Abstract
Alcohol use is a contributor in the premature deaths of approximately 3 million people annually. Among the risk factors for alcohol misuse is circadian rhythm disruption; however, this connection remains poorly understood. Inhibition of the circadian nuclear receptor REV-ERBα is known to disrupt molecular feedback loops integral to daily oscillations, and impact diurnal fluctuations in the expression of proteins required for reward-related neurotransmission. However, the role of REV-ERBα in alcohol and substance use-related phenotypes is unknown. Herein, we used a Rev-erbα knockout mouse line and ethanol two-bottle choice preference testing to show that disruption of Rev-erbα reduces ethanol preference in male and female mice. Rev-erbα null mice showed the lowest ethanol preference in a two-bottle choice test across all genotypes, whereas there were no ethanol preference differences between heterozygotes and wildtypes. In a separate experiment, alcohol-consuming wildtype C57Bl/6N mice were administered the REV-ERBα/β inhibitor SR8278 (25 mg/kg or 50 mg/kg) for 7 days and alcohol preference was evaluated daily. No differences in alcohol preference were observed between the treatment and vehicle groups. Our data provides evidence that genetic variation in REV-ERBα may contribute to differences in alcohol drinking.
Keywords: alcohol, circadian rhythms, REV-ERBα, NR1D1, addiction
1. Introduction
Alcohol use is a contributing factor in the premature deaths of approximately 3 million people annually [1]. It is estimated that 23.9% of US citizens engage in binge drinking (4–5 or more drinks during one occasion) and 5.8% in heavy drinking (binge drinking for 5 or more days in a 30-day period), thus contributing to profound morbidity and mortality levels [2]. Those with perturbed circadian cycles, such as shift workers [3], adolescents [4], and frequent trans-meridian travelers [5,6], have greater odds of hazardous alcohol use than the general population. Similarly, chronic alcohol users and those in remission experience circadian rhythm and sleep disturbances, which may also contribute to the risk of relapse [7]. Collectively, these observations suggest that circadian desynchrony shares a reciprocal relationship with adverse alcohol consumption patterns and is a risk factor in problematic alcohol consumption.
Clinical and preclinical studies illustrate that alcohol use behaviors are linked with variations in genes of the cellular circadian mechanism. Briefly, the positive arm of the circadian mechanism involves Complex Circadian Locomotor Output Cycles Kaput (CLOCK) or its paralogue Neuronal Per-Arnt-Sim Domain Protein 2 (NPAS2), which heterodimerize with Brain and Muscle Arnt-like proteins (BMAL1, BMAL2) to drive transcriptional regulation of gene expression through binding enhancer elements. This in turn drives the expression of proteins that act as feedback regulators of CLOCK-NPAS2/BMAL, including the primary negative feedback loop proteins Cryptochrome (CRY) and Period (PER1, 2, and 3), and the accessory proteins REV-ERBα (otherwise known as Nuclear Receptor Subfamily 1 Group D Member 1). The 24 h day and night cycling of the circadian mechanism and its effects on organ pathophysiology have been well reviewed (e.g., [8,9,10,11,12]). Important to this study, polymorphisms of CLOCK/NPAS2 [13,14], BMAL [15,16], PER genes [15,17,18,19,20,21,22], and REV-ERBα [23] predict alcohol use phenotypes in humans independently or co-morbidly with other conditions (e.g., depression, stress). Mechanistically, elimination or downregulation of Clock [24], Per1 [21,25], and Per2 [22,25,26,27] is associated with changes in ethanol consumption patterns and/or sensitivity in rodent models of drinking behavior. Meanwhile, global Clock [24], Per1 [21,25], and Per2 [22,25] deficiency increases alcohol self-administration and preference; a triple knockdown of Per1, Per2, and Clock localized to the nucleus accumbens reduces alcohol consumption in a mouse model of binge drinking [28]. Together, these findings suggest that circadian biology and circadian mechanism gene mutations play an important role in predicting alcohol consumption phenotypes.
Although many preclinical studies have sought to characterize the role of the core circadian genes within the primary feedback loop on ethanol drinking and response [21,22,24,25,26,27], there is a lack of understanding of how key accessory circadian proteins influence alcohol-related behaviors. Factors such as REV-ERBα are of particular interest because they could be important contemporary targets of drug therapies. Given REV-ERBα gene-based correlation with alcohol use [23] and that agonist administration impairs cocaine reward sensitivity [29], it would seem to be an important target in the treatment of alcohol use disorders as well. Moreover, several studies suggest that REV-ERBα directly regulates dopaminergic neurotransmission, which is key to reward response [30]. In wildtype mice, striatal levels of dopamine are lower at subjective dusk than dawn [31] and the nucleus accumbens exhibits daily fluctuations in its expression of tyrosine hydroxylase (TH) and dopamine transporter, which are required for dopamine synthesis and clearance from the synaptic cleft, respectively [32]. However, genetic or pharmacological inhibition of REV-ERBα in mice results in striatal hyperdopaminergia and ablation of Th mRNA transcript periodicity in the ventral midbrain [31]. Rev-erbα knockout mice also demonstrate circadian-independent behavioral phenotypes indicative of hippocampal dysfunction and elevated dopamine synthesis, turnover, and dopamine receptor D1 mRNA transcript levels in the hippocampus [33]. Upregulation of genes involved with dopaminergic signaling in Rev-erbα null mice is consistent with phenotypes in Clock mutant mice that also show hyperdopaminergic behavioral phenotypes and elevated TH levels, as well as enhanced drug reward [34]. Considering the critical role of dopaminergic activity in drug response and addiction, it seems likely that the circadian mechanism factor REV-ERBα is a critical modulator of alcohol-related phenotypes; thus, being a druggable target, it is a compelling focus of investigation.
Given the role of REV-ERBα in the circadian feedback loop [35], that agonist administration impairs drug reward sensitivity [29], and the similarities between REV-ERBα-depleted rodents and Clock mutants in the altered expression of genes related dopaminergic signaling [33,34], we hypothesized that functional disruption of Rev-erbα would elevate ethanol consumption and preference. In the first phase of this study, we investigated ethanol two-bottle choice preference in Rev-erbα null male and female mice. This experiment was complemented with pharmacological manipulation of REV-ERB via daily injections of the REV-ERBα/β antagonist SR8278 [36].
2. Results
2.1. Genetic Deletion of Rev-erbα Decreases Ethanol Preference and Consumption
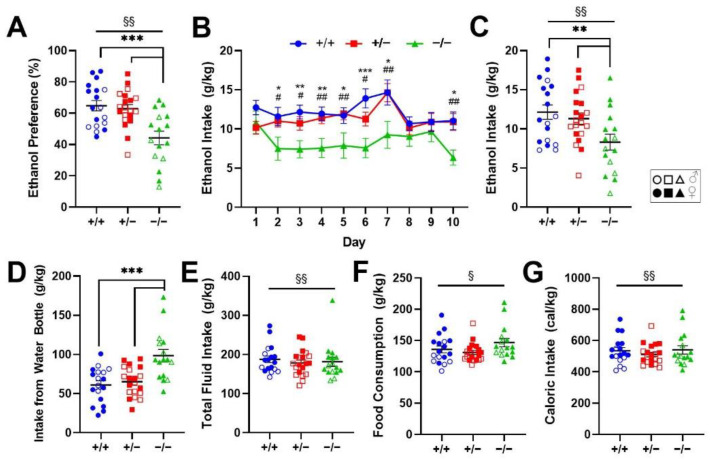
To evaluate the role of Rev-erbα in voluntary ethanol intake, we employed the two-bottle choice paradigm in a line of Rev-erbα knockout mice and compared ethanol preference and consumption across male and female wildtype (Rev-erb+/+; n = 7 males, 11 females), heterozygous (Rev-erb+/−; n = 10 males, 10 females), and knockout (Rev-erb−/−; n = 5 males, 11 females) mice. In the 10-day period following development of stable ethanol preference (less than 15% variance in ethanol preference across 3 consecutive days), a significant effect of genotype was observed on ethanol preference (F2, 48 = 11.96, p = 6.10 × 10−5). Post hoc analysis revealed a significant reduction in ethanol preference in Rev-erb−/− (44.18 ± 4.34%) compared to Rev-erb+/+ (64.83 ± 3.26%, p = 2.32 × 10−4) and Rev-erb+/− counterparts (62.89 ± 2.65%, p = 2.10 × 10−4; Figure 1A). Consistent with prior research [10,37,38], a significant main effect of sex was also observed (F1, 48 = 5.55, p = 0.02): ethanol preference was greater in females compared to males regardless of genotype (60.56 ± 3.03% vs. 54.26 ± 3.35%; Figure 1A). However, there were no significant sex × genotype (F2, 48 = 0.78, p = 0.46), sex × day (F3.80, 182.51 = 1.14, p = 0.17), or sex × genotype × day (F7.61, 182.51 = 0.66, p = 0.72) interactions in ethanol preference.
Figure 1.
Genetic deletion of Rev-erbα reduces ethanol preference and consumption in male and female mice. Preference for 10% ethanol is (A) reduced in Rev-erb−/− mice relative to Rev-erb+/+ and Rev-erb+/− mice (p < 0.001 ***) and elevated in females compared to males (p < 0.01 §§). Ethanol intake normalized to body mass is also (B) reduced in Rev-erb−/− mice compared to the other groups on most testing days (p < 0.05 *, < 0.01 **, < 0.001 *** vs. Rev-erb+/+, p < 0.05 #, <0.01 ## vs. Rev-erb+/−) and (C) greater in females compared to males according to daily average consumption (p < 0.01 §§). (D) Water consumption normalized to body mass was greater in Rev-erb−/− compared to the Rev-erb+/+ and Rev-erb+/− groups (p < 0.001 ***). Average daily (E) total fluid intake (p < 0.01 §§), (F) food intake (p < 0.05 §), (G) calorie intake (p < 0.01 §§) normalized to body mass was greater in females than males.
Daily ethanol consumption across the 10-day period was also evaluated according to genotype and sex. Again, a significant main effect of genotype was detected in both daily ethanol (F2, 48 = 6.537, p = 3.08 × 10−3) and water intake from the water-only bottle (F2, 48 = 9.857, p = 2.59 × 10−4), with the knockouts varying significantly from the wildtype and heterozygous groups in both parameters. Average daily ethanol consumption was significantly reduced in Rev-erb−/− mice (8.31 ± 1.01 g/kg) compared to Rev-erb+/+ (12.13 ± 0.92 g/kg, p = 5.17 × 10−3) and Rev-erb+/− (11.31 ± 0.73 g/kg, p = 0.01) mice. A significant day × genotype interaction was observed (F9.26, 222.28 = 2.34, p = 0.01), with post hoc testing revealing lower ethanol consumption in the Rev-erb−/− group compared to Rev-erb+/+ and Rev-erb+/− on most testing days (Figure 1B,C). As expected, there was a significant effect of sex on ethanol intake (F1,48 = 10.94, p = 1.79 × 10−3): females voluntarily consumed more ethanol than males (11.81 ± 0.71 g/kg vs. 9.07 ± 0.73 g/kg; Figure 1C). In contrast, water consumption from the water-only bottle was highest in the Rev-erb−/− group (98.34 ± 8.06 g/kg), followed by the Rev-erb+/− group (65.10 ± 4.15 g/kg, p = 9.49 × 10−4) and the Rev-erb+/+ group (60.99 ± 5.27 g/kg, p = 6.72 × 10−4; Figure 1D). Consistent with these observations, there was a significant main effect of sex on total fluid consumption: females consumed more liquid than males (F2, 48 = 9.42, p = 3.53 × 10−3; 163.35 ± 8.01 g/kg vs. 194.81 ± 6.39 g/kg; Figure 1E). We also observed an effect of day on total fluid consumption (F4.42, 211.91 = 6.82, p = 3.44 × 10−9; data not shown) and greater consumption observed on day 1 compared to days 2, 3, and 4 (p < 0.05), and on day 7 compared to most other days (p < 0.05), possibly explained by cage cleaning on those days. However, we did not observe any significant main effect of genotype or any interactions on total fluid consumed.
Body mass was also recorded daily and analyzed. There was a significant effect of sex on body mass: weight was lower in females (F1, 48 = 218.43, p = 1.72 × 10−19; data not shown). While there was no significant main effect of genotype on body mass (F2,48 = 0.41, p = 0.67), there were also significant day × genotype (F6.37,152.99 = 3.52, p = 2.22 × 10−3) and sex × genotype (F2, 48 = 4.72, p = 0.01) interactions, though neither survived multiple comparison post hoc testing (data not shown). Average daily chow consumption and caloric intake across the 10 days significantly differed according to sex, with females consuming more chow (F1, 48 = 5.05, p = 0.03; 143.12 ± 4.13 g/kg vs. 128.71 ± 3.60 g/kg) and more calories (F1,48 = 8.28, p = 5.96 × 10−3; 554.78 ± 15.87 Cal/kg vs. 488.25 ± 13.15 Cal/kg) per day on average (Figure 1F,G). There were no significant main effects of genotype (chow consumption: F2,48 = 2.00, p = 0.15; caloric intake: F2,48 = 0.23, p = 0.80) or sex × genotype (chow consumption: F2,48 = 1.24, p = 0.30; caloric intake: F2,48 = 0.33, p = 0.33) interaction on food consumption patterns, suggesting no effect of genotype on appetitive behavior.
2.2. Pharmacological Inhibition of REV-ERBα/β Does Not Affect Ethanol Preference or Intake in Male and Female Mice
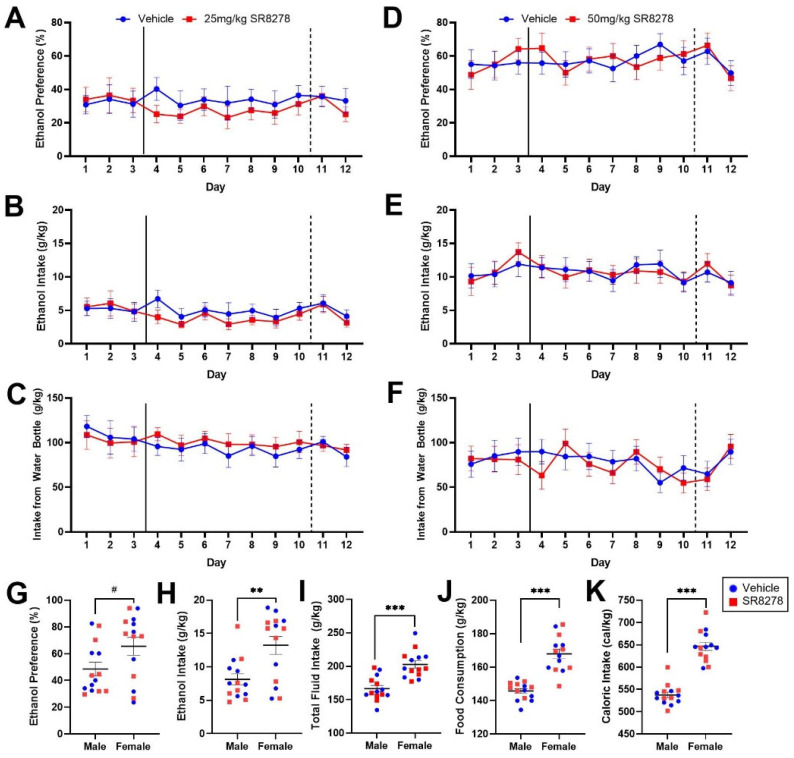
To further evaluate the role of REV-ERBα in voluntary ethanol consumption and ethanol preference, we attempted to recapitulate the findings in the Rev-erbα null mouse line using pharmacological means. Using a cohort of male C57Bl/6N mice (n = 8/group) with stable ethanol preference in the two-bottle choice paradigm, we administered a vehicle solution or the REV-ERBα/β antagonist SR8278 daily and assessed changes to ethanol preference, liquid consumption, body mass, and food intake. Mice were habituated to vehicle injection for 3 days prior to drug administration, after which vehicle solution or 25 mg/kg of SR8278 was administered for 7 days; recordings continued for 2 post-injection days. On each day, SR8278 or vehicle were administered at zeitgeber time (ZT) 06, when REV-ERBα levels are highest [39]. Overall, there were no significant effects of treatment on ethanol preference (F4.97, 69.53 = 1.04, p = 0.40), ethanol consumption (F4.90, 68.62 = 1.153, p = 0.34), water consumption from the water-only bottle (F4.90, 68.63 = 0.60, p = 0.70), or total fluid intake (F11, 154 = 0.317, p = 0.98) across the experimental period (Figure 2A–C).
Figure 2.
Pharmacological inhibition of REV-ERBα/β with 25 mg/kg (A–C) or 50 mg/kg (D–K) of SR8278 did not alter ethanol preference, ethanol intake, or water intake from the water-only bottle in mice. In male mice, 25 mg/kg of SR8278 did not alter (A) ethanol preference, (B) ethanol intake normalized to body mass, or (C) water intake from the water-only bottle normalized to body mass compared to vehicle treatment. In male and female mice, 50 mg/kg of SR8278 did not alter (D) ethanol preference, (E) ethanol intake normalized to body mass, or (F) water intake from the water-only bottle normalized to body mass compared to vehicle treatment. Irrespective of treatment (50 mg/kg SR8278 or vehicle), there was a (G) trend toward a significant effect of sex on ethanol preference (p = 0.06), with females showing greater average daily (H) ethanol intake (p < 0.01), (I) total fluid intake (p < 0.001), (J) food intake (p < 0.001), and (K) caloric intake (p < 0.001) compared to males when normalized to body mass. p = 0.06 #, p < 0.01 **, p < 0.001 ***. (A–F) Solid lines indicate time of onset of SR8278 injection; dashed lines indicate time of cessation of daily injections.
Although not significant, ethanol preference and consumption were marginally lower in male mice treated with 25 mg/kg of SR8278 than the vehicle group across the injection period (Figure 2A,B). We therefore extended these investigations with a naïve cohort of male and female C57Bl/6N mice (n = 7/group/sex) injected with 50 mg/kg of SR8278 to assess whether there might be an effect of SR8278 dose or a potential interaction with sex on alcohol drinking. Comparable with our previous results, we observed no significant differences between vehicle and treatment groups in ethanol preference (F6.17, 148.14 = 0.99, p = 0.46), ethanol intake (F6.41, 153.87 = 0.45, p = 0.85), or water intake from the water-only bottle (F5.72, 137.35 = 1.26, p = 0.28) during the injection period (Figure 2D–F). Females consumed more ethanol (F1, 24 = 9.66, p = 4.79 × 10−3; 13.22 ± 1.34 g/kg vs. 8.13 ± 0.83 g/kg), chow (F1,24 = 27.72, p = 2.12 × 10−5; 166.90 ± 2.99 g/kg vs. 146.74 ± 2.34 g/kg), total fluid (F1,24 = 23.82, p = 5.63 × 10−5; 202.78 ± 5.26 g/kg vs. 166.45 ± 5.26 g/kg), and calories (F1, 24 = 78.09, p = 5.19 × 10−9, 644.40 ± 8.26 kcal/g vs. 539.20 ± 8.62 kcal/g) per gram of body weight than males, and there was a trend toward a significant difference in ethanol preference according to sex (F1, 24 = 3.881, p = 0.06, 65.58 ± 6.62% vs. 48.58 ± 5.02%; Figure 2G–K). In addition, there was a sex × day interaction on total fluid intake (F11, 264 = 2.96, p = 1.02 × 10−3), with females consuming more fluid than males on most days regardless of treatment (p < 0.05; data not shown).
3. Discussion
Excessive alcohol use is a major public health concern and is especially prevalent among those experiencing circadian disruptions, such as frequent travelers, shift workers, and adolescents [3,4,5,6]. Findings from the present study indicate that expression of Rev-erbα (a gene encoding an auxiliary circadian constituent) in mice might promote ethanol preference and self-administration. To our knowledge, this is the first study to causally suggest a function for Rev-erbα in patterns of ethanol use. The finding that Rev-erbα deletion reduces ethanol self-administration in male and female mice is surprising considering prior observations of REV-ERB modulation on reward response; notably, treatment of mice with the REV-ERBα/β agonist SR9011 was previously shown to attenuate wheel running without affecting overall locomotion [40], and impair cocaine-conditioned place preference [29]. Together with our findings, this may suggest that aberrant REV-ERBα activity in either direction suppresses reward-related behavior, or that the unique actions of dual REV-ERBα/β modulation may differ from the actions of either auxiliary circadian constituent alone.
We hypothesized that Rev-erbα knockout mice would consume and prefer ethanol to a greater degree than wildtype mice considering that Rev-erbα deficiency results in a hyperdopaminergic profile in reward-related brain regions, an observation consistent with Clock mutant mice that also demonstrate greater drug intake, drug self-administration, and reductions in Rev-erbα mRNA transcripts [41,42]. However, our results suggest that Rev-erbα knockout mice may find ethanol less rewarding or are less motivated to consume ethanol despite their behavioral and neurobiological similarities to Clock mutants in other domains. REV-ERBs participate in negative feedback of their expression by suppressing their own transcription [43] as well as transcription of Bmal1 [35], whose protein product heterodimerizes with CLOCK to transcriptionally upregulate components of the circadian loop, including Rev-erbs. Presumably, the core circadian loop would remain intact in Rev-erbα-/- mice, unlike Clock mutants. While both genetic lines demonstrate some level of disrupted circadian rhythmicity in activity and clock machinery, Clock mutants have severe circadian arrhythmia under constant darkness [44] and reduced expression of circadian mRNA transcripts [45,46,47] in the suprachiasmatic nucleus, whereas Rev-erbα knockouts show a more modest effect on periodicity [48] and elevated circadian clock machinery transcript levels [35,49,50], including Bmal1. Interestingly, whole-brain BMAL1 expression in male mice is negatively correlated with genetic vulnerability to consuming ethanol in a drinking in the dark paradigm [51], and striatal deletion of Bmal1 enhances daily ethanol consumption in males while decreasing consumption in females [52]. These findings support that reduced ethanol consumption and preference by Rev-erbα deletion could be caused by elevated BMAL1 expression. Although more investigations must be conducted to determine the scope of its impact on downstream effector proteins, our findings in combination with others suggest that REV-ERBα could be a potential candidate for the development of novel pharmacotherapies targeting ethanol misuse.
Drug response and addiction liability are principally mediated by dopaminergic signaling through mesocorticolimbic pathways. Dopaminergic dysregulation in the rodent brain by REV-ERBα inhibition has been observed through irregular expression of dopamine-related behaviors, elevated dopamine turnover, higher expression of mRNA and protein related to dopamine metabolism, and disrupted dopaminergic cyclicity in the ventral midbrain; these characteristics culminate in hyperdopaminergia in Rev-erbα−/− mice [31,33]. Notably, elevating tonic dopamine neurotransmission by optogenetic stimulation of the VTA is known to attenuate self-administration in ethanol-drinking rats [53,54]. However, alcohol response, sensitivity, and reward are influenced directly by multiple neurotransmitter systems, notably at GABA receptors where ethanol acts to produce some of its acute and chronic effects [55]. Two recent studies revealed that Rev-erbs affect oscillatory GABAergic patterns in the brain, with Rev-erbα disruption elevating tonic GABA currents in the hippocampus and impairing the oscillatory patterns of GABA reuptake proteins in the hippocampus and cortex [56,57]. It is currently unclear if decreased ethanol preference by Rev-erbα deletion is primarily due to impaired dopamine activity affecting reward response or by ancillary systems that participate in ethanol response directly, such as GABAergic neurotransmission. Future investigations of this mouse line and vulnerability to other drugs and naturally rewarding substances will help elucidate whether Rev-erbα contributes to ethanol vulnerability specifically, or whether expression of this gene is relevant to generalized drug reward.
In our attempt to establish the effect of one such pharmacotherapeutic strategy, treatment of C57Bl/6N mice with 25 mg/kg or 50 mg/kg of the REV-ERBα/β antagonist SR8278 did not recapitulate the decreased ethanol intake and preference observed in our genetic knockout model. Considering the rapid metabolism of SR8278 in the brain following 25 mg/kg i.p. injection [57], it is likely that a once daily injection of SR8278 is insufficient to affect reward-related functions over the course of a day despite administration during peak Rev-erbα expression. Moreover, since we did not assess any other behavioral end-points to assess the effects of SR8278, it is possible that the doses tested here were not behaviorally relevant. It is notable that most studies of Rev-erbα on behavior and reward exclusively use genetic models, with only one study comparing the genetic model with bidaily intraperitoneal REV-ERBα/β agonist injections [29], and another microinfusing SR8278 directly into the ventral midbrain 4 h prior to behavioral testing [31]. While our expectation that pharmacological inhibition of REV-ERBs would recapitulate the findings of Rev-erbα deletion, future investigations may benefit from the use of the agonist previously observed to affect behavior, or utilizing more invasive, direct methods of antagonist delivery. However, it is also possible that Rev-erbα expression during development may affect future ethanol drinking patterns, which would not be apparent with pharmacological inhibition restricted to adulthood. In addition, the role of Rev-erbβ in circadian circuitry has been relatively underexplored compared to Rev-erbα, and while Rev-erbβ is considered functionally redundant to Rev-erbα, we cannot discount a unique function of Rev-erbβ in ethanol intake vulnerability, warranting specific pharmacological manipulation of each target alone in future studies. Lastly, we did not assess drinking microstructure (e.g., bout duration, licks, etc.), which may be assessed in future studies to establish the effects of REV-ERBα/β pharmacological manipulation on alcohol drinking [58].
It is also possible that decreased ethanol preference in our knockout model is due to an intermediate behavioral phenotype. For instance, there could be enhanced sensitivity to the effects of ethanol within the knockout mice, which may mediate intake. As preference for bitter tasting substances has not been characterized in this line, we also cannot discount a potential difference in taste perception according to genotype. However, one previous study found that Rev-erbα knockout mice show enhanced consumption of high caloric sweet food but no differences in the intake of the non-caloric sweetener saccharine [59]. This may suggest no differences in sweet palatability according to genotype, although further preference studies that include high caloric sweet alcohol, low caloric sweet alcohol, and unsweetened alcohol could provide insights into genetic vulnerability to consuming high caloric sweet-tasting (e.g., cocktails, coolers) versus bitter-tasting (e.g., hard liquors, beer) alcoholic beverages. Moreover, it would be important to assess whether REV-ERBα disruption impacts the effects of alcohol on righting reflex and other alcohol-related effects. Previous findings using transgenic mice with alterations in other diurnal function-related genes have some effects on alcohol loss of righting reflex (e.g., Clock), whereas Per1 and Per2 disruption did not impact loss of righting [24,25]. Finally, multiple studies suggest that Rev-erbα affects mood, and REV-ERBα disruption diminishes depression- and anxiety-like phenotypes in mice [31,60]. A positive relationship between mood disorders and alcohol use has been well-documented [61], suggesting that Rev-erbα knockout mice could show lower ethanol preference than wildtype counterparts due to greater mood resilience.
In summary, this work establishes a novel causative association between Rev-erbα and ethanol preference in a genetic model of alcohol use vulnerability. These findings provide support for previous correlative human studies highlighting REV-ERBα polymorphisms in the risk for developing alcohol use disorders. This is the first study to provide evidence that Rev-erbα expression may alter drug use vulnerability, possibly by affecting the expression of core clock components, suggesting that REV-ERBα could be a future pharmacological target for mitigating problematic substance use.
4. Materials and Methods
4.1. Ethics Statement
All experiments herein were approved by the University of Guelph Institutional Animal Care and Use Committee (AUP#3922, 3858) and were performed in accordance with the guidelines set forth by the Canadian Council on Animal Care (1993).
4.2. Two-Bottle Choice Test
All mice were given ad libitum access to tap water, 10% ethanol, and standard food chow, as in previous studies [42,62,63]. Water and ethanol bottles had stainless steel drinking spouts inserted through grommets on the cage lids. A two-bottle choice paradigm was implemented by rotating the water bottle and ethanol bottle positions daily to prevent a positional preference. Ethanol, water, food consumption, and body weight were measured and recorded daily at ZT06.
4.3. Drug Administration
The REV-ERBα/β inhibitor SR8278 was dissolved in a 5:5:90 solution of DMSO:Cremophor:PBS as previously described (Welch et al., 2017). Injection administration began when there was less than 15% variance in ethanol preference across 3 consecutive days (approximately 2–3 weeks of access). Mice were pseudo-randomly assigned to treatment groups such that there were an equal number of low and high ethanol preferers in each group. The mice were habituated to a 10mL/kg intraperitoneal vehicle injection once daily for 3 days prior to drug administration to mitigate the effects of handling and injection stress on ethanol drinking. REV-ERBα/β inhibitor SR8278 was delivered at 25 mg/kg or 50 mg/kg at ZT06. One group continued to receive vehicle injections and the other were given the REV-ERBα/β inhibitor once daily for 7 days. Measurements were continued for 2 days post-injection.
4.4. Experiment 1: Genetic Disruption
Adult (approximately 8 weeks old) male and female Rev-erb+/+, Rev-erb+/−, and Rev-erb−/− mice (n = 5–11/sex/genotype) were obtained from a breeding colony maintained by Dr. Tami Martino at the University of Guelph. Heterozygous founders (B6.Cg-Nr1d1tm1Ven/LazJ, strain no. #018447) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Mice were pair-housed in cages divided by perforated metal dividers to ensure mice only accessed their own food, water, and ethanol while also avoiding the confounding effects of social isolation [42,64]. A constant environment was maintained, with an ambient temperature of 21 ± 2 °C, circulating air, and a constant humidity of 50 ± 10%. Mouse cages were housed in custom-built cabinets under controlled conditions to maintain mice on a standard 12 h light (L):12-h dark (D) cycle, with lights on at 10 am (ZT0) and lights off at 10 pm (ZT12). All mice were habituated to the 12:12 LD cycle for 1 week prior to the start of the study period.
4.5. Experiment 2: Pharmacological Disruption 25 mg/kg
C57Bl/6N male mice (n = 8/group) were obtained from Charles River Laboratories (Saint Constant, QC, CA) at approximately 8 weeks old. Mice were housed in pairs with perforated metal dividers. They were habituated to the vivarium and handled by experimenters for 1 week before experiment commencement. A constant environment was maintained, with an ambient temperature of 21 ± 2 °C, circulating air, a constant humidity of 50 ± 10% and a 12:12 LD cycle beginning at 7 am (ZT0). SR8278 was administered daily as described above.
4.6. Experiment 3: Pharmacological Disruption, 50 mg/kg
C57Bl/6N male and female mice (n = 7/group/sex) were obtained from Charles River Laboratories (Saint Constant, QC, CA, USA) at approximately 8 weeks old. In adherence with recently updated housing guidelines by the Canadian Council on Animal Care, mice were singly housed. Otherwise, experimental conditions and drug administration were the same as for Experiment 2.
4.7. Statistics
All data were analyzed using SPSS 26 (IBM, Armonk, NY, USA), and graphs were generated in GraphPad Prism version 9 (GraphPad Software Inc., La Jolla, CA, USA). Ethanol preference, ethanol consumption, water consumption, and body mass data were analyzed across recorded days, whereas daily food consumption and daily caloric intake were averaged across the total experimental period and within experimental phases when appropriate. Ethanol preference was calculated as % Preference = Ethanol intake (g)/Total Liquid Intake (g) × 100%. Ethanol consumption was reported as the amount of absolute ethanol consumed in the 10% ethanol solution. Caloric intake was calculated as the sum of calories from food chow (3.3 kcal/g) and calories from 100% ethanol (7 kcal/g). Daily ethanol intake, water intake, food intake, and caloric intake were normalized to daily body mass in kilograms. All data are reported at the onset of stable ethanol preference (<15% variance across 3 consecutive days) and are presented as group means ± standard error of the mean. Where appropriate, a univariate ANOVA or repeated measures ANOVA was conducted followed by Bonferroni post hoc testing, with significance determined at p < 0.05.
Author Contributions
Conceptualization, T.A.M. and J.Y.K.; investigation, Y.A.-S., B.W.J., C.J.R., S.H., M.A.T., C.B.S. and H.H.A.T.; formal analysis, H.H.A.T.; visualization, H.H.A.T.; writing—original draft preparation, H.H.A.T., C.B.S. and B.W.J.; writing—review and editing, J.Y.K.; supervision, B.W.J., C.J.R., S.H., H.H.A.T., T.A.M. and J.Y.K.; project administration, H.H.A.T.; resources, M.R., T.A.M. and J.Y.K.; funding acquisition, T.A.M. and J.Y.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocols were approved by the University of Guelph Institutional Animal Care and Use Committee (protocol code: AUP 3922; protocol code AUP 4731) and were performed in accordance with the guidelines set forth by the Canadian Council on Animal Care (1993).
Data Availability Statement
The data generated and presented in this study are openly available in Open Science Framework at https://osf.io/uynf6/?view_only=1e7721ea2eee4708ab703524d0a79927.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by CIHR Project Grants awarded to J.Y.K. (PJT-436591) and T.A.M. (PJT-450152, 450171).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization . Global Status Report on Alcohol and Health 2018. WHO; Geneva, Switzerland: 2018. [Google Scholar]
- 2.Substance Abuse and Mental Health Services Administration . Key Substance Use and Mental Health Indicators in the United States: Results from the 2019 National Survey on Drug Use and Health. Substance Abuse and Mental Health Services Administration; Rockville, MD, USA: 2020. [Google Scholar]
- 3.Richter K., Peter L., Rodenbeck A., Weess H.G., Riedel-Heller S.G., Hillemacher T. Shiftwork and Alcohol Consumption: A Systematic Review of the Literature. Eur. Addict. Res. 2021;27:9–15. doi: 10.1159/000507573. [DOI] [PubMed] [Google Scholar]
- 4.Hasler B.P., Soehner A.M., Clark D.B. Sleep and Circadian Contributions to Adolescent Alcohol Use Disorder. Alcohol. 2015;49:377–387. doi: 10.1016/j.alcohol.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkholder J.D., Joines R., Cunningham-Hill M., Xu B. Health and Well-Being Factors Associated with International Business Travel. J. Travel Med. 2010;17:329–333. doi: 10.1111/j.1708-8305.2010.00441.x. [DOI] [PubMed] [Google Scholar]
- 6.Rogers H.L., Reilly S.M. A Survey of the Health Experiences of International Business Travelers. Part One--Physiological Aspects. AAOHN J. 2002;50:449–459. doi: 10.1177/216507990205001006. [DOI] [PubMed] [Google Scholar]
- 7.Meyrel M., Rolland B., Geoffroy P.A. Alterations in Circadian Rhythms Following Alcohol Use: A Systematic Review. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2020;99:109831. doi: 10.1016/j.pnpbp.2019.109831. [DOI] [PubMed] [Google Scholar]
- 8.Parekh P.K., Ozburn A.R., McClung C.A. Circadian Clock Genes: Effects on Dopamine, Reward and Addiction. Alcohol. 2015;49:341–349. doi: 10.1016/j.alcohol.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alibhai F.J., Tsimakouridze E.V., Reitz C.J., Pyle W.G., Martino T.A. Consequences of Circadian and Sleep Disturbances for the Cardiovascular System. Can. J. Cardiol. 2015;31:860–872. doi: 10.1016/j.cjca.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Aziz I.S., McMahon A.M., Friedman D., Rabinovich-Nikitin I., Kirshenbaum L.A., Martino T.A. Circadian Influence on Inflammatory Response During Cardiovascular Disease. Curr. Opin. Pharmacol. 2021;57:60–70. doi: 10.1016/j.coph.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Khaper N., Bailey C.D.C., Ghugre N.R., Reitz C., Awosanmi Z., Waines R., Martino T.A. Implications of Disturbances in Circadian Rhythms for Cardiovascular Health: A New Frontier in Free Radical Biology. Free Radic. Biol. Med. 2018;119:85–92. doi: 10.1016/j.freeradbiomed.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Mistry P., Duong A., Kirshenbaum L., Martino T.A. Cardiac Clocks and Preclinical Translation. Heart Fail. Clin. 2017;13:657–672. doi: 10.1016/j.hfc.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Gamble K.L., Motsinger-Reif A.A., Hida A., Borsetti H.M., Servick S.V., Ciarleglio C.M., Robbins S., Hicks J., Carver K., Hamilton N., et al. Shift Work in Nurses: Contribution of Phenotypes and Genotypes to Adaptation. PLoS ONE. 2011;6:e18395. doi: 10.1371/journal.pone.0018395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sjoholm L.K., Kovanen L., Saarikoski S.T., Schalling M., Lavebratt C., Partonen T. Clock Is Suggested to Associate with Comorbid Alcohol Use and Depressive Disorders. J. Circadian Rhythms. 2010;8:1. doi: 10.1186/1740-3391-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banach E., Pawlak J., Kapelski P., Szczepankiewicz A., Rajewska-Rager A., Skibinska M., Czerski P., Twarowska-Hauser J., Dmitrzak-Weglarz M. Clock Genes Polymorphisms in Male Bipolar Patients with Comorbid Alcohol Abuse. J. Affect. Disord. 2018;241:142–146. doi: 10.1016/j.jad.2018.07.080. [DOI] [PubMed] [Google Scholar]
- 16.Kovanen L., Saarikoski S.T., Haukka J., Pirkola S., Aromaa A., Lonnqvist J., Partonen T. Circadian Clock Gene Polymorphisms in Alcohol Use Disorders and Alcohol Consumption. Alcohol Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- 17.Baranger D.A., Ifrah C., Prather A.A., Carey C.E., Corral-Frias N.S., Drabant Conley E., Hariri A.R., Bogdan R. Per1 Rs3027172 Genotype Interacts with Early Life Stress to Predict Problematic Alcohol Use, but Not Reward-Related Ventral Striatum Activity. Front. Psychol. 2016;7:464. doi: 10.3389/fpsyg.2016.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blomeyer D., Buchmann A.F., Lascorz J., Zimmermann U.S., Esser G., Desrivieres S., Schmidt M.H., Banaschewski T., Schumann G., Laucht M. Association of Per2 Genotype and Stressful Life Events with Alcohol Drinking in Young Adults. PLoS ONE. 2013;8:e59136. doi: 10.1371/journal.pone.0059136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brower K.J., Wojnar M., Sliwerska E., Armitage R., Burmeister M. Per3 Polymorphism and Insomnia Severity in Alcohol Dependence. Sleep. 2012;35:571–577. doi: 10.5665/sleep.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comasco E., Nordquist N., Gokturk C., Aslund C., Hallman J., Oreland L., Nilsson K.W. The Clock Gene Per2 and Sleep Problems: Association with Alcohol Consumption among Swedish Adolescents. Ups. J. Med. Sci. 2010;115:41–48. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L., Bilbao A., Laucht M., Henriksson R., Yakovleva T., Ridinger M., Desrivieres S., Clarke T.K., Lourdusamy A., Smolka M.N., et al. Effects of the Circadian Rhythm Gene Period 1 (Per1) on Psychosocial Stress-Induced Alcohol Drinking. Am. J. Psychiatry. 2011;168:1090–1098. doi: 10.1176/appi.ajp.2011.10111579. [DOI] [PubMed] [Google Scholar]
- 22.Spanagel R., Pendyala G., Abarca C., Zghoul T., Sanchis-Segura C., Magnone M.C., Lascorz J., Depner M., Holzberg D., Soyka M., et al. The Clock Gene Per2 Influences the Glutamatergic System and Modulates Alcohol Consumption. Nat. Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- 23.Dalvie S., King A., Fein G., Ramesar R., Stein D.J. Possible Involvement of the Circadian Pathway in Alcohol Use Disorder in a South African Adolescent Cohort. Metab. Brain Dis. 2016;31:75–80. doi: 10.1007/s11011-015-9744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozburn A.R., Falcon E., Mukherjee S., Gillman A., Arey R., Spencer S., McClung C.A. The Role of Clock in Ethanol-Related Behaviors. Neuropsychopharmacology. 2013;38:2393–2400. doi: 10.1038/npp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamsby J.J., Templeton E.L., Bonvini L.A., Wang W., Loros J.J., Dunlap J.C., Green A.I., Gulick D. The Circadian Per1 and Per2 Genes Influence Alcohol Intake, Reinforcement, and Blood Alcohol Levels. Behav. Brain Res. 2013;249:15–21. doi: 10.1016/j.bbr.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brager A.J., Prosser R.A., Glass J.D. Circadian and Acamprosate Modulation of Elevated Ethanol Drinking in Mper2 Clock Gene Mutant Mice. Chronobiol. Int. 2011;28:664–672. doi: 10.3109/07420528.2011.601968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perreau-Lenz S., Zghoul T., de Fonseca F.R., Spanagel R., Bilbao A. Circadian Regulation of Central Ethanol Sensitivity by the Mper2 Gene. Addict. Biol. 2009;14:253–259. doi: 10.1111/j.1369-1600.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- 28.Sharma R., Puckett H., Kemerling M., Parikh M., Sahota P., Thakkar M. Antisense-Induced Downregulation of Clock Genes in the Shell Region of the Nucleus Accumbens Reduces Binge Drinking in Mice. Alcohol Clin. Exp. Res. 2021;45:530–542. doi: 10.1111/acer.14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee S., Wang Y., Solt L.A., Griffett K., Kazantzis M., Amador A., El-Gendy B.M., Huitron-Resendiz S., Roberts A.J., Shin Y., et al. Pharmacological Targeting of the Mammalian Clock Regulates Sleep Architecture and Emotional Behaviour. Nat. Commun. 2014;5:5759. doi: 10.1038/ncomms6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkow N.D., Morales M. The Brain on Drugs: From Reward to Addiction. Cell. 2015;162:712–725. doi: 10.1016/j.cell.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 31.Chung S., Lee E.J., Yun S., Choe H.K., Park S.B., Son H.J., Kim K.S., Dluzen D.E., Lee I., Hwang O., et al. Impact of Circadian Nuclear Receptor Rev-Erbalpha on Midbrain Dopamine Production and Mood Regulation. Cell. 2014;157:858–868. doi: 10.1016/j.cell.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 32.Sleipness E.P., Sorg B.A., Jansen H.T. Diurnal Differences in Dopamine Transporter and Tyrosine Hydroxylase Levels in Rat Brain: Dependence on the Suprachiasmatic Nucleus. Brain Res. 2007;1129:34–42. doi: 10.1016/j.brainres.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 33.Jager J., O’Brien W.T., Manlove J., Krizman E.N., Fang B., Gerhart-Hines Z., Robinson M.B., Klein P.S., Lazar M.A. Behavioral Changes and Dopaminergic Dysregulation in Mice Lacking the Nuclear Receptor Rev-Erbalpha. Mol. Endocrinol. 2014;28:490–498. doi: 10.1210/me.2013-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClung C.A., Sidiropoulou K., Vitaterna M., Takahashi J.S., White F.J., Cooper D.C., Nestler E.J. Regulation of Dopaminergic Transmission and Cocaine Reward by the Clock Gene. Proc. Natl. Acad. Sci. USA. 2005;102:9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The Orphan Nuclear Receptor Rev-Erbalpha Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 36.Kojetin D., Wang Y., Kamenecka T.M., Burris T.P. Identification of Sr8278, a Synthetic Antagonist of the Nuclear Heme Receptor Rev-Erb. ACS Chem. Biol. 2011;6:131–134. doi: 10.1021/cb1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Middaugh L.D., Kelley B.M., Bandy A.L., McGroarty K.K. Ethanol Consumption by C57bl/6 Mice: Influence of Gender and Procedural Variables. Alcohol. 1999;17:175–183. doi: 10.1016/S0741-8329(98)00055-X. [DOI] [PubMed] [Google Scholar]
- 38.Yoneyama N., Crabbe J.C., Ford M.M., Murillo A., Finn D.A. Voluntary Ethanol Consumption in 22 Inbred Mouse Strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reitz C.J., Alibhai F.J., Khatua T.N., Rasouli M., Bridle B.W., Burris T.P., Martino T.A. Sr9009 Administered for One Day after Myocardial Ischemia-Reperfusion Prevents Heart Failure in Mice by Targeting the Cardiac Inflammasome. Commun. Biol. 2019;2:353. doi: 10.1038/s42003-019-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solt L.A., Wang Y., Banerjee S., Hughes T., Kojetin D.J., Lundasen T., Shin Y., Liu J., Cameron M.D., Noel R., et al. Regulation of Circadian Behaviour and Metabolism by Synthetic Rev-Erb Agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triqueneaux G., Thenot S., Kakizawa T., Antoch M.P., Safi R., Takahashi J.S., Delaunay F., Laudet V. The Orphan Receptor Rev-Erbalpha Gene Is a Target of the Circadian Clock Pacemaker. J. Mol. Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizk A.A., Jenkins B.W., Al-Sabagh Y., Hamidullah S., Reitz C.J., Rasouli M., Martino T.A., Khokhar J.Y. The Impact of Sex, Circadian Disruption, and the Clockδ19/Δ19 Genotype on Alcohol Drinking. Genes. 2022;13:701. doi: 10.3390/genes13040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelmant G., Begue A., Stehelin D., Laudet V. A Functional Rev-Erb Alpha Responsive Element Located in the Human Rev-Erb Alpha Promoter Mediates a Repressing Activity. Proc. Natl. Acad. Sci. USA. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitaterna M.H., King D.P., Chang A.M., Kornhauser J.M., Lowrey P.L., McDonald J.D., Dove W.F., Pinto L.H., Turek F.W., Takahashi J.S. Mutagenesis and Mapping of a Mouse Gene, Clock, Essential for Circadian Behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin X., Shearman L.P., Weaver D.R., Zylka M.J., de Vries G.J., Reppert S.M. A Molecular Mechanism Regulating Rhythmic Output from the Suprachiasmatic Circadian Clock. Cell. 1999;96:57–68. doi: 10.1016/S0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 46.Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. Mcry1 and Mcry2 Are Essential Components of the Negative Limb of the Circadian Clock Feedback Loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 47.Oishi K., Fukui H., Ishida N. Rhythmic Expression of Bmal1 Mrna Is Altered in Clock Mutant Mice: Differential Regulation in the Suprachiasmatic Nucleus and Peripheral Tissues. Biochem Biophys Res. Commun. 2000;268:164–171. doi: 10.1006/bbrc.1999.2054. [DOI] [PubMed] [Google Scholar]
- 48.Cho H., Zhao X., Hatori M., Yu R.T., Barish G.D., Lam M.T., Chong L.W., Di Tacchio L., Atkins A.R., Glass C.K., et al. Regulation of Circadian Behaviour and Metabolism by Rev-Erb-Alpha and Rev-Erb-Beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delezie J., Dumont S., Sandu C., Reibel S., Pevet P., Challet E. Rev-Erbalpha in the Brain Is Essential for Circadian Food Entrainment. Sci. Rep. 2016;6:29386. doi: 10.1038/srep29386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeda R., Tsuchiya Y., Koike N., Umemura Y., Inokawa H., Ono R., Inoue M., Sasawaki Y., Grieten T., Okubo N., et al. Rev-Erbalpha and Rev-Erbbeta Function as Key Factors Regulating Mammalian Circadian Output. Sci. Rep. 2019;9:10171. doi: 10.1038/s41598-019-46656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanderlinden L.A., Saba L.M., Bennett B., Hoffman P.L., Tabakoff B. Influence of Sex on Genetic Regulation of “Drinking in the Dark” Alcohol Consumption. Mamm. Genome. 2015;26:43–56. doi: 10.1007/s00335-014-9553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Zavalia N., Schoettner K., Goldsmith J.A., Solis P., Ferraro S., Parent G., Amir S. Bmal1 in the Striatum Influences Alcohol Intake in a Sexually Dimorphic Manner. Commun. Biol. 2021;4:1227. doi: 10.1038/s42003-021-02715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bass C.E., Grinevich V.P., Gioia D., Day-Brown J.D., Bonin K.D., Stuber G.D., Weiner J.L., Budygin E.A. Optogenetic Stimulation of Vta Dopamine Neurons Reveals that Tonic but Not Phasic Patterns of Dopamine Transmission Reduce Ethanol Self-Administration. Front. Behav. Neurosci. 2013;7:173. doi: 10.3389/fnbeh.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Budygin E.A., Bass C.E., Grinevich V.P., Deal A.L., Bonin K.D., Weiner J.L. Opposite Consequences of Tonic and Phasic Increases in Accumbal Dopamine on Alcohol-Seeking Behavior. iScience. 2020;23:100877. doi: 10.1016/j.isci.2020.100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberto M., Varodayan F.P. Synaptic Targets: Chronic Alcohol Actions. Neuropharmacology. 2017;122:85–99. doi: 10.1016/j.neuropharm.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding G., Li X., Hou X., Zhou W., Gong Y., Liu F., He Y., Song J., Wang J., Basil P., et al. Rev-Erb in Gabaergic Neurons Controls Diurnal Hepatic Insulin Sensitivity. Nature. 2021;592:763–767. doi: 10.1038/s41586-021-03358-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T., Yu F., Xu H., Chen M., Chen X., Guo L., Zhou C., Xu Y., Wang F., Yu J., et al. Dysregulation of Rev-Erbalpha Impairs Gabaergic Function and Promotes Epileptic Seizures in Preclinical Models. Nat. Commun. 2021;12:1216. doi: 10.1038/s41467-021-21477-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frie J.A., Khokhar J.Y. An Open Source Automated Two-Bottle Choice Test Apparatus for Rats. HardwareX. 2019;5:e00061. doi: 10.1016/j.ohx.2019.e00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feillet C.A., Bainier C., Mateo M., Blancas-Velazquez A., Salaberry N.L., Ripperger J.A., Albrecht U., Mendoza J. Rev-Erbalpha Modulates the Hypothalamic Orexinergic System to Influence Pleasurable Feeding Behaviour in Mice. Addict. Biol. 2017;22:411–422. doi: 10.1111/adb.12339. [DOI] [PubMed] [Google Scholar]
- 60.Zhao C., Gammie S.C. The Circadian Gene Nr1d1 in the Mouse Nucleus Accumbens Modulates Sociability and Anxiety-Related Behavior. Eur. J. Neurosci. 2018;48:1924–1943. doi: 10.1111/ejn.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castillo-Carniglia A., Keyes K.M., Hasin D.S., Cerda M. Psychiatric Comorbidities in Alcohol Use Disorder. Lancet. Psychiatry. 2019;6:1068–1080. doi: 10.1016/S2215-0366(19)30222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruffolo J., Frie J.A., Thorpe H.H.A., Talhat M.A., Khokhar J.Y. Alcohol and Vaporized Nicotine Co-Exposure During Adolescence Contribute Differentially to Sex-Specific Behavioral Effects in Adulthood. Nicotine. Tob. Res. 2021:ntab250. doi: 10.1093/ntr/ntab250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamidullah S., Lutelmowski C.D., Creighton S.D., Luciani K.R., Frie J.A., Winters B.D., Khokhar J.Y. Effects of Vapourized Thc and Voluntary Alcohol Drinking During Adolescence on Cognition, Reward, and Anxiety-Like Behaviours in Rats. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2021;106:110141. doi: 10.1016/j.pnpbp.2020.110141. [DOI] [PubMed] [Google Scholar]
- 64.Lesscher H.M., Spoelder M., Rotte M.D., Janssen M.J., Hesseling P., Lozeman-van’t Klooster J.G., Baars A.M., Vanderschuren L.J. Early Social Isolation Augments Alcohol Consumption in Rats. Behav. Pharmacol. 2015;26:673–680. doi: 10.1097/FBP.0000000000000165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated and presented in this study are openly available in Open Science Framework at https://osf.io/uynf6/?view_only=1e7721ea2eee4708ab703524d0a79927.