Abstract
We quantified the diversity of oxygenic phototrophic microorganisms present in eight hypersaline microbial mats on the basis of three cultivation-independent approaches. Morphological diversity was studied by microscopy. The diversity of carotenoids was examined by extraction from mat samples and high-pressure liquid chromatography analysis. The diversity of 16S rRNA genes from oxygenic phototrophic microorganisms was investigated by extraction of total DNA from mat samples, amplification of 16S rRNA gene segments from cyanobacteria and plastids of eukaryotic algae by phylum-specific PCR, and sequence-dependent separation of amplification products by denaturing-gradient gel electrophoresis. A numerical approach was introduced to correct for crowding the results of chromatographic and electrophoretic analyses. Diversity estimates typically varied up to twofold among mats. The congruence of richness estimates and Shannon-Weaver indices based on numbers and proportional abundances of unique morphotypes, 16S rRNA genes, and carotenoids unveiled the underlying diversity of oxygenic phototrophic microorganisms in the eight mat communities studied.
Due to their physiological diversity, microorganisms play major roles in the cycling of chemical elements within the biosphere, but this relevance for environmental processes is only fragmentarily reflected in our current knowledge about microbial diversity (39, 40) because the small size and morphological simplicity of microorganisms have hampered the study of their diversity. While microbial physiology and genetics can be investigated in great detail in cultivated isolates, the majority of microorganisms have so far resisted cultivation efforts (40). From most habitats studied, with only a few exceptions (47), less than 1% of the microorganisms observed by microscopy have been brought into culture (1). It is clear, then, that current isolation procedures will fail to adequately investigate the microbial diversity extant in natural environments (1, 10). Molecular biological techniques, and particularly the study of small-subunit rRNAs and the respective genes, have provided new insights into the phylogenetic diversity of microorganisms (68). Microbial nucleic acids extracted directly from environmental samples are amenable to comparative analyses of nucleotide sequences (18, 41, 64). Numerous publications based on this approach have reported the exploration of uncultivated microbial diversity in the last decade (10, 40). However, our understanding of forces that shape and sustain microbial diversity in the environment and of the impact that microbial diversity may have on ecosystem processes is as yet very limited (25, 39). Theoretically, empirical investigations of such interdependencies should lead to considerable progress in the field of microbial ecology, but such investigations depend unavoidably on the evaluation of biodiversity in quantitative terms. This quantification has not yet been achieved on the basis of the new molecular methodologies (39), but in principle it is possible and probably desirable (17).
The quantification of diversity requires the grouping of individual elements into nonoverlapping classes according to a differentiating criterion (26). If the study is to be restricted to certain organisms, which usually will be the case, individuals to be excluded from the analysis need to be identified as such. Ecological diversity is considered a function of the number of different classes (richness) and the relative distribution of individual elements among these classes (evenness) (3, 65). Various indices have been proposed as measures of diversity that incorporate both aspects, richness and evenness (30). The Shannon-Weaver index is the most common diversity index used by ecologists (65); it weights individual classes by their relative abundances. It can be understood as an estimator of the degree of uncertainty attached to the identity of any individual randomly selected from a community, which increases with richness as well as with evenness (29). Optimally, individual elements in a class should be uniform with respect to their ecology. However, functional diversity, the actual ecologically relevant parameter, cannot be directly determined, and some deviation from this ideal must be expected when single criteria are used as bases for diversity determinations. Exposed to environmental selection, ecological units are also evolutionary units (43, 63), and the use of evolutionarily coherent entities as classes for diversity estimates is desirable. For practical reasons identification procedures should be as little time-consuming as possible, since often large numbers of organisms need to be investigated. Ecologists studying macroscopic plants and animals commonly use taxonomic species as classes for grouping individual organisms and assess species richness and species diversity accordingly (3). The delineation of species on the basis of morphologies is the most common practice (8), but it does not necessarily result in evolutionarily and ecologically coherent entities, particularly when applied to microorganisms. The determination of prokaryotic species richness and diversity in nature is impracticable because the current bacteriological species concept applies exclusively to organisms in pure cultures (66). The value of available species concepts for the quantification of diversity probably will depend on the group of microorganisms considered and on the habitats to be studied; it may be necessary to replace species with some other appropriate units of biodiversity (25).
We studied microbial mats from hypersaline waters in evaporation ponds of the saltern in Guerrero Negro, Baja California, Mexico, as well as from salt marshes in its proximity (9, 22). The biomass of these benthic laminated ecosystems is almost exclusively composed of microorganisms, most of which are prokaryotes. We focused our investigations on communities of oxygenic phototrophs, whose activities are the basis for the existence of these mats. These microorganisms, namely cyanobacteria, diatoms, and a small proportion of green microalgae, traditionally have been classified on the basis of their morphologies (2, 7, 49). Diversity studies of microalgae that were restricted to morphotype analyses alone (e.g., reference 60) have been reported in the past. However, the phycological systems of classification do not necessarily reflect evolutionary relationships (35, 46, 67) and may underestimate diversity (11, 69). Therefore, we investigated the diversity of oxygenic phototrophic microorganisms by applying three cultivation-independent approaches in parallel. Classes for grouping the respective elements analyzed were defined by unique cell and colony morphologies, rRNA gene sequences, and carotenoid molecule structures. Cells and molecules from organisms other than oxygenic phototrophs were excluded from the analyses. The relative morphological complexity of oxygenic phototrophic microorganisms and their content of unique pigments allowed us to evaluate the diversity reflected in rRNA genes. The congruence of results obtained for eight different mat communities demonstrated that microbial diversity can be meaningfully quantified. The numbers and proportional abundances of unique morphotypes, carotenoids, and 16S rRNA genes could describe the diversity of oxygenic phototrophic microorganisms.
MATERIALS AND METHODS
Sampling.
Microbial mats were sampled during the second to fourth weeks of April 1996 (mats P2, P4, P6, NC2, and NC3) and 1997 (mats P3/4, P5, and NC52). Sampling sites were located in evaporation ponds of the saltern in Guerrero Negro, Baja California Sur, Mexico, and in the salt marsh of Ojo de Liebre Lagoon. Detailed descriptions of these sites can be found elsewhere (9, 22). The characteristics of the microbial mats investigated are summarized in Table 1. Measurements of photosynthesis were performed in a field laboratory with microsensor techniques (48). Except for giving information about the thickness of the photic zone, based on the maximum depth where gross photosynthesis was detectable, we do not address the results of these supplementary studies in this report (but see reference 14). Three cores 10 to 20 cm apart were taken as samples from each mat. For light microscopy, the mat samples (core diameter, 4 mm) were fixed in 5% (wt/vol) formaldehyde and stored at 4°C. For extractions of nucleic acids and carotenoids, the mat samples (core diameter, 25 mm) were frozen on site, transported to the laboratory in liquid nitrogen, and stored at −80°C until processed.
TABLE 1.
Characteristics of the microbial mats studied
| Mat | Field conditions
|
Photic zone characteristics
|
|||
|---|---|---|---|---|---|
| Salinity (%)a | Water depth (m) | Origin | Thickness (mm) | Texture | |
| P2 | 6 | 0.5 | Evaporation pond | 2.0 | Soft |
| P3/4 | 6 | 0.5 | Evaporation pond | 1.5 | Cohesive |
| P4 | 9 | 1.0 | Evaporation pond | 3.0 | Cohesive, rubbery |
| P5 | 11 | 1.0 | Evaporation pond | 3.0 | Cohesive, rubbery |
| P6 | 14 | 1.0 | Evaporation pond | 4.8 | Gelatinous, translucent |
| NC2 | 5–8 | 0.2–0.5 | Tidal channel | 1.5 | Soft |
| NC52 | 5–11 | 0.2–0.5 | Tidal channel | 1.6 | Soft |
| NC3 | 6–21 | 0.3–0.6 | Tidal channel | 1.7 | Very cohesive, rubbery, compact |
Measured in the field site at time of collection. Salinities in evaporation ponds typically vary by less than 5% per year. The salinities and water depths in tidal channels vary with tidal cycles.
Microscopy and morphotype quantification.
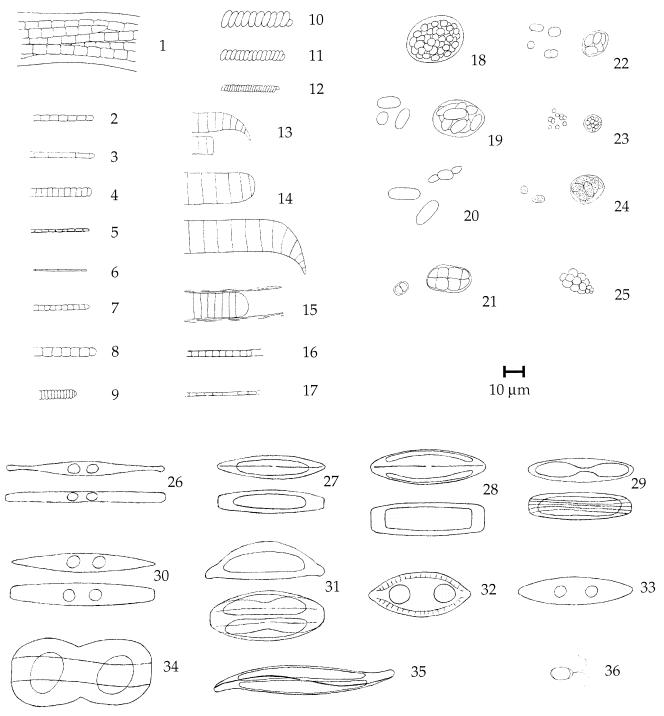
The layers corresponding to photic zones were cut from formaldehyde-fixed mat samples with scalpel blades and sectioned vertically into subcores of approximately 0.5- by 0.5-mm mat surface area. These pieces were placed on glass slides in 1 drop of water and chopped and stirred to achieve even distribution. Sketches of morphotypes were prepared for illustration (Fig. 1). For each subcore, 25 to 40 randomly chosen phase-contrast microscopic fields were photographed at 400-fold magnification. Counts and size measurements were performed on projections of the resulting slides, exclusively taking into account focused cells of oxygenic phototrophs. Diatom valves without plastids were omitted from the analyses. For unicellular organisms, cell numbers were counted. For filamentous organisms, total filament lengths were divided by the respective mean cell lengths to calculate cell numbers. Final cell numbers or total filament lengths were converted into biovolumes with geometric formulae (15). Rounded cells were considered spheres, and rod-shaped and filamentous organisms were considered cylinders. Naviculoid and nitzschoid diatom cells (Fig. 1, morphotypes 26 to 30, 32, and 33) were considered flat elongated cylinders or prisms with elliptical or rhombic surface areas (valve view), respectively.
FIG. 1.
Morphotypes of oxygenic-phototrophic microorganisms from eight mat communities as observed by phase-contrast light microscopy. Morphotypes 1 to 25 are cyanobacteria, of which 1 to 17 can be assigned to the order Oscillatoriales and 18 to 25 can be assigned to the order Chroococcales (7). Morphotypes 26 to 35 are diatoms, to which the following generic assignments can be made (49): Nitzschia, 26, 30, 33; Brachysira, 27; Navicula, 28; Amphora, 31; Mastogloia, 32; Entomoneis, 34; and Gyrosigma, 35. Morphotype 36 is a green alga (Dunaliella sp.).
DNA extraction.
The layers corresponding to photic zones were aseptically cut from mat cores (100 to 400 mg, representing approximately 60 mm2 of mat surface) and homogenized in Dounce tissue homogenizers (Novodirect, Kehl, Germany). Cell lysis and DNA extraction were performed as described previously (38). Briefly, the suspensions were repeatedly frozen and thawed and subsequently incubated in the presence of sodium dodecyl sulfate and proteinase K. Cell lysis was controlled microscopically. DNA was extracted by applying hexadecylmethylammonium bromide, phenol, chloroform, and isoamyl alcohol and precipitated by the addition of isopropyl alcohol.
PCR.
The oligonucleotide primers CYA359F and CYA781R were applied to selectively amplify 16S rRNA gene segments from cyanobacteria and plastids (38). The numbers in the primer designations refer to the 5′ ends of target signature sites in 16S rRNA genes (Escherichia coli nucleotide numbering [5]). A 40-nucleotide GC-rich sequence was attached to the 5′ end of the primer CYA359F to improve the detection of sequence variation in amplified DNA fragments by subsequent denaturing-gradient gel electrophoresis (DGGE [38]). As templates for amplifications, 10 ng of DNAs extracted from mat samples was added to each 100-μl reaction mixture.
DGGE and digital image analysis.
Amplification products generated by duplicate PCRs with the same template DNAs were pooled and subsequently purified and concentrated by using the QIAquick PCR purification kit (Diagen, Düsseldorf, Germany). DNA concentrations in the resulting solutions were determined by comparison to the Gibco low-DNA-mass standard (Gibco, Eggenstein, Germany) after agarose gel electrophoresis; 500 ng of DNA was applied to denaturing-gradient gels. DGGE was performed as described previously (38). Briefly, polyacrylamide gels with a denaturant gradient from 20 to 60% were used, and electrophoreses were run for 3.5 h at 200 V. Subsequently, the gels were incubated for 30 min in 1× TAE (40 mM Tris-HCl [pH 8.3], 20 mM acetic acid, 1 mM EDTA) containing 20 mg of ethidium bromide/ml. Fluorescence of dye bound to DNA was excited by UV irradiation by applying a UV transilluminator and was photographed with a digital image gel documentation system (Cybertech, Berlin, Germany). The intensities of gel band fluorescences were measured on digital images by applying the gel-plotting macro implemented in the NIH-Image software package version 1.62 (National Institutes of Health, Bethesda, Md.). To enable the transformation of fluorescence values into amounts of DNA in individual bands, we designed and applied on each gel a DNA mass calibration standard for DGGE, which was a mixture of PCR products with known concentrations (see Fig. 4).
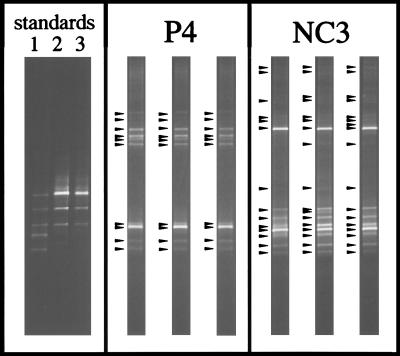
FIG. 4.
Composite figure of ethidium bromide-stained DGGE separation patterns of PCR-amplified segments of 16S rRNA genes. Mixtures of PCR products derived from five cyanobacterial strains were applied to each gel as standards (in lanes 1 to 3 [top to bottom] are Scytonema sp. strain B-77-Scy.jav., Synechococcus leopoliensis SAG 1402-1, Microcoleus chthonoplastes MPI-NDN-1, Geitlerinema sp. strain PCC 9452 [“Microcoleus” sp. strain 10 mfx], and Cyanothece sp. strain PCC 7418). The standard in lane 1 allows gel-to-gel comparisons. The DNA mass calibration standard in lanes 2 and 3 enables the transformation of measured band fluorescence values into amounts of DNA (in lane 2, the amounts of DNA in individual bands are (top to bottom) 528, 176, 59, 20, and 7 ng; in lane 3, half of the amount of the standard in lane 2 was applied). The gels labelled P4 and NC3 show separation patterns of PCR products derived from triplicate sampling cores of those microbial mats. The arrowheads indicate the bands included in the subsequent analyses.
Carotenoid extraction and analysis.
Frozen samples of mat layers (100 to 400 mg, representing approximately 60 mm2 of mat surface) corresponding to photic zones were ground in a mortar while cooled by liquid nitrogen. The ground samples were extracted in 10 to 15 ml of degassed acetone for 24 h in the dark at 4°C. Extracts were clarified by filtration on Whatman GF/F glass fiber filters and subsequently concentrated under a stream of N2 gas. Concentrated pigment extracts were separated and analyzed by high-pressure liquid chromatography (HPLC), with on-line detection by diode array-based spectroscopy between 350 and 700 nm, allowing for the detection of typical carotenoid spectra. The details of chromatographic conditions and equipment were essentially as previously described (24).
Estimation of richness and diversity.
The richness and diversity of morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophic microorganisms were estimated. Classes for grouping organisms and molecules were defined by unique cell and colony morphologies, nucleotide sequences, and pigment molecule structures. Cells and molecules from organisms other than oxygenic phototrophs were excluded from the analyses.
Morphotype analysis was performed by light microscopy. Diatoms and cyanobacteria could be distinguished from most other microorganisms due to their sizes and characteristic morphologies. Epifluorescence microscopy was used to identify putative members of the family Chloroflexaceae. These green filamentous bacteria occur in marine microbial mats (45) and may be mistakenly identified as cyanobacteria, but they were distinguished by their lack of visible fluorescence. Proportional abundances of morphotypes were calculated on the basis of cell numbers and, alternatively, cell volumes. 16S rRNA gene fragments were amplified by PCR from cyanobacterial and plastid DNA after nucleic acid extraction from mat samples (38). Phylum-specific amplification enabled the exclusion from the analyses of DNA from organisms other than oxygenic phototrophs (38). Numbers and proportional abundances of the unique rRNA gene segments amplified were estimated after DGGE analysis of PCR products. Carotenoids were extracted directly from mat samples, and their analysis by HPLC enabled the determination of the numbers and abundances of unique pigments. Quantification of the relative abundance of specific carotenoids was based on area integration of peaks resulting from absorption at 470 nm, and no attempts were made to introduce corrections for differences in extinction coefficients. Peak delimitation and area integration were carried out automatically by the intrument’s software. Absorption peaks in these chromatograms corresponding to pigments other than carotenoids (chlorophyll a, pheophytins, diverse bacteriochlorophylls, and scytonemin) were identified from the corresponding absorption spectra (350 to 700 nm) and deleted from the automated analysis a posteriori (for an example of this procedure, see Fig. 5). The assumption was made that the contribution to total carotenoids of pigments from heterotrophic and chemotrophic bacteria was negligible, as phototrophs make up the bulk of the biomass in the photic zone and they contain much higher specific amounts of carotenoids than nonphototrophic bacteria. In those samples displaying measurable amounts of bacteriochlorophylls, a correction for carotenoids stemming from anoxygenic phototrophic bacteria was carried out. These were identified by the comparison of retention times and spectra to carotenoid standards from purple sulfur bacteria and green filamentous nonsulfur bacteria. However, these corrections were seldom necessary. The majority of carotenoids detected could be assigned to typical cyanobacterial and diatom standards from cultivated strains.
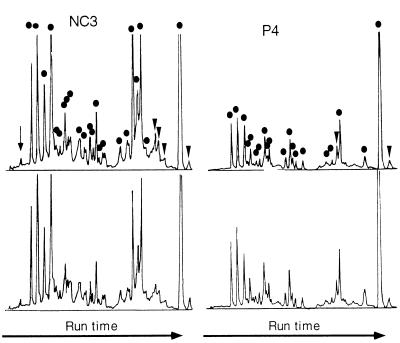
FIG. 5.
Duplicate carotenoid analysis by HPLC in two mats, P4 and NC3. Two independent analyses for each mat are shown. Each peak in the 470-nm chromatrogram was assigned to either a carotenoid (solid circles), a tetrapyrrol (chlorophylls and phaeophytin) (arrowheads), or scytonemin (arrow) on the basis of absorption spectra obtained on-line.
Using the approaches described above for triplicate samples from each oxygenic phototrophic mat community, the numbers, D, and proportional abundances, ai, of classes i were determined. Shannon-Weaver indices (H′) (30, 52) were calculated as
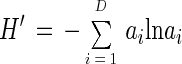 |
In the following discussion, subscripts refer to the respective analyses of morphotypes (M), rRNA genes (R), and carotenoids (C). Corrections for crowding of bands and peaks in electrophoretic and chromatographic analyses transformed DR and DC into richness estimates SR and SC (see below). DM was used directly as an estimate of the richness of the morphotypes, SM. Arithmetic means and standard errors were calculated from the results of triplicate analyses. Statistical analysis was performed by using SPSS version 6.1 software (SPSS, Erkrath, Germany).
Correction for crowding of the number of classes measured by electrophoresis and chromatography.
With increasing numbers of bands or peaks detected in electrophoretic or chromatographic analyses, respectively, the probability increases that classes cannot be discerned because they run at identical positions in the gel or chromatogram. In the following paragraph, we describe a way to approximate this probability distribution and, on that basis, to estimate how many classes are likely to have been missed in a given analysis due to crowding.
Let S be the number of specific classes present in a sample, D the number of classes actually detected in the analysis, and Dmax the maximum number of classes that the analytical procedure can detect. Due to crowding within the chromatogram or electrophoretic gel, for any given S there exists a certain probability to detect only D classes, where D ≤ S and D ≤ Dmax. A distribution, PS(D), describing this probability can be used to correct measured values for crowding. Assuming that there are no preferred sites of occurrence for the classes within the chromatogram or electrophoresis gel, i.e., that crowding is homogeneous, and that all classes can be detected with equal resolution, then the probability, p, that two classes in a sample with S = 2 cannot be discerned is p = P2(1) = (Dmax)−1. The probability that we measure D classes in a sample with S + 1 specific classes is PS + 1 (D) = PS(D) Dp + PS (D − 1) [1 − (D − 1)p]. Because P1(1) = 1, if p is known, one can inductively calculate any value of PS(D). Alternatively, PS(D) can be computed directly as
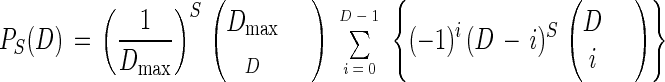 |
Once the values of PS(D) are computed, a simple correction consists of calculating expectation values for D, Dexp, for any given S by using PS(D), as
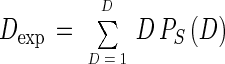 |
Values of S that yield a Dexp corresponding to a measured value of D can be taken as corrected estimates of richness.
Values for Dmax were estimated, on the basis of the average peak width and total distance of separation for our experiments, to be 200 for DGGE and 96 for HPLC. Accordingly, crowding was likely to affect the results if DR was ≥20 and DC was ≥14, respectively. None of our DGGE results exceeded this threshold, but values of carotenoid richness in most cases needed to be raised by one to three classes (i.e., by up to 12%). We did not attempt to take into account the effect of varying class frequencies on crowding or to correct the calculation of Shannon-Weaver indices, as such corrections can be extremely complicated. Thus, our values for H′C tend to underestimate carotenoid diversity.
RESULTS
Diversity of morphotypes.
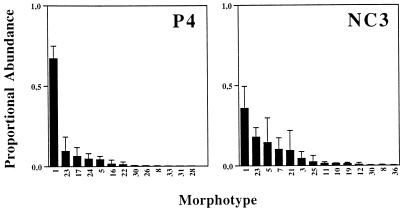
Morphotypes that were distinguished are illustrated in Fig. 1. In total, 36 different morphotypes were observed, of which 25 were cyanobacteria, 10 were diatoms, and 1 was a green alga. The number of morphotypes detected in each of the specimens is considered an estimate of the morphotype richness, SM, of the respective oxygenic-phototrophic community. Proportional abundances based on cell counts and, alternatively, estimated cell volumes were used to calculate the Shannon-Weaver indices, H′M (Table 2). Figure 2 illustrates the proportional abundances of morphotypes in two of the eight mats on the basis of cell counts performed on triplicate randomly drawn subcores.
TABLE 2.
Numbers of classes, richnesses, and Shannon-Weaver indices determineda
| Mat | Morphotypes
|
16S rRNA genes
|
Carotenoids
|
|||||
|---|---|---|---|---|---|---|---|---|
| SM | H′M | H′V | SR | H′R | DC | SC | H′C | |
| P2 | 10.67 ± 0.33 | 1.45 ± 0.09 | 1.82 ± 0.12 | 11.33 ± 0.66 | 1.38 ± 0.08 | 20.67 ± 0.66 | 22.67 ± 0.66 | 2.28 ± 0.03 |
| P3/4 | 7.33 ± 0.33 | 0.98 ± 0.09 | 1.42 ± 0.17 | 4.00 ± 0.00 | 0.35 ± 0.02 | 9.33 ± 0.88 | 9.33 ± 0.88 | 1.62 ± 0.03 |
| P4 | 10.33 ± 0.88 | 1.10 ± 0.14 | 1.07 ± 0.17 | 10.00 ± 0.00 | 1.75 ± 0.02 | 22.00 ± 1.15 | 24.00 ± 1.16 | 2.28 ± 0.02 |
| P5 | 7.00 ± 0.58 | 1.02 ± 0.12 | 1.31 ± 0.03 | 6.67 ± 0.33 | 1.31 ± 0.09 | 17.00 ± 0.58 | 18.00 ± 0.58 | 1.97 ± 0.02 |
| P6 | 9.67 ± 0.33 | 1.39 ± 0.05 | 1.72 ± 0.10 | 10.00 ± 0.00 | 1.90 ± 0.02 | 14.67 ± 0.33 | 15.67 ± 0.33 | 2.20 ± 0.03 |
| NC2 | 12.00 ± 0.58 | 1.41 ± 0.21 | 1.68 ± 0.08 | 12.00 ± 0.00 | 2.09 ± 0.01 | 22.00 ± 1.00 | 24.00 ± 1.00 | 2.44 ± 0.02 |
| NC52 | 12.00 ± 0.58 | 1.18 ± 0.13 | 1.65 ± 0.06 | 12.33 ± 0.33 | 1.74 ± 0.05 | 13.67 ± 0.66 | 14.00 ± 1.00 | 1.90 ± 0.03 |
| NC3 | 12.67 ± 0.33 | 1.67 ± 0.04 | 1.29 ± 0.03 | 16.00 ± 1.00 | 2.08 ± 0.11 | 24.00 ± 1.00 | 26.67 ± 1.34 | 2.53 ± 0.02 |
D, number of classes detected; S, richness; H′, Shannon-Weaver index. The subscripts refer to the respective analyses of morphotypes (M), volumes of morphotypes (V), 16S rRNA genes (R), and carotenoids (C). The means and standard errors from triplicate analyses are shown.
FIG. 2.
Proportional abundances of morphotypes based on cell counts. The morphotype numbers refer to Fig. 1. The data are from analyses of communities in mats P4 and NC3. Because sets of morphotypes found in triplicate subcores from the same mat do not necessarily completely coincide, the cumulative number of morphotypes observed in a community may exceed the mean richness, SM (Table 2).
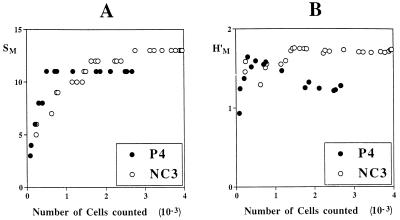
A marked patchiness in mat community structure was observed at the scale of cyanobacterial colonies and filaments (10 to 100 μm). Therefore, a significant dependence of SM and H′M on the number of microscopic fields investigated had to be expected. The minimum number of cells needed to achieve representative subsamples was determined for each of the mats by randomly removing slides (microscopic fields) one by one from the respective analyses and subsequently determining richness and Shannon-Weaver indices for the resulting samples of reduced sizes. For a low number of cells included in an analysis both parameters increased (not necessarily monotonically) concomitantly with increasing sample size and then, provided that a large enough number of cells was investigated, leveled off. A subsample was assumed to be sufficiently representative if its further enlargement caused no further increase of the determined diversity (44, 60). Figure 3 shows plots of these analyses for two specimens. The sample sizes needed to detect all rare and localized morphotypes (Fig. 3A) and to weight them according to their actual abundances (Fig. 3B) were estimated from such plots to be 2,000 to 3,000 cells. Significant heterogeneity of the distribution of organisms at the scale of millimeters caused major differences among microscopic analyses of triplicate preparations from the same mat. For example, the surface of the mat from pond 5 consisted of a loose film of diatoms (Nitzschia sp.; morphotype 30) forming irregularly scattered tufts up to 1 mm in height and diameter. Concomitantly, depending on the exact site of sampling, the relative proportion of diatoms in a randomly drawn subcore, representing only a 0.5- by 0.5-mm mat surface, varied from 1.4 to 9.5% of the total cell numbers of oxygenic phototrophs. This level of patchiness resulted in large variances of estimates of both abundances of individual morphotypes (Fig. 2) and morphotype diversity as reflected in H′M (see Fig. 7), whereas SM was less affected. In all specimens, the rarest morphotypes detected accounted for less than 1% of the total number of cells and total biovolumes. Richness estimates and Shannon-Weaver indices varied almost twofold among mats, whereas coefficients of variation for each mat amounted to at most 16%.
FIG. 3.
Relationship of SM (A) and H′M (B [based on cell counts]) to the number of cells encountered. The data are from analyses of communities in mats P4 and NC3.
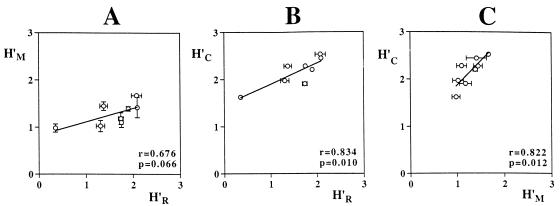
FIG. 7.
Relationships among Shannon-Weaver indices for communities of oxygenic-phototrophic microorganisms in eight microbial mats based on analyses of morphologies (H′M), 16S rRNA genes (H′R), and carotenoids (H′C). The arithmetic means and standard errors from triplicate analyses are shown. The Pearson correlation coefficient, r, and its statistical significance, P, have been calculated.
Diversity of 16S rRNA genes.
Total nucleic acids were extracted from triplicate mat samples, and 16S rRNA gene segments were amplified from cyanobacterial and plastid DNA by applying a phylum-specific PCR that had recently been developed on the basis of published nucleotide sequences (38). The sequence-dependent separation of the resulting amplification products by DGGE (36) generated band patterns that were characteristic for each of the mats (Fig. 4). The number of bands visible in denaturant gradient polyacrylamide gels (corrected for crowding) provides an estimate of the richness, SR. Fluorescence measurements and comparisons to a DNA mass calibration standard (Fig. 4) allowed the calculation of the amounts of DNA in individual gel bands, proportional abundances of sequence-defined populations, and Shannon-Weaver indices, H′R (Table 2). An exceptionally low diversity was detected in mat community P3/4. Among the other communities the variation of SR was more than twofold, and Shannon-Weaver indices varied from 1.31 to 2.09, with coefficients of variation derived from triplicate analyses smaller than 7%. The lower detection limit for ethidium bromide-stained DNA in these gels was approximately 10 ng per band, which is 2% of the total amount of PCR product applied to each gel lane. To date, we have been able to extract, reamplify, and sequence DNA from the majority of gel bands. This analysis should allow the determination of phylogenetic relationships among organisms forming the investigated mats and cultivated reference strains, which is outside the scope of the present paper. However, it may be significant to note that no “heteroduplex bands” (12) could be detected and that all determined sequences were derived from cyanobacteria or plastids, so that most probably, none of the bands represents any undesired amplification product.
Diversity of carotenoids.
According to carotenoid pigment analysis, more than twofold differences in richness could be observed among mats. Richness SC was estimated to range from 9.33 ± 0.88 in mat P3/4 to 24.00 ± 1.00 in mat NC3 (Table 2). The corresponding estimates of the Shannon-Weaver diversity index, measured from the relative abundance of each carotenoid, varied between 1.62 and 2.53, almost twofold. The crowding of peaks was likely to have affected the results of HPLC analyses (Fig. 5). While estimates of carotenoid richness could be corrected accordingly, this was not attempted for Shannon-Weaver indices. Thus, the latter may underestimate diversity. There may have been some misidentification events with carotenoids, particularly with the less abundant ones, as the spectra from small peaks were not always clear. These may have been degradation products of original phototrophic carotenoids, or they may have stemmed from other bacteria.
DISCUSSION
Units of biodiversity.
Biologists studying the ecology of communities of plants or animals usually choose species as basic units for diversity estimates. For prokaryotes this is currently impossible because of the bacteriological practice of delineating species on the basis of cultivated strains (66). At present, probably far less than 5% of extant cyanobacterial species are successfully cultivated (6) and only a minority of these cultures are axenic (7). This disproportion reflects general problems of obtaining, within a scientist’s lifetime, a collection of strains that represents the microbial richness extant in an environmental sample. In addition, the current bacteriological species concept yields groupings that are not equivalent to species of larger organisms (54) and that do not necessarily correspond to real ecological units (43, 63). In contrast, the traditional phycological taxonomy enables the identification of morphological species without the need for cultivation, which is advantageous for ecological studies (2). But especially for organisms with less complex morphologies, such as unicellular or simple filamentous cyanobacteria, the power of this system of classification to identify evolutionarily and ecologically meaningful clusters may be rather limited (6, 27). However, the lack of any appropriate species concept does not hinder the estimation of microbial diversity. In fact, the predominant use of species as units of diversity in ecology is mainly due to practical reasons and may not always be the best solution possible (31, 59).
We compared estimates of the diversity of oxygenic phototrophic microorganisms as reflected in morphotypes, carotenoids, and 16S rRNA gene segments. All three approaches have limitations and potential drawbacks. Morphological groupings of mat-forming cyanobacteria may (13) or may not (11) represent phylogenetically coherent entities, and sibling morphological species have also been described for eukaryotic microalgae (34, 69). The potential variability of morphological traits with the conditions and state of growth may cause additional difficulties for identification in the field (16). rRNA genes are strongly conserved in function and structure, and their information content is therefore limited. Strains of bacteria with considerably different physiologies were reported to contain identical 16S rRNA genes (43). On the other hand, slightly different rRNA gene sequences detected in an environmental sample do not prove the presence of several microbial populations but may instead be derived from a single organism (37). Similarly, a single microalga or cyanobacterium usually produces a multiplicity of carotenoid types, and conversely, identical pigments may be extracted from several different algae.
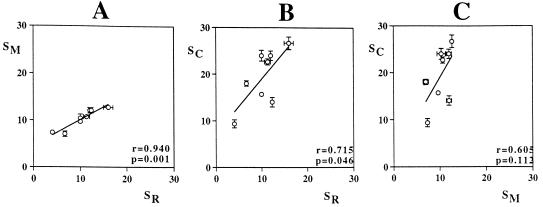
None of the approaches we applied enables the indisputable determination of absolute numbers of ecologically distinct populations forming a community. Yet the congruity of the results obtained indicates that all of the characteristics analyzed provide valuable information on the diversity of the organisms of interest. Therefore, their simultaneous investigation allows a meaningful comparison of the different communities in relative terms. Richness data obtained for the eight mat communities by applying three independent approaches are positively linearly correlated (Fig. 6), meaning that, on average, an increase in the number of unique 16S rRNA genes is accompanied by an increase in the number of morphotypes and types of carotenoids. The correlation of morphotype and 16S rRNA richness is particularly strong and highly significant. Interestingly, for six of the eight mats the ratio of SM to SR is 1.0 (mean values of triplicate analyses). However, in the communities with the highest (NC3) and lowest (P3/4) richnesses according to carotenoid and genetic data, these ratios are 0.8 and 1.8, causing the slope of the respective regression curve to deviate from one. The richness determined on the basis of carotenoids for all mats is higher than the number of either morphotypes or rRNA gene sequence types, which is due to the fact that all phototrophs produce more than a single type of carotenoid (19). The ratio of SC to SR ranges from 1.1 to 2.7, causing a less significant positive correlation of these data and possibly indicating that the number of pigments synthesized depends on the specific identity of the organism. Thus, for some communities richness estimates based on different approaches can be rather contradictory, which strongly confirms the notion that the analysis of any single characteristic can be misleading. But, very importantly, the simultaneous application of three independent approaches unveils an underlying trend of richness among the various communities investigated, which may now be related to further mat characteristics or other environmental parameters.
FIG. 6.
Relationships among estimates of the richness of oxygenic-phototrophic microorganisms in eight microbial mats based on triplicate analyses of morphologies (SM), 16S rRNA genes (SR), and carotenoids (SC). The arithmetic means and standard errors from triplicate analyses are shown. The Pearson correlation coefficient, r, and its statistical significance, P, have been calculated.
Proportional abundances.
The determination of abundances of microorganisms depends on meaningful units of counting, the choice of which is difficult and may to some extent depend on research objectives. Since microorganisms are clonal organisms, some authors have argued that colonies or trichomes instead of cells should be considered individuals (23, 60). However, in the densely packed communities within microbial mats, the boundaries of colonies often could not be recognized. Therefore, we simply counted cells and, as an alternative measure, weighted them by their respective individual volumes. When we applied these alternative approaches, drastically variable cell sizes caused significantly divergent estimates of the proportional abundances of certain morphotypes, seriously affecting Shannon-Weaver indices. While the abundances of cell components may reflect the relative proportions of organisms, the number of marker molecules per cell depends on the cell’s specific identity and physiological state. The production of carotenoids depends on environmental conditions, with the quantity and spectral quality of incident irradiation being especially important (42). The number of rRNA genes per cell varies with the number of copies per genome as well as the number of identical chromosomes (and replicated parts thereof) per cell. The latter may change considerably with growth conditions (4, 62).
Because of practical problems with all of the methodologies applied, the relative abundance of any population detected may deviate from that actually present in a sample. Morphotype-based abundance determinations are biased in favor of large organisms, since they are more likely to be encountered in focused microscopic planes. Procedures for extracting nucleic acids and pigments from mat samples may be selective in principle. However, the application of alternative protocols employing mechanical disruption (bead beating) or enzymatic digestion (lysozyme treatment) of cells resulted in no detectable differences in DGGE band patterns (data not shown), and microscopic observations indicated that complete lysis was achieved by the protocol that was finally applied. The quantitative use of PCR may be compromised when the reactions reach a plateau phase of amplification. This typically occurs at a product concentration of 10−8 M (50, 56), which is approximately the final molarity yielded in our experiments. By performing amplification reactions with a series of template dilutions, we could confirm that above this limit the integrated amplification efficiency decreased; however, the proportional abundances of DNA molecules with different sequences remained unaffected (data not shown). Other potential sources of bias cannot be excluded, however. Primer degeneracies and greatly varying G+C contents of amplified DNA molecules have been suspected to cause differential amplification efficiencies (61). Unknown target organisms may exist, the 16S rRNA genes of which do not contain the signature sites necessary for efficient amplification (38). Hybridization techniques may allow us to determine abundances of environmental nucleic acids more accurately than by PCR. However, their application depends on the availability of suitable nucleic acid probes, which either require prior knowledge of the sequences to be detected (synthetic oligonucleotide probes [1, 53]) or need to be prepared from available reference DNA (polynucleotide probes [21, 28]).
Despite the inherent difficulties of population abundance determinations, overall congruent estimates of diversity were obtained based on the analyses of 16S rRNA genes, morphotypes, and carotenoids. Shannon-Weaver indices calculated for the eight mat communities on the basis of the different approaches are positively linearly correlated (Fig. 7). Mainly because of the relatively low 16S rRNA gene diversity detected in community P3/4, the slopes of the regression lines deviate from one. Similar to the richness results, carotenoid analyses for all mats yielded higher absolute values for Shannon-Weaver indices than did other measurements. Shannon-Weaver indices based on proportional volumes of morphotypes (not shown in Fig. 7) are exceptional, since they are only weakly and insignificantly correlated with those based on cell number proportions (r = 0.35; P = 0.39) and not at all correlated with indices calculated on the basis of 16S rRNA genes (r = 0.09) and carotenoids (r = 0.02). Since individual cell volumes differed by up to 3 orders of magnitude, the transformation of cell counts into biovolumes significantly changed the proportional abundances calculated for certain morphotypes. As was the case for estimates of richness, binary comparisons of Shannon-Weaver indices for particular communities are also contradictory when based on different methodologies. This again reflects the limitations inherent in all of the identifying traits and their analyses.
Quantification of microbial diversity.
Facing the numerous limitations inherent in the various methodologies currently available, many authors conclude that an ecological evaluation of microbial diversity has not yet been convincingly reported (39, 57) or that it would be—at present—impossible (55). The identification of basic units of microbial diversity is difficult, and the determination of the proportional abundances of microbial populations can be very ambiguous. However, the latter may be of interest because, generally, rare species in a community have little effect on the overall flux of energy and matter but may instead become important under changing environmental conditions (51). Therefore, if any current activities of communities or ecosystems are investigated, diversity indices weighting populations by their proportional abundances may be more relevant than the number of distinct populations. Such indices also are less sensitive to the detection limits of the respective methodologies applied. This feature may be especially useful for the study of habitats such as soil, with difficult-to-determine and tremendously high microbial richness.
Many of the basic problems discussed in this paper are not specific to the exploration of the microbial world. For the majority of ecological collections the diversity can be estimated and expressed only in relative terms. A comprehensive census usually is not achievable, and in many cases even random samples cannot be drawn (44). Species concepts of larger organisms may themselves be as controversial as is their relevance for diversity estimates (32). Depending on research objectives, it may be more fruitful to take into account the organisms’ specific identities and their ecologically relevant properties (31, 59). However, diversity is an inherent aspect of community structure, and it has been reported to be related to ecosystem functioning and predictability (33, 58), the study of which is considered to be a grand challenge of ecological research (20). Our report demonstrates that the study of biomarker molecules enables the quantification of microbial richness and diversity in natural habitats. The analysis of pigments and morphotypes cannot be generally applied or will be less informative for microorganisms other than phototrophs. However, other identifying features may be investigated, such as cell wall components, fatty acids, enzyme activities, or other traits that can be related to a functional group of interest. Our approach can be considered analogous to the determination of species diversity for macroscopic organisms and makes achievable future synecological research on microbial diversity beyond purely descriptive studies.
ACKNOWLEDGMENTS
We gratefully acknowledge S. Koch and C. Wawer for numerous extractions of carotenoids and nucleic acids and Exportadora de Sal, S. A. de C. V., Baja California Sur, Mexico, for their continued logistic support. We thank E. Clavero for her introduction to the classification of the diatoms, R. Amann for use of the facilities, and T. Richter for the explicit formula for calculating PS(D).
The research described in this paper was financially supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostidis K, Komárek J. Modern approach to the classification system of cyanophytes. 1. Introduction. Arch Hydrobiol (Suppl) 1985;71:291–302. [Google Scholar]
- 3.Begon M, Harper J L, Townsend C R. Ecology—individuals, populations, communities. Oxford, United Kingdom: Blackwell Scientific Publications; 1990. [Google Scholar]
- 4.Birky C W, Jr, Walsh J B. Biased gene conversion, copy number, and apparent mutation rate differences within chloroplast and bacterial genomes. Genetics. 1992;130:677–683. doi: 10.1093/genetics/130.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius M, Dull T, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Castenholz R W. Species usage, concept, and evolution in the Cyanobacteria (blue-green algae) J Phycol. 1992;28:737–745. [Google Scholar]
- 7.Castenholz R W, Waterbury J B. Oxygenic phototrophic bacteria, group I. Cyanobacteria. In: Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams and Wilkins Co.; 1989. pp. 1710–1728. [Google Scholar]
- 8.Claridge M F, Dawah H A, Wilson M R. Practical approaches to species concepts for living organisms. In: Claridge M F, Dawah H A, Wilson M R, editors. Species: the units of biodiversity. London, United Kingdom: Chapman and Hall; 1997. pp. 1–15. [Google Scholar]
- 9.Des Marais D J. The biogeochemistry of hypersaline microbial mats. Adv Microb Ecol. 1995;14:251–274. doi: 10.1007/978-1-4684-7724-5_6. [DOI] [PubMed] [Google Scholar]
- 10.Embley T M, Stackebrandt E. Species in practice: exploring uncultured prokaryote diversity in natural samples. In: Claridge M F, Dawah H A, Wilson M R, editors. Species: the units of biodiversity. London, United Kingdom: Chapman and Hall; 1997. pp. 1–15. [Google Scholar]
- 11.Ferris M J, Ruff Roberts A L, Kopczynski E D, Bateson M M, Ward D M. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl Environ Microbiol. 1996;62:1045–1050. doi: 10.1128/aem.62.3.1045-1050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Pichel F, Prufert-Bebout L, Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Pichel, F., M. Kühl, U. Nübel, and G. Muyzer. Salinity-dependent limitation of photosynthesis and oxygen exchange in microbial mats. J. Phycol., in press.
- 15.Garcia-Pichel F, Mechling M, Castenholz R W. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl Environ Microbiol. 1994;60:1500–1511. doi: 10.1128/aem.60.5.1500-1511.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Pichel F, Nübel U, Muyzer G. The phylogeny of unicellular, extremely halotolerant cyanobacteria. Arch Microbiol. 1998;169:469–482. doi: 10.1007/s002030050599. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Pichel, F., U. Nübel, G. Muyzer, and M. Kühl. On cyanobacterial community diversity and its quantification. In C. Bell (ed.), Trends in microbial ecology, in press.
- 18.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin T W. The biochemistry of carotenoids. 2nd ed. Vol. 1. London, United Kingdom: Chapman and Hall; 1981. [Google Scholar]
- 20.Hanski I. Be diverse, be predictable. Nature. 1997;390:440–441. [Google Scholar]
- 21.Holben W E, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javor B. Hypersaline environments. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 23.Jordan T L, Staley J T. Electron microscopic study of succession in the periphyton community of Lake Washington. Microb Ecol. 1976;2:241–251. doi: 10.1007/BF02011645. [DOI] [PubMed] [Google Scholar]
- 24.Karsten U, Garcia Pichel F. Carotenoids and mycosporine-like amino acid compounds in members of the genus Microcoleus (cyanobacteria): a chemosystematic study. Syst Appl Microbiol. 1996;19:285–294. [Google Scholar]
- 25.Klug M J, Tiedje J M. Response of microbial communities to changing environmental conditions: chemical and physiological approaches. In: Guerrero R, Pedrós-Alió C, editors. Trends in microbial ecology. Madrid, Spain: Spanish Society for Microbiology; 1993. pp. 371–374. [Google Scholar]
- 26.Kolasa J, Biesiadka E. Diversity concept in ecology. Acta Biotheoretica. 1984;33:145–162. [Google Scholar]
- 27.Komárek J. Towards a combined approach for the taxonomy and species delimitation of picoplanktic cyanoprokaryotes. Arch Hydrobiol Suppl. 1996;117:377–401. [Google Scholar]
- 28.Lanoil B D, Giovannoni S J. Identification of bacterial cells by chromosomal painting. Appl Environ Microbiol. 1997;63:1118–1123. doi: 10.1128/aem.63.3.1118-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legendre L, Legendre P. Numerical ecology. Amsterdam, The Netherlands: Elsevier; 1982. [Google Scholar]
- 30.Ludwig J A, Reynolds J F. Statistical ecology. New York, N.Y: John Wiley and Sons; 1988. [Google Scholar]
- 31.May R. Conceptual aspects of the quantification of the extent of biological diversity. In: Hawksworth D L, editor. Biodiversity—measurement and estimation. London, United Kingdom: Chapman and Hall; 1995. pp. 13–20. [Google Scholar]
- 32.Mayden R. A hierarchy of species concepts: the denouement in the saga of the species problem. In: Claridge M F, Dawah H A, Wilson M R, editors. Species: the units of biodiversity. London, United Kingdom: Chapman and Hall; 1997. pp. 381–424. [Google Scholar]
- 33.McNaughton S J. Biodiversity and function of grazing ecosystems. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem function. Berlin, Germany: Springer-Verlag; 1993. pp. 361–382. [Google Scholar]
- 34.Medlin L K. Can molecular techniques change our ideas about the species concept? In: Joint I, editor. Molecular ecology of aquatic microbes. G38. Berlin, Germany: Springer-Verlag; 1995. pp. 133–152. [Google Scholar]
- 35.Medlin L K, Kooistra W H C F, Gersonde R, Wellbrock U. Evolution of the diatoms (Bacillariophyta). II. Nuclear-encoded small-subunit rRNA sequence comparisons confirm a paraphyletic origin for the centric diatoms. Mol Biol Evol. 1996;13:67–75. doi: 10.1093/oxfordjournals.molbev.a025571. [DOI] [PubMed] [Google Scholar]
- 36.Muyzer G, De Waal E D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nübel U, Garcia Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Donnell A G, Goodfellow M, Hawksworth D L. Theoretical and practical aspects of the quantification of biodiversity among microorganisms. In: Hawksworth D L, editor. Biodiversity—measurement and estimation. London, United Kingdom: Chapman and Hall; 1995. pp. 65–73. [DOI] [PubMed] [Google Scholar]
- 40.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 41.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 42.Paerl H W. Cyanobacterial carotenoids: their roles in maintaining optimal photosynthetic production among aquatic bloom-forming genera. Oecologia. 1984;61:143–149. doi: 10.1007/BF00396752. [DOI] [PubMed] [Google Scholar]
- 43.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 44.Pielou E C. The measurement of diversity in different types of biological collections. J Theor Biol. 1966;13:131–144. [Google Scholar]
- 45.Pierson B K, Valdez D, Larsen M, Morgan E, Mack E E. Chloroflexus-like organisms from marine and hypersaline environments: distribution and diversity. Photosynth Res. 1994;41:35–52. doi: 10.1007/BF02184144. [DOI] [PubMed] [Google Scholar]
- 46.Pinevich A V, Averina S G, Velichko N V. Another view on the role of photosynthetic pigments in taxonomy of oxygenic-phototrophic bacteria: proposed rejection of the order Prochlorales Florenzano, Balloni, and Materassi 1986 (Emend. Burger-Wiersma, Stal, and Mur 1989), the family Prochloraceae Florenzano, Balloni, and Materassi 1986, and the family Prochlorotrichaceae Burger-Wiersma, Stal, and Mur 1989. Int J Syst Bacteriol. 1997;47:1264–1267. [Google Scholar]
- 47.Pinhassi J, Zweifel U L, Hagström A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. Adv Microb Ecol. 1986;9:293–352. [Google Scholar]
- 49.Round F E, Crawford R M, Mann D G. The diatoms: morphology and biology of the genera. Cambridge, United Kingdom: Cambridge University Press; 1990. [Google Scholar]
- 50.Sardelli A D. Plateau effect—understanding PCR limitations. Amplifications. 1993;9:1–3. [Google Scholar]
- 51.Schulze E D, Mooney H A. Ecosystem function and biodiversity: a summary. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem function. Berlin, Germany: Springer-Verlag; 1993. pp. 497–510. [Google Scholar]
- 52.Shannon C E, Weaver W. The mathematical theory of communication. Urbana, Ill: University of Illinois Press; 1949. [Google Scholar]
- 53.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staley J T. Biodiversity: are microbial species threatened? Curr Opin Biotechnol. 1997;8:340–345. doi: 10.1016/s0958-1669(97)80014-6. [DOI] [PubMed] [Google Scholar]
- 55.Steinberg C E W, Geller W. Biodiversity and interactions within pelagic nutrient cycling and productivity. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem function. Berlin, Germany: Springer-Verlag; 1993. pp. 497–510. [Google Scholar]
- 56.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiedje J M. Approaches to the comprehensive evaluation of prokaryote diversity of a habitat. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function. Oxon, United Kingdom: CAB International; 1995. pp. 73–87. [Google Scholar]
- 58.Tilman D, Downing J A. Biodiversity and stability in grasslands. Nature. 1994;367:363–365. [Google Scholar]
- 59.Tilman D, Knops J, Wedin D, Reich P, Ritchie M, Siemann E. The influence of functional diversity and composition on ecosystem processes. Science. 1997;277:1300–1302. [Google Scholar]
- 60.Tinnberg L. Phytoplankton diversity in Lake Norvikken 1961–1975. Holarctic Ecol. 1979;2:150–159. [Google Scholar]
- 61.Von Wintzingerode F, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 62.Wagner R. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch Microbiol. 1994;161:100–109. doi: 10.1007/BF00276469. [DOI] [PubMed] [Google Scholar]
- 63.Ward D M. A natural species concept for prokaryotes. Curr Opin Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. [DOI] [PubMed] [Google Scholar]
- 64.Ward D M, Weller R, Bateson M M. 16S ribosomal RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 65.Washington H G. Diversity, biotic and similarity indices. Water Res. 1984;18:653–694. [Google Scholar]
- 66.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 67.Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–25. [Google Scholar]
- 68.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood A M, Leatham T. The species concept in phytoplankton ecology. J Phycol. 1992;28:723–729. [Google Scholar]