Abstract
We constructed and characterized three stress probe plasmids which utilize a green fluorescent protein as a noninvasive reporter in order to elucidate Escherichia coli cellular stress responses in quiescent or resting cells. Cellular stress levels were easily detected by fusing three heat shock stress protein promoter elements, those of the heat shock transcription factor ς32, the protease subunit ClpB, and the chaperone DnaK, to the reporter gene gfpuv. When perturbed by a chemical or physical stress (such as a heat shock, nutrient [amino acid] limitation, or addition of IPTG [isopropyl-β-d-thiogalactopyranoside], acetic acid, ethanol, phenol, antifoam, or salt [osmotic shock]), the E. coli cells produced GFPuv, which was easily detected within the cells as emitted green fluorescence. Temporal and amplitudinal mapping of the responses was performed, and the results revealed regions where quantitative delineation of cell stress was afforded.
When cells are exposed to chemical or physical stress, they undergo many changes, including alterations in the patterns of gene expression, as well as protein stability. For example, when bacterial cells are exposed to a high temperature, a set of heat shock proteins are transcriptionally upregulated by a transcription factor, ς32 (21, 37, 58). These heat shock proteins are evolutionarily conserved, and many play an important role in the folding, assembly, degradation, and translocation of proteins, not only under stress conditions but also during normal cell growth (16, 17, 25). In addition to heat shock, a number of other stresses induce the synthesis of heat shock proteins in many organisms; these stresses include viral infection (42), oxygen limitation, the presence of abnormal proteins (5, 18, 23), overexpression of heterologous proteins (4, 19, 34, 53), nutrient limitation (carbon source limitation, amino acid source limitation, etc.) (20, 33, 45), and exposure to various chemicals, including ethanol, phenol, hydrogen peroxide, and heavy metals (6, 28, 50–52, 54).
The proteins that are produced include transcription factors, chaperones, proteases, and other proteins that confer a survival advantage on the stressed organism, particularly when the levels of the stresses are subinhibitory (21, 51). Monitoring the heat shock response has resulted in detection of toxic compounds and other environmental insults (51), as well as conditions which lead to suboptimal cell growth and suboptimal yields of recombinant proteins (2, 39). A common method for determining cellular regulation has been transcriptional and/or translational fusion of a reporter gene (e.g., lacZ, cat) to endogenous regulatory elements (27, 35, 36). Recently, Van Dyk et al. showed that bioluminescence could be used to detect the heat shock response after fusion of the dnaK and grpE heat shock promoters to the luxCDABE bioluminescence operon from Vibrio fischeri (51). This method obviated the need for centrifugation, cell lysis, pH adjustment, and subsequent kinetic enzyme activity measurements.
In this study, we utilized a green fluorescent protein (GFP), GFPuv (13), as a reporter of cellular stress responses in Escherichia coli. When GFPuv is the reporter, ATP or other cofactors are not necessary for fluorescence. Also, the metabolic requirements for generation of this GFP are minimal as GFPuv is relatively small (27 kDa). Stress was detected by transcriptional fusion of three heat shock stress protein promoter elements (promoter elements of the heat shock transcription factor ς32 [9, 57], the protease subunit ClpB [30, 44], and the chaperone DnaK [3, 12, 14]) to the reporter gene gfpuv. Because the native stress promoters were amplified from genomic DNA and incorporated into a plasmid so that they controlled the expression of GFPuv, we were able to quantify the cellular stress responses of E. coli by simply measuring GFP fluorescence intensity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli HB101 [supE44 hsdS20(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1] (8) was used to construct a recombinant plasmid containing the GFPuv gene, gfpuv, and heat shock protein promoters. E. coli JM105 [supE endA sbcB15 hsdR4 rpsL thiΔ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] (56) was the host used to investigate the cellular stress response under several stress conditions. Plasmid pGFPuv (Clontech Laboratories, Inc., Palo Alto, Calif.) was used for excision of the gfpuv gene. Plasmid pBR322 was the parent plasmid used for insertion of the gfpuv fusions.
Construction of recombinant plasmids.
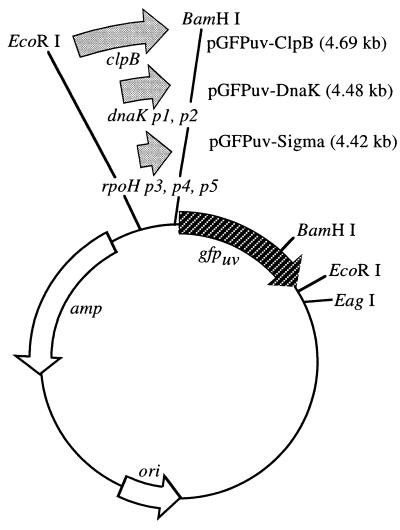
Three heat shock promoters, specifically those of the heat shock transcription factor ς32, the protease subunit ClpB, and the chaperone DnaK, were obtained by performing PCR amplification with a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) from genomic DNA obtained from E. coli K-12. The E. coli K-12 promoter-operator sequences for rpoH (for ς32) (9, 57), clpB (30, 44), and dnaK (3, 12, 14) were obtained from GenBank (http://www.ncbi.nlm.nih.gov/web/search). The −35 and −10 (TATA box) regions, transcription initiation site, ribosome binding site (RBS), and translation initiation site (codon, ATG) of the native proteins were used for regulation of GFPuv; that is, since the length between the RBS and the translation initiation site is an important factor for the translational efficiency of foreign protein expression (46), we used the translation initiation sites of the original stress promoters. In the case of rpoH, a sequence spanning 121 bases prior to the ATG was targeted. This region includes three rpoH promoters (p3, recognized by the RNA polymerase EςE; and p4 and p5, recognized by the RNA polymerase Eς−0 [17]). With clpB and dnaK, the total target sequences were 361 and 181 bases prior to the native ATG, respectively. In the case of dnaK, this region contains promoters p1 and p2 (58). To adjust the open reading frame, two extra bases, GG, were added during design of the PCR primer sequences (rpoH, 5′-CCGGAATTCAAGCTTGCATTGAACTTAGTGG-3′ and 5′-CGCGGATCCCCCATTCAAATCCTCTCAATCGATATC-3′; clpB, 5′-CCGGAATTCCCGGCAATTGGTCCACGCGCG-3′ and 5′-CGCGGATCCCCCATAACTCCTCCCATAACGGATC-3′; dnaK, 5′-CCGGAATTCCGAAATTTCTGCGCAAAAGCAC-3′ and 5′-CGCGGATCCCCCATCTAAACGTCTCCACTATATATTC-3′), which allowed cloning of the 142-, 410-, and 204-bp EcoRI- and BamHI-digested amplified products into the EcoRI and BamHI sites of pBR322. The gfpuv gene was excised from the pGFPuv plasmid by BamHI (partial) and EagI (full) digestion and was inserted into the BamHI and EagI sites of three pBR322 plasmids, each of which contained a heat shock stress protein promoter. The recombinant plasmids, pGFPuv-Sigma, pGFPuv-ClpB, and pGFPuv-DnaK, contained the heat shock stress protein promoters rpoH, clpB, and dnaK, respectively, and the gfpuv structural gene (Fig. 1).
FIG. 1.
Schematic diagram of recombinant plasmids pGFPuv-Sigma, pGFPuv-ClpB, and pGFPuv-DnaK. pGFPuv-Sigma, pGFPuv-ClpB, and pGFPuv-DnaK are pBR322-based plasmids containing three heat shock stress protein promoters from the heat shock transcription factor ς32, the protease ClpB, and the chaperone DnaK, respectively, and the reporter gene gfpuv.
Culture media, growth conditions, and chemicals.
E. coli strains were grown to the early log phase at 30°C in 100 ml of Luria broth (5 g of yeast extract [Sigma Chemical Co., St. Louis, Mo.] per liter, 10 g of Bacto Tryptone [Difco Laboratories, Detroit, Mich.] per liter, 10 g of NaCl per liter) supplemented with 50 μg of ampicillin (Sigma) per ml from 30°C overnight cultures grown in the same medium. After the cells were harvested, they were washed with phosphate-buffered saline (1.44 g of Na2HPO4 per liter, 0.24 g of KH2PO4 per liter, 0.2 g of KCl per liter, 8 g of NaCl per liter; pH 7.4). The cells were then resuspended in 20 ml of phosphate-buffered saline and were in a resting (nongrowth) state; these cells were used immediately for stress induction tests.
Stresses.
Heat shock was induced by raising the temperature from 30 to 42°C by immersion in a water bath. Chemical pollutants (ethanol, methanol, phenol, heavy metals, etc.) are known to induce cellular stresses in E. coli (41, 51, 54); in this study the responses of cells to ethanol and phenol were tested by adding 4% (vol/vol) ethanol and 1 g of phenol per liter, respectively. The response of cells to high osmolarity (7, 55) was investigated by adding 40 g of NaCl per liter (osmotic pressure, 17 atm). A stringent response is induced by a lack of aminoacyl-tRNA at the ribosomes during translation (10, 15), which in turn induces expression of heat shock proteins (20). The stringent stress was induced in this study by adding 100 mg of serine hydroxamate per liter, which inhibited the aminoacylation of seryl-tRNA without inhibiting serine synthesis or guanosine 5′-diphosphate 3′-diphosphate decay (38). Antifoam agents are widely used to reduce foaming in highly agitated and sparged fermentors, particularly fermentors containing complex media. Selected antifoam agents have previously been shown to inhibit cell growth. In this study, 0.2 ml of antifoam agent (food grade; Dow Corning Corp.) per liter was added. Finally, 10 g of acetic acid per liter and 1, 3, and 5 mM isopropyl β-d-thiogalactopyranoside (IPTG) (Sigma) were added to resting cells. The presence of acetate has been linked to low productivity in E. coli cultures overexpressing recombinant proteins (32), and it has been reported that IPTG (which is used to derepress lac-based promoters) stresses cells independent of overexpression (31).
Analytical methods.
Cell growth was monitored by determining optical density at 600 nm with a spectrophotometer (Spectronic 21D; Milton Roy Company). The GFP assay was performed by measuring fluorescence intensity with a fluorescence spectrometer (model LS-3B; Perkin-Elmer Ltd., Beaconsfield, Buckinghamshire, England) at an excitation wavelength of 395 nm and an emission wavelength of 509 nm. Duplicate measurements were obtained for each sample; the average values of the duplicate measurements are reported below. The data reported below include the specific fluorescence intensity (SFI) (the raw fluorescence intensity divided by the optical density at 600 nm) and the relative fluorescence intensity (RFI), which was obtained by subtracting the specific fluorescence value for E. coli JM105 containing unmodified plasmid pBR322 (a stress probe plasmid without the gfpuv gene and stress promoters) from the fluorescence value for the stressed gfpuv-containing cells: RFI = (SFI|t − SFI|t0)stress probe − (SIF|t − SFI|t0)pBR322 where SFI|t and SFI|t0 are the SFIs at time t and time zero, respectively. This allowed us to compare insults, as cultures had different optical densities at time zero. The results shown below were recorded at 8 h unless indicated otherwise. Also, by using parent plasmid pBR322 as a fluorescence control, the baseline transcriptional activity of all cultures could be determined. This is reported below as the SFI at time zero.
Western blot analysis of DnaK.
A total protein assay was performed with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.) by using bovine serum albumin as the standard. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed by mixing a sample with sample buffer (0.5 M Tris-HCl [pH 6.8], 10% glycerol, 5% SDS, 5% β-mercaptoethanol, 0.25% bromophenol blue), incubating the preparation at 100°C for 3 min, centrifuging it for 1 min, and loading it onto a 15% slab gel. After electrophoresis, the gel was transferred onto a nitrocellulose membrane (Bio-Rad) by using a Bio-Rad Mini-Trans Blot Cell in Bjerrum–Schafer-Nielsen transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol; pH 9.2) for 40 min at 20 V. The nitrocellulose membrane was probed with a 1:1,000 dilution of polyclonal anti-DnaK antibody (StressGen Biotechnologies Corp., Victoria, British Columbia, Canada) and detected with a 1:5,000 dilution of goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) and 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium color development reagent (Sigma). Also, pure serially diluted (two times each) DnaK (StressGen) was loaded onto the membrane as a standard; the amounts used ranged from 1.0 to 0.008 μg. The stained membranes were photographed with a digital camera (Strategene Eagle Eye) and were analyzed with National Institutes of Health image software (NIH Image, written by Wayne Rasband of the National Institutes of Health and available on the Internet by anonymous ftp from zippy.nimh .nih.gov).
RESULTS
Cellular stress response by heat shock.
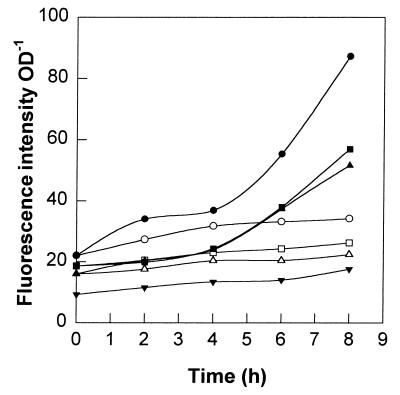
An increase in the SFI was observed for the ς32, ClpB, and DnaK plasmids in E. coli JM105 cells that were heat shocked by a temperature shift from 30 to 42°C (Fig. 2). Also, there was significant prestress fluorescence, which revealed the following pattern of intensity: pGFPuv-Sigma > pGFPuv-ClpB > pGFPuv-DnaK > pBR322. Since the RBS and promoter elements were included in the regulatory sequences of the fusions, increasing the fluorescence intensity (pre- and poststress) demonstrated that there was increased transcription and/or translation of the ς32 transcription factor, the protease subunit ClpB, and the chaperone DnaK. In the case of the strain containing the pGFPuv-Sigma plasmid, the levels of fluorescence increased within the first 2 h after the temperature shift. For the pGFPuv-ClpB and pGFPuv-DnaK plasmids, the levels of fluorescence increased most after 4 h. The level of fluorescence of rpoH::gfpuv was always at least 50% greater than the levels of fluorescence of the clpB::gfpuv and dnaK::gfpuv plasmids under heat shock conditions. Also, because the raw fluorescence data were very similar to the specific fluorescence data, we concluded that there was virtually no interference due to the intrinsic fluorescence of the cells.
FIG. 2.
GFPuv (cellular stress) response profiles under heat shock conditions. Symbols: •, pGFPuv-Sigma; ○, nonstressed pGFPuv-Sigma; ■, pGFPuv-ClpB; □, nonstressed pGFPuv-ClpB; ▴, pGFPuv-DnaK; ▵, nonstressed pGFPuv-DnaK; ▾, pBR322 (control). The heat shock consisted of transferring initially resting cells from 30 to 42°C. OD, optical density.
Stress probe sensitivity.
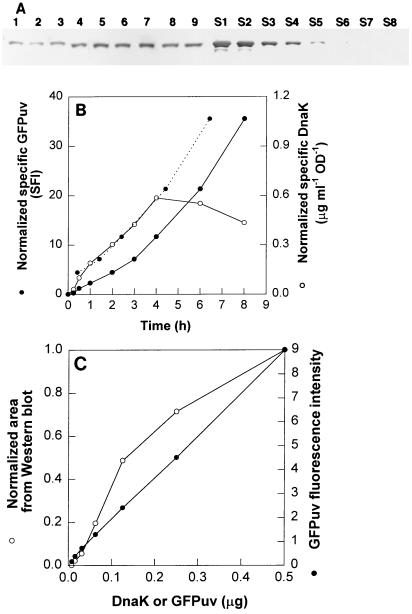
To evaluate the sensitivity of GFPuv monitoring, the pGFPuv-DnaK strain was heat shocked. In addition, serially diluted GFP and DnaK were analyzed by fluorescence and Western blotting in order to establish detection limits and regions of linearity. The results of the DnaK Western blot and GFP fluorescence analyses are shown in Fig. 3. Although less sensitive, the Western blot analyses confirmed that there was an increase in DnaK synthesis (Fig. 3A). The normalized specific DnaK levels and the normalized specific GFPuv SFIs during heat shock are shown in Fig. 3B. The DnaK profile was almost identical to the GFPuv fluorescence profile until 4 h, after which the amount of DnaK leveled off and the GFP SFI increased. The difference was probably due to DnaK proteolytic sensitivity. A smaller band was typically found below DnaK on immunoblots, while GFPuv is stable in E. coli (11). Note that the time lag for GFPuv chromophore cyclization is known to be constant, 95 min, so the fluorescence data were also shifted (as shown) to more accurately track the appearance of the protein (1). An improved correlation was obtained for the shifted data. Figure 3C shows the linearity of the fluorescence intensity down to 0.1 ± 0.01, which corresponded to 0.01 μg of protein, and outward to 1.0 μg of protein. The results for an analogous dilution of DnaK, detected and quantified by Western blotting (Fig. 3A), revealed a limited linear region from 0.03 to 0.15 μg of protein (Fig. 3C).
FIG. 3.
(A) Western blot analysis of DnaK expression after heat shock. Lanes 1 to 9, samples taken 0, 0.25, 0.5, 1, 2, 3, 4, 6, and 8 h, respectively, after stress; lanes S1 to S8, serial dilutions of standard DnaK (lane S1 contained 1.0 μg, and lane S8 contained 0.007813 μg; the preparation was serially diluted twofold per lane for quantification of the linear range and detection limits). (B) Increase in DnaK after heat shock (Western blot) correlated with GFPuv (SFI). The heat shock consisted of changing the temperature from 30 to 42°C. The dotted line shows the results for a 95-min shift in the GFPuv fluorescence profile. OD, optical density. (C) Fluorescence intensity of serially diluted E. coli JM105 containing GFPuv. The linearity of the fluorescence was demonstrated to be accurate to 0.1 fluorescence unit (r2 [from 0 to 1 μg] = 0.999). The DnaK data are from the Western blot shown in panel A; the results were linear for amounts ranging from 0.03 to 0.15 μg (r2 = 0.92).
Dose-response curve for ethanol shock.
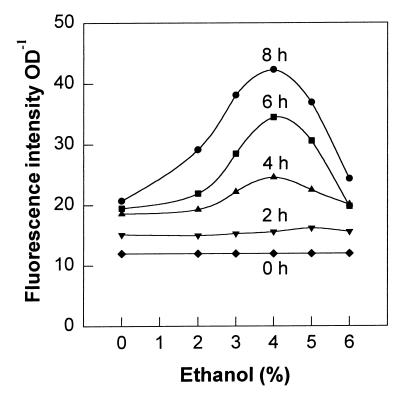
Ethanol is an efficient inducer of heat shock proteins (37), and we evaluated the ethanol dose-response profile for the pGFPuv-DnaK plasmid (Fig. 4). At early times after ethanol addition, the fluorescence response was nonlinear for low ethanol concentrations (<4%, vol/vol). The fluorescence was linear at all times with ethanol concentrations ranging from 2 to 4% (vol/vol), and at later times the response was linear for a wider range of ethanol concentrations (the r2 value was 0.989 at 8 h for 0 to 4% [vol/vol] ethanol). Since the slopes of fluorescence-versus-ethanol concentration plots for concentrations less than 4% (vol/vol) and of fluorescence-versus-time plots were positive, the best sensitivity for backward discrimination of ethanol levels was at 8 h or at the end of the experiment. Interestingly, as the ethanol concentration was increased to levels greater than 4% (vol/vol), an apparent toxicity threshold was reached, and DnaK-promoted GFPuv expression started to decrease at high ethanol concentrations.
FIG. 4.
Dose-response curves for dnaK::gfpuv under ethanol-imposed stress conditions. Samples were taken every 30 min. Symbols: ⧫, samples taken at time zero; ▾, ▴, ■, and •, samples taken 2, 4, 6, and 8 h, respectively, after ethanol was added. OD, optical density.
Detection of several environmental insults.
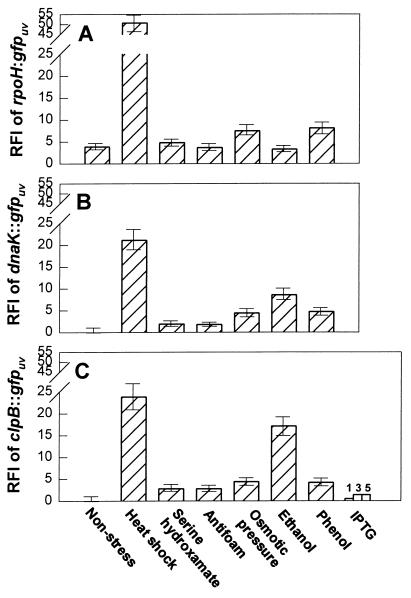
The levels of fluorescence of each strain subjected to several stresses were also plotted (Fig. 5). A positive response was observed for all probes and for all but one stress. In the outlier case, IPTG was added at three concentrations to the ClpB strain. E. coli JM105 cells are lacIq, and there was no plasmid-encoded lac-based promoter; in the absence of mRNA amplification (which might have led to stress), there was only a slight increase in fluorescence. This finding was consistent with the results of a previous study in which there were small increases in GroEL, DnaK, and GroES when IPTG was added to a lac− strain (31). With the exception of ethanol addition to the rpoH::gfpuv strain, the response to each insult was greatest for the rpoH::gfpuv strain and smallest for the dnaK::gfpuv strain. Figure 5 also shows that the increases observed with the rpoH::gfpuv strain for the serine hydroxamate, antifoam agent, and ethanol insults were not appreciably different than the increases observed with the unstressed control. Figure 5 does not show temporal results which revealed that ς32 always led the chaperone and protease subunit. Also, addition of acetic acid (10 g liter−1) quenched fluorescence of the ς32 and DnaK plasmids (data not shown).
FIG. 5.
Detection of stress after several insults. (A) rpoH::gfpuv. (B) dnaK::gfpuv. (C) clpB::gfpuv. The controls were not stressed. Other resting cell preparations were subjected to the following stresses at time zero: heat shock, 100 mg of serine hydroxamate per liter, 0.2 ml of antifoam agent (Dow Corning) per liter, 40 g of NaCl per liter, 4% (vol/vol) ethanol, and 1 g of phenol per liter. An IPTG test was performed by using 1, 3, and 5 mM IPTG and clpB::gfpuv. The RFIs were determined at 8 h.
DISCUSSION
Heat shock stress proteins can for the most part be divided into the following three categories: effector molecules, such as the heat shock transcription factor ς32, which is a signal for heat shock protein synthesis; proteases, which play a role in degrading both foreign and endogenous proteins; and chaperones, which catalyze the native protein folding processes but also may be “unfoldases” that hold other proteins in slightly altered configurations while proteases attack and digest them (35, 43, 48). We constructed three noninvasive stress probe plasmids to examine the regulatory response of one protein belonging to each of these broad categories. The proteins studied were three well-studied heat shock proteins, the heat shock transcription factor ς32, the protease subunit ClpB, and the chaperone DnaK. The endogenous promoter sequences of the genes encoding these proteins were used to drive expression of the novel reporter gene gfpuv. Sensitive detection of cellular stress was accomplished by simply measuring green fluorescence.
We investigated the responses to several imposed stresses (heat shock; addition of serine hydroxamate, which was a stringent stress; several environmental stresses, including IPTG, acetic acid, antifoam agent, and osmotic pressure; and the chemical pollutants ethanol and phenol) under resting cell conditions. We employed the resting cell state in this work for two reasons. First, it was simple; that is, the resting cell state can be considered a pseudo-steady state much like the steady state in a continuous culture, in which a perturbation is easily detected as a transient response from the steady state. Second, based on the recent work of Rupani et al. (41), we might have expected a minimal response or no response in resting cells, which would have provided a more stringent test of GFPuv sensitivity. In their work performed with the grpE promoter used to drive luciferase, Rupani et al. observed no response in cells entering the stationary phase of a batch culture or in cells cultivated in a chemostat at dilution rates less than 0.25 h−1 (41). They also found that the dnaK-regulated luciferase responses were typically 1 order of magnitude lower than the grpE-regulated responses. Our results, therefore, are noteworthy in that significant responses for ς32, DnaK, and ClpB were obtained with resting cells. Like grpE, the genes which we studied normally respond most to stress when the cells are in a growth phase (21, 26, 58). One possible explanation for the results is that the GFP system requires expression of only one protein instead of the five lux genes of the V. fischeri operon. In addition, luciferase requires ATP for bioluminescence, while GFP requires only molecular oxygen (24). Thus, at low growth rates or in resting cells, there may be less available ATP, while molecular oxygen should be abundant. Also, there was little confounding increase in fluorescence due to increasing cell mass, as would be the case in growing cultures.
A negative aspect of the resting cell state was the relatively long response time. In part, however, the delayed response was due to the delay in fluorescence following GFP expression (1, 13). The first significant increase in the fluorescence of the ς32 strain was roughly 2 h after the heat shock. Interestingly, a more dramatic increase followed after 4 h (Fig. 2A). This biphasic profile was similar to the profile obtained by Tomoyasu et al. for a mutant strain lacking FtsH (HflB), the membrane-bound protease that degrades ς32 (49). These authors observed an initial increase in the level of ς32 within 5 min after the heat shock, which was followed later (∼20 min) by another increase and ultimately by a sustained threefold accumulation. In addition to demonstrating that FtsH was involved in destabilizing ς32, Tomoyasu et al. showed that ς32 expression occurred for at least 1 h after the heat shock. Our rpoH::gfpuv results are qualitatively similar except for the time scale. Interestingly, by shifting the GFP fluorescence data by ∼95 min, we improved the agreement between our results and those of Tomoyasu et al. (49). Similar results were obtained previously for GFP (1, 40), and in this study a ∼95-min shift improved the correlation between GFPuv fluorescence and the DnaK level (Fig. 3B).
It was also interesting that two chemical pollutants, ethanol and phenol, were efficient inducers of the heat shock proteins (Fig. 5), suggesting the stress probe strains may be useful as noninvasive biological sensors for chemical detection and tests for toxicity of specific pollutants (51). Naturally, the responses might be dependent on the dose of the imposed stress. In order for a compound to be useful as a biological sensor, a significant response should occur at a dose that is less than the dose which is toxic to E. coli or otherwise inhibits cell growth (e.g., 4% [vol/vol] ethanol [41]). Conversely, there may be a minimum threshold level needed to elicit a response (41, 51). For example, there was no dnaK::gfp response at ethanol concentrations less than 1% soon after addition (Fig. 4). However, the response was always linear at concentrations ranging from 2 to 4%, and excellent results were obtained at the last sample times examined (8 h). Indeed, in each case, the fluorescence of the stressed cells was greater than the fluorescence of the controls, and the difference increased with time because of both the relatively long cyclization time of the GFP chromophore and the stability of GFPuv.
Although different stress response profiles were obtained for the three plasmids after different stresses were imposed, several general trends were evident. First, the fluorescence intensity of the ς32 heat shock transcription factor was greatest, followed by the fluorescence intensity of the chaperone DnaK and the fluorescence intensity of the protease subunit ClpB. Furthermore, only the ethanol shock resulted in a significant difference between DnaK fluorescence and ClpB fluorescence. The order of the relative cellular GFPuv levels under normal cell growth conditions was similar to the order under stressed conditions (ς32 > ClpB > DnaK [data not shown]). Also, we found that the fluorescence followed a similar trend in the prestress samples (ς32 > ClpB > DnaK) (Fig. 2). These results indicated that there was more constitutive or background transcription and/or translation of ς32, which perhaps reflected the numbers of promoter elements within the upstream regulatory regions of the plasmids used (p3, p4, and p5 of rpoH for rpoH::gfp versus p1 and p2 of dnaK for dnak::gfp).
We also found that the heat shock transcription factor ς32 generally responded fastest to stress, which is consistent with the role of this factor as the principal regulator of heat shock protein synthesis (36, 58) and the specific regulator of DnaK and ClpB (2, 28). Our ς32 results were consistent with the results of Nagai et al. (36) and other workers (22, 47), who demonstrated that ς32 is upregulated upon heat shock and that rapid accumulation of ς32 is not just due to increased stability (through sequestration of DnaJ, DnaK, and GrpE in protease pathways) (29). Interestingly, the principal regulatory component for upregulation (the translational control region) was not in our plasmid construct. This ς32 result was observed in experiments in which highly affected sources of stress, such as heat shock, osmotic pressure, ethanol, and phenol, were used.
In summary, one of the three stress probes which we constructed, the ς32 plasmid, responded more rapidly and to a larger extent than the others, suggesting that it has greater utility for discriminating between elicitors of the heat shock response. Also, although the GFP-based fluorescence method described here is rather slow, much greater sensitivity was obtained than could be obtained by Western analysis, which requires cell disruption, SDS-PAGE, electrotransfer, blotting, and imaging.
ACKNOWLEDGMENTS
This work was supported by grant BCS-9157852 from National Science Foundation to G.R., by grant BCS-9319366 from the National Science Foundation to W.E.B., by U.S. Army ERDEC contract DAAM01-96-C-0037 to W.E.B., and by a grant from the Korea Science and Engineering Foundation, Republic of Korea, to H.J.C.
REFERENCES
- 1.Albano C R, Randers-Eichhorn L, Chang Q, Bentley W E, Rao G. Quantitative measurement of green fluorescent protein expression and chromophore cyclization. Biotechnol Tech. 1996;10:953–958. [Google Scholar]
- 2.Andersson L, Yang S J, Neubauer P, Enfors S O. Impact of plasmid presence and induction on cellular responses in fed batch cultures of Escherichia coli. J Biotechnol. 1996;46:255–263. doi: 10.1016/0168-1656(96)00004-1. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell J C, Craig E A. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci USA. 1984;81:848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley W E, Davis R H, Kompala D S. Dynamics of induced CAT expression in E. coli. Biotechnol Bioeng. 1991;38:749–760. doi: 10.1002/bit.260380709. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum S, Bailey J E. Plasmid presence changes the relative levels of many host cell proteins and ribosome components in recombinant Escherichia coli. Biotechnol Bioeng. 1991;37:736. doi: 10.1002/bit.260370808. [DOI] [PubMed] [Google Scholar]
- 6.Blom A, Harder W, Matin A. Unique and overlapping pollutant stress proteins of Escherichia coli. Appl Environ Microbiol. 1992;58:331–334. doi: 10.1128/aem.58.1.331-334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botsford J L. Analysis of protein expression in response to osmotic stress in Escherichia coli. FEMS Microbiol Lett. 1990;72:355–360. doi: 10.1111/j.1574-6968.1990.tb03916.x. [DOI] [PubMed] [Google Scholar]
- 8.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–672. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 9.Calendar R, Erickson J W, Halling C, Nolte A. Deletion and insertion mutations in the rpoH gene of Escherichia coli that produce functional sigma-32. J Bacteriol. 1988;170:3479–3484. doi: 10.1128/jb.170.8.3479-3484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashel M, Rudd K E. The stringent response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1410–1438. [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Cowing D W, Bardwell J C, Craig E A, Woolford C, Hendrix R W, Gross C A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci USA. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crameri A, Whitehorn E A, Tate E, Stemmer W P C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 14.Ezaki B, Ogura T, Mori H, Niki H, Hiraga S. Involvement of DnaK protein in mini-F plasmid replication: temperature-sensitive seg mutations are located in the dnaK gene. Mol Gen Genet. 1989;218:183–189. doi: 10.1007/BF00331267. [DOI] [PubMed] [Google Scholar]
- 15.Gallant J A. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- 16.Georgopoulos C, Ang D, Liberek K, Zylicz M. Properties of the Escherichia coli heat shock proteins and their role in bacteriophage growth. In: Motimoto R I, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 191–221. [Google Scholar]
- 17.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Motimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock protein and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 18.Goff S A, Goldberg A L. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 21.Grossman A D, Taylor W E, Burton Z F, Burgess R R, Gross C A. Stringent response in Escherichia coli induces expression of heat shock proteins. J Mol Biol. 1985;186:357–365. doi: 10.1016/0022-2836(85)90110-x. [DOI] [PubMed] [Google Scholar]
- 22.Grossman A D, Straus D B, Walter W A, Gross C A. ς32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- 23.Harcum S W, Bentley W E. Response dynamics of 26, 34, 39, 54, and 80 kDa proteases in induced cultures of recombinant Escherichia coli. Biotechnol Bioeng. 1993;42:675–685. doi: 10.1002/bit.260420602. [DOI] [PubMed] [Google Scholar]
- 24.Heim R, Tsien R Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 25.Hendrick J P, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 26.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 27.Herman C, Thevenet D, D’Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins D E, Auger E A, Matin A. Role of RpoH, a heat shock regulator protein, in Escherichia coli: effect of alternations of the outer membrane permeability on sensitivity of environmental toxicants. Toxic Assess. 1991;5:253–264. [Google Scholar]
- 29.Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of ς32 in Escherichia coli. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock ς factor (ς32) J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosinski M J, Rinas U, Bailey J E. Isopropyl-β-d-thiogalactopyranoside influences the metabolism of Escherichia coli. Appl Microbiol Biotechnol. 1992;36:782–784. [Google Scholar]
- 32.Majewski R A, Domach M M. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol Bioeng. 1990;35:732–738. doi: 10.1002/bit.260350711. [DOI] [PubMed] [Google Scholar]
- 33.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 34.Maurizi M R. Proteases and protein degradation in Escherichia coli. Experimentia. 1992;48:178–200. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- 35.Muffler A, Barth M, Marshall C, Hengge-Aronis R. Heat shock regulation of ςs turnover: a role for DnaK and relationship between stress responses mediated by ςs and ς32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai H, Yuzawa H, Yura T. Interplay of two cis-acting mRNA regions in translational control of ς32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt F C, VanBogelen R A. Positive regulatory gene for temperature-controlled proteins in Escherichia coli. Biochem Biophys Res Commun. 1981;100:894–900. doi: 10.1016/s0006-291x(81)80257-4. [DOI] [PubMed] [Google Scholar]
- 38.Pizer L I, Merlie J P. Effects of serine hydroxamate on phospholipid synthesis in Escherichia coli. J Bacteriol. 1973;114:980–987. doi: 10.1128/jb.114.3.980-987.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez D M, Bentley W E. Fed-batch feeding and induction policies that improve foreign protein synthesis and stability by avoiding stress response. Biotechnol Bioeng. 1995;47:596–608. doi: 10.1002/bit.260470512. [DOI] [PubMed] [Google Scholar]
- 40.Randers-Eichhorn L, Albano, C. R. C R, Sipior J, Bentley W E, Rao G. On-line green fluorescent protein sensor with LED excitation. Biotechnol Bioeng. 1997;55:921–927. doi: 10.1002/(SICI)1097-0290(19970920)55:6<921::AID-BIT9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 41.Rupani S, Gu M B, Konstantinov K B, Dhurjati P S, Van Dyk T K, LaRossa R A. Characterization of the stress response of a bioluminescent biological sensor in batch and continuous cultures. Biotechnol Prog. 1996;12:387–392. doi: 10.1021/bp960015u. [DOI] [PubMed] [Google Scholar]
- 42.Sedger L, Ruby J. Heat-shock response to vaccinia virus infection. J Virol. 1994;68:4685–4689. doi: 10.1128/jvi.68.7.4685-4689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman M Y, Goldberg A L. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 1992;11:71–78. doi: 10.1002/j.1460-2075.1992.tb05029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Squires C L, Pedersen S, Ross B M, Squires C L. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St. John A C, Goldberg A L. Effects of starvation for potassium and other inorganic ions on protein degradation and ribonucleic acid synthesis in Escherichia coli. J Bacteriol. 1980;143:1223–1233. doi: 10.1128/jb.143.3.1223-1233.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stormo G D. Translation initiation. In: Reznikoff W, Gold L, editors. Maximizing gene expression. Stoneham, Mass: Butterworths Publishers; 1986. pp. 201–206. [Google Scholar]
- 47.Straus D B, Walter W A, Gross C A. The heat shock response of E. coli is regulated by changes in the concentration of ς32. Nature (London) 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 48.Straus D B, Walter W A, Gross C A. Escherichia coli heat shock gene mutants are defective in proteolysis. Genes Dev. 1988;2:1851–1858. doi: 10.1101/gad.2.12b.1851. [DOI] [PubMed] [Google Scholar]
- 49.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H J, Rutman A B, Oppenheim A, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanBogelen R A, Kelly P M, Neidhardt F C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati, P. S. P S, Larossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Dyk T K, Reed T R, Vollmer A C, Larossa R A. Synergistic induction of the heat-shock response in Escherichia coli by simultaneous treatment with chemical inducers. J Bacteriol. 1995;177:6001–6004. doi: 10.1128/jb.177.20.6001-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M Y, Pulliam T R, Valle M, Vakharia V N, Bentley W E. Kinetic analysis of alkaline protease activity, recombinant protein production and metabolites for infected insect (Sf9) cells under different DO levels. J Biotechnol. 1996;46:243–254. [Google Scholar]
- 54.Welch W. The mammalian stress response: cell physiology and biochemistry of stress proteins. In: Motimoto R I, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 223–278. [Google Scholar]
- 55.Yancey P H, Clark M E, Hand S C, Bowlus R D, Somero G N. Living with water stress: evolution of osmolytic systems. Science. 1982;217:1214–1217. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 57.Yura T, Tobe T, Ito K, Osawa T. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc Natl Acad Sci USA. 1984;81:6803–6807. doi: 10.1073/pnas.81.21.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yura T, Nagai H, Mori H. Regulation of the heat shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]