 was the nuclear internal reference, GAPDH was the cytoplasmic internal reference. The results showed that in SW620 cells and LOVO cells, circ_0085315 was preferentially localized in the cytoplasm, and EIF3E was preferentially localized in the nucleus (figure 1f). Then, CC patients were divided into two groups, including the group with high circRNA_0085315 expression (n = 38) and the group with low circRNA_0085315 expression (n = 32), according to the median circ_0085315 mRNA level (median value = 3). Then, Kaplan–Meier survival curves showed that patients in circ_0085315 Low group had a longer survival time than those in circ_0085315 High group (Figure 1g). After clinicopathological features were compared among CC patients, we found that the high expression of circ_0085315 was significantly related to lymph node metastasis (P = 0.016) (Table 1).
Figure 1.
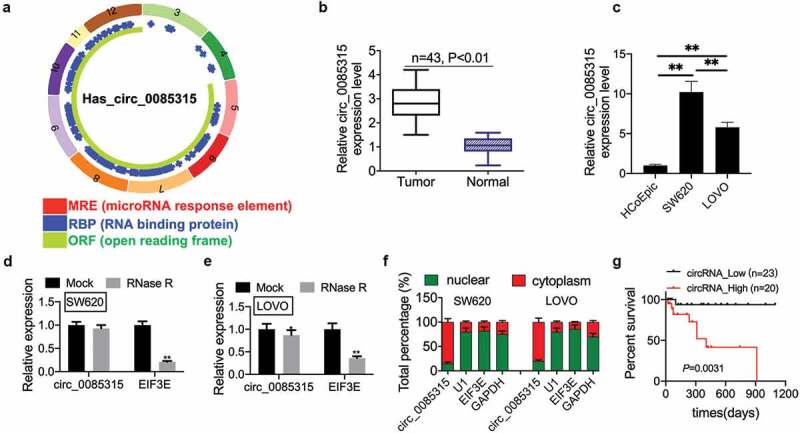
CircRNA_0085315 is highly expressed in CC tissues and closely related to survival status of CC patients.
The structure of hsa_circ_0085315 was shown in (A) based on the Cancer-Specific CircRNA Database (CSCD) database. (B) Relative circ_0085315 expression in CC tumor tissues compared with adjacent normal tissues (n = 43) by qRT-PCR analysis. (C) Relative circ_0085315 expression in CC cell lines compared with HCoEpic by qRT-PCR analysis. (D and E) qRT-PCR helped to determine circ_0085315 expression after RNase R treatment in CC cell line. (F) Nuclear RNA fractionation and cytoplasmic experiments analysis of circ_0085315ʹs location in CC cells. (G) The overall survival (OS) probability of CC patients with low- and high-circRNA_0085315 expression (**P < 0.01). CC, colon cancer.
Circ_0085315-silencing suppressed cell proliferation, invasion, and migration in CC
To comprehend the biological function of circ_0085315 in CC, circ_0085315 siRNA#1 or circ_0085315 siRNA#2 was transfected into SW620 cells to silence the circ_0085315 expression. The results showed that transfection with circ_0085315 siRNA significantly reduced circ_0085315 expression in SW620 cells (Figure 2a). However, transfection with circ_0085315 siRNA did not inhibit the linear_EIF3E expression in SW620 and LOVO cells (Figure 2b). In circ_0085315-knockout SW620 cells, the viability (Figure 2c), proliferation (Figure 2 D), invasion (Figure 2e), and migration (figure 2f) of cells were remarkably inhibited compared to those in untreated SW620 cells. Next, the expression of Ki67, MMP2, and N-cadherin was weakened by circ_0085315-silencing, while the E-cadherin expression was promoted by circ_0085315-overexpression in CC cells (Figure 2g and h).
Figure 2.
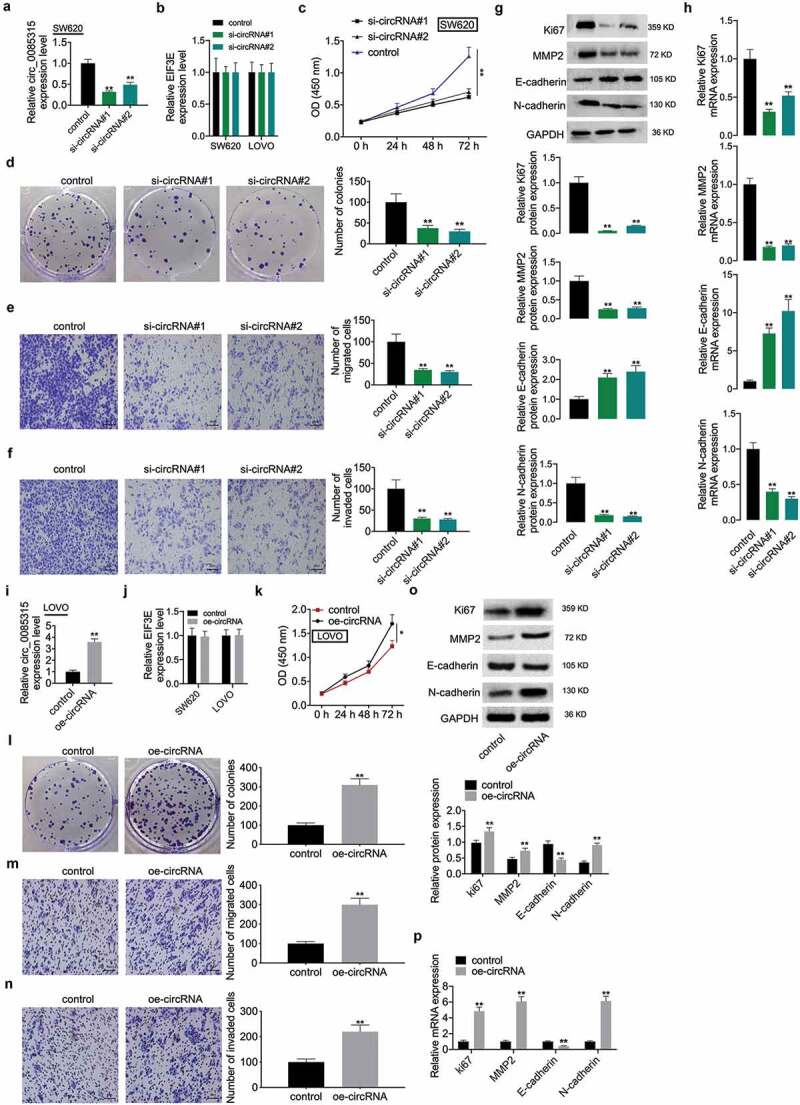
Circ_0085315 regulated cell proliferation, invasion, and migration as well as the expression change of Ki67, MMP2, E-cadherin, and N-cadherin in CC.
(A) circRNA_0085315 level was detected by qRT-PCR in si-con or si-circ_0085315 (Number 1 and 2) transfected SW620 cells. (B) The linear EIF3E level was detected by qRT-PCR in si-con or si-circ_0085315 (Number 1 and 2) transfected cells. (C) Evaluation of the viability exhibited was performed via the CCK-8 assay in cells transfected by si-con or si-circ_0085315. (D) Colony Formation Assay helped determine the impact of circ_0085315 silencing on colony formation ability. The transwell migration assay (E) and transwell invasion assay (F) helped determine the migration and invasion of SW620 cells and LOVO cells following the siRNA transfection. Western blot assay (G) and qRT-PCR assay (H) aimed at analyzing the expression of Ki67, MMP2, E-cadherin, and N-cadherin in CC cells under the transfection of si-con or si-circ_0085315 (Number 1 and 2). (B) In vector-con or vector-circ_0085315 transfected LOVO cells, circRNA_0085315 level (I), linear EIF3E level (J), cell viability (K), cell proliferation (L), cell invasion (M) and migration (N), and the expression of Ki67, MMP2, E-cadherin, and N-cadherin at protein level (O) and mRNA level (P) were detected by qRT-PCR, CCK-8, colony formation, Transwell, and Western blot assays. *P < 0.05; **P < 0.01.
Circ_0085315-overexpression enhanced cell proliferation, invasion, and migration in CC
As circ_0085315-silencing mediated the inhibition on cell proliferation, invasion, and migration, this study also explored the effect of high circ_0085315 expression on these biological behaviors and the expression of related proteins. To this end, vectors expressing circ_0085315 were transfected into LOVO cells with lower circRNA expression, so as to increase circ_0085315 expression (Figure 2i). Not surprisingly, transfection with vectors expressing circ_0085315 did not affect the linear_EIF3E expression in LOVO and SW620 cells (Figure 2j). Next, circ_0085315-overexpression distinctly reduced the abilities of cell proliferation ((Figure 2k and L), invasion (Figure 2m), and migration (2 N), down-regulated the expression of Ki67, MMP2, and N-cadherin, and up-regulated that of E-cadherin (Figure 2o and P).
CircRNA_0085315 functioned as a ceRNA to sponge miR-1200 to affect cell proliferation and invasion in CC
Firstly, the results of the qRT-PCR assay showed that the biotinylated circRNA_0085315 probe selectively reduced the circRNA_0085315 level in SW620 and LOVO cells, but it did not affect the expression of EIF3E, the linear counterpart RNA of circRNA_0085315 (Figure 3a). Based on the above research results, it was the top priority to screen the target miRNAs of circ_0085315 in CC that affected cell proliferation and metastasis. There were 57 target miRNAs of circ_0085315 in the CircInteractome database (Figure 3b). RNA pull-down assay demonstrated that seven miRNAs (including miR-1200, miR-1257, miR-604, miR-634, miR-936, miR-659, and miR-217) might interact with circ_0085315. Of them, the enrichment level of miR-1200 caused by circ_0085315 was the highest in SW620 and LOVO cells (Figure 3c-e). Then, we found that the biotinylated circRNA_0085315 probe also significantly reduced the circ_0085315 enrichment level (figure 3f). Similarly, miR-1200 expression in CC cell lines and tumor tissues was significantly lower than that in HCoEpic and normal tissues (Figure 3g and 3 H). Furthermore, we determined the negative correlation between the expression level of circ_0085315 and miR-1200 among 43 paired CC tumor tissues (Figure 3i).
Figure 3.
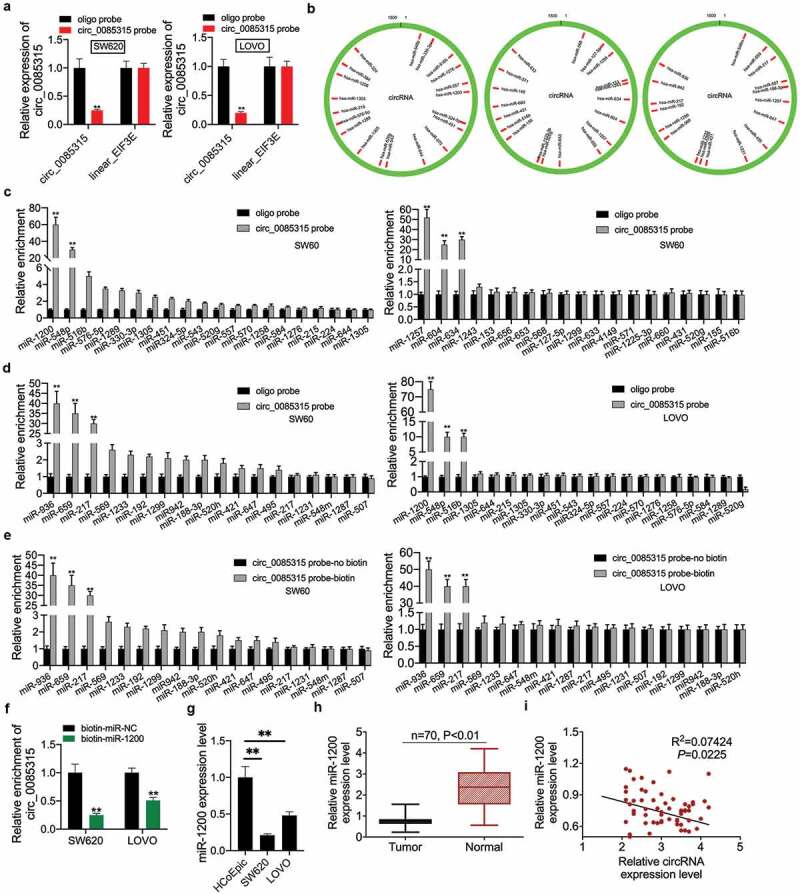
CircRNA_0085315 functioned as a ceRNA sponge to miR-1200.
(A) qRT-PCR assay was used to determine that the biotinylated circRNA_0085315 probe selectively brings down circRNA_0085315 level in SW620 and LOVO cells and not the expression of linear EIF3E. A total of 57 target miRNAs of circ_0085315 were shown in B based on Circular RNA Interactome database. (C, D, and E) RNA pulldown assays were conducted to examine the binding abilities between candidate miRNAs and circ_0085315 in SW620 and LOVO cells. (F) qRT-PCR assay was used to determine the biotinylated miR-1200 probe effectively reduced circRNA_0085315 level in SW620 and LOVO cells. (G) Relative miR-1200 expression in CC cell lines compared with HCoEpic by qRT-PCR analysis. (H) Relative miR-1200 expression in CC tumor tissues compared with adjacent normal tissues (n = 43) by qRT-PCR analysis. (I) Pearson correlation analysis of between circ_0085315 and miR-1200. *P < 0.05; **P < 0.01.
The full-length sequence of circ_0085315 was shown on this website: https://circinteractome.nia.nih.gov/api/v2/circgenomic?circular_rna_query=hsa_circ_0085315. To determine whether circ_0085315 affected cell proliferation and invasion by adsorbing miR-1200, the site that bound to miR-1200 located on the full-length sequence of circ_0085315 was mutated (Figure 4a-C). Then, the effects of circ_0085315 mutant (mut) on the biological behavior of CC cells and the expression of related proteins were examined. As a result, the biotinylated circRNA_0085315-mut probe no longer affected the miR-1200 enrichment level (Figure 4d). After the transfection of the vector expressing circ_0085315-mut into LOVO cells, the circ_0085315 mRNA level still increased in the group with overexpression of circ_0085315-mut (Figure 4e). Further experimental results showed that the overexpression of circ_0085315-mut in LOVO cells had basically no effect on cell proliferation, invasion, and migration, or the expression of Ki67, MMP2, E-cadherin, and N-cadherin (figure 4f-L).
Figure 4.
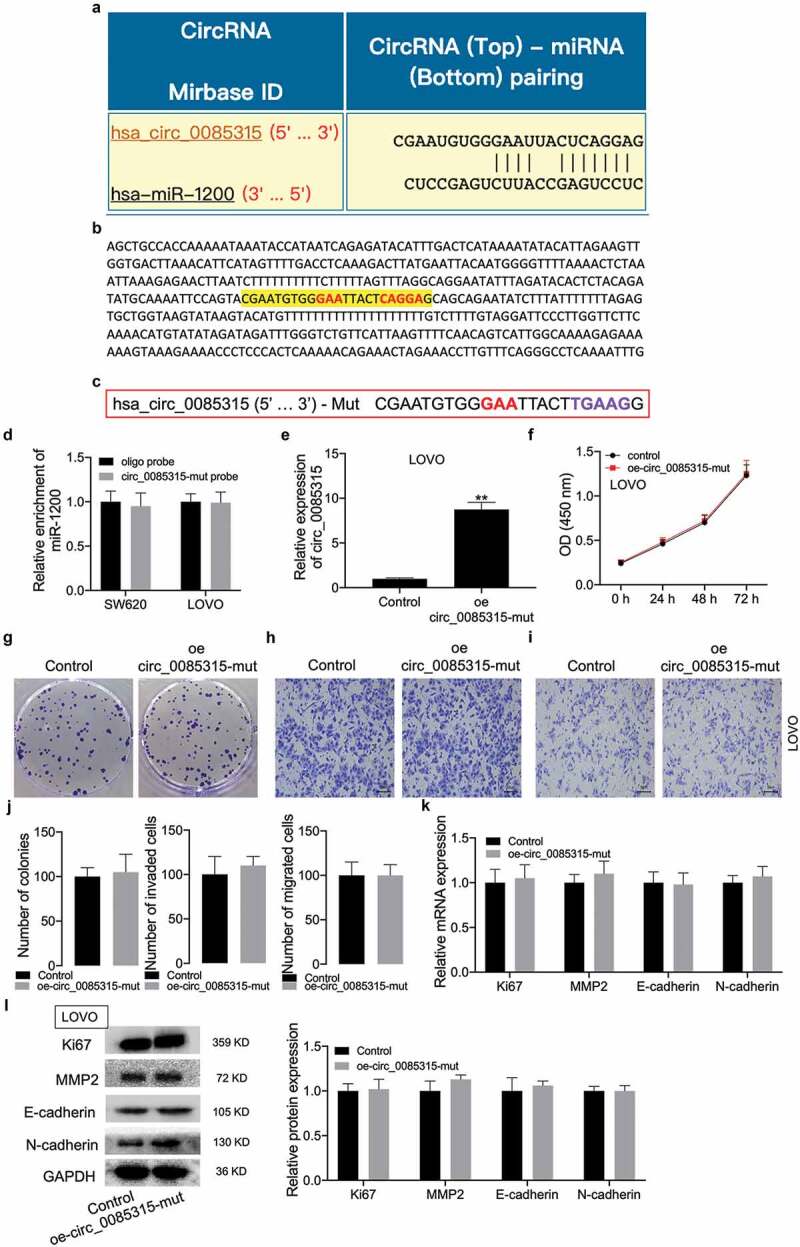
CircRNA_0085315 mut had no affect cell proliferation, invasion, and migration in CC.
(A) The binding sequence of miR-1200 on circRNA_0085315 is predicted from the Circular RNA Interactome database. (B) the site that can bind to miR-1200 locating on the full-length sequence of circ_0085315. (C) The sequence of circRNA_0085315 mut. (D) qRT-PCR assay was used to determine that the biotinylated circRNA_0085315-mut probe had no effect on the enrichment level of miR-1200. A vector expressing circRNA_0085315 mut was transfected into LOVO cells, circRNA_0085315 level (E), cell viability (F), proliferation (G), invasion (H), and migration (I) were detected by qRT-PCR, CCK-8, colony formation, Transwell assays. The statistical diagrams were shown in J. qRT-PCR assay (K) and Western blot assay (L) aimed at analyzing the expression of Ki67, MMP2, E-cadherin, and N-cadherin in CC cells under the transfection of oe-circ_0085315 mut. *P < 0.05; **P < 0.01.
CircRNA_0085315 sponged miR-1200 to up-regulate the MAP3K1 expression in CC cells
According to the ceRNAs theory, circ_0085315 and the target gene of miR-1200 should share miR-1200, thereby removing mRNA repression through competitively binding to miR-1200. Therefore, we screened 4779 target genes of miR-1200 from the TargetScan database and 918 target genes of miR-1200 from the miRDB database. A total of 845 overlapping target genes among the above databases are shown in Figure 5a. MAP3K1, MAP3K2, and MAP3K3 belonged to such intersected target genes. To determine the complementary-binding association between miR-1200 and MAP3K1, MAP3K2, or MAP3K3, luciferase reporter assay and qRT-PCR assay were carried out. To this end, miR-1200 mimics and miR-1200 inhibitors were transfected separately into SW620 and LOVO cells, so as to inhibit or increase the miR-1200 level (Figure 5b). The MAP3K1 expression was the most significantly repressed by miR-1200 mimics (Figure 5c). Thus, the TargetScan database was used to predicate the target genes of miR-1200 as well as their binding sites. The results suggest that MAP3K1 had a binding site of miR-1200 (Figure 5d). miR-1200 mimics significantly inhibited the luciferase activity of MAP3K1 (wt), but it had no effect on that of MAP3K1 (mut) (Figure 5e). In addition, the MAP3K1 expression was significantly inhibited by miR-1200 mimics (figure 5f) and up-regulated by miR-1200 inhibitors (Figure 5g). It also found that its expression equally increased in tumor tissues of CC patients (Figure 5h) and was negatively correlated with miR-1200 expression (Figure 5i). Next, we found that the up-regulated circ_0085315 also significantly increased the luciferase activity of MAP3K1 (wt) (Figure 5j), while the down-regulated circ_0085315 remarkably inhibited that of MAP3K1 (wt) (Figure 5k), but they had no effect on that MAP3K1 (mut). Then, the miR-1200 mimics-induced enhanced luciferase activity of MAP3K1 (wt) was abolished by the up-regulated circ_0085315. However, the up-regulated circ_0085315-mut had no impact on this effect (Figure 5l). In addition, circ_0085315 expression in CC patients was negatively correlated with miR-1200 expression (Figure 5m).
Figure 5.
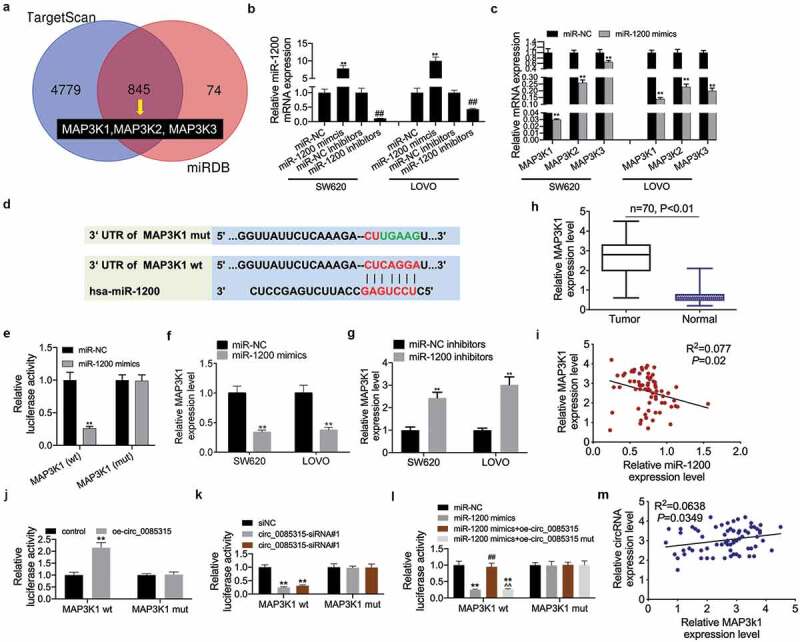
CircRNA_0085315 sponges to miR-1200 to upregulate MAP3K1 expression in CC cells.
(A) Venn analysis of 845 overlapping target genes between TargetScan and miRDB database. (B) The transfection efficiency of miR-1200 mimics or inhibitors was verified using qRT-PCR. (C) The expression of MAP3K1, MAP3K2, and MAP3K3 was detected using qRT-PCR in SW620 and LOVO cells transfected by miR-1200 mimics. (D) The binding sequence of miR-1200 on MAP3K1 is predicted from TargetScan. (E) The interaction between MAP3K1 and miR-1200 was assessed by luciferase reporter assay. (F and G) The expression of MAP3K1 was detected using qRT-PCR in SW620 and LOVO cells transfected by miR-1200 mimics or miR-1200 inhibitors. (H) Relative MAP3K1 expression in CC tumor tissues compared with adjacent normal tissues (n = 43) by qRT-PCR analysis. (I) Pearson correlation analysis of between MAP3K1 and miR-1200. (J and K) The interaction between MAP3K1 and circ_0085315 was assessed by luciferase reporter assay. (L) The interaction among MAP3K1, miR-1200, and circ_0085315 was assessed by luciferase reporter assay. *P < 0.05; **P < 0.01.
MAP3K1 regulated miR-1200 to affect the biological behavior of CC cells and the activation of the MAPK signaling pathway
This study further examined whether miR-1200 overexpression inhibited the proliferation, migration, and invasion of CC cells via MAP3K1. First, MAP3K1 mRNA expression significantly increased in MAP3K1-overexpression CC cells (Figure 6a). It was then found that miR-1200 mimics inhibited cell viability; however, this effect was reversed by MAP3K1-overexpression in SW620 cells (Figure 6b). Similarly, miR-1200 mimics-mediated inhibition on the proliferation and metastasis in SW620 cells was abolished by MAP3K1-overexpression (Figure 6c-E). The phosphorylation levels of JNK, p38, and ERK1/2 were inhibited by miR-1200 mimics; however, the up-regulated MAP3K1 reversed the above effects (figure 6f).
Figure 6.
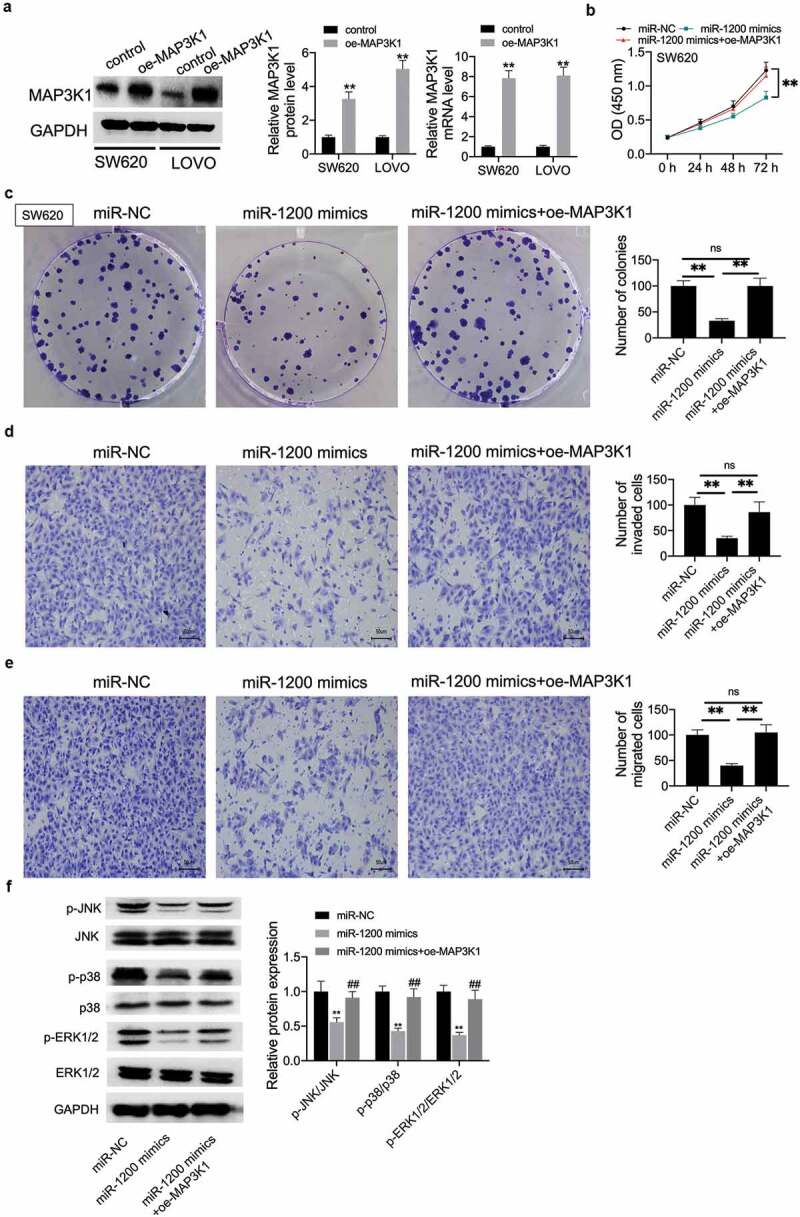
MAP3K1 regulated miR-1200 impacting the biological behavior of CC cells and the activation of the MAPK signaling pathway.
(A) The transfection efficiency of MAP3K1 was verified using qRT-PCR and Western blot assays in SW620 and LOVO cells transfected by vector-con or vector-MAP3K1. Then, vector-MAP3K1 was transfected into miR-1200 mimics transfected cells, cell viability (B), proliferation (C), cell invasion (D), migration (E), and the phosphorylation level of JNK/p38/ERK1/2 were detected by CCK-8, colony formation, transwell, and Western blot assays. *P < 0.05; **P < 0.01.
The regulatory network of circRNA_0085315/miR-1200/MAP3K1 exerted its function by the activation of the MAPK signaling pathway
To understand the interplay among circRNA_0085315, miR-1200, and MAP3K1, we used SW620 cells with the higher circRNA_0085315 expression as research objects to explore the effects of miR-1200 and MAP3K1 on its proliferation, migration, and invasion. The results showed that both miR-1200 mimics and MAP3K1 silencing could inhibit the abilities of proliferation, invasion, and migration of SW620 cells (Figure 7a-G). Next, circRNA_0085315-overexpressing plasmids were transfected into LOVO cells with the lower circRNA_0085315 expression, thereby analyzing the effects of miR-1200 mimics or si-MAP3K1 on the above cell proliferation, invasion, and migration. Notwithstanding the presence of circRNA_0085315 promoted the proliferation, invasion, and migration of cells, the additional expression of miR-1200 mimics or si-MAP3K1 reversed the above phenomenon (Figure 7h-N).
Figure 7.
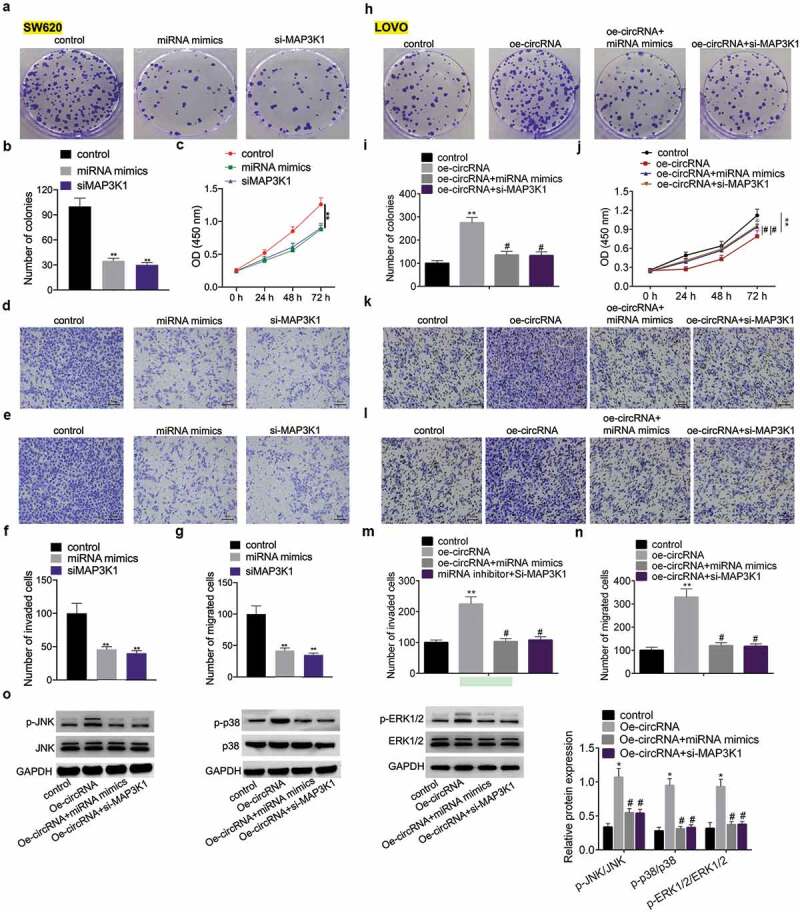
The regulatory network of circRNA_0085315/miR-1200/MAP3K1 functioned via the activation of the MAPK signaling pathway.
In SW620 cells transfected by miR-1200 mimics or si-MAP3K1, cell proliferation, invasion, migration, and the phosphorylation level of JNK/p38/ERK1/2 were detected by colony formation assay (A, B), CCK-8 (C), transwell invasion and migration assays (D, E, F, G). In LOVO cells transfected by vector-circ_0085315 and miR-1200 mimics/or si-MAP3K1, cell proliferation, invasion, migration, and the phosphorylation level of JNK/p38/ERK1/2 were detected by colony formation assay (H, I), CCK-8 (J), transwell invasion and migration assays (K, L, M, N), and Western blotting (O). *P < 0.05; **P < 0.01.
MAP3K1 is a signaling pathway protein of MAPK. It turned out that the overexpression of circRNA_0085315 indeed remarkably increased the phosphorylation levels of JNK, p38, and ERK in SW620 cells (Figure 7o). More importantly, the miR-1200 mimics or MAP3K1 silencing blocked the activation of the MAPK pathway by circRNA_0085315 overexpression (Figure 7o). According to the above data, circRNA_0085315 altered the activation of the MAPK signaling pathway by affecting the MAP3K1 expression. This suggests that the feedback loop of circRNA_0085315-miR-1200-MAP3K1 served as the key positive regulator for the signal transduction of MAPK/ERK.
Discussion
Abnormal circRNA expression in tumor cells affects cancer progression [8]. Zhang Q reported that circ_PIP5K1A promoted CC development and exerted the oncogenic effects, which might be regulated by miR-1273a [15]. Additionally, the results from Daishi Chen et al. showed that circ_100395 suppressed the malignant behavior of lung cancer by virtue of regulating the miR-1228/TCF21 axis [16]. Hence, some circRNAs contain microRNA (miRNA) response elements, Which interact with miRNAs to adsorb miRNAs, thereby terminating the translation inhibition of miRNAs on their target genes. Furthermore, Wang YG et al. reported that circ_0091570 acted as the ceRNA to sponge miR-1307, and it played functional roles to regulate ISM1 expression in liver cancer [17]. However, many unreported circRNAs may also play important roles in tumorigenesis [18]. Due to the limitation of time and space, it is impossible to explore the role of every circRNA that has not been reported in the development of CC. Therefore, how to quickly screen out a circRNA related to CC progression is an urgent problem to be solved. Based on the ceRNA’s theory [19], the known cancer-related miRNAs were selected as the penetrative points to select their target circRNAs, thereby shortening and reducing the time and difficulty in screening CC-related circRNAs. Here, circRNA_0085315 was a circRNA originating from EIF3E. EIF3, as a eukaryotic cell translation initiation factor with the largest relative molecular weight, plays an important regulatory role in the transcription and translation of eukaryotic cells, which is abnormally expressed in tumors to affect the occurrence and development of tumors [20–22]. EIF3E is a subunit of EIF3, which promotes tumorigenesis by mediating the mRNA circulation to enhance gene translation [23]. Therefore, more attention should be paid to the expression pattern and role of EIF3E-derived circ_0085315 in CC. We discovered that the upregulation of circRNA_0085315 in CC tumor tissues was positively correlated with lymph node metastasis and stage, suggesting that circRNA_0085315 might be a predictor of poor prognosis for CC patients. According to the ceRNA hypothesis, RNA transcripts from circRNAs and mRNA can use the same miRNA response elements, which scramble for binding to miRNAs to regulate the expression of each other [12,24]. In this study, the Circular RNA Interactome database indicated that circ_0085315 showed that many target miRNAs of circ_0085315. However, RNA pull-down assay was conducted, which suggested that miR-1200 was the miRNA with the highest enrichment level in CC cells treated with the circRNA_0085315 probe. Meanwhile, it was discovered that transfection with circ_0085315-mut-overexpressing plasmid without the binding site of miR-1200 suppressed the promotion of CC cell proliferation and metastasis. In addition, the expression pattern of miR-1200 in CC tumor tissues and cell lines was downregulation, which was contrary to the expression pattern of circ_0085315. Based on these data, it is basically authentic for us to consider that circRNA_0085315 is a ceRNA that binds to miR-1200.
Specificity of the circRNA mechanism by acting as a ceRNA, the circRNA-miRNA-mRNA regulatory network for circRNA_0085315 was explored. It was important to find out that the target genes of miR-1200 are closely related to circ_0085315 expression. However, among the numerous target genes of miR-1200, three attracted our attention, including MAP3K1, MAP3K2, and MAP3K3, respectively [25]. All three genes belong to the MAPK signaling pathway-related proteins. Our results show that, compared with the inhibition of miR-1200 mimics on the expression of MAP3K2 and MAP3K3, the inhibition of MAP3K1 expression induced by miR-1200 mimics was much stronger. MAP3K1 can express MEK kinase 1 (MEKK1, an important kinase that links Ras activation to MAPK signaling) [26], which is tagged as a marker of the MAPK signaling pathway. In this regard, the continuous activation of the MAPK signaling in many cancers has a close involvement in cell proliferation, invasion, and migration [25,27–29]. Furthermore, the role of MAP3K1-overexpression in promoting the proliferation and metastasis of tumor cells has been reported in many studies [30–32]. Our study indicated that miR-1200 mimics inhibited luciferase activity of MAP3K1 (wt). However, the role of circ_0085315-overexpression in enhancing the luciferase activity of MAP3K1 (wt) was abolished by miR-1200 mimics, and the role of miR-1200 mimics in decreasing the luciferase activity of MAP3K1 (wt) was not affected by the overexpression of circ_0085315-mut. The above results suggested that circ_0085315 sponged to miR-1200 to up-regulate the MAP3K1 expression.
In order to further understand the relationship between miRNA and MAPK signaling pathways, miR-1200 mimics and MAP3K1-overexpression are transfected into SW620 cells by the manner of solely or together so that to detect cell proliferation, invasion, and migration as well as the expression change of mark proteins of MAPK signaling pathway. Our results showed that miR-1200 mimics reduced cell proliferation, invasion, and migration, which was consistent with the results reported in previous studies [12–14]. These effects of miR-1200 in CC cells were reversed by MAP3K1-overexpression. These data indicate that miR-1200 regulates the biological behavior of CC cells by targeting MAP3K1. JNK, p38, and ERK, the markers of the MAPK signaling pathway, can be activated by their respective high level of phosphorylation in many cancers [33,34].
Our previous experimental results showed that circ_0085315 had higher expression in SW620 cells, hence we used miR-1200 mimics or MAP3K1 silencing to treat SW620 cells, thereby studying the effects of miR-1200 and MAP3K1 on cell proliferation, invasion, and migration of SW620 cells with the high level of circ_0085315. Our results show that both miR-1200 mimics and MAP3K1 silencing can inhibit the abilities of cell proliferation, invasion, and migration. Next, circ_0085315 overexpression was transfected into LOVO cells with a low level of circ_0085315 to observe the effects of miR-1200 mimics and MAP3K1 silencing on the proliferation, invasion, and migration of this cell. Then, we found that both miR-1200 mimics and MAP3K1 silencing also could reverse circ_0085315-overexpression induced cell proliferation, invasion, and migration. Furthermore, miR-1200-mediated suppressed activation of the MAPK signaling pathway was reversed by MAP3K1-overexpression. Likewise, the circ_0085315-mediated promotion on cell proliferation and metastasis and the activation of the MAPK signaling pathway were reversed by miR-1200 mimics or MAP3K1-inhibition. Combined with the above results, the miR-1200/MAP3K1 pathway mediated the effect of circ_0085315 on promoting cell proliferation and metastasis of CC cancer.
In conclusion, we find that circ_0085315 is significantly up-regulated in human CC tissues, which can successfully sponge miR-1200 to promote the proliferation, migration, and invasion of CC cells. We also demonstrate that circ_0085315 can up-regulate the MAP3K1 expression and activate the MAPK signaling pathway by adsorbing miR-1200. The above data help to confirm that the circ_0085315/miR-1200/MAP3K1 regulatory network exists in CC, and this may provide a new strategy to treat CC.
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics approval and consent to participate
Prior informed consent from patients and approval from the Ethics Committee of Ningbo First Hospital were obtained for the study. The study was performed following the guidelines of the Committee and the Declaration of Helsinki. All of the operations were carried out strictly following the applicable guidelines, laws and regulations.
Data availability statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
References
- [1].Cappell MS. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol Clin North Am. 2008;37(1):1–24. [DOI] [PubMed] [Google Scholar]
- [2].Sharma R. An examination of colorectal cancer burden by socioeconomic status: evidence from GLOBOCAN 2018. EPMA Journal. 2020;11(1):95–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ko J-H, Um J-Y, Lee S-G, et al. Conditioned media from adipocytes promote proliferation, migration, and invasion in melanoma and colorectal cancer cells. J Cell Physiol. 2019;234(10):18249–18261. [DOI] [PubMed] [Google Scholar]
- [4].Xi X, Teng M, Zhang L, et al. Retracted: microRNA-204-3p represses colon cancer cells proliferation, migration, and invasion by targeting HMGA2. J Cell Physiol. 2020;235(2):1330–1338. [DOI] [PubMed] [Google Scholar]
- [5].Zeng M, Zhu L, Li L, et al. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xiong W, Qin J, Cai X, et al. Overexpression LINC01082 suppresses the proliferation, migration and invasion of colon cancer. Mol Cell Biochem. 2019;462(1–2):33–40. [DOI] [PubMed] [Google Scholar]
- [7].Chen -L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. [DOI] [PubMed] [Google Scholar]
- [8].Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xia L, Song M, Sun M, et al. Circular RNAs as biomarkers for cancer. Adv Exp Med Biol. 2018;1087:171–187. [DOI] [PubMed] [Google Scholar]
- [10].Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- [11].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li S, Pei Y, Wang W, et al. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. 2019;18(11):1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pan B, Zhao M, Wang N, et al. LncRNA RGMB-AS1 promotes glioma growth and invasion through miR-1200/HOXB2 Axis. Onco Targets Ther. 2019;37:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Z, Yu X, Zhou B, et al. Circular RNA circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. Biomed Res Int. 2020;2020:5103272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang Q, Zhang C, Ma J-X, et al. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25(35):5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen D, Ma W, Ke Z, et al. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Y-G, Wang T, Ding M, et al. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–138. [DOI] [PubMed] [Google Scholar]
- [18].Guarnerio J, Zhang Y, Cheloni G, et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29(8):628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. [DOI] [PubMed] [Google Scholar]
- [20].Gomes-Duarte A, Lacerda R, Menezes J, et al. eIF3: a factor for human health and disease. RNA Biol. 2018;15(1):26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yin Y, Long J, Sun Y, et al. The function and clinical significance of eIF3 in cancer. Gene. 2018;673:130–133. [DOI] [PubMed] [Google Scholar]
- [22].Hershey JWB. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta. 2015;1849(7):792–800. [DOI] [PubMed] [Google Scholar]
- [23].Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peng P, Zhang B, Huang J, et al. Identification of a circRNA-miRNA-mRNA network to explore the effects of circRNAs on pathogenesis and treatment of spinal cord injury. Life Sci. 2020;257:118039. [DOI] [PubMed] [Google Scholar]
- [25].Slattery ML, Lundgreen A, Wolff RK. Dietary influence on MAPK-signaling pathways and risk of colon and rectal cancer. Nutr Cancer. 2013;65(5):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suddason T, Anwar S, Charlaftis N, et al. T-Cell-Specific Deletion of Map3k1 Reveals the Critical Role for Mekk1 and Jnks in Cdkn1b -Dependent Proliferative Expansion. Cell Rep. 2016;14(3):449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Slattery ML, Lundgreen A, Wolff RK. MAP kinase genes and colon and rectal cancer. Carcinogenesis. 2012;33(12):2398–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xue Z, Vis DJ, Bruna A, et al. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. Cell Res. 2018;28(7):719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Okumura K, Shirasawa S, Nishioka M, et al. Activated Ki-Ras suppresses 12-O-tetradecanoylphorbol-13-acetate-induced activation of the c-Jun NH2-terminal kinase pathway in human colon cancer cells. Cancer Res. 1999;59(10):2445–2450. [PubMed] [Google Scholar]
- [30].Avivar-Valderas A, McEwen R, Taheri-Ghahfarokhi A, et al. Functional significance of co-occurring mutations in PIK3CA and MAP3K1 in breast cancer. Oncotarget. 2018;9(30):21444–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pham TT, Angus SP, Johnson GL. MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer. 2013;4(11–12):419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bi CL, Liu JF, and Zhang MY, et al. LncRNA NEAT1 promotes malignant phenotypes and TMZ resistance in glioblastoma stem cells by regulating let-7g-5p/MAP3K1 axis. Biosci Rep. 2020;40(10);1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim EK, Choi E-J. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89(6):867–882. [DOI] [PubMed] [Google Scholar]
- [34].Liu M-G, Wang -R-R, Chen X-F, et al. Differential roles of ERK, JNK and p38 MAPK in pain-related spatial and temporal enhancement of synaptic responses in the hippocampal formation of rats: multi-electrode array recordings. Brain Res. 2011;1382:57–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.


