Abstract
Extracellular vesicles (EVs) are rounded vesicles enclosed by a lipid bilayer membrane, released by eukaryotic cells and by bacteria. They carry various types of bioactive substances, including nucleic acids, proteins, and lipids. Depending on their cargo, EVs have a variety of well‐studied functions in mammalian systems, including cell‐to‐cell communication, cancer progression, and pathogenesis. In contrast, EVs in plant cells (which have rigid walls) have received very little research attention for many decades. Increasing evidence during the past decade indicates that both plant cells and plant pathogens are able to produce and secrete EVs, and that such EVs play key roles in plant–pathogen interactions. Plant EVs contains small RNAs (sRNAs) and defence‐related proteins, and may be taken up by pathogenic fungi, resulting in reduced virulence. On the other hand, EVs released by gram‐negative bacteria contain a wide variety of effectors and small molecules capable of activating plant immune responses via pattern‐recognition receptor‐ and BRI1‐ASSOCIATED RECEPTOR KINASE‐ and SUPPRESSOR OF BIR1‐mediated signalling pathways, and salicylic acid‐dependent and ‐independent processes. The roles of EVs in plant–pathogen interactions are summarized in this review, with emphasis on important molecules (sRNAs, proteins) present in plant EVs.
Keywords: effector‐triggered immunity, exosome, extracellular vesicles, pathogen‐associated molecular patterns‐triggered immunity, plant, plant disease, systemic acquired resistance
Extracellular vesicles, the rounded vesicles released by both plants and plant‐pathogenic pathogens, play multifaceted roles in plant–microbe interactions (fungi, viruses, and bacteria).
1. INTRODUCTION
Plants are typically exposed to a variety of potential pests and pathogens throughout their lifespan. During the course of evolution, plants have developed numerous defensive mechanisms against infection by pathogens. Defensive responses at infection sites are initiated by recognition of pathogen‐associated molecular patterns (PAMPs) that are conserved in pathogenic microbes; such responses are collectively termed PAMP‐triggered immunity (PTI) (Zhang et al., 2018). In a typical case of plant–pathogen co‐evolution, microbes have developed effector proteins that they release inside host cells and which interfere with PTI. Plants detect such effector proteins through resistance (R) genes and activate a more robust and faster type of defensive response, termed effector‐triggered immunity (ETI) (Naveed et al., 2020; Qi et al., 2011). In some cases, mobile signalling molecules such as glycerol‐3‐phosphate, azelaic acid, pipecolic acid, and N‐hydroxy‐pipecolic acid are synthesized in infected tissues and translocated to pathogen‐free systemic tissues, where they induce defensive responses (Chanda et al., 2011; Hartmann et al., 2018; Jung et al., 2009; Návarová et al., 2012). This process is termed systemic acquired resistance (SAR). Direct communication between plants and pathogens involves transport of substances (e.g., effector proteins, antimicrobial peptides) across cellular boundaries from pathogens to host cells, or vice versa (Cai et al., 2021).
Studies during the past decade have documented the crucial roles of extracellular vesicles (EVs) in the exchange of molecules between plants and pathogenic microbes. EVs are membranous, rounded vesicles that are produced and secreted by prokaryotic and eukaryotic cells under both normal and pathophysiological conditions. The EVs secreted by eukaryotic cells can be classified into three major groups with distinct biogenesis mechanisms: (a) apoptotic bodies (1000–5000 nm diameter), which are generated by membrane blebbing and released during late stages of programmed cell death; (b) microvesicles (100–1000 nm diameter), which are shed directly from the plasma membrane (PM) surface through external budding; and (c) exosomes (30–150 nm diameter), which are released following fusion of a multivesicular body with the PM (Akers et al., 2013; György et al., 2011; Liebana‐Jordan et al., 2021; van der Pol et al., 2012).
In early studies, exosomes were viewed as structures involved in the disposal of cellular wastes. However, more recent studies have revealed their essential roles in cell communication, developmental processes, tissue repair, fertilization, and immune responses (Théry et al., 2002). Exosomes contain a variety of bioactive substances, including nucleic acids, proteins, and lipids, and can be taken up by target cells for delivery of their cargo (Doyle & Wang, 2019; Macia et al., 2019; Mu et al., 2014; Rodrigues et al., 2018; Valadi et al., 2007).
The presence and functions of EVs in mammalian systems have been well documented. Most exosome research to date has been performed using animal body fluids and cell cultures. Although Jensen's group observed EVs in carrot cells in 1967 (Halperin & Jensen, 1967), studies on exosome function are far fewer, and slower to progress, for plants than for animals. Recent reports from Jin's group indicate that both plants and plant pathogens can release EVs, which play key roles in plant–pathogen communication (Cai et al., 2018; Cai, He & Jin, 2019). In this review, we summarize recent advances in the roles of EVs in research on plant–microbe interactions (fungi, viruses, and bacteria), and on the important molecules (small RNAs [sRNAs], proteins) found in plant EVs.
2. BOTH PLANT AND PLANT‐PATHOGENIC MICROBES CAN RELEASE EVS
Jensen's group observed EVs in carrot cells by electron microscopy in 1967 (Halperin & Jensen, 1967). However, the presence of a cell wall in plant cells continued to be viewed as an obstacle to the production or perception of vesicles, and almost no research on plant EVs was performed in the following decades. In 2009, de la Canal's group isolated EVs from sunflower apoplastic fluids and demonstrated that plant cells, like animal cells, can secrete exosomes into the intercellular space (Regente et al., 2009). Studies during the past decade have shown that plant EVs are derived from multivesicular body endosomes (MVBs) and exocyst‐positive organelles (EXPO), contain distinctive sets of RNAs, proteins, lipids, and metabolites (Figure 1), and are functionally involved in defensive responses against pathogens (Cai et al., 2018; He et al., 2021; Liu et al., 2020; Regente et al., 2017; Rutter & Innes, 2017).
FIGURE 1.
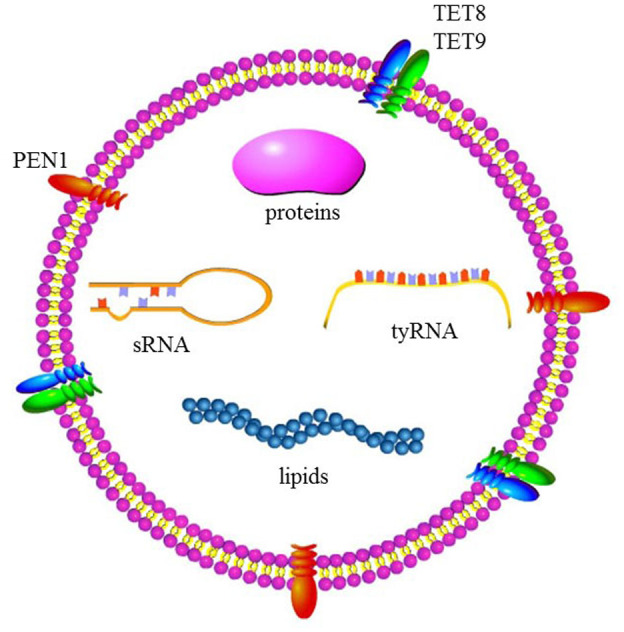
Cargo of plant extracellular vesicles (EVs). PEN1, plant‐specific penetration 1; sRNA, small RNA; TET, tetraspanin; tyRNA, tiny RNA
Bacterial pathogens are also capable of secreting EVs. Bacterial EVs were first described by Work's group in 1966 (Knox et al., 1966). Gram‐positive bacteria have thick cell walls and were previously considered to be incapable of producing EVs. However, recent studies have demonstrated that some of them do have such a capability, although the biological functions of the secreted EVs remain unclear (Bose et al., 2020). Gram‐negative bacteria can produce EVs by pinching off part of their outer membrane (Kulp & Kuehn, 2010); the resulting structures are known as outer membrane vesicles (OMVs). The OMVs are surrounded by a lipid bilayer, ranging from 20 to 400 nm in size (da Rocha et al., 2020), and contain various types of macromolecules, including signalling compounds, nucleic acids, virulence factors, toxins, and lipopolysaccharides (Bose et al., 2020). They are involved in numerous bacterial functions, including cell‐to‐cell communication, biofilm formation, tolerance to physiological stress factors, adherence to hosts, and modulation of host immunity (Bose et al., 2020; Manning & Kuehn, 2011). Our knowledge of OMVs secreted by plant‐pathogenic bacteria is fragmentary, but recent studies on plant–bacteria interactions suggest that OMVs play roles in biofilm formation, virulence, and modulation of plant immunity (Bose et al., 2020; Katsir & Bahar, 2017; McMillan et al., 2021).
Our knowledge of EVs produced by pathogens is based mainly on studies of those secreted by human‐pathogenic gram‐negative bacteria (Pathirana & Kaparakis‐Liaskos, 2016). Relatively few studies have focused on fungal EVs. Hu's group presented the first evidence that fungi secrete EVs into the extracellular microenvironment (Rodrigues et al., 2007). They demonstrated that extracellular capsular polysaccharide, the major virulence factor of Cryptococcus neoformans, is transported via EVs during growth in vitro and during infection in vivo. Later studies revealed that fungal EVs contain various cargo (proteins, nucleic acids, lipids, and polysaccharides) (Silva et al., 2013). The EVs from Candida albicans elicit a notable biological response in immune cells and can be internalized by macrophages and dendritic cells (Vargas et al., 2015). Recent studies have reported that plant‐pathogenic fungi, such as Fusarium oxysporum f. sp. vasinfectum and Zymoseptoria tritici (Bleackley et al., 2020; Hill & Solomon, 2020), can also release EVs. Some research progress has been made regarding functions of EVs during interactions between pathogenic fungi and mammalian cells, but much less is known about the roles of EVs in plant–fungus interactions.
Plants and plant‐pathogenic fungi and bacteria can secrete EVs. However, the existence of the cell wall might prevent the passage of EVs. Thus, some fundamental issues need to be elucidated: (a) How do EVs pass through the cell walls of plant cells and fungi? Focused ion beam‐scanning electron microscopy analyses by Movahed et al. (2019) revealed that plant EVs are capable of penetrating the cell wall. Plant EVs contain abundant cell wall‐modifying proteins (Movahed et al., 2019; Regente et al., 2017; Rutter & Innes, 2017). Therefore, one possible mechanism is that plant EVs degrade a part of the cell wall to allow their passage. (b) Are plant cells able to take up EVs from plants/bacteria/fungi? The EVs mediate communication between cells by transferring signal molecules after they are taken up by the target cells. Therefore, in further research on the function of EVs in plant–microbe interactions, it will be interesting to determine whether plant cells in suspension culture can take up EVs produced by plants/bacteria/fungi.
3. ROLES OF EVS IN PLANT–FUNGUS INTERACTIONS
3.1. Plant EVs inhibit fungal growth and virulence by transferring their cargo into fungal cells
A proteomic analysis of the EVs isolated from sunflower seedlings revealed that they contain numerous defence‐related proteins (pathogenesis‐related [PR] proteins [PR4, PR5, PR9, and PR14], disease resistance dirigent protein, Gnk2 antifungal protein), and also GDSL (consensus amino acid sequence motif of Gly, Asp, Ser, and Leu around the active site Ser) lipase acyl hydrolases, lectins, and germin‐like proteins (Regente et al., 2017). Tucci's group found that EVs released by tomato root cells contain a wide variety of proteins involved in perception and transduction of plant–pathogen interactions and typical defence‐related proteins, and strongly inhibit spore germination of several fungal pathogens, including F. oxysporum, Botrytis cinerea, and Alternaria alternata (Palma et al., 2020). These findings suggest that plant EVs play an essential role in communication between host plants and fungal pathogens.
Small RNAs, which include microRNAs (miRNAs) and small interfering RNAs (siRNAs), are short regulatory RNAs that silence genes with complementary sequences (Baulcombe, 2004). The miRNAs are processed by Dicer‐like (DCL) proteins from single‐stranded stem‐loop‐forming RNA precursors, while siRNAs are processed by DCL proteins from double‐stranded RNA (dsRNA) precursors (Cai, He, Weiberg, et al., 2019). The expression of sRNAs targeting fungal transcripts in barley and wheat during host‐induced gene silencing was found to affect the development of Blumeria graminis (powdery mildew fungus) (Mu et al., 2014), indicating that RNA molecules can be transferred from plants to the invading fungus, where they silence the expression of target genes. Interestingly, transmission electron microscopy (TEM) analyses revealed exosome‐like vesicles in the extrahaustorial matrix (the space between the symbiont's plasma membrane and the extrahaustorial membrane of the host) (Micali et al., 2011). Such exosome‐like vesicles may mediate the transfer of sRNAs between the plant and the fungus, although it is unclear from which organism they originate. The EVs from sunflower seedlings can be taken up by the fungal pathogen Sclerotinia sclerotiorum and inhibit its growth, suggesting that they may transport sRNA molecules from the host plant to the fungus (Regente et al., 2017). Hailing's group isolated pure fungal cells from infected Arabidopsis tissues and identified 42 Arabidopsis sRNAs from those cells (Cai et al., 2018). Among them, 32 sRNAs, including TAS1c‐siR483, TAS2‐siR453, and IGN‐siR1 (whose levels were higher than those of TAS1c‐siR585, TAS2‐siR710, and IGN‐siR107), were also identified in EVs isolated from extracellular apoplastic fluids from fungus‐infected leaves. A nuclease protection assay demonstrated that the transferred host sRNAs localized inside the EVs because they were protected from nuclease digestion. B. cinerea cells can take up plant EVs, indicating that EVs secreted by plant cells can transfer sRNAs into fungal cells. Knockout of an sRNA synthesis gene in Arabidopsis rendered it more susceptible to B. cinerea infection, and knockout of Arabidopsis sRNA target genes in B. cinerea reduced its virulence (Cai et al., 2018). These findings indicate that plant EVs affect plant–pathogen interactions by transferring sRNAs into pathogens, thereby reducing their virulence (Figures 1 and 2a).
FIGURE 2.
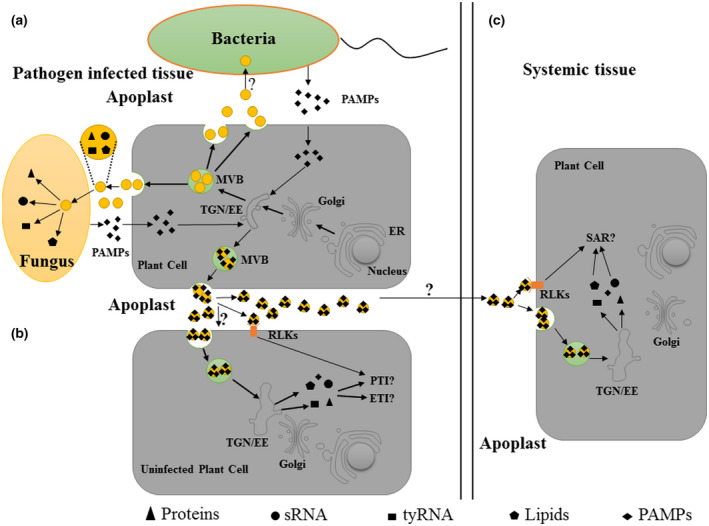
Function of plant extracellular vesicles (EVs) in plant–microbe interactions. (a) Plant EVs can be taken up by pathogens and can inhibit the virulence of pathogens by releasing their cargos (small RNAs [sRNA], proteins, tiny RNAs [tyRNA], and lipids). Meanwhile, pathogen‐associated molecular patterns (PAMPs) from pathogens can be loaded into plant EVs in pathogen‐infected cells. (b) Plant EVs may be functionally related to the signal transmission of PAMP‐triggered immunity (PTI) and effector‐triggered immunity (ETI). First, PAMPs in plant EVs released by infected cells are recognized by receptor‐like kinases (RLKs) located on the membrane to activate PTI in adjacent uninfected cells. Second, plant EVs contain abundant proteins involved in PTI and ETI. When plant cells perceive pathogens, the contents of plant EVs secreted by these cells may change. These EVs can be taken up by adjacent pathogen‐free cells and induce them to produce PTI and ETI downstream signals. (c) Plant EVs may be functionally related to systemic acquired resistance (SAR) signal transmission. Plant EVs contain SAR‐related proteins and PAMPs. During SAR, EVs released by pathogen‐infected cells may be transported to systemic tissues via the apoplast to transmit SAR signals when they are taken up by target cells or induce an immune response by the recognition of PAMPs and RLKs in systemic tissues. ER, endoplasmic reticulum; MVB, multivesicular bodies endosomes; TGN/EE, trans‐Golgi network/early endosome
An EV‐sRNA profiling analysis revealed that numerous sRNAs were less abundant in EVs than in Arabidopsis. The EVs isolated from B. cinerea‐infected leaves were found to be enriched with trans‐acting siRNA locus 1C‐derived siRNA483 (TAS1c‐siR483), but not TAS1c‐siR585, even though the two sRNAs are generated from the same TAS1c mRNA precursor and are among the 20 most abundant sRNAs in Arabidopsis (Cai et al., 2018). This finding suggests that sRNAs are selectively loaded by host plants into EVs during pathogen infection. In eukaryotes, Argonaute (AGO) proteins bind selectively to specific sRNAs and then guide the RNA‐induced silencing complex to corresponding target genes by complementary base pairing between the target mRNA and the sRNA guide strand (Krishnatreya et al., 2021; Liu et al., 2017). To elucidate the mechanism underlying selective loading of RNAs into EVs by plants, He et al. (2021) used proteomic analysis to identify various RNA‐binding proteins in Arabidopsis EVs; these included AGO1, DEAD‐box RNA helicases (RH11, RH37), and annexins (ANN1, ANN2). In their analyses, AGO1 bound selectively to EV‐enriched sRNAs but not to non‐EV‐associated sRNAs, indicating its involvement in selective loading of sRNAs into EVs. In addition, the sRNA content was lower in EVs isolated from ago1 plants than in those isolated from wild‐type plants. The Arabidopsis genome encodes 10 AGOs, which are divided into three distinct phylogenetic clades (AGO1/5/10, AGO4/6/8/9, AGO2/3/7) based on their functional domains, indicating potential functional redundancy within these clades (Jullien et al., 2020). Each AGO binds to a distinct set of sRNAs; however, only AGO1 and its associated sRNAs were detected in EVs (He et al., 2021). These findings suggest a crucial role of AGO1 in sRNA loading and/or stabilization in EVs.
Meyers' group observed that Arabidopsis EVs are highly enriched in sRNAs with a length of 10–17 nucleotides, which they termed tiny RNAs (tyRNAs) (Figure 1) (Baldrich et al., 2019). The tyRNAs may be derived from degradation of mRNAs, primary miRNAs, siRNAs, tasiRNAs, or hcRNAs. The cellular functions of tyRNAs are unknown. In mammalian cells, EVs mediate long‐distance transport of mRNAs and noncoding RNAs, in addition to sRNAs. The RNAs protected within the EV lumen may be transported to distant cells and retain their function (Mittelbrunn et al., 2011; Ratajczak et al., 2006). Whether these types of RNA are present in plant EVs and whether they are involved in plant–pathogen interactions are yet to be determined.
The composition and function of exosomes secreted by different animal cells are distinct. Different types of exosomes are able to specifically interact with a target cell or microbe (Théry et al., 2009). The EVs secreted by plants contain a variety of stress‐related substances and toxic compounds. Thus, plant cells must be able to avoid taking up these EVs for successful growth and development. Like animal cells, plant cells may be able to secrete different types of EVs that have distinct cargo and target cells, for example specific plant cells or pathogen cells. This speculation is supported by the results of a comparative proteomic analysis of virus‐induced and bacteria‐induced EVs in Arabidopsis (Movahed et al., 2019; Rutter & Innes, 2017), where only about 41% proteins were shared by both proteomes. In the mammalian system, ligand–receptor interactions are an important mechanism for the uptake of exosomes by target cells (Escrevente et al., 2011). Exosome uptake was reduced when exosomes or cancer cells were treated with proteinase K, suggesting that protein recognition is an important part of this process (Escrevente et al., 2011). Regente et al. (2017) and Rutter and Innes (2017) identified many receptor proteins in Arabidopsis EVs. Further research is required to determine whether these proteins are involved in the specific uptake of exosomes by target pathogens.
3.2. Fungal EVs induce a phytotoxic response in plants
Although many studies have explored the functions of plant EVs in interactions between pathogenic fungi and plant cells, fewer have focused on the roles of fungal EVs in plant–fungus interactions. Anderson's group observed that EVs from F. oxysporum f. sp. vasinfectum (cotton wilt pathogen) induced a phytotoxic response in cotton and Nicotiana benthamiana plants (Bleackley et al., 2020). Interestingly, the EVs isolated from F. oxysporum f. sp. vasinfectum were coloured deep purple. These findings, combined with the results of a proteomic analysis of F. oxysporum f. sp. vasinfectum EVs, suggested a possible role of EVs as sites of pigment and/or toxin biosynthesis. Similarly, a proteomic analysis of EVs isolated from the wheat‐pathogenic fungus Z. tritici revealed that they contained a set of putative virulence‐associated proteins (Hill & Solomon, 2020). The above findings provide further evidence that fungal EVs have important functions in host plant–fungus interactions.
4. ROLES OF EVS IN PLANT–BACTERIA INTERACTIONS
4.1. Functions of plant EVs in plant–bacteria interactions
So far, there have been no reports on the functions of plant EVs in plant–bacteria interactions. In mammalian systems, EVs released by bacteria‐infected cells contain PAMPs and abundant host proteins, and are able to induce an immune response in noninfected adjacent cells (Anand et al., 2010; Bhatnagar & Schorey, 2007). These findings demonstrate that exosomes released by bacteria‐infected cells play a role in promoting the immune response on bacterial infection. Similarly, EVs from Pseudomonas syringae‐ and mock‐infected Arabidopsis were found to contain numerous signal transduction proteins related to biotic stress (Rutter & Innes, 2017), suggesting that plant EVs may also transmit immune signals during bacterial infection. The possible signal transduction roles of plant EVs during the immune response will be discussed later in this review. In addition, mammalian cells can deliver sRNA to bacterial cells via EVs. These sRNAs are able to decrease the formation of antibiotic‐resistant biofilms by suppressing the expression of several genes essential for biofilm formation (Koeppen et al., 2021). The secretion of EVs in P. syringae‐infected Arabidopsis was approximately double that in uninfected controls (Rutter & Innes, 2017), indicative of the functional involvement of plant EVs in resistance to bacterial pathogens. Because plant EVs contain sRNAs and pathogenesis‐related proteins (Cai et al., 2018; Regente et al., 2017), it is reasonable to speculate that they can inhibit bacterial growth and virulence by delivering their cargo to bacterial cells.
4.2. OMVs activate plant immune responses
Proteomic analyses of OMVs secreted by plant‐pathogenic bacteria have revealed that they contain effectors and various types of virulence factors (Bose et al., 2020; Chowdhury & Jagannadham, 2013; Kulkarni et al., 2015; Sidhu et al., 2008), suggesting that they help modulate plant immune responses. Ronald's group was the first to report such a function (Bahar et al., 2016). In an Arabidopsis model, OMVs from the aerobic, gram‐negative bacteria Xanthomonas campestris pv. campestris and Xanthomonas oryzae pv. oryzae rapidly induced a burst of reactive oxygen species (ROS) and the expression of defence‐related genes. The presence of the immune elicitor EF‐Tu (elongation factor‐thermo unstable) in OMVs was detected by western blot analysis. In plants, PTI is mediated by surface pattern‐recognition receptors (PRRs) that detect PAMPs in the extracellular microenvironment. In the Arabidopsis model, flagellin sensitive 2 (FLS2) and elongation factor receptor (EFR) are the best‐characterized PRRs. Recognition of flagellin by FLS2, or of EF‐Tu by EFR, induces PTI (Gómez‐Gómez & Boller, 2000; Zipfel et al., 2006). efr‐1 plants are insensitive to OMVs, suggesting that the ROS burst induced by OMVs is mediated by EFR (Bahar et al., 2016). The immune co‐receptors BRI1‐ASSOCIATED RECEPTOR KINASE (BAK1) and SUPPRESSOR OF BIR1 (SOBIR1) are involved in perception of OMVs and downstream signalling (Figure 3). Notably, many other Arabidopsis PRR mutant lines are able to respond to OMVs, suggesting that OMVs contain other immune elicitors besides EF‐Tu (Bahar et al., 2016).
FIGURE 3.
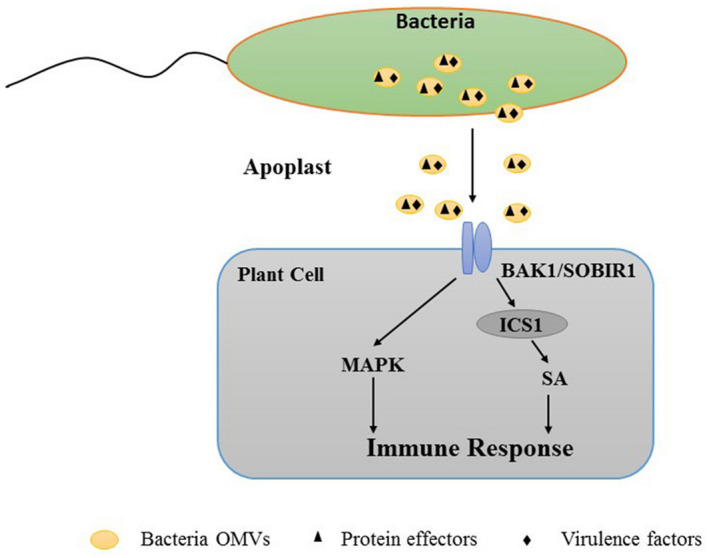
Bacterial outer membrane vesicles (OMVs), which contain protein effectors, can elicit plant protective immune responses. The immune co‐receptors BAK1 and SOBIR1 mediate the perception of, and the response to, OMVs. The OMVs induce immune responses via salicylic acid (SA)‐dependent/‐independent processes and MAPK pathways
Kuehn's group observed that OMVs from P. syringae and Pseudomonas fluorescens activate plant immune responses against bacterial and oomycete pathogens (McMillan et al., 2021). ETI is induced by recognition of bacterial type III secretion system (T3SS) effector‐mediated modifications of plant proteins (Jones & Dangl, 2006; Spoel & Dong, 2012). Both effectors and components of the T3SS machinery have been identified in association with OMVs (Kulkarni & Jagannadham, 2014). It is reasonable to speculate that immune responses induced by OMVs are related to the effectors they contain. However, in a recent study by McMillan et al. (2021), OMVs from T3SS mutants that do not express effector proteins induced a normal immune response, indicating that T3SS effectors are not responsible for the induction of protective immunity by OMVs (McMillan et al., 2021). The protective effect of OMVs was only slightly affected by proteinase K treatment or boiling, indicating that OMV‐mediated resistance to bacterial infection is related not to OMV protein components, but rather to other OMV‐associated molecules (metabolites, lipids, small protein epitopes). A variety of salicylic acid (SA)‐dependent and ‐independent immune responses were found to be induced by OMVs. Plants treated with P. syringae OMVs showed increased ICS1 expression and accumulated SA, whereas those treated with P. fluorescens OMVs did not, reflecting important differences in plant immune responses activated by OMVs from different bacteria. The P. syringae OMVs induced MAPK phosphorylation in a much more sustained manner than did the well‐characterized PAMP signalling elicited by flg22, indicating that OMVs activate different—probably more complex—plant immune responses than does a single PAMP signal (Figure 3). Further research is required to explore the detailed mechanisms of OMV‐mediated activation of plant immunity. Of particular interest is the identification of OMV‐associated components that activate the plant immune response. Such information will greatly facilitate the development of novel, more effective biopesticides.
4.3. OMVs facilitate infection by pathogenic bacteria
Many studies have explored the functions of OMVs in mammalian systems, where they promote the secretion efficiency of virulence factors (Bonnington & Kuehn, 2014; Lloubes et al., 2013). The direct uptake of OMVs by plant cells has not yet been documented, but it is reasonable to assume that plant‐pathogenic bacteria deliver a variety of cargo to plant cells via EVs. Plant cell walls, composed mainly of pectin, cellulose, and hemicellulose, are barriers to the transport of factors from bacteria to plant cells. To overcome this obstacle, bacteria have evolved a variety of cell wall‐degrading enzymes using type II secretion systems (T2SSs), for example polygalacturonases, cellulases, amylases, and xylanases. These enzymes degrade the rigid plant cell wall, thereby facilitating the transport of virulence factors into the cell (Faulkner & Robatzek, 2012; Gibson et al., 2011; Hématy et al., 2009; Rajeshwari et al., 2005; Szczesny et al., 2010). Büttner's group demonstrated that T2SS substrates such as xylanases were secreted by X. campestris pv. vesicatoria via OMVs; T2SS mutants did not produce or secrete xylanases, and the presence of xylanases in OMVs in wild‐type X. campestris pv. vesicatoria was confirmed by immunoelectron microscopy (Solé et al., 2015). These findings indicate that OMVs provide an alternative transport route to the extracellular microenvironment for a variety of virulence factors, including T2SS substrates, and play a functional role in pathogen infection.
Lindow's group found that OMVs were involved in the migration ability and virulence of the aerobic, gram‐negative bacterium Xylella fastidiosa (Ionescu et al., 2014). The secretion of OMVs was found to be suppressed by a diffusible signal factor‐dependent quorum‐sensing system. A ΔrpfF strain with disrupted quorum signalling showed greater OMV production and stronger virulence to plants, relative to the wild‐type strain, but lower adhesion to glass or plant surfaces. The adhesion of X. fastidiosa cells to xylem vessels was strongly suppressed by OMV treatment, and the OMVs facilitated cell movement in the xylem and enhanced virulence. It remains to be determined whether OMVs from other plant‐pathogenic bacteria perform similar functions.
5. ROLES OF EVS IN PLANT–VIRUS INTERACTIONS
The EVs generated in mammals during virus infection have been extensively studied. Exosomes isolated from hepatitis C virus (HCV)‐infected human hepatoma Huh7.5.1 cells were found to contain full‐length viral RNAs, viral proteins, and particles, and successfully transmitted infection to naive cells and established productive infection (Ramakrishnaiah et al., 2013), suggesting that exosome‐mediated viral transmission plays a role in immune‐evasive properties of the virus. In contrast, exosomes may inhibit viral transmission through various mechanisms. PAMPs, such as viral nucleic acids, are recognized by pattern recognition receptors (PRRs) that mobilize innate immune responses (Kawai & Akira, 2009). Chisari's group observed that exosomes from HCV‐infected cells transferred viral PAMP (RNA) to plasmacytoid dendritic cells (pDCs) and triggered host innate immune responses such as production of antiviral type 1 interferons (Dreux et al., 2012). Therefore, exosomes secreted by virus‐infected cells play dual roles in animal cells: they can both enhance and inhibit viral infection (Jia et al., 2021). However, information about the function of plant EVs in plant–virus interactions is rather limited. Turnip mosaic virus (TuMV) is a positive‐sense, single‐stranded RNA virus belonging to the order Picornavirales. Zheng's group reported, based on immunogold labelling and confocal microscopy analyses, that plant EVs in infected N. benthamiana leaves contain TuMV replication complexes (proteins and RNA) (Movahed et al., 2019). Proteomic analyses of EVs isolated from TuMV‐infected plants revealed the presence of numerous proteins, including those involved in TuMV replication (putative poly(A)‐binding protein, PABP) and in TuMV infection (S‐adenosylhomocysteine hydrolase, AGO2), as well as a protein involved in the plant immunity response (14‐3‐3 protein), and transcription factors found exclusively in TuMV‐enriched EVs. These findings suggest a possible dual role of EVs in plant–virus interactions: (a) plant EVs containing viral replication complexes facilitate the spread of viruses from infected cells to adjacent cells or systemic tissues via the phloem. Further studies should focus on whether EVs isolated from phloem sap of virus‐infected tissues contain viral replication complexes, although it is challenging to isolate a large volume of phloem sap; (b) EVs act as carriers of PAMPs or signal molecules to mediate the transmission of immune signals against viruses from infected cells to adjacent cells. It will be interesting to determine whether the viral components in plant EVs are able to function as PAMPs to induce plant immune responses (Figure 4).
FIGURE 4.
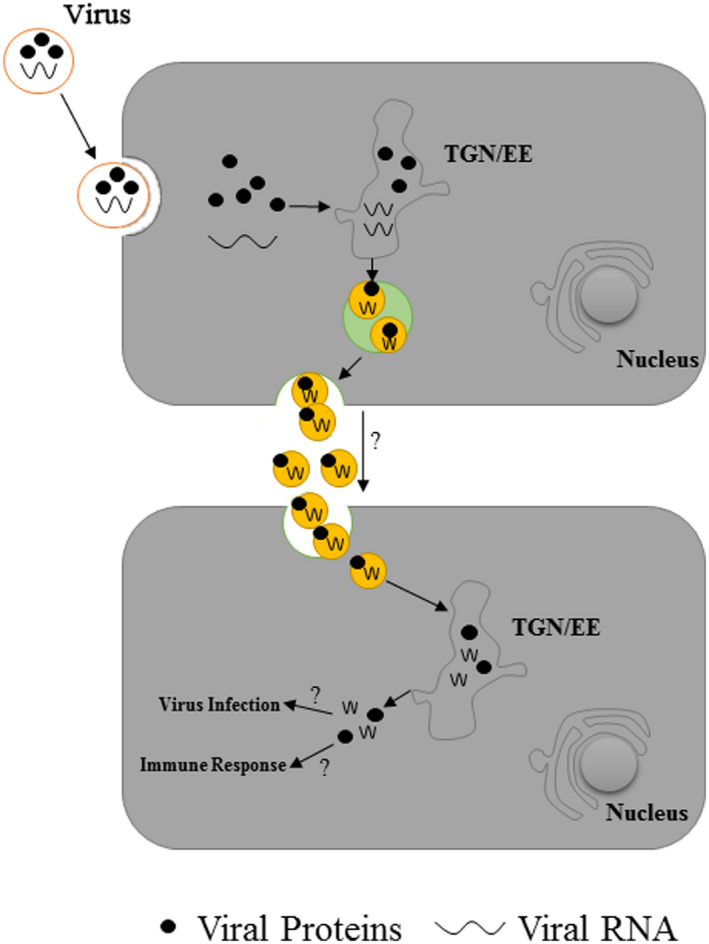
Plant extracellular vesicles (EVs) are involved in the virus‐induced immune response. Plant EVs released by virus‐infected cells contain viral proteins and RNA. These EVs may facilitate the spread of viruses from infected cells to adjacent cells, or act as pathogen‐associated molecular pattern (PAMP) carriers to induce plant immune responses
6. ROLES OF PLANT EVS IN PTI, ETI, AND SAR
Multivesicular bodies proliferate in plant cells under pathogen challenge, indicating increased exosome secretion (An et al., 2006; Wang et al., 2014). The secretion of EVs was higher in P. syringae‐infected Arabidopsis than in uninfected controls (Rutter & Innes, 2017), suggesting functional involvement of plant EVs in resistance to bacterial pathogens. In plants, SA is an important phytohormone that plays key roles in various defence responses, including SAR, basal defence, and resistance gene‐mediated defence (Lu et al., 2016). Rutter and Innes (2017) observed that SA treatment promoted EV secretion in Arabidopsis, suggesting that plant EVs are involved in intercellular transmission of immune signals. Along this line, a wide variety of proteins associated with immune signal transmission, ROS signalling, and defence‐related proteins have been identified in EVs using proteomic analyses (Movahed et al., 2019; Regente et al., 2017; Rutter & Innes, 2017).
Plants have a two‐tiered innate immune system comprising PTI and ETI, of which PTI is the first tier. On infection of plants by pathogens, PRRs located on the cell membrane recognize PAMPs released by the pathogens and activate PTI (Li et al., 2018). The PRRs comprise receptor‐like kinases (RLKs) and receptor‐like proteins (RLPs) (Wu et al., 2019). The proteins CARD1, KIN7, LRR‐RLK 2, and SIF3 are among the RLKs identified in Arabidopsis EVs isolated from both P. syringae‐infected and control plants (Movahed et al., 2019; Rutter & Innes, 2017). Plant EVs are localized mainly in extracellular spaces, and conceivably play some role in PTI (Figure 2b).
Through co‐evolution, pathogens have developed effectors to suppress PTI and promote efficient infection. In turn, plants have developed R proteins that respond to a particular T3SS effector by activating specific immune responses (an example of ETI). In Arabidopsis, RPM1‐INTERACTING PROTEIN 4 (RIN4) is phosphorylated by P. syringae effectors, and consequently activates R protein signalling and initiates an immune response (Mackey et al., 2002, 2003). RIN4 and numerous other proteins that interact with it are present in plant EVs (Rutter & Innes, 2017), indicating functional involvement of EVs in ETI. The EV proteome contains many calcium‐dependent protein kinases (CDPKs) (Movahed et al., 2019; Rutter & Innes, 2017), for example CPK3 and CPK21. The CDPKs phosphorylate distinct substrates in PTI and ETI, and help regulate a variety of plant immune responses, including ROS production, transcriptional reprogramming of genes involved in immunity, and the hypersensitive response (Kadota et al., 2014). Plant EVs are thereby able to communicate with neighbouring cells to transmit or amplify immune signals (Figure 2b).
Inducers of SAR, including azelaic acid, glycerol‐3‐phosphate, pipecolic acid, and N‐hydroxy‐pipecolic acid, are generated in pathogen‐infected tissues and subsequently transferred via the phloem to systemic tissues where SAR is established (Chanda et al., 2011; Hartmann et al., 2018; Jung et al., 2009; Návarová et al., 2012). Various SAR‐related proteins, such as DEFECTIVE IN INDUCED RESISTANCE 1 (DIR1), THIOREDOXIN h3 (TRXm3), and PATHOGENESIS‐RELATED 1 (PR1), show increased abundance in phloem exudates from SAR‐induced leaves (Carella et al., 2016; Champigny et al., 2013; Chanda et al., 2011). Both azelaic acid and glycerol‐3‐phosphate are transported via a symplastic route, which involves specialized channels, the plasmodesmata (Lim et al., 2016). In mammalian systems, EVs are secreted by most cells into various bodily fluids (e.g., urine, blood, milk, saliva) under certain physiological or pathological conditions. It is therefore conceivable that plant EVs are transported via the phloem from locally infected tissues to distant leaves and deliver their cargo to cells of systemic leaves. Carella et al. (2016) identified a large number of putative mobile proteins from phloem exudates collected during SAR induction. Eighteen of the differentially accumulated proteins, including the SAR‐related protein TRXh3 (AT5G42980; a member of the m‐type family of plastid‐targeted thioredoxins), have been detected in EVs (Table 1) (Rutter & Innes, 2017). The abundance of TRXh3 was found to be increased in phloem exudates from leaves infected with either virulent or avirulent P. syringae strains (Carella et al., 2016). TRXh3 plays essential roles in SA‐dependent local resistance and in SAR by catalysing an SA‐induced NPR1 oligomer‐to‐monomer reaction (Tada et al., 2008). These findings suggest that plant EVs mediate the transport of SAR signals via the phloem (Figure 2c). Future studies should investigate whether pathogen‐induced plant EVs can promote disease resistance and induce SAR.
TABLE 1.
Summary of proteins in proteome of both phloem and extracellular vesicles
| Locus | Gene symbol | Description | Accumulation level in PEXPsm versus PEXMock |
|---|---|---|---|
| AT3G16470 | JAL35 | Jacalin‐related lectin 35 | Decreased |
| AT1G65930 | CICDH | Cytosolic isocitrate dehydrogenase (NADP) | Decreased |
| AT4G09000 | GF14 | 14‐3‐3‐like protein GF14 chi | Decreased |
| AT1G18210 | CML27 | Probable calcium‐binding protein | Increased |
| AT3G53420 | PIP2‐1 | Aquaporin PIP2‐1 | Decreased |
| AT4G23600 | CORI3 | Cystine lyase | Decreased |
| AT2G44060 | – | Late embryogenesis abundant protein, group 2 | Increased |
| AT1G22300 | GRF10 | 14‐3‐3‐like protein GF14 epsilon | Decreased |
| AT1G67090 | RBCS1A | Ribulose bisphosphate carboxylase small chain 1A | Decreased |
| AT1G70890 | MLP43 | MLP‐like protein 43 | Increased |
| AT1G72150 | PALT1 | Patellin 1 | Decreased |
| AT1G76180 | – | Dehydrin | Decreased |
| AT1G22530 | PATL2 | Patellin 2 | Decreased |
| AT1G35720 | ANN1 | Annexin D1 | Decreased |
| AT2G21660 | CCR2 | Glycine‐rich RNA‐binding protein 7 | Decreased |
| AT1G13440 | GAPC2 | Glyceraldehyde‐3‐phosphate dehydrogenase | Decreased |
| AT2G45470 | FLA8 | Fasciclin‐like arabinogalactan protein 8 | Decreased |
| AT2G44790 | – | Uclacyanin‐2 | Increased |
In mammalian systems, exosomes released by bacteria‐infected cells contain PAMPs and specific bacterial proteins, which can induce an immune response in their target cells (Schorey et al., 2015; Wang et al., 2018). For example, exosomes from macrophages infected with Mycobacterium tuberculosis contain PAMPs and are capable of activating immune responses in their target cells (Singh et al., 2015). The plant PTI is activated by the recognition of PAMPs and PRRs, which are mainly RLKs (Wu et al., 2019). In our recent study, we found the phosphorylation levels of 17 RLKs were significantly changed in systemic leaves of SAR‐induced plants (Zhou et al., 2021), suggesting that these proteins are also involved in SAR establishment in cells of systemic leaves. Therefore, we can speculate that plant EVs also contain PAMPs released by the pathogens, which contribute to the induction of local resistance and SAR (Figure 2). Further research to clarify whether plant EVs contain PAMPs or other pathogen‐derived components will be important to better understand the functions of EVs in plant immunity. Furthermore, sRNAs in exosomes from Helicobacter pylori‐infected macrophages are able to induce an inflammatory response in target cells by promoting the production of cytokines such as IL‐23, TNF‐α, and IL‐6. Cai et al. (2018) observed that plant EVs contain a variety of sRNAs that can inhibit fungal virulence by reducing the expression of virulence‐related genes (Cai et al., 2018). In further research, it will be interesting to explore the involvement of these sRNAs in plant EVs in plant immunity by manipulating the expression of their target genes. The signalling functions of EVs are determined by its cargo. In mammalian cells, the composition and functions of exosomes can change during stress (Beninson & Fleshner, 2014). Numerous studies have shown that plant EVs contain abundant cargo (nucleic acids, proteins, and lipids) (Cai et al., 2018; He et al., 2021; Liu et al., 2020; Regente et al., 2017; Rutter & Innes, 2017); however, such cargo has been analysed only qualitatively. Therefore, application of ‐omics methods to elucidate differentially accumulated cargo (specific sRNAs, mRNAs, lncRNAs, proteins, and lipids) in pathogen‐induced plant EVs will improve our understanding of how EVs function in local resistance and SAR. Analyses of plant EV proteomes have revealed that they contain various signalling proteins (kinases, calmodulin, etc.) in the absence of pathogen infection. Are these signal proteins in an activated state, or do they become activated after being recognized by pathogens? Posttranslational modification (phosphorylation, glycosylation, cetylation, etc.) of proteins plays important roles in regulating protein activity, stability, and protein–protein interactions (Li et al., 2015). The application of proteomics methods to identify EV proteins with posttranslational modifications on pathogen infection will shed further light on the functions of EVs in plant immunity.
7. CONCLUSION
Both plant cells and plant pathogens release EVs, analogously to animal cells. Plant EVs contain a wide variety of defence‐related cargoes, including proteins, nucleic acids (sRNA, tyRNA), and lipids, and play key roles in plant–pathogen interactions. Plant EVs can be taken up by fungi and inhibit their virulence by transferring sRNAs/defence‐related proteins into pathogens. Plant EVs contain a wide variety of proteins involved in perception and transduction of plant–pathogen interactions, and are involved in signal transduction in PTI, ETI, and SAR.
EVs from plant‐pathogenic bacteria and fungi also mediate interkingdom communication. Bacterial EVs contain various immune elicitors and can induce plant immune responses via PRR‐ and BAK1/SOBIR1‐mediated signalling pathways. Bacterial EVs also activate plant immune responses via SA‐dependent and ‐independent processes. This ability is related not to immune elicitors in EVs, but to the metabolites, lipids, and small protein epitopes they contain. Bacterial EVs facilitate cell movement in host plants and enhance virulence. Fungal EVs can induce phytotoxic responses in host plants and appear to play key roles in some infection processes.
Our knowledge of the involvement of EVs in plant cell–microbe interactions is still limited and fragmentary; much more is known about the roles of EVs in mammalian cell–microbe interactions. The precise functions of EVs remain to be clearly elucidated. More work is clearly needed on isolating plant EVs specifically related to the plant immune response, defining their composition, and documenting their function. Such studies will be essential for understanding the functions of EVs in plant–microbe interactions and will greatly facilitate the development of improved methods for sustainable agriculture.
ACKNOWLEDGEMENTS
The authors are grateful to Dr S. Anderson for English editing of the manuscript. This work was supported by the Key Cultivation Project of Shangqiu Normal University (7001700235).
Zhou, Q. , Ma, K. , Hu, H. , Xing, X. , Huang, X. & Gao, H. (2022) Extracellular vesicles: their functions in plant–pathogen interactions. Molecular Plant Pathology, 23, 760–771. 10.1111/mpp.13170
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Akers, J.C. , Gonda, D. , Kim, R. , Carter, B.S. & Chen, C.C. (2013) Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus‐like vesicles, and apoptotic bodies. Journal of Neuro‐Oncology, 113, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, Q. , Ehlers, K. , Kogel, K.H. , van Bel, A.J. & Hückelhoven, R. (2006) Multivesicular compartments proliferate in susceptible and resistant MLA12‐barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytologist, 172, 563–576. [DOI] [PubMed] [Google Scholar]
- Anand, P.K. , Anand, E. , Bleck, C.K. , Anes, E. & Griffiths, G. (2010) Exosomal Hsp70 induces a pro‐inflammatory response to foreign particles including mycobacteria. PLoS One, 5, e10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar, O. , Mordukhovich, G. , Luu, D.D. , Schwessinger, B. , Daudi, A. , Jehle, A.K. et al. (2016) Bacterial outer membrane vesicles induce plant immune responses. Molecular Plant‐Microbe Interaction, 29, 374–384. [DOI] [PubMed] [Google Scholar]
- Baldrich, P. , Rutter, B.D. , Karimi, H.Z. , Podicheti, R. , Meyers, B.C. & Innes, R.W. (2019) Plant extracellular vesicles contain diverse small RNA species and are enriched in 10‐ to 17‐nucleotide "tiny" RNAs. The Plant Cell, 31, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Beninson, L.A. & Fleshner, M. (2014) Exosomes: an emerging factor in stress‐induced immunomodulation. Seminars in Immunopathology, 26, 394–401. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, S. & Schorey, J.S. (2007) Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. Journal of Biological Chemistry, 282, 25779–25789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleackley, M.R. , Samuel, M. , Garcia‐Ceron, D. , McKenna, J.A. , Lowe, R.G.T. , Pathan, M. et al. (2020) Extracellular vesicles from the cotton pathogen fusarium oxysporum f. sp. vasinfectum induce a phytotoxic response in plants. Frontiers in Plant Science, 10, 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington, K.E. & Kuehn, M.J. (2014) Protein selection and export via outer membrane vesicles. Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research, 1843, 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, S. , Aggarwal, S. , Singh, D.V. & Acharya, N. (2020) Extracellular vesicles: an emerging platform in gram‐positive bacteria. Microbial Cell, 7, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , He, B. & Jin, H. (2019) A safe ride in extracellular vesicles‐small RNA trafficking between plant hosts and pathogens. Current Opinion in Plant Biology, 52, 140–148. [DOI] [PubMed] [Google Scholar]
- Cai, Q. , He, B. , Wang, S. , Fletcher, S. , Niu, D. , Mitter, N. et al. (2021) Message in a bubble: shuttling small RNAs and proteins between cells and interacting organisms using extracellular vesicles. Annual Review of Plant Biology, 72, 497–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , He, B. , Weiberg, A. , Buck, A.H. & Jin, H. (2019) Small RNAs and extracellular vesicles: new mechanisms of cross‐species communication and innovative tools for disease control. PLoS Pathogens, 15, e1008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , Qiao, L. , Wang, M. , He, B. , Lin, F.‐M. , Palmquist, J. et al. (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science, 360, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella, P. , Merl‐Pham, J. , Wilson, D.C. , Dey, S. , Hauck, S.M. , Vlot, A.C. et al. (2016) Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiology, 171, 1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champigny, J.C. , Isaacs, M. , Carella, P. , Faubert, J. , Fobert, P.R. & Cameron, R.K. (2013) Long distance movement of DIR1 and investigation of the role of DIR1‐like during systemic acquired resistance in Arabidopsis . Frontiers in Plant Science, 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda, B. , Xia, Y.E. , Mandal, M.K. , Yu, K. , Sekine, K.‐T. , Gao, Q.‐M. et al. (2011) Glycerol‐3‐phosphate is a critical mobile inducer of systemic immunity in plants. Nature Genetics, 43, 421–427. [DOI] [PubMed] [Google Scholar]
- Chowdhury, C. & Jagannadham, M.V. (2013) Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochimica et Biophysica Acta (BBA) ‐ Proteins and Proteomics, 1834, 231–239. [DOI] [PubMed] [Google Scholar]
- De Palma, M. , Ambrosone, A. , Leone, A. , Del Gaudio, P. , Ruocco, M. , Turiák, L. et al. (2020) Plant roots release small extracellular vesicles with antifungal activity. Plants, 9, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, L.M. & Wang, M.Z. (2019) Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells, 8, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux, M. , Garaigorta, U. , Boyd, B. , Décembre, E. , Chung, J. , Whitten‐Bauer, C. et al. (2012) Short‐range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host and Microbe, 12, 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrevente, C. , Keller, S. , Altevogt, P. & Costa, J. (2011) Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer, 11, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner, C. & Robatzek, S. (2012) Plants and pathogens: putting infection strategies and defence mechanisms on the map. Current Opinion in Plant Biology, 15, 699–707. [DOI] [PubMed] [Google Scholar]
- Gibson, D.M. , King, B.C. , Hayes, M.L. & Bergstrom, G.C. (2011) Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Current Opinion in Microbiology, 14, 264–270. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. & Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Molecular Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- György, B. , Szabó, T.G. , Pásztói, M. , Pál, Z. , Misják, P. , Aradi, B. et al. (2011) Membrane vesicles, current state‐of‐the‐art: emerging role of extracellular vesicles. Cellular and Molecular Life Sciences, 68, 2667–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin, W. & Jensen, W.A. (1967) Ultrastructural changes during growth and embryogenesis in carrot cell cultures. Journal of Ultrastructure Research, 18, 428–443. [DOI] [PubMed] [Google Scholar]
- Hartmann, M. , Zeier, T. , Bernsdorff, F. , Reichel‐Deland, V. , Kim, D. , Hohmann, M. et al. (2018) Flavin monooxygenase‐generated N‐hydroxypipecolic acid is a critical element of plant systemic immunity. Cell, 173, 456–469. [DOI] [PubMed] [Google Scholar]
- He, B. , Cai, Q. , Qiao, L. , Huang, C.‐Y. , Wang, S. , Miao, W. et al. (2021) RNA‐binding proteins contribute to small RNA loading in plant extracellular vesicles. Nature Plants, 7, 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy, K. , Cherk, C. & Somerville, S. (2009) Host–pathogen warfare at the plant cell wall. Current Opinion in Plant Biology, 12, 406–413. [DOI] [PubMed] [Google Scholar]
- Hill, E.H. & Solomon, P.S. (2020) Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici . Fungal Biology and Biotechnology, 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu, M. , Zaini, P.A. , Baccari, C. , Tran, S. , da Silva, A.M. & Lindow, S.E. (2014) Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proceedings of the National Academy of Sciences of the United States of America, 111, E3910–E3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, X. , Yin, Y. , Chen, Y. & Mao, L. (2021) The role of viral proteins in the regulation of exosomes biogenesis. Frontiers in Cellular and Infection Microbiology, 11, 671625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jullien, P.E. , Grob, S. , Marchais, A. , Pumplin, N. , Chevalier, C. , Bonnet, D.M.V. et al. (2020) Functional characterization of Arabidopsis ARGONAUTE 3 in reproductive tissues. The Plant Journal, 103, 1796–1809. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. , Tschaplinski, T.J. , Wang, L. , Glazebrook, J. & Greenberg, J.T. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Molecular Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Katsir, L. & Bahar, O. (2017) Bacterial outer membrane vesicles at the plant–pathogen interface. PLoS Pathogens, 13, e1006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T. & Akira, S. (2009) The roles of TLRs, RLRs and NLRs in pathogen recognition. International Immunology, 21, 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, K. , Vesk, M. & Work, E. (1966) Relation between excreted lipopolysaccharide complexes and surface structures of a lysine‐limited culture of Escherichia coli . Journal of Bacteriology, 92, 1206–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen, K. , Nymon, A. , Barnaby, R. , Bashor, L. , Li, Z. , Hampton, T.H. et al. (2021) Let‐7b‐5p in vesicles secreted by human airway cells reduces biofilm formation and increases antibiotic sensitivity of P. aeruginosa . Proceedings of the National Academy of Sciences of the United States of America, 118, e2105370118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnatreya, D.B. , Baruah, P.M. , Dowarah, B. , Chowrasia, S. , Mondal, T.K. & Agarwala, N. (2021) Genome‐wide identification, evolutionary relationship and expression analysis of AGO, DCL and RDR family genes in tea. Scientific Reports, 11, 8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, H.M. & Jagannadham, M.V. (2014) Biogenesis and multifaceted roles of outer membrane vesicles from Gram‐negative bacteria. Microbiology, 160, 2109–2121. [DOI] [PubMed] [Google Scholar]
- Kulkarni, H.M. , Swamy, C.V.B. & Jagannadham, M.V. (2015) The proteome of the outer membrane vesicles of an Antarctic bacterium Pseudomonas syringae Lz4W. Data in Brief, 4, 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp, A. & Kuehn, M.J. (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Review of Microbiology, 64, 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Silva‐Sanchez, C. , Zhang, T. , Chen, S. & Li, H. (2015) Phosphoproteomics technologies and applications in plant biology research. Frontiers in Plant Science, 6, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K.E. , Xiong, X.I. , Zhu, S. , Liao, H. , Xiao, X. , Tang, Z. et al. (2018) MeBIK1, a novel cassava receptor‐like cytoplasmic kinase, regulates PTI response of transgenic Arabidopsis . Functional Plant Biology, 45, 658–667. [DOI] [PubMed] [Google Scholar]
- Liebana‐Jordan, M. , Brotons, B. , Falcon‐Perez, J.M. & Gonzalez, E. (2021) Extracellular vesicles in the fungi kingdom. International Journal of Molecular Sciences, 22, 7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, G.‐H. , Shine, M.B. , de Lorenzo, L. , Yu, K. , Cui, W. , Navarre, D. et al. (2016) Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host and Microbe, 19, 541–549. [DOI] [PubMed] [Google Scholar]
- Liu, N. , Wang, N. , Bao, J.J. , Zhu, H.X. , Wang, L.J. & Chen, X.Y. (2020) Lipidomic analysis reveals the importance of GIPCs in Arabidopsis leaf extracellular vesicles. Molecular Plant, 13, 1523–1532. [DOI] [PubMed] [Google Scholar]
- Liu, S.R. , Zhou, J.J. , Hu, C.G. , Wei, C.L. & Zhang, J.Z. (2017) MicroRNA‐mediated gene silencing in plant defense and viral counter‐defense. Frontiers in Microbiology, 8, 1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloubes, R. , Bernadac, A. , Houot, L. & Pommier, S. (2013) Nonclassical secretion systems. Research in Microbiology, 164, 655–663. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Greenberg, J.T. & Holuigue, L. (2016) Editorial: salicylic acid signaling networks. Frontiers in Plant Science, 7, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia, L. , Nanan, R. , Hosseini‐Beheshti, E. & Grau, G.E. (2019) Host‐ and microbiota‐derived extracellular vesicles, immune function, and disease development. International Journal of Molecular Sciences, 21, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alonso, J.M. , Ecker, J.R. & Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. , Wiig, A. & Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis . Cell, 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Manning, A.J. & Kuehn, M.J. (2011) Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiology, 11, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, H.M. , Zebell, S.G. , Ristaino, J.B. , Dong, X. & Kuehn, M.J. (2021) Protective plant immune responses are elicited by bacterial outer membrane vesicles. Cell Reports, 34, 108645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, C.O. , Neumann, U. , Grunewald, D. , Panstruga, R. & O'Connell, R. (2011) Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria . Cell Microbiology, 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Mittelbrunn, M. , Gutiérrez‐Vázquez, C. , Villarroya‐Beltri, C. , González, S. , Sánchez‐Cabo, F. , González, M.Á. et al. (2011) Unidirectional transfer of microRNA‐loaded exosomes from T cells to antigen‐presenting cells. Nature Communications, 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahed, N. , Cabanillas, D.G. , Wan, J. , Vali, H. , Laliberté, J.F. & Zheng, H. (2019) Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiology, 180, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, J. , Zhuang, X. , Wang, Q. , Jiang, H. , Deng, Z.‐B. , Wang, B. et al. (2014) Interspecies communication between plant and mouse gut host cells through edible plant derived exosome‐like nanoparticles. Molecular Nutrition and Food Research, 58, 1561–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová, H. , Bernsdorff, F. , Döring, A.C. & Zeier, J. (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. The Plant Cell, 24, 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed, Z.A. , Wei, X. , Chen, J. , Mubeen, H. & Ali, G.S. (2020) The PTI to ETI continuum in Phytophthora–plant interactions. Frontiers in Plant Science, 11, 593905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana, R.D. & Kaparakis‐Liaskos, M. (2016) Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell Microbiology, 18, 1518–1524. [DOI] [PubMed] [Google Scholar]
- van der Pol, E. , Böing, A.N. , Harrison, P. , Sturk, A. & Nieuwland, R. (2012) Classification, functions, and clinical relevance of extracellular vesicles. Pharmacological Reviews, 64, 676–705. [DOI] [PubMed] [Google Scholar]
- Qi, Y. , Tsuda, K. , Glazebrook, J. & Katagiri, F. (2011) Physical association of pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI) immune receptors in Arabidopsis . Molecular Plant Pathology, 12, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. & Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Molecular Plant‐Microbe Interactions, 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Ramakrishnaiah, V. , Thumann, C. , Fofana, I. , Habersetzer, F. , Pan, Q. , de Ruiter, P.E. et al. (2013) Exosome‐mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proceedings of the National Academy of Sciences of the United States of America, 110, 13109–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, J. , Miekus, K. , Kucia, M. , Zhang, J. , Reca, R. , Dvorak, P. et al. (2006) Embryonic stem cell‐derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia, 20, 847–856. [DOI] [PubMed] [Google Scholar]
- Regente, M. , Corti‐Monzón, G. , Maldonado, A.M. , Pinedo, M. , Jorrín, J. & de la Canal, L. (2009) Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Letters, 583, 3363–3366. [DOI] [PubMed] [Google Scholar]
- Regente, M. , Pinedo, M. , Clemente, H.S. , Balliau, T. , Jamet, E. & de la Canal, L. (2017) Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. Journal of Experimental Botany, 68, 5485–5495. [DOI] [PubMed] [Google Scholar]
- da Rocha, I.F.M. , Amatuzzi, R.F. , Lucena, A.C.R. , Faoro, H. & Alves, L.R. (2020) Cross‐kingdom extracellular vesicles EV–RNA communication as a mechanism for host–pathogen interaction. Frontiers in Cellular and Infection Microbiology, 10, 593160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, M. , Fan, J. , Lyon, C. , Wan, M.H. & Hu, Y. (2018) Role of extracellular vesicles in viral and bacterial infections: pathogenesis, diagnostics, and therapeutics. Theranostics, 8, 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, M.L. , Nimrichter, L. , Oliveira, D.L. , Frases, S. , Miranda, K. , Zaragoza, O. et al. (2007) Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans‐cell wall transport. Eukaryotic Cell, 6, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, B.D. & Innes, R.W. (2017) Extracellular vesicles isolated from the leaf apoplast carry stress‐response proteins. Plant Physiology, 173, 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey, J.S. , Cheng, Y. , Singh, P.P. & Smith, V.L. (2015) Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO Reports, 16, 24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu, V.K. , Vorhölter, F.‐J. , Niehaus, K. & Watt, S.A. (2008) Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris . BMC Microbiology, 8, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, B.M.A. , Prados‐Rosales, R. , Espadas‐Moreno, J. , Wolf, J.M. , Luque‐Garcia, J.L. , Goncalves, T. et al. (2013) Characterization of Alternaria infectoria extracellular vesicles. Sabouraudia, 52, 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P.P. , Li, L. & Schorey, J.S. (2015) Exosomal RNA from Mycobacterium tuberculosis infected cells is functional in recipient macrophages. Traffic, 16, 555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé, M. , Scheibner, F. , Hoffmeister, A.‐K. , Hartmann, N. , Hause, G. , Rother, A. et al. (2015) Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps Type II secretion system and outer membrane vesicles. Journal of Bacteriology, 197, 2879–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. & Dong, X. (2012) How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews Immunology, 12, 89–100. [DOI] [PubMed] [Google Scholar]
- Szczesny, R. , Jordan, M. , Schramm, C. , Schulz, S. , Cogez, V. , Bonas, U. et al. (2010) Functional characterization of the Xps and Xcs type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria . New Phytologist, 187, 983–1002. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. et al. (2008) Plant immunity requires conformational charges of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Duban, L. , Segura, E. , Véron, P. , Lantz, O. & Amigorena, S. (2002) Indirect activation of naïve CD4+ T cells by dendritic cell‐derived exosomes. Nature Immunology, 3, 1156–1162. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Ostrowski, M. & Segura, E. (2009) Membrane vesicles as conveyors of immune responses. Nature Reviews Immunology, 9, 581–593. [DOI] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J.J. & Lotvall, J.O. (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vargas, G. , Rocha, J.D.B. , Oliveira, D.L. , Albuquerque, P.C. , Frases, S. , Santos, S.S. et al. (2015) Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans . Cell Microbiology, 17, 389–407. [DOI] [PubMed] [Google Scholar]
- Wang, F. , Shang, Y. , Fan, B. , Yu, J.Q. & Chen, Z. (2014) Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen‐responsive MAPK cascade in plant basal defense. PLoS Pathogens, 10, e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Yao, Y. , Chen, X. , Wu, J. , Gu, T. & Tang, X. (2018) Host derived exosomes‐pathogens interactions: potential functions of exosomes in pathogen infection. Biomedicine and Pharmacotherapy, 108, 1451–1459. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Reca, I.B. , Spinelli, F. , Lironi, D. , Lorenzo, G.D. , Poltronieri, P. et al. (2019) An EFR‐Cf‐9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1‐ and BAK1‐dependent recognition of eIF18. Molecular Plant Pathology, 20, 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Zhao, F. , Jiang, L. , Chen, C. , Wu, L. & Liu, Z. (2018) Different pathogen defense strategies in Arabidopsis: more than pathogen recognition. Cells, 7, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. , Meng, Q.I. , Tan, X. , Ding, W. , Ma, K. , Xu, Z. et al. (2021) Protein phosphorylation changes during systemic acquired resistance in Arabidopsis thaliana . Frontiers in Plant Science, 12, 748287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. et al. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


