Abstract
When glucose is the carbon source, the white rot fungus Pycnoporus cinnabarinus produces a characteristic red pigment, cinnabarinic acid, which is formed by laccase-catalyzed oxidation of the precursor 3-hydroxyanthranilic acid. When P. cinnabarinus was grown on media containing cellobiose or cellulose as the carbon source, the amount of cinnabarinic acid that accumulated was reduced or, in the case of cellulose, no cinnabarinic acid accumulated. Cellobiose-dependent quinone reducing enzymes, the cellobiose dehydrogenases (CDHs), inhibited the redox interaction between laccase and 3-hydroxyanthranilic acid. Two distinct proteins were purified from cellulose-grown cultures of P. cinnabarinus; these proteins were designated CDH I and CDH II. CDH I and CDH II were both monomeric proteins and had apparent molecular weights of about 81,000 and 101,000, respectively, as determined by both gel filtration and sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The pI values were approximately 5.9 for CDH I and 3.8 for CDH II. Both CDHs used several known CDH substrates as electron acceptors and specifically adsorbed to cellulose. Only CDH II could reduce cytochrome c. The optimum pH values for CDH I and CDH II were 5.5 and 4.5, respectively. In in vitro experiments, both enzymes inhibited laccase-mediated formation of cinnabarinic acid. Oxidation intermediates of 3-hydroxyanthranilic acid served as endogenous electron acceptors for the two CDHs from P. cinnabarinus. These results demonstrated that in the presence of a suitable cellulose-derived electron donor, CDHs can regenerate fungal metabolites oxidized by laccase, and they also supported the hypothesis that CDHs act as links between cellulolytic and ligninolytic pathways.
The cellobiose dehydrogenases (CDHs) are a group of oxidative enzymes that are widespread in fungi that can utilize cellulose (1, 2, 20). Even though a number of fungi belonging to different ecological groups have been shown to produce CDH-like activity, only a few of the corresponding enzymes have been characterized biochemically. CDHs have been purified from the white rot fungi Phanerochaete chrysosporium (Sporotrichum pulverulentum) (6, 47), Heterobasidion annosum (27), Schizophyllum commune (22), and Trametes versicolor (38), the nonligninolytic fungi Myceliophthora thermophila (9), Neurospora sitophila (12), and Chaetomium cellulolyticum (21), the brown rot fungus Coniophora puteana (28, 41), and, recently, the thermophilic soft rot fungus Humicola insolens (42).
All of the CDHs that have been characterized are extracellular proteins that consist of two prosthetic groups, a heme group and a flavin adenine dinucleotide (FAD) moiety. They preferentially oxidize cellobiose and use a variety of substrates as electron acceptors; the substrates used as electron acceptors include quinones, phenoxy radicals (reviewed in references 1 and 20), triiodide (5), Fe (24, 29), and Mn (6, 39). Each CDH may be split into a heme domain and an FAD domain by proteolytic cleavage (15, 23, 25). The FAD moiety provides most of the catalytic activity of the CDH (48) and has long been regarded as a separate cellobiose-oxidizing enzyme, which is also known as cellobiose:quinone oxidoreductase (46). CDH activity has been detected only in the presence of cellulose or cellodextrins (45). The recent cloning and sequencing of two CDH genes from the white rot fungus P. chrysosporium (32, 33, 36), as well as one CDH gene from T. versicolor (13), allowed molecular analyses of the genes to be performed and should provide better tools for studying the regulation of CDHs.
The catalytic features of CDH suggest that it functions in both lignin degradation and cellulose degradation, two processes which are usually studied as if they took place independently. Consequently, synergistic effects between the two pathways are poorly understood. The functions of CDH may include generating the active hydroxyl radicals which initiate Fenton’s reactions to degrade wood components (30, 49) and preventing the repolymerization of radicals generated during the oxidation of lignin by phenol oxidases (1, 20). There is increasing evidence that CDH plays an important role in lignocellulose degradation (3, 4, 11, 24, 34, 44). However, the exact function(s) of the enzyme remains to be identified.
Our studies have focused on the white rot fungus Pycnoporus cinnabarinus, which has an unusual set of ligninolytic phenol oxidases (16, 19). P. cinnabarinus degrades lignin very efficiently with laccase as the only phenol oxidase. Laccase also mediates the formation of a pigment, cinnabarinic acid (CA), which imparts a characteristic orange-red color to the fruiting bodies of the fungus and serves as an antimicrobial agent (14, 18). The precursor of CA, 3-hydroxyanthranilic acid (3-HAA), could aid laccase during oxidation of recalcitrant lignin structures (17). In this study our objectives were (i) to determine if CA levels are dependent on CDH activities, (ii) to identify, purify, and characterize CDHs from P. cinnabarinus, and (iii) to determine whether CDHs prevent laccase-catalyzed oxidation of 3-HAA to CA.
MATERIALS AND METHODS
Biological material and reagents.
P. cinnabarinus ATCC 200478, which was isolated from decaying wood in New South Wales, Australia, was maintained as previously described (19). P. cinnabarinus laccase was purified as described previously (19). All chemicals were obtained from either Sigma (Deisenhofen, Germany) or Aldrich (Steinheim, Germany) and were at least analytical grade. Cytochrome c (catalog no. C-2037; Sigma) was obtained from bovine heart material. Bacterial cellulose purified from Acetobacter xylinum cultures was provided by L. Einfeldt (Institute of Macromolecular Chemistry, Friedrich Schiller University Jena, Jena, Germany).
Culture conditions.
P. cinnabarinus conidial spore suspensions were prepared from rice-grown cultures as described previously (19). The concentrations of the spore suspensions were adjusted to about 1.5 × 108 spores/ml. Portions (2.5 ml) of spore suspensions were used to inoculate 400 ml of culture medium in 1,000-ml Erlenmeyer flasks containing modified Dodson medium (19) supplemented with 2,2-dimethylsuccinic acid (pH 5.0). The carbon source used was either glucose (5 g/liter), cellobiose (5 g/liter), or cellulose (Avicel; Sigma) (5 g/liter). All incubations were done at 24°C on a rotary shaker (125 rpm). For the time course study of production of CA, fungal cultures were grown in triplicate.
Quantification of CA and cellobiose.
Production of the phenoxazinone derivative CA was monitored spectrophotometrically by determining the increase in absorbance at 450 nm (18). Cellobiose concentrations were quantified by an isocratic high-performance liquid chromatography analysis performed with an ion-exchange column (Aminex ion-exclusion HPX-87P; 300 by 7.8 mm; Bio-Rad, Richmond, Calif.) maintained at 80°C (31). A model 2700 solvent delivery system (Bio-Rad) was equipped with a model 1755 refractive index detector (Bio-Rad). Centrifuged culture samples were separated by using water as the eluant at a flow rate of 0.5 ml/min, and cellobiose concentrations were quantified by comparison with an external cellobiose standard.
Purification of P. cinnabarinus CDHs.
For CDH production, P. cinnabarinus was cultivated in modified Dodson medium containing Avicel cellulose (5 g/liter). When maximum CDH activity was observed (usually after 10 days), the mycelia and cellulose fibers were separated from 8 liters of culture broth by filtration through glass fiber filters (Whatman, Wiesloch, Germany) and 0.45-μm-pore-size nylon membrane filters (Millipore, Eschborn, Germany). The culture filtrate was concentrated by ultrafiltration with a 10-kDa membrane filter (type PM10; Amicon, Witten, Germany) and was excessively rebuffered with 50 mM sodium acetate (pH 5.0) by using the same ultrafiltration system. After concentration to a volume of approximately 10 ml, the crude protein was loaded onto a DEAE-M column (15 by 250 mm; Toyopearl, Tokyo, Japan). The column was washed with 1 volume of 50 mM sodium acetate buffer (pH 5.0), and the proteins were eluted with a linear 0 to 500 mM NaCl gradient in 50 mM sodium acetate buffer (pH 5.0) at a flow rate of 1 ml/min; the total volume used was 1,200 ml. A Bio-Rad Econo system was used to control the chromatographic steps. CDH-containing fractions were pooled, concentrated by ultrafiltration, and separated on a gel filtration column (Bio-Gel P 100; 10 by 1,200 mm; Bio-Rad) by using 50 mM sodium acetate (pH 5.0) at a flow rate of 0.2 ml/min. The final purification step involved high-performance liquid chromatography performed with the model 2700 solvent delivery system equipped with a Bio-Dimension UV-visible monitor (Bio-Rad) and a UNO anion-exchange column (volume, 1 ml; Bio-Rad). The CDH was eluted with a linear 0 to 500 mM NaCl gradient in 50 mM sodium acetate buffer (pH 5.0) at a flow rate of 1 ml/min; the total volume used was 35 ml.
Gel electrophoresis and staining.
The isoelectric points of CDH I and CDH II in crude (cell-free) culture filtrates and pure enzyme preparations were determined with isoelectric focusing-polyacrylamide gel electrophoresis (PAGE) gels pH 3 to 10 gradients (125 by 65 mm; thickness, 0.4 mm; Bio-Lyte; Bio-Rad). After PAGE, duplicate lanes were stained for protein with Serva Blue W (Serva) or for CDH activity with 2 mM 2,6-dichlorophenolindophenol (DCPIP) in 50 mM sodium tartrate buffer (pH 4.0) containing 2 mM cellobiose. The isoelectric points of the two proteins were determined by comparison with a protein standard mixture (Sigma) containing amyloglycosidase (pI 3.6), trypsin inhibitor (pI 4.6), β-lactoglobulin A (pI 5.1), carbonic anhydrase II (pI 5.9), carbonic anhydrase I (pI 6.6), myoglobin (pI 6.8 and 7.2), lectin (pI 8.2 to 8.8), and trypsinogen (pI 9.3). Sodium dodecyl sulfate (SDS)-PAGE was performed to determine the purity and the molecular weights of the CDHs with a Mini-Protean system (Bio-Rad). Protein bands were visualized by silver staining and were compared to the bands produced by molecular weight markers (broad range; Bio-Rad). For activity staining of CDHs, proteins were isolated by nondenaturing PAGE without prior boiling of the samples, and the gels were stained with DCPIP as described above. To monitor the time course of CDH activity in P. cinnabarinus cultures, 1-ml samples were removed at the times indicated below and concentrated 10-fold by ultrafiltration with a 10-kDa membrane filter (Nanospin; Millipore). Twenty-microliter aliquots of the concentrated samples were separated by nondenaturing PAGE and subsequently stained to determine DCPIP-reducing activity. The molecular weights of purified CDH I and CDH II were determined with a calibrated (molecular weight markers; broad range, Sigma) gel filtration column (Bio-Gel P 100; 10 by 1,200 mm; Bio-Rad) by using 50 mM sodium acetate (pH 5.0) as the eluant. Protein concentrations were determined by using the Bradford reagent (Bio-Rad) and bovine serum albumin as the standard.
Enzyme assays.
Laccase activity was determined spectrophotometrically by monitoring the oxidation of 500 μM 2,2′-azino-bis-(3-ethylthiazoline-6-sulfonate) (ABTS) in 50 mM sodium tartrate buffer (pH 4.0) at 420 nm (molar extinction coefficient, 3.6 × 104 M−1 cm−1). CDH activity was routinely assayed by monitoring the DCPIP-reducing activity in 50 mM sodium acetate buffer (pH 4.5) containing 2 mM cellobiose at 600 nm (molar extinction coefficient, 2.7 M−1 cm−1). One unit of activity was defined as the amount of enzyme that reduced 1 μmol of substrate/min · ml.
Kinetic measurements.
The rates of reduction of test substrates by CDH I and CDH II were determined spectrophotometrically at 25°C in 50 mM sodium acetate buffer (pH 4.5) containing 2 mM cellobiose at the wavelength at which the absorbance for each compound was maximal. With the exception of 3,5-di-tert-butylbenzoquinone (TBBQ), substrates were added from 10-fold-concentrated stock solutions to a final concentration of 500 μM in a total reaction volume of 1,000 μl. TBBQ (1.2 mM) was dissolved in 96% ethanol and added to the enzyme assay reaction mixture to a final concentration of 300 μM. Mn(III) malonate was prepared and quantified as described by Roy et al. (39). The pH optima of P. cinnabarinus CDH I activity and CDH II activity were determined with 500 μM TBBQ and 2 mM cellobiose by using 50 mM sodium citrate (pH 2.5 to 3.25), 50 mM sodium acetate (pH 4.0 to 5.0), 50 mM sodium succinate (pH 5.5), and 50 mM sodium phosphate (pH 6.0 to 7.0).
Effect of P. cinnabarinus CDHs on oxidation of 3-HAA by laccase.
To study the interaction of laccase and CDH during oxidation of 3-HAA by laccase, reactions were started in 1,000-μl (total volume) reaction mixtures containing 50 mM sodium tartrate buffer (pH 4.5), 500 μM 3-HAA, 0.15 U of laccase, and 0.12 U of CDH II, and changes in absorbance at 450 nm were monitored spectrophotometrically. After 2 and 133 min, 10 μl of cellobiose (final concentration, 500 μM) was added to each reaction mixture. Control experiments were performed under the same conditions without cellobiose. In a second set of experiments, the increase in absorbance at 450 nm was monitored in the presence of different concentrations of purified CDH II (0.015 to 0.3 U). The reaction conditions were as follows: 500 μM 3-HAA, 2 mM cellobiose, and 0.15 U of P. cinnabarinus laccase in 1,000 μl (total volume) of 50 mM sodium tartrate buffer (pH 4.5).
Cellulose binding of CDH I and CDH II.
To study the ability of CDH I and CDH II to bind to cellulose, each protein (0.5 U/ml) was incubated with 1 mg of Avicel cellulose, 1 mg of Whatman CF 11 cellulose, 1 mg of bacterial cellulose, 1 mg of birch wood xylan, 1 mg of starch, or 1 mg of chitin, in 1,000 μl of 50 mM sodium acetate (pH 4.5). The reaction mixtures were incubated for 1 h at room temperature with constant agitation on a rocking table. Then samples were centrifuged, the supernatants were collected, and the activities were measured by using the standard DCPIP assay. The remaining activities were compared to the activities of control reaction mixtures without polysaccharide. All values reported below are means based on duplicate values from three independent experiments; the maximal sample mean deviation was ±5%.
Spectroscopy.
Enzyme kinetics, as well as protein spectra, were recorded with a Shimadzu model UV-1601PC spectrophotometer equipped with the Hyper UV software package (Shimadzu, Duisburg, Germany). Spectra of purified CDH II were recorded in 25 mM sodium acetate buffer (pH 5.0). To reduce the enzyme, cellobiose was added to a final concentration of 5 mM.
RESULTS
Influence of carbon source on CA accumulation and CDH production.
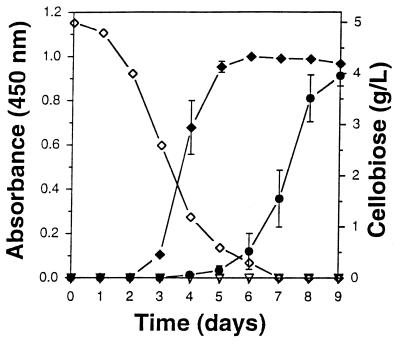
P. cinnabarinus was cultivated in defined liquid medium containing glucose, cellobiose, or cellulose as the carbon source. In the presence of glucose, accumulation of a characteristic red pigment, identified previously as CA (18), was observed after 48 h (Fig. 1). CA formation is catalyzed by laccase-mediated oxidation of the tryptophan metabolite 3-HAA (18). The maximal levels of CA were reached within 5 to 6 days. However, when P. cinnabarinus was grown on cellobiose as the carbon source, the accumulation of CA was considerably delayed and proceeded at a much slower rate than it did in glucose-containing cultures, even though the specific laccase activities were similar under both conditions. CA began to accumulate when cellobiose levels were low (approximately <0.8 g/liter). The final CA levels, which were reached after 10 days of cultivation, were as high as the levels reached in glucose-grown cultures (Fig. 1). When cellulose was the carbon source, no CA was formed during 10 days of cultivation.
FIG. 1.
Time course of CA accumulation in P. cinnabarinus cultures grown on basal media supplemented with glucose (⧫), cellobiose (•), and cellulose (▿). Depletion of the carbon source was monitored in the cellobiose-grown cultures (◊).
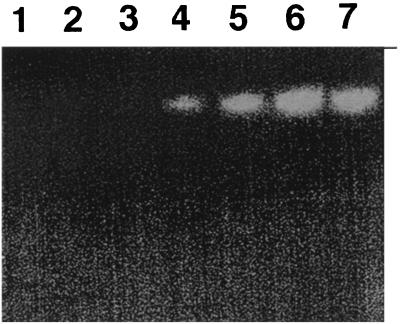
Detection of CDH activity in the P. cinnabarinus culture filtrate was masked by high levels of laccase, which interfered with all common CDH substrates (e.g., DCPIP and cytochrome c). To circumvent this problem, CDH activities were monitored by separating laccase from CDH-like proteins by nondenaturing PAGE and subsequent activity staining of the gels with DCPIP. No CDH activity was found in glucose-grown cultures, but CDH activity was detected after 4 days in cellulose-grown cultures and this activity peaked between days 7 and 9 (Fig. 2). A similar time course of CDH activity was observed when P. cinnabarinus was grown on cellobiose as the carbon source.
FIG. 2.
CDH activity in filtrates of P. cinnabarinus cultures grown on cellulose. Twenty-microliter aliquots of 10-fold-concentrated culture samples obtained from days 1 to 7 were separated by native PAGE (lanes 1 to 7, respectively). After electrophoresis, the gel was stained for CDH activity by using 2 mM DCPIP in 50 mM sodium tartrate (pH 4.0) and containing 2 mM cellobiose. Bands corresponding to CDH activity appear as clearing zones. The gel was scanned (Power Lock II; UMAX) and was labeled in Photoshop (Adobe) by using an Apple Power PC 9500 computer.
Purification of CDH from P. cinnabarinus.
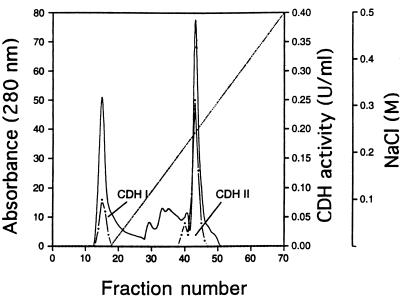
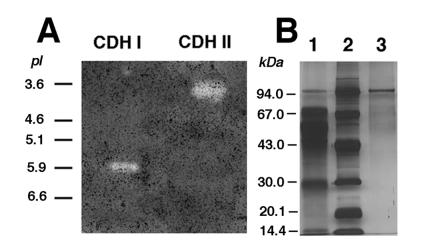
For partial characterization and in vitro studies, the CDHs of P. cinnabarinus were purified. Even though cellobiose is a potent inducer of P. cinnabarinus CDHs, purification of the enzymes from cellobiose-grown cultures was hampered by the accumulation of high concentrations of extracellular glucan. Therefore, cellulose (Avicel) was the carbon source used for P. cinnabarinus CDH production. Ten-day-old cultures grown on cellulose were harvested and used for enzyme isolation and purification. The time of harvest was less critical than that previously reported for T. versicolor CDHs (38). In P. cinnabarinus, CDH activity remained constant for at least 2 days. The two CDHs secreted by P. cinnabarinus CDH I and CDH II, were purified from 8 liters of culture fluid in two and three chromatographic steps, respectively. After the culture filtrate was concentrated by ultrafiltration, the initial separation was carried out by using a DEAE column, and this resulted in two major CDH activity peaks (Fig. 3). Fractions containing the first CDH activity peak, which was found in the flowthrough of the column and was designated CDH I, were pooled and partially purified by subsequent gel filtration chromatography. CDH I was unstable during purification, and, therefore, only partial purification was attempted. CDH I is a monomeric protein with a molecular mass of approximately 81 kDa, as determined by gel filtration and SDS-PAGE (data not shown), and an isoelectric point of about 5.9 (Fig. 4A).
FIG. 3.
Separation of concentrated culture filtrate from cellulose-grown cultures of P. cinnabarinus by DEAE-M ion-exchange chromatography. Proteins were eluted with a linear 0 to 500 mM NaCl gradient in 25 mM sodium acetate (pH 5.0) (·····). The protein (——) and CDH activity (·——·) in the eluate were monitored.
FIG. 4.
(A) Isoelectric focusing (pH 3 to 10; Bio-Rad) electrophoresis of CDH I and CDH II after the final chromatographic step. (Only part of the gel is shown.) The gel was stained with 2 mM DCPIP in 50 mM sodium tartrate buffer (pH 4.0) containing 2 mM cellobiose. Bands corresponding to CDHs were detected by clearing zones, which developed only in the presence of cellobiose. The positions of standard proteins with known pI values are indicated. The protein standard was electrophoresed on a portion of the same gel which was stained for protein. The gel was scanned (Power Lock II; UMAX) and was labeled in Photoshop (Adobe) by using an Apple Power PC 9500 computer. (B) SDS-PAGE (total acrylamide concentration, 10%; bisacrylamide cross-linker concentration, 3%; 0.1% SDS) of P. cinnabarinus CDH II after different purification steps. Protein was visualized by silver staining. Lane 1, DEAE-M column eluate (3.5 μg of protein); lane 2, protein marker; lane 3, anion-exchange (UNO) column eluate (0.3 μg of protein). Each lane contained approximately 0.05 CDH activity unit.
The second CDH activity peak, referred to as CDH II, consisted of a minor portion and a major portion and contributed approximately 75% of the total CDH activity in the P. cinnabarinus culture filtrate. CDH II-containing fractions were pooled and subjected to further purification procedures. Gel filtration of CDH II followed by a final anion-exchange step yielded a red-brown protein that corresponded to a single band on SDS-PAGE gels (Fig. 4B). The molecular mass of CDH II, as determined by gel filtration and SDS-PAGE, was approximately 101 kDa. The pI of CDH II was approximately 3.9, as determined by isoelectric focusing (Fig. 4A).
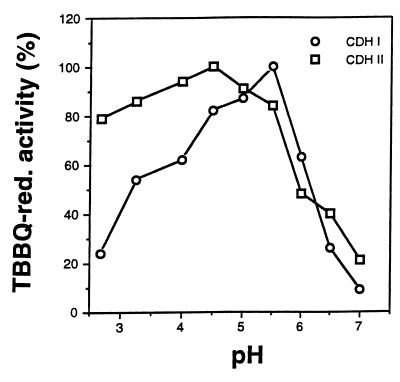
The specific activity of apparently homogeneous CDH II was 58 μmol/min · mg of protein when TBBQ in 50 mM sodium acetate (pH 4.5) was the substrate. In addition to the differences in size and isoelectric point, CDH I and CDH II also differed with respect to their pH optima. When TBBQ was the reducing substrate, CDH I exhibited maximal activity at pH 5.5 and CDH II exhibited maximal activity at pH 4.5 (Fig. 5).
FIG. 5.
pH activity profiles of CDH I and CDH II determined with 500 μM TBBQ as the electron acceptor in the presence of 2 mM cellobiose in 50 mM sodium citrate (pH 2.5 to 3.25), 50 mM sodium acetate (pH 4.0 to 5.0), 50 mM sodium succinate (pH 5.5), and 50 mM sodium phosphate (pH 6.0 to 7.0). The values are means based on triplicate measurements; the maximal sample mean deviation from the values shown was ±6%. red., reducing.
Several CDH substrates were reduced by both enzymes (Table 1), and both CDHs catalyzed reduction of the ABTS cation radical. In general, the substrate specificities were similar for the two enzymes, but CDH I had a higher Km value for all of the substrates tested, and this difference was particularly pronounced in the case of ferricyanide ions. Cytochrome c is a suitable substrate for distinguishing between intact CDH and its FAD fragment (40). CDH II could reduce cytochrome c at significant rates, but CDH I could not, suggesting that P. cinnabarinus CDH II resembles the intact CDH protein, whereas CDH I corresponds to the FAD fragment.
TABLE 1.
Reductive activities of P. cinnabarinus CDH I and CDH II
When DCPIP was the electron acceptor, both CDHs exhibited the greatest affinity for cellobiose as the reductive partner (Table 2). Besides cellobiose, only lactose and mannitol could serve as electron donors for CDH I and CDH II even though the DCPIP reduction rates were lower than for cellobiose. No reduction of DCPIP was detected with the monomeric sugars d-glucose, d-xylose, and d-mannose. Furthermore, none of the cellulose preparations tested was a suitable substrate for the oxidative half-reactions of both CDHs.
TABLE 2.
Oxidative activities of P. cinnabarinus CDH I and CDH IIa
| Substrate | Sp act (U/mg)
|
|
|---|---|---|
| CDH I | CDH II | |
| Cellobiose | 4.1 ± 0.5 | 15 ± 0.4 |
| Lactose | 1.2 ± 0.2 | 13 ± 0.3 |
| Mannitol | 0.3 ± 0.01 | 0.1 ± 0.02 |
| Sorbose | 0.3 ± 0.01 | 0.1 ± 0.01 |
Each reaction mixture (total volume, 1,000 μl) contained 50 mM sodium acetate (pH 4.5), 500 μM DCPIP, a water-soluble sugar substrate at a concentration of 2 mM, and 50 mg of a cellulose preparation. The assay mixtures were incubated at 25°C for 15 min. The results are means ± standard deviations based on triplicate measurements. No activity (DCPIP reduction after 12 h of incubation) was detected when any of the following compounds was the putative electron-donating substrate: bacterial cellulose, Whatman CF 11 cellulose, Avicel cellulose, d-glucose, d-xylose, and d-mannose.
Both P. cinnabarinus CDHs selectively bound to cellulose (Table 3). The binding of CDH II was more efficient than the binding of CDH I, but the affinities for the cellulose preparations tested were similar for the two enzymes. Both enzymes adsorbed best to Avicel cellulose, followed by bacterial cellulose and Whatman CF 11 cellulose.
TABLE 3.
Adsorption of P. cinnabarinus CDH I and CDH II to polysaccharides
| Polysaccharide | Remaining activity in the supernatant (%)a
|
|
|---|---|---|
| CDH I | CDH II | |
| Avicel cellulose | 66 | 41 |
| Bacterial cellulose | 61 | 37 |
| Whatman CF 11 cellulose | 81 | 90 |
| Xylan | 102 | 96 |
| Starch | 103 | 99 |
| Chitin | 100 | 98 |
| Control | 100 | 100 |
CDH I (0.5 U/ml) or CDH II (0.5 U/ml) sodium was incubated with 1 mg of a polysaccharide in 50 mM sodium acetate (pH 4.5) in a 1,000-μl (total volume) reaction mixture at 25°C. After 1 h of incubation with constant agitation, the polysaccharides were removed by centrifugation, and the CDH activities in the supernatant were determined by using the standard DCPIP assay. Control reaction mixtures contained no polysaccharide. The values are the means based on duplicate values from three independent experiments; the maximum sample mean deviation from the values shown was 5%.
Spectrophotometric analysis of CDH II reduction.
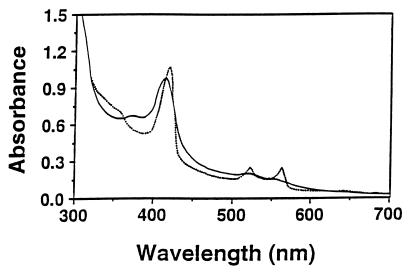
P. cinnabarinus CDH II had absorbance peaks at 420 nm (which indicated that heme was present) and at 520 and 553 nm (Fig. 6). When the enzyme was reduced to its ferrous form by adding cellobiose, the absorbance peaks at 530 and 562 nm increased and there was a concurrent decrease in absorbance at wavelengths between 440 and 500 nm. The absorbance at 420 nm/absorbance at 280 nm ratio of highly purified, oxidized CDH II was about 0.60 (±0.05).
FIG. 6.
Visible spectrum of purified CDH II in its oxidized (——) and reduced (·····) forms. The spectrum was recorded in 25 mM sodium acetate buffer (pH 5.0), and the enzyme was reduced by adding cellobiose (final concentration, 5 mM).
In vitro experiments performed with laccase and CDH.
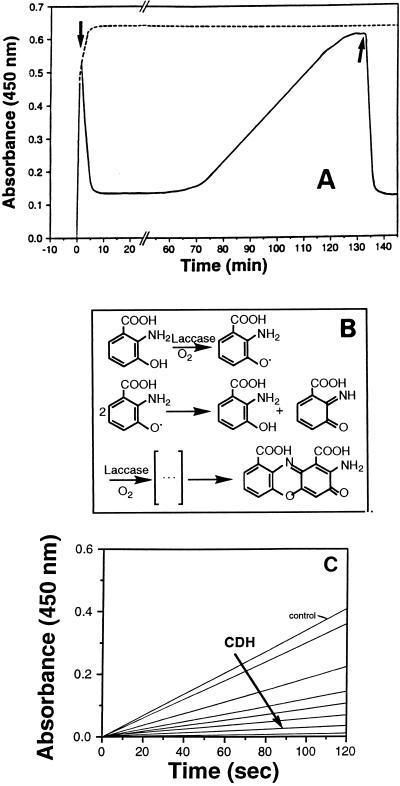
Christen et al. (10) and Toussaint and Lerch (43) proposed that 3-HAA is converted to CA via a six-electron oxidation reaction (Fig. 7B). Oxidation of 3-HAA (500 μM) by laccase (0.15 U) in a reaction mixture containing CDH II (0.12 U) was inhibited when cellobiose (final concentration, 500 μM) was added (Fig. 7A). In the presence of cellobiose, reduction of reaction intermediates by CDH II resulted in a decrease in absorbance until a basal level was reached, and this basal level remained constant for approximately 45 min. After 45 min, 3-HAA oxidation was again detected, but this reaction proceeded at a much lower rate than it did in the control to which no cellobiose was added. When fresh cellobiose (final concentration, 500 μM) was added, CDH II-catalyzed reduction of 3-HAA oxidation products began again. Increasing the concentration of CDH II resulted in a significant decrease in the rate of 3-HAA oxidation (Fig. 7). Inhibition of laccase-catalyzed CA formation was also observed when CDH I was added to the reaction mixture. Thus, both CDHs are able to reduce reaction intermediates produced during oxidative dimerization of 3-HAA. CA itself was not a substrate for either of the P. cinnabarinus CDHs, suggesting that the final step in the dimerization of 3-HAA is irreversible. These results indicate that in P. cinnabarinus cultures, CA accumulation depends on the interaction of laccase and CDHs.
FIG. 7.
Inhibition of laccase-catalyzed 3-HAA oxidation in the presence of purified P. cinnabarinus CDH II and cellobiose. (A) 3-HAA oxidation by laccase in a reaction mixture containing 500 μM 3-HAA, 0.15 U of laccase, and 0.12 U of CDH II in 1,000 μl of 50 mM sodium tartrate (pH 4.5) was monitored by determining changes in absorbance at 450 nm (——). The times at which 10 μl of cellobiose (final concentration, 500 μM) was added to the reaction mixture are indicated by arrows. Control experiments were performed without cellobiose (– – – –). (B) Hypothetical reaction mechanism for laccase-mediated oxidation of 3-HAA to CA (adapted from references 10 and 43 with permission from the publisher). (C) Effects of different concentrations of CDH II on the oxidation of 3-HAA (500 μM) by laccase (0.15 U) in 50 mM sodium tartrate (pH 4.5) containing 2 mM cellobiose. The control contained no CDH II and the amounts of CDH II that were added to the reaction mixtures were 0.015, 0.03, 0.06, 0.12, 0.18, 0.25, and 0.3 U.
DISCUSSION
We purified CDHs from P. cinnabarinus and used them for in vitro studies performed with 3-HAA and laccase in order to show that CDHs were responsible for the repression of laccase-catalyzed CA formation in P. cinnabarinus cultures grown on cellobiose or cellulose. Two proteins, designated CDH I and CDH II, used several known CDH substrates as electron acceptors; these substrates included quinones, the ABTS cation, and metal ions. Cellobiose was the most efficient electron donor for both proteins. Moreover, like other CDHs, both proteins specifically bound to cellulose. Biochemical data, as well as DNA sequence data, suggest that CDHs harbor a unique cellulose binding domain that is different from the cellulose binding domains of hydrolytic cellulases (26).
The molecular mass of P. cinnabarinus CDH II (approximately 101 kDa) is very similar to the molecular masses of CDH 4.2 purified from T. versicolor (97 kDa) (38), the CDH of S. commune (102 kDa) (22), and the CDH of P. chrysosporium (90 kDa) (5, 45). The UV-visible spectra of the oxidized and reduced states of CDH II indicate that heme and flavin cofactors are present. The spectrophotometric characteristics are very similar to those reported for CDH 4.2 of T. versicolor (38) and the heme-containing CDH of P. chrysosporium (35). In P. chrysosporium, the decrease in absorbance at wavelengths between 460 and 500 nm after reduction of the CDH has been associated with reduction of the flavin cofactor (4), and a similar response was observed for CDH 4.2 of T. versicolor (38). Based on its size, its isoelectric properties and its inability to reduce cytochrome c, CDH I resembles CDH 6.4 of T. versicolor and the FAD-containing CDH (cellobiose:quinone oxidoreductase) of P. chrysosporium. CDH I was less active on all of the substrates tested and was not able to reduce cytochrome c.
In P. chrysosporium, the smaller, FAD-containing CDH, also known as cellobiose:quinone oxidoreductase, is a proteolytic cleavage product of the FAD- and heme-containing CDH (23, 25). We can only speculate that this is also the case for CDH I of P. cinnabarinus and cannot exclude the possibility that CDH I is encoded by a separate gene.
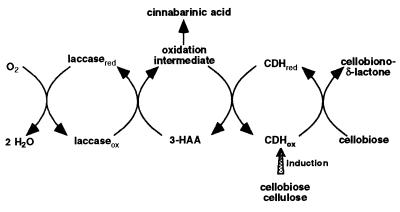
Archibald et al. (4) reported that oxidation of guaiacol by laccase in the presence of T. versicolor CDH prevented the accumulation of stable end products and resulted in indefinite consumption of oxygen. Our results suggest that CDH plays a similar role in vivo. Laccase oxidation products of the tryptophan metabolite 3-HAA were identified as in vivo electron acceptors for both P. cinnabarinus CDHs. 3-HAA is the precursor of the laccase-mediated formation of CA, the characteristic phenoxazinone pigment in P. cinnabarinus (18). We hypothesize that laccase and CDH interact during pigment synthesis in P. cinnabarinus (Fig. 8). In the absence of cellulose, laccase oxidizes 3-HAA to CA. In the presence of cellulose or cellodextrins, CDH is produced, which catalyzes the reduction of 3-HAA oxidation intermediates. As long as a suitable electron donor, preferably cellobiose, is available for the oxidative half-reaction, this cycle is maintained indefinitely, which prevents accumulation of CA. This model is consistent with the observation that wood colonized by P. cinnabarinus has the typical white rot appearance. CA and other related phenoxazinones do not accumulate until fruiting bodies are formed.
FIG. 8.
Model of the in vivo interaction of CDHs and laccase during CA formation in P. cinnabarinus. Reaction intermediates of the laccase-catalyzed oxidation of 3-HAA have not been identified yet.
In recent years, the substrate range of laccases has been expanded to include nonphenolic lignin structures, which are not oxidized by laccase alone. These nonphenolic substrates can be oxidized by laccases in the presence of certain cooxidants, which are often referred to as mediators (7, 8). Previously, we identified 3-HAA as a possible endogenously produced cooxidant that could aid laccase in oxidizing lignin (17). If such compounds were used only in catalytic amounts in a natural system, (re)cycling of cooxidants would be a prerequisite. In addition to its importance in vivo, (re)cycling of cooxidants has great significance for biotechnical applications of laccase-cooxidant systems.
P. cinnabarinus has proven to be an interesting model organism for studying new mechanisms of lignocellulose degradation by white rot fungi. The novel interaction between CDH and laccase described in this paper is evidence that CDHs have a physiological function, which supports the hypothesis that CDHs are involved not only in cellulose degradation but also in lignin degradation.
ACKNOWLEDGMENTS
We thank R. Machmerth for technical assistance and Karl-Erik L. Eriksson for valuable discussions.
REFERENCES
- 1.Ander P. The cellobiose-oxidizing enzymes CBQ and CBO as related to lignin and cellulose degradation—a review. Enzyme Microbiol Technol. 1994;15:1002–1007. [Google Scholar]
- 2.Ander P, Eriksson K-E L. Selective degradation of wood components by white-rot fungi. Physiol Plant. 1977;41:239–248. [Google Scholar]
- 3.Ander P, Marzullo L. Sugar oxidoreductases and veratryl alcohol oxidase as related to lignin degradation. J Biotechnol. 1996;53:115–131. doi: 10.1016/s0168-1656(97)01680-5. [DOI] [PubMed] [Google Scholar]
- 4.Archibald F S, Bourbonnais R, Jurasek L, Paice M G, Reid I D. Kraft pulp bleaching and delignification by Trametes versicolor. J Biotechnol. 1997;53:215–236. [Google Scholar]
- 5.Bao W, Renganathan V. Triiodide reduction by cellobiose:quinone oxidoreductase of Phanerochaete chrysosporium. FEBS Lett. 1991;279:30–32. doi: 10.1016/0014-5793(91)80242-u. [DOI] [PubMed] [Google Scholar]
- 6.Bao W, Usha S N, Renganathan V. Purification and characterization of cellobiose dehydrogenase, a novel hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys. 1993;300:705–713. doi: 10.1006/abbi.1993.1098. [DOI] [PubMed] [Google Scholar]
- 7.Bourbonnais R, Paice M G. Oxidation of nonphenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-w. [DOI] [PubMed] [Google Scholar]
- 8.Call H P, Mücke I. History, overview and applications of mediated ligninolytic systems, especially laccase-mediator-systems. J Biotechnol. 1997;53:163–202. [Google Scholar]
- 9.Canevascini G. Cellobiose dehydrogenase from Sporotrichum thermophile. Methods Enzymol. 1988;160:443–448. [Google Scholar]
- 10.Christen S, Southwell-Keely P, Stocker R. Oxidation of 3-hydroxyanthranilic acid to the phenoxazinone cinnabarinic acid by peroxyl radicals and by compound I of peroxidases or catalases. Biochemistry. 1992;31:8090–8097. doi: 10.1021/bi00149a045. [DOI] [PubMed] [Google Scholar]
- 11.Costa-Ferreira M, Ander P, Duarte J. On the relationship between cellobiose dehydrogenase and cellobiose:quinone oxidoreductase under conditions where [14C]DHP is mineralized by whole cultures of Phanerochaete chrysosporium. Enzyme Microb Technol. 1994;16:771–776. [Google Scholar]
- 12.Dekker R F H. Induction and characterization of a cellobiose dehydrogenase produced by a species of Monilia. J Gen Microbiol. 1980;120:309–316. [Google Scholar]
- 13.Dumonceaux T J, Bartholomew K A, Charles T C, Moukha S M, Archibald F S. Cloning and sequencing of a gene encoding cellobiose dehydrogenase from Trametes versicolor. Gene. 1998;210:211–219. doi: 10.1016/s0378-1119(98)00084-5. [DOI] [PubMed] [Google Scholar]
- 14.Eggert C. Laccase is responsible for antimicrobial activity of Pycnoporus cinnabarinus. Microbiol Res. 1997;152:315–318. doi: 10.1016/S0944-5013(97)80046-8. [DOI] [PubMed] [Google Scholar]
- 15.Eggert C, Habu N, Temp U, Eriksson K-E L. Cleavage of Phanerochaete chrysosporium cellobiose dehydrogenase (CDH) by three endogenous proteases. In: Srebotnik E, Messner K, editors. Biotechnology in the pulp and paper industry. Recent advances in applied and fundamental research. Vienna, Austria: Fakultas-Universitätsverlag; 1996. pp. 551–554. [Google Scholar]
- 16.Eggert C, LaFayette P R, Temp U, Eriksson K-E L, Dean J F D. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1998;64:1766–1772. doi: 10.1128/aem.64.5.1766-1772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggert C, Temp U, Dean J F D, Eriksson K-E L. A fungal metabolite mediates oxidation of non-phenolic lignin model compounds and synthetic lignin by laccase. FEBS Lett. 1996;391:144–148. doi: 10.1016/0014-5793(96)00719-3. [DOI] [PubMed] [Google Scholar]
- 18.Eggert C, Temp U, Dean J F D, Eriksson K-E L. Laccase-mediated formation of the phenoxazinone derivative, cinnabarinic acid. FEBS Lett. 1995;376:202–204. doi: 10.1016/0014-5793(95)01274-9. [DOI] [PubMed] [Google Scholar]
- 19.Eggert C, Temp U, Eriksson K-E L. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus:purification and characterization of the laccase. Appl Environ Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson K-E L, Habu N, Samejima M. Recent advances in fungal cellobiose oxidoreductases. Enzyme Microb Technol. 1993;15:1001–1008. [Google Scholar]
- 21.Fähnrich P, Irrgang K. Conversion of cellulose to sugars and cellobionic acid by the extracellular enzyme system of Chaetomium cellulolyticum. Biotechnol Lett. 1982;12:775–780. [Google Scholar]
- 22.Fang J, Liu W, Gao P J. Cellobiose dehydrogenase from Schizophyllum commune:purification and study of some catalytic, inactivation and cellulose binding properties. Arch Biochem Biophys. 1998;353:37–46. doi: 10.1006/abbi.1998.0602. [DOI] [PubMed] [Google Scholar]
- 23.Habu N, Samjima M, Eriksson K-E L. Release of the FAD domain from cellobiose oxidase by proteases from cellulolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1993;327:161–164. doi: 10.1016/0014-5793(93)80162-n. [DOI] [PubMed] [Google Scholar]
- 24.Henriksson G, Ander P, Pettersson B, Pettersson G. Cellobiose dehydrogenase (cellobiose oxidase) from Phanerochaete chrysosporium as a wood-degrading enzyme. Studies on cellulose, xylan, and synthetic lignin. Appl Microbiol Biotechnol. 1995;42:790–796. [Google Scholar]
- 25.Henriksson G, Petterson G, Johansson G, Ruiz A, Uzcategui E. Cellobiose oxidase can be cleaved by papain into two domains. Eur J Biochem. 1991;196:101–106. doi: 10.1111/j.1432-1033.1991.tb15791.x. [DOI] [PubMed] [Google Scholar]
- 26.Henriksson G, Salumets A, Divne C, Pettersson G. Studies of cellulose binding by cellobiose dehydrogenase and a comparison with cellobiohydrolase I. Biochem J. 1997;324:833–838. doi: 10.1042/bj3240833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hüttermann A, Noelle A. Characterization and regulation of cellobiose dehydrogenase in Fomes annosus. Holzforschung. 1982;36:283–286. [Google Scholar]
- 28.Hyde S M, Wood P M. Kinetic and antigenic similarities for cellobiose dehydrogenase from the brown-rot fungus Coniophora putanea and the white-rot fungus Phanerochaete chrysosporium. FEMS Lett. 1996;145:439–444. [Google Scholar]
- 29.Kremer S M, Wood P M. Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe(III) reductase. Eur J Biochem. 1992;205:133–138. doi: 10.1111/j.1432-1033.1992.tb16760.x. [DOI] [PubMed] [Google Scholar]
- 30.Kremer S M, Wood P M. Production of Fenton’s reagent by cellobiose oxidase from cellulolytic cultures of Phanerochaete chrysosporium. Eur J Biochem. 1992;208:807–814. doi: 10.1111/j.1432-1033.1992.tb17251.x. [DOI] [PubMed] [Google Scholar]
- 31.Lesage-Meessen L, Haon M, Delattre M, Thibault J-F, Ceccaldi B C, Asther M. An attempt to channel the transformation of vanillic acid into vanillin by controlling methoxyquinone formation in Pycnoporus cinnabarinus with cellobiose. Appl Microbiol Biotechnol. 1997;47:393–397. [Google Scholar]
- 32.Li B, Nagalla S R, Renganathan V. Cellobiose dehydrogenase from Phanerochaete chrysosporium is encoded by two allelic variants. Appl Environ Microbiol. 1997;63:796–799. doi: 10.1128/aem.63.2.796-799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Nagalla S R, Renganathan V. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:1329–1335. doi: 10.1128/aem.62.4.1329-1335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansfield S D, de Jong E, Saddler J N. Cellobiose dehydrogenase, an active agent in cellulose depolymerization. Appl Environ Microbiol. 1997;63:3804–3809. doi: 10.1128/aem.63.10.3804-3809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morpeth F, Jones G. Resolution, purification and some properties of multiple forms of cellobiose dehydrogenase from the white-rot fungus Sporotrichum pulverulentum. Biochem J. 1986;236:221–226. doi: 10.1042/bj2360221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raices M E, Paifer E, Cremata J, Montesino M, Stahlberg J, Divne C, Szabo I J, Henriksson G, Pettersson G. Cloning and characterization of a cDNA encoding a cellobiose dehydrogenase from the white-rot fungus Phanerochaete chrysosporium. FEBS Lett. 1995;369:233–238. doi: 10.1016/0014-5793(95)00758-2. [DOI] [PubMed] [Google Scholar]
- 37.Roy B P, Archibald F S. An indirect free radical-based assay for the enzyme cellobiose:quinone oxidoreductase. Anal Biochem. 1994;216:291–298. doi: 10.1006/abio.1994.1044. [DOI] [PubMed] [Google Scholar]
- 38.Roy B P, Dumonceaux T, Koukoulas A A, Archibald F S. Purification and characterization of cellobiose dehydrogenase from the white rot fungus Trametes versicolor. Appl Environ Microbiol. 1996;62:4417–427. doi: 10.1128/aem.62.12.4417-4427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy B P, Paice M G, Archibald F S, Misra S K, Misiak J E. Creation of metal-complexing agents, reduction of manganese dioxide, and promotion of manganese peroxidase-mediated Mn(III) production by cellobiose:quinone oxidoreductase from Trametes versicolor. J Biol Chem. 1994;269:19745–19750. [PubMed] [Google Scholar]
- 40.Samejima M, Eriksson K-E L. A comparison of the catalytic properties of cellobiose:quinone oxidoreductase and cellobiose oxidase from Phanerochaete chrysosporium. Eur J Biochem. 1992;207:103–107. doi: 10.1111/j.1432-1033.1992.tb17026.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmidthalter D R, Canevascini G. Isolation and characterization of the cellobiose dehydrogenase from the brow-rot fungus Coniophora putanea (Schum ex Fr.) Karst. Arch Biochem Biophys. 1993;300:559–563. doi: 10.1006/abbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 42.Schou C, Christensen M H, Schulein M. Characterization of a cellobiose dehydrogenase from Humicola insolens. Biochem J. 1998;330:565–571. doi: 10.1042/bj3300565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toussaint O, Lerch K. Catalytic oxidation of 2-aminophenols and ortho hydroxylation of aromatic amines by tyrosinase. Biochemistry. 1987;26:8568–8571. doi: 10.1021/bi00400a011. [DOI] [PubMed] [Google Scholar]
- 44.Vallim M A, Janse B J H, Gaskell J, Pizzirani-Kleiner A A, Cullen D. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl Environ Microbiol. 1998;64:1924–1928. doi: 10.1128/aem.64.5.1924-1928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westermark U, Eriksson K-E L. Carbohydrate dependent enzymic quinone reduction during lignin degradation. Acta Chem Scand B. 1974;28:204–208. [Google Scholar]
- 46.Westermark U, Eriksson K-E L. Cellobiose:quinone oxidoreductase, a new wood-degrading enzyme from white-rot fungi. Acta Chem Scand B. 1974;28:209–214. [Google Scholar]
- 47.Westermark U, Eriksson K-E L. Purification and properties of cellobiose:quinone reductase from Sporotrichum pulverulentum. Acta Chem Scand B. 1975;29:419–424. [PubMed] [Google Scholar]
- 48.Wood D, Wood P M. Evidence that cellobiose:quinone oxidoreductase from Phanerochaete chrysosporium is a breakdown product of cellobiose oxidase. Biochim Biophys Acta. 1992;1119:90–96. doi: 10.1016/0167-4838(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 49.Wood P M. Pathways of production of Fenton’s reagent by wood-rotting fungi. FEMS Microbiol Rev. 1994;13:313–320. [Google Scholar]