Abstract
We evaluated the use of the PCR for detection of enteric viruses in groundwater. To do this, we used an improved sample-processing technique and a large-volume amplification protocol. The objective of this study was to use advanced molecular techniques to develop a rapid and simple method which can be used by the water industry for detection of viral contamination in a variety of water samples. The strategy described here fulfills the water industry’s need for a rapid, reliable, easily performed method for analyzing groundwater for virus contamination. Viruses were detected after concentration from at least 400 gallons (1,512 liters) of water by a filter adsorption and elution method, which resulted in a concentrate containing viruses. A total of 150 samples were analyzed by performing cell culture assays for enteroviruses and by performing reverse transcription PCR (RT-PCR) analyses for enteroviruses, hepatitis A virus, and rotavirus. Thirteen samples (8.7%) produced cellular cytopathic effects when the Buffalo green monkey cell line was used. When primers specific for enteroviruses were used in RT-PCR, 40 of 133 samples (30.1%) tested positive for the presence of enterovirus RNA. When hepatitis A virus-specific primers were used, 12 of 139 samples (8.6%) were considered positive for the presence of hepatitis A viral RNA. The RT-PCR analysis performed with rotavirus-specific primers identified 18 of 130 samples (13.8%) that were positive for rotavirus RNA sequences. Our sample-processing technique and large-volume PCR protocol (reaction volume, 300 μl) resulted in sufficient removal or dilution of inhibitors so that more than 95% of the samples could be assayed by PCR. Because of its sensitivity for detecting viral nucleic acid sequences, PCR analysis should produce more positive results than cell culture analysis. Since either cell culture analysis or PCR can reveal only a “snapshot” of the quality of the groundwater being sampled, PCR seems to be a desirable rapid initial screening tool.
Human enteric viruses are excreted in feces of infected individuals and may directly or indirectly contaminate water intended for drinking. These viruses are excreted in high numbers (108 to 1010 particles per g of feces) by infected individuals. The enteric viruses include the enteroviruses, rotaviruses, Norwalk and Norwalk-like viruses, adenoviruses, reoviruses, and others.
The enteroviruses (poliovirus, coxsackie A and B viruses, and echovirus) can cause a variety of illnesses ranging from gastroenteritis to myocarditis and aseptic meningitis (8). Numerous studies have documented the presence of enteroviruses in raw and treated drinking water (6, 7), wastewater (11), and sludge (2). Enteroviruses in the environment create a public health risk because they can be transmitted via the fecal-oral route through contaminated water (2) and low numbers are able to initiate infections in humans.
The PCR (9, 14) can be used to enzymatically amplify to detectable levels nucleic acid sequences that are present in low copy numbers in water samples. The speed, specificity, low cost, and ease of use of this procedure have led to its use in environmental science.
The advantages of PCR are several. Compared to techniques such as cell culturing for detection of viruses, the time required for the assay can be reduced from days or weeks to hours. Both the initial and recurring costs of PCR are substantially less than the costs of cell culture techniques, and PCR is easily performed. In addition, PCR can be used to identify a specific pathogen found in water. It cannot, however, be used to determine the infectious state of an organism; it can only determine the presence or absence of pathogen-specific DNA or RNA sequences. PCR assays have been used to detect enterovirus nucleic acid sequences in clinical (5, 13) and environmental samples (1, 3, 12).
The primary objective of this research was to use advanced molecular techniques to develop a rapid, simple, inexpensive assay which could be used by the water industry for detection of viral contamination in water. Molecular techniques are now used in environmental research and monitoring, and the necessary tools and techniques are available from a variety of sources. We developed a comprehensive research plan to evaluate a “universal” sample-processing and large-volume PCR method for detection of viruses in groundwater and to investigate the applicability of the method for detection of enteroviruses, hepatitis A virus, and rotavirus in groundwater sources. The approach which we used included laboratory studies in which we developed and optimized the PCR method and determined the specificity of the PCR primers used for detection of viruses, followed by a field evaluation of the method performed with groundwater obtained from different geographical locations in a variety of physical, chemical, and geological settings. The specific objectives were to develop and evaluate a simple, rapid, inexpensive method for detecting human viruses in groundwater samples with the reverse transcription PCR (RT-PCR); to optimize the PCR method for detecting low concentrations of viruses in groundwater as an alternative to cell culture assays; to develop a sample treatment protocol for removing reaction inhibitors from groundwater concentrates; to develop a method for assaying a larger equivalent sample volume of each water concentrate by the PCR technique; and to conduct a field evaluation of the optimized method for detection of enteroviruses, hepatitis A virus, and rotavirus by using 150 groundwater samples.
MATERIALS AND METHODS
Sample collection and site selection.
A total of 150 water samples were obtained from groundwater from different geographical locations in a variety of physical, chemical, and geological settings. The site selection process provided a variety of samples in order to ensure the best possible field evaluation of the applicability of the PCR method for detection of viruses in groundwater. The samples were collected by passing at least 400 gallons (1,512 liters) of raw groundwater (prior to any treatment) through a 1MDS filter (CUNO Inc., Meriden, Conn.) at a flow rate of no more than 4 gallons per min. The filters remained in the filter housing and were shipped overnight to our laboratory at 4°C, and they were processed within 48 h after completion of the sample collection process.
In order to obtain consistent sampling, 30 identical sampling kits were assembled. Each kit contained all of the equipment needed to collect a sample, including all hoses and connectors, a filter and a filter housing, protective gloves, reusable ice packs, sample bottles, a sample data sheet, and a detailed written protocol. Each kit also included a water meter, which allowed the sampler to record the volume of water sampled, and a in-line flow-restricting device to limit the filtration rate to 4 gallons per min. In addition, to help ensure that the water-sampling procedure was consistent, a 10-min VHS video describing and illustrating all of the procedures was professionally produced. The training video was provided to samplers before they collected samples.
Filter elution.
The filters were eluted by using an autoclaved solution containing 1.5% beef extract (Becton Dickinson, Cockeysville, Md.) and 0.05 M glycine (U.S. Biochemical Corp., Cleveland, Ohio) (pH 9.4). One liter of the beef extract solution was poured into the filter housing containing the 1MDS filter and left for 15 min. The solution was then forced from the filter housing into a sterile 2-liter beaker by using nitrogen (N2) gas. The eluant was then poured back into the filter housing and again forced out into the same beaker by using N2. The pH of the solution was then lowered to 7.0 to 7.4 by using 1 M HCl, and the solution was stirred for 15 min. Forty milliliters of this eluant was mixed with 4 ml of glycerol and stored at −80°C until the phage assay was performed. Another 100 ml was stored at −80°C for archival purposes.
Virus flocculation and reconcentration.
Each elution solution was either stored at −20°C or immediately adjusted to pH 3.5 and stirred for 15 min. Each stirred solution was then centrifuged for 30 min at 4,000 × g at 4°C. The resulting pellet was initially resuspended in 9 ml of 0.15 M Na2HPO4 buffer (pH 9.4) and transferred to a fresh 50-ml centrifuge tube. Three milliliters of the buffer was then used to rinse both centrifuge bottles and combined with the initial 9 ml. The pH was adjusted to 7.2, and the volume was brought to 15 ml with 0.15 M Na2HPO4 (pH 7.2). The solution was then mixed with an equal volume of Freon (Fisher Scientific Co., Pittsburgh, Pa., or Aldrich Chemical Co., Milwaukee, Wis.), vortexed for 2 min, and centrifuged at 2700 × g for 10 min. The upper, aqueous portion was removed and transferred to a fresh 15-ml tube, and the volume was brought to 15 ml with 0.15 M Na2HPO4 (pH 7.2). One-half of this 15-ml preparation (i.e., 7.5 ml) was stored at −80°C until it was used in the PCR analysis. The other half (containing 50% of the original pellet) was brought to 15 ml with 0.15 M Na2HPO4 (pH 7.2) and was stored at −80°C until it was used in the cell culture analysis.
Cell culture assay.
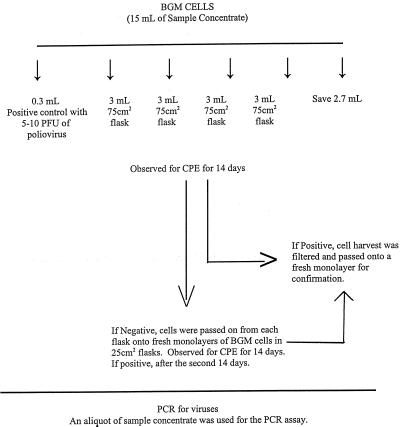
Buffalo green monkey (BGM) kidney cells were grown until they produced confluent monolayers in 25- and 75-cm2 plastic flasks by using Eagle’s minimum essential medium with Earle’s salts (Irving Scientific, Irving, Calif.) containing 10% fetal bovine serum (Sigma Chemical Co., St. Louis, Mo.). Included in the maintenance medium was an antibiotic-antimycotic solution (BRL Life Technologies, Gaithersburg, Md.) containing 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.25 μg of amphotericin B per ml. Prior to the actual assay, 1 ml of the concentrate that was being tested was placed on BGM cells in a 25-cm2 flask, and the monolayer was observed for 1 week for toxicity or bacterial contamination. If toxicity was observed, the sample was diluted 1:3 in 0.15 M sodium phosphate (pH 7.0 to 7.5) for the actual assay. If bacterial contamination was observed, prior to the actual sample assay the concentrate was filtered through a 0.22-μm-pore-size filter (Millipore Corp., Bedford, Mass.) through which 10 ml of 1.5% beef extract had been passed. Before exposure to the sample, the growth medium was poured off and the cell monolayer was washed twice with a Tris (Sigma Chemical Co.)-buffered saline solution. For each sample, a 3-ml portion of the final concentrate was inoculated into each of four 75-cm2 flasks. A total of 12 ml of the final concentrate was assayed for each sample. The flasks were incubated at 37°C for 60 min and rocked every 15 min to facilitate virus adsorption to the cells. Twenty milliliters of maintenance medium consisting of Eagle’s minimum essential medium (Irvine Scientific) supplemented with 2% fetal bovine serum (Sigma Chemical Co.) and one ml of gentamicin (50 μg/ml; BRL Life Technologies) was added to each flask. The flasks were incubated at 37°C and examined daily for 14 days for viral cytopathic effects (CPE). Suspected viral CPE were confirmed by inoculating the medium onto a fresh monolayer of BGM cells and observing the cells for CPE. The method is summarized in Fig. 1.
FIG. 1.
Cell culture assay.
All of the samples that did not exhibit CPE after the first passage were passed a second time on BGM cells; all of the samples that exhibited CPE were confirmed to be virus positive by two additional passages on BGM cells.
Physiochemical analysis.
The samplers measured the water temperature and pH at the time of sampling at each collection site. Turbidity and UV absorbance at 254 nm were measured when samples were received. One liter of each water sample was used to determine the general chemical characteristics (U.S. Environmental Protection Agency [EPA] method 300.0 for minerals and U.S. EPA methods 200.7, 200.8, and 200.9 for metals) and the total organic carbon (TOC) content.
To determine whether a water sample contained a high or low level of aquatic humic materials, the specific UV absorbance (SUVA) was determined. The SUVA was defined as the absorbance at 254 nm (expressed per meter of absorbance) divided by the dissolved organic carbon (DOC) concentration (in milligrams per liter). A SUVA of more than 4 indicated that the DOC in a water sample consisted largely of aquatic humic material. A SUVA of less than 3 indicated that the DOC consisted largely of nonhumic material (4). Because the samples were from groundwater sources, the TOC values were used to calculate the SUVAs. The SUVAs for 119 samples ranged from 0.15 to 25.13, with a mean of 3.07 (standard deviation, 3.79) and a median of 1.90. The SUVAs for this set of samples did not correlate with inhibition of PCR assays performed with water samples. It seems that aquatic humic and nonhumic materials both inhibit PCR amplification.
TOC assay.
TOC analyses were performed in triplicate at the Belleville Laboratory of the American Water Works Service Company, Inc. Three 5-ml portions of each raw water sample were analyzed with a model TOC-5000 TOC analyzer (Shimadzu, Columbia, Md.).
UV analysis for SUVA determinations.
Two 2-ml portions of each raw water sample were analyzed to determine spectral absorbance at 254 nm by using a Milton Roy Spectronic 21-D spectrophotometer. The water sample was placed in a quartz cuvette (Spectrocell Corp., Oreland, Pa.), and duplicate readings were taken. The instrument was zeroed by using double-distilled H2O.
Statistical analyses.
The analyses performed in this study included some basic statistics and assessment of distributions for each of the variables. Exploratory analyses were also performed to determine correlations between the variables. Normality tests were performed with the set of data by using SAS’s PPROC UNIVARIABLE. Correlation analyses were performed both as numeric analyses (by using Pearson correlations) and as rank correlations (by using Spearman correlations).
Analyses of variance for cell culture and PCR values by well depth, by distance from surface water sources, and by distance from sewage sources were performed. The results indicated that the mean distances for cell culture-positive and -negative values were not significantly different for any of the distance parameters tested. The mean distances for overall PCR-positive and -negative values were not significantly different by well depth or by distance from surface water sources but were significantly different for PCR-positive and -negative values by distance from sewage sources.
Primers for virus detection.
The following primers used for detection of enteroviruses in the sample concentrates, as previously described by DeLeon et al. (3), produced a 196-bp product: 5′-CCT CCG GCC CCT GAA TG-3′ and 5′-ACC GGA TGG CCA ATC CAA-3′.
The hepatitis A virus primers (5′-CAG CAC ATC AGA AAG GTG AG-3′ and 5′-CTC CAG AAT CAT CTC CAA C-3′) produced a 192-bp product (3).
The upstream primer used for rotavirus (CON 1; 5′-TTG CCA CCA ATT CAG AAT AC-3′) and the downstream primer (CON 2; 5′-ATT TCG GAC CAT TTA TAA CC-3′) produced a 211-bp product. The rotavirus primer sequences were kindly provided by Jon Gentsch of The Centers for Disease Control and Prevention Viral Gastroenteritis Unit, Atlanta, Ga.
Large-volume PCR.
Most manufacturers of the enzymes needed for PCR describe reaction protocols in which the total reaction volume is 30 to 100 μl. These volumes are also the most commonly described reaction volumes in the scientific literature, although reaction volumes of 10 μl or less are not uncommon (10). There has been a trend to minimize the reaction volume in order to conserve reagents and make it easier to perform a large number of reactions simultaneously. The drawback to this approach when environmental samples are analyzed is that smaller portions of potentially diluted sources are examined.
We initially used our previously described protocol (1), in which 10 μl of a sample was used in a 30-μl RT reaction mixture. This 10 μl represented 0.5 liter of the original sample. As viruses may be present at very low concentrations and still present health problems, we wished to maximize the sample size. We accomplished this by increasing the sample size to 100 μl (representing 10 liters of sample or 15 ml of concentrate) without increasing the reaction mixture size or amounts of reagents 10-fold. The amount of RNase inhibitor used was 3.3 times the amount used in the smaller reaction mixture, and the amount of reverse transcriptase used was only two times the amount used in the smaller reaction mixture, so the reaction volume was increased 10-fold but the cost increased only about 2.5-fold. In addition, we observed that the sensitivity of the reaction was greater and the results were more consistent with the larger reaction volumes than with the smaller reaction volumes.
Pre-PCR sample treatment.
Prior to PCR analysis, each sample concentrate was extracted once with phenol-chloroform (5:1, pH 4.7) (Amresco Inc., Solon, Ohio) and once with chloroform (Amresco) as follows. The concentrate was combined 1:1 with the phenol-chloroform mixture and vortexed for 3 min. The sample was then centrifuged for 15 min at 14,000 × g. The aqueous portion was removed and combined with an equal volume of chloroform, and the resulting preparation was vortexed for 1 min and centrifuged for 5 min at 14,000 × g. The resulting aqueous portion (500 to 750 μl) was applied to the top of a column consisting of 5 ml of autoclaved DNA grade Sephadex G-100 (Pharmacia Biotech AB, Uppsala, Sweden), equilibrated in high-performance liquid chromatography (HPLC) grade water, in a 5-ml syringe plugged at the bottom with a 1-in.-square piece of sterile Kim-Wipe tissue (Kimberly-Clark Corp., Roswell, Ga.). The initial column eluant was discarded. Three successive 750-μl aliquots of HPLC grade H2O were applied, and the column was allowed to drain between applications. The first two column eluants were discarded. The final 750-μl eluant was collected in a 1.5-ml microcentrifuge tube containing approximately 50 μl of autoclaved Chelex 100 resin (Bio-Rad Laboratories, Hercules, Calif.) and was kept at −20°C until the RT-PCR analysis was performed.
RT Reaction.
Two RT-PCR were performed with each sample concentrate; in one reaction virus-seeded concentrate was used as part of quality control to evaluate reaction inhibition, and in the other reaction unseeded water concentrate was used to determine whether there was any viral contamination of the sample. One 50-μl reaction mixture was seeded with 10 PFU of poliovirus, 10 PFU of hepatitis A virus, or 10 50% tissue culture infective doses (TCID50) of rotavirus, while the other reaction mixture (final volume, 300 μl) contained only sample.
The small-volume, seeded RT reactions were performed as follows. A 10-μl portion of sample was combined with 5 μl of sterile, nuclease-free water containing either 10 PFU of virus or 10 TCID50 of virus and 0.7 μl of a solution containing random hexamers (250 μM stock solution) in a 500-μl microcentrifuge tube. The mixture was heated at 99°C for 4 min and then placed on ice. Then a 33-μl reaction cocktail was added; this cocktail was prepared by mixing 18.5 μl of sterile, nuclease-free water, 6 μl of 10× buffer (35 mM MgCl2, 750 mM KCl, 100 mM Tris; pH 9.5), 6 μl 0.1 M dithiothreitol, 1.3 μl of a deoxynucleoside triphosphate (dNTP) mixture containing each dNTP at a concentration of 10 mM, 0.8 μl of RNasin (40 U per μl; Promega, Madison, Wis.), and 0.4 μl of SuperScript II reverse transcriptase (200 U per μl; BRL Life Technologies). The RT reaction mixture (total volume 48.5 μl) was then incubated at 25°C for 15 min, at 42°C for 45 min, and at 99°C for 5 min. The reaction mixtures were stored at 4°C until the amplification reaction was performed.
For the large-volume, unseeded reaction, 50 μl of sample and 50 μl of sterile, nuclease-free water were combined with 4 μl of a solution containing random hexamers (250 μM stock solution; Pharmacia Biotech) in a 500-μl microcentrifuge tube. The mixture was heated at 99°C for 4 min and then placed on ice. Then a 186-μl reaction cocktail was prepared by combining 110.5 μl of HPLC grade H2O, 30 μl of 10× buffer, 30 μl of 0.1 M dithiothreitol, 8 μl of a dNTP mixture containing each dNTP at a concentration of 10 mM (Pharmacia Biotech), 5 μl of RNasin (40 U per μl; Promega), and 2.5 μl of SuperScript reverse transcriptase (200 U per μl; BRL Life Technologies) and added to the tube containing the sample. The RT reaction mixture (total volume, 290 μl) was then incubated at 25°C for 15 min, at 42°C for 45 min, and at 99°C for 5 min. The reaction mixtures were stored at 4°C until the amplification reaction was performed.
cDNA amplification by PCR.
A PCR in which the entire RT reaction mixture was used was performed by adding a reaction cocktail consisting of primers and AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.).
For each small-volume, virus-seeded reaction, a 1.5-μl cocktail consisting of 0.3 μl of each primer (concentration, 75 μM), 0.3 μl of AmpliTaq DNA polymerase (1.5 U), and 0.6 μl of water was added to the RT reaction mixture. The reaction mixture was incubated for 3 min at 96°C and then subjected to 35 cycles consisting of 45 s at 94°C, 30 s at 55°C, and 45 s at 72°C. The final annealing step was performed for 7 min at 72°C. The reaction mixtures were stored at 4°C until they were analyzed by agarose gel electrophoresis.
For each large-volume (300-μl) reaction, a 10-μl cocktail consisting of 2 μl of each primer (concentration, 75 μM), 2 μl of AmpliTaq DNA polymerase, and 4 μl of water was added to the RT reaction mixture. The reaction mixture was incubated for 4 min at 96°C and then subjected to 35 cycles consisting of 75 s at 94°C, 60 s at 55°C, and 75 s at 72°C. The final annealing step was performed at 72°C for 7 min. The reaction mixtures were stored at 4°C until they were analyzed by agarose gel electrophoresis.
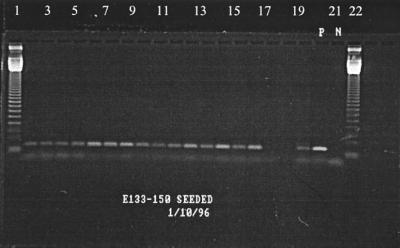
Agarose gel electrophoresis was performed in 1.6% agarose gels (Amresco) containing 15 μg of ethidium bromide per ml. The gels were electrophoresed for 2 h at a constant voltage of 100 V and were analyzed by photographing them as they were exposed to UV light (UVP Inc., Upland, Calif.). Typical gel photographs are shown in Fig. 2 and 3.
FIG. 2.
Enterovirus-seeded reactions. Lanes 1 and 22, 123-bp marker; lanes 2 to 19, samples 133 to 150; lane 20, positive control (10 PFU of poliovirus); lane 21, negative control.
FIG. 3.
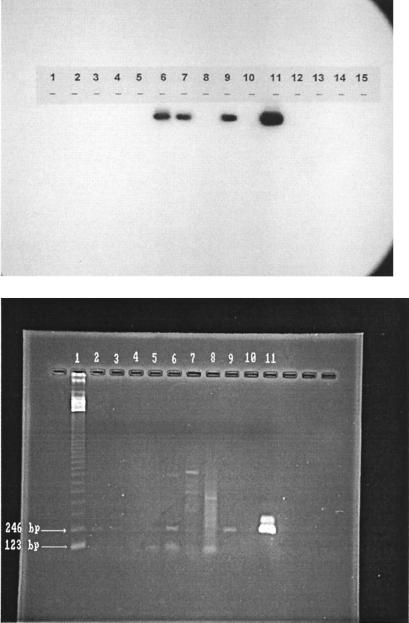
Rotavirus-seeded and nonseeded reactions. Lane 1, 123-bp marker; lanes 2 to 6, seeded reactions (samples 80 to 84); lanes 7 to 11, nonseeded reactions (samples 80 to 84); lane 12, positive control (10 TCID50 of rotavirus); lane 13, negative control.
Hybridization with radiolabeled DNA probes.
Following electrophoresis, agarose gels were soaked in 0.4 M HCl for 15 min, rinsed in double-distilled H2O, and then soaked in 0.4 M NaOH for 15 min to denature the double-stranded PCR product. The DNA was then transferred (15) to a charged nylon membrane (GeneScreen Plus; DuPont NEN Research, Boston, Mass.) by using a vacuum blotter (model 785; Bio-Rad Laboratories). The membrane was soaked for 30 min in 10× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate, pH 7.0) and then placed on a piece of blotting paper on the vacuum blotter surface. The vacuum blotter surface was then overlaid with a plastic sheet in which a window slightly smaller than the membrane was cut. The gel was placed over the membrane, 1 liter of 10× SSC was added to the blotter chamber, and a vacuum (5 in. of Hg) was applied for 90 min. Following transfer, the membrane was soaked for 1 min in 0.4 M NaOH to completely denature the DNA on the membrane and then soaked for 1 min in 1.0 M Tris-HCl (pH 7.5)–5× SSC to neutralize the NaOH. The membrane was placed between two pieces of blotting paper to remove the excess moisture and then either air dried overnight at room temperature or placed in a UV light chamber (model UVC 500 UV Crosslinker; Pharmacia, Piscataway, N.J.) and exposed to 120,000 μJ of 254-nm light per cm2. Both air drying and UV exposure resulted in permanent fixation of the transferred DNA to the nylon membrane, as recommended by the membrane manufacturer.
Following fixation of the DNA to the membrane, the membrane was placed in glass roller bottle (Robbins Scientific, Sunnyvale, Calif.), and enough hybridization buffer (Rapib-Hyb; Amersham Life Sciences, Arlington Heights, Ill.) that had been prewarmed to 42°C was added to the tube to completely soak the membrane. The tube was placed in a hybridization incubator (model 400; Robbins Scientific) equipped with a rotating tube holder and was rotated for 30 min at 42°C, after which 5 μl of radiolabeled DNA probe was added to the buffer in the tube. The tube was returned to the incubator and rotated for an additional 120 min at 42°C.
Following hybridization, the hybridization buffer was poured off, 30 ml of 2× SSC was added to the bottle, and the bottle was gently shaken by hand for 10 min at room temperature. The wash solution was then poured off, and an additional 30 ml of 2× SSC was added the tube, which was again gently shaken for 10 min at room temperature. The wash solution was discarded, approximately 30 ml of 2× SSC–1% sodium dodecyl sulfate that had been prewarmed to 42°C was added to the bottle, and the bottle was rotated in the hybridization incubator for 20 min at 42°C. After 20 min, the wash solution was poured off, 30 ml of 0.2× SSC–1% sodium dodecyl sulfate that had been prewarmed to 42°C was added to the bottle, and the bottle was rotated in the incubator for 20 min at 42°C.
The membrane was blotted to remove the excess moisture and placed in a sealable plastic envelope (Kapak Corp., Minneapolis, Minn.). The envelope was placed in a photographic exposure cassette and allowed to expose a sheet of X-ray film (X-OMAT AR; Eastman Kodak Co., Rochester, N.Y.) overnight at −80°C. Depending on the intensity of the signal observed on the film, some exposures were repeated for as little as 2 h or as long as 48 h. The film was developed according to the film manufacturer’s directions. An typical autoradiograph is shown in Fig. 4.
FIG. 4.
Gel photograph and autoradiograph of the same samples. Lanes 6, 7, and 9 contained rotavirus-positive samples. Lane 1, 123-bp marker; lanes 2 to 9, samples 1, 6, 16, 31, 32, 46, 47, and 54; lane 10, negative control; lane 11, positive control (10 TCID50 of rotavirus).
Radiolabeling of DNA probes.
DNA probes were 3′ end labeled with [32P]dATP (Amersham Life Sciences) by using a DNA 3′ end labeling system kit (Promega). For each 20-μl reaction mixture, 4 μl of 5× terminal transferase buffer (supplied with the kit), 1 μl (2 pmol) of DNA probe, 1 μl of terminal transferase (10 to 20 U/μl), 1.6 μl of 32P-labeled dATP (800 Ci/mmol), and 12.4 μl of water were added to a 50-μl microcentrifuge tube. The tube was incubated for 60 min at 37°C, and the reaction was stopped by heating the reaction mixture for 10 min at 70°C. The labeled probe was stored at −20°C for 10 days or less until it was used.
RESULTS
Cell culture analysis of environmental concentrates.
A total of 150 samples were analyzed for enteroviruses by cell culture techniques by using BGM cells; 13 of the 150 samples (8.7%) exhibited cellular CPE in both the initial phase and the confirmation phase of the analysis.
RT-PCR analysis of environmental concentrates.
A total of 150 samples were analyzed by RT-PCR for enteroviruses, hepatitis A virus, and rotavirus. Each sample was assayed twice for each virus, once by using concentrate alone as a template for RT-PCR and once by using concentrate which had been seeded with either 10 PFU of poliovirus, 10 PFU of hepatitis A virus, or 10 TCID50 of rotavirus.
When primers specific for enterovirus were used in RT-PCR, 17 samples (11.3%) failed to exhibit amplification when they were seeded. A total of 40 of the 133 samples which could be assayed (30.1%) were deemed positive for the presence of enterovirus RNA.
When primers specific for hepatitis A virus were used in the RT-PCR, 11 samples (7.3%) failed to exhibit amplification when they were seeded. Twelve of the 139 samples which could be assayed (8.6%) were deemed positive for the presence of hepatitis A viral RNA.
In the RT-PCR analysis performed with rotavirus-specific primers there were 20 samples (13.3%) that could not be assayed. Eighteen of the remaining 130 samples (13.8%) were positive for rotavirus RNA.
The cell culture and RT-PCR results are summarized in Table 1, and the results of a comparative analysis are shown in Table 2.
TABLE 1.
Summary of viral analyses
| Type of assay | No. of samples analyzeda | No. of samples
|
|||
|---|---|---|---|---|---|
| Positive | Negative | Sample did not precipitate | Reaction failed | ||
| Cell cultureb | 150 | 13 (8.7)c | 136 (91.3) | 1 (0.1) | |
| Enterovirus PCR | 150 | 40 (26.7, 30.1) | 93 (62.0, 69.9) | 17 (11.3) | |
| Hepatitis A virus PCR | 150 | 12 (8.0, 8.6) | 127 (84.7, 91.4) | 11 (7.3) | |
| Rotavirus PCR | 150 | 8 (12.0, 13.8) | 112 (74.7, 86.2) | 20 (13.3) | |
Number of samples for which the analysis was completed.
The virus assayed for in the cell culture analysis was enterovirus.
The values in parentheses are percentages. Where there are two values, the first value is the percentage of the total samples and the second value is the percentage of the samples assayed.
TABLE 2.
Comparison of enterovirus assays
| Reaction in cell culture assay | No. of samples with the following reactions in the RT-PCR assay:
|
||
|---|---|---|---|
| Positive | Negative | Unknown | |
| Positive | 6 | 6 | 1 |
| Negative | 34 | 89 | 14 |
Summary of physicochemical characteristics of groundwater sites.
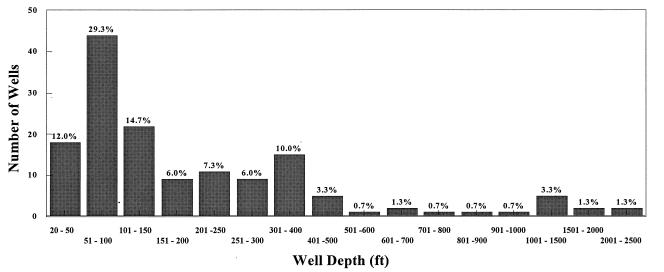
The average depth of the wells surveyed was 269 ft, and the depths ranged from 19 to 2,247 ft. The average well depth at the sites that were positive as determined by cell culture analysis was 418 ft, while the average depth of the wells at the sites that were positive as determined by RT-PCR was 213 ft. The average pH was 7.16 for all wells, and the pH ranged from 4.83 to 9.20. The average pH values were 7.18 for cell culture-positive samples and 7.10 for RT-PCR-positive samples. The average temperature for all wells was 14.8°C, and the range of temperatures was 7.0 to 34.0°C. The average temperatures were 12.9°C for cell culture-positive samples and 13.5°C for RT-PCR-positive samples. The average turbidity was 1.4 nephelometric turbidity units, and the turbidities ranged from 0.039 to 15.6 nephelometric turbidity units. The average TOC content was 0.97 mg/liter, and the TOC contents ranged from 0.12 to 5.21 mg/liter. The physicochemical characteristics and geological formations of the groundwater sites are summarized in Table 3 and Fig. 5.
TABLE 3.
Summary of physicochemical characteristics
| Category | Depth (m) | pH | Turbidity (NTU)a | Temp (°C) | TOC concn (mg/liter) |
|---|---|---|---|---|---|
| Avg | 269 | 7.16 | 2.20 | 14.8 | 0.97 |
| Minimum | 19 | 4.83 | 0.039 | 7.0 | 0.12 |
| Maximum | 2,247 | 9.20 | 15.6 | 34.0 | 5.21 |
| Avg for cell culture-positive samples | 418 | 7.18 | 1.75 | 12.9 | 1.18 |
| Avg for enterovirus PCR-positive samples | 213 | 7.10 | 1.02 | 13.5 | 0.97 |
| Avg for rotavirus PCR-positive samples | 235 | 7.27 | 1.60 | 14.2 | 0.95 |
| Avg for hepatitis A virus-positive samples | 167 | 6.81 | 0.37 | 14.2 | 1.18 |
NTU, nephelometric turbidity units.
FIG. 5.
Numbers of groundwater wells sampled at different depths. More than 85% of the wells were 51 to 500 ft deep.
DISCUSSION
The objective of this work was to develop a simple, rapid, low-cost PCR-based assay that could be used by water utilities to monitor viruses in groundwater samples. Molecular techniques are now widely used in environmental research and monitoring, and the necessary tools and techniques are available from a variety of sources. The strategy described here fulfills the water industry’s need for a rapid, reliable, inexpensive, easily performed technique for analyzing groundwater for virus contamination.
Strategy for detection of viruses by PCR.
The PCR is a powerful technique for detecting organism-specific nucleic acid sequences and can differentiate types of enteric viruses, such as enterovirus, rotavirus, hepatitis A virus, and Norwalk virus. The strategy developed in this study involved removing or inactivating PCR-inhibiting substances, using a large-volume PCR which allowed a larger equivalent volume of a water sample to be used, seeding water concentrates with viruses to test the applicability of the PCR for each sample, and using assay controls.
PCR cannot be performed with most concentrated water samples unless interfering substances are removed prior to RT and/or the amplification reaction (PCR). Selection of the sample treatment method must be based on applicability and the efficiency of the protocol. The fact that the specific enterovirus, hepatitis A virus, and rotavirus sequences were amplified in more than 98% of the groundwater concentrates and the fact that the results were confirmed by Southern hybridization suggest that our sample treatment protocol and the RT-PCR assay can be used to detect enteroviruses in environmental samples.
Removal of potentially inhibiting material from a sample, either by chemical means (such as phenol-chloroform treatment) or by physical means (such as dilution and chromatographic separation), is critical to virus detection by RT-PCR. Although the virus concentration in groundwater may be very low, RT-PCR techniques can potentially reveal the presence of viral RNA molecules. We found, however, that some untreated environmental samples mask detection of the viral RNA by this technique, even when they are seeded with virus at high concentrations. In dilution experiments, a 100-μl environmental sample which had been seeded with 102 PFU was subjected to RT-PCR and failed to show any DNA amplification. The same sample diluted 10 times and 100 times exhibited increasing amplification with each dilution (data not shown).
(i) Positive and negative controls for PCR assays.
We believe that for each environmental sample, both a positive virus-seeded control and a negative control sample need to be tested simultaneously with the environmental sample to allow reasonable interpretation of data. The lack of amplification from a sample does not necessarily mean that no human enteroviruses are present in the sample. Controls must be subjected to the same procedures as the samples to ensure that PCR inhibition does not occur.
(ii) Confirmation and sensitivity.
The sensitivity of the PCR technique and confirmation of PCR amplification should be evaluated by techniques such as Southern hybridization and seminested PCR assays. Using non-PCR-based confirmation, such as Southern transfer and hybridization, is more desirable. This technique allows an increase in the detection limit following PCR and provides confirmation of the PCR assay results.
Sample inhibition of RT-PCR.
Several samples assayed by the PCR resisted amplification when they were seeded with virus. Many of the same samples, however, could be assayed by PCR when steps were taken to neutralize inhibitors and isolate the viruses from the samples. Some of the samples exhibited amplification after the initial phenol-chloroform and Sephadex treatments described above, while other samples required a second PCR in which a seminested primer (a third primer located between the initial two primers) was used. Interestingly, the nucleic acids in several samples which resisted amplification with the primers specific to enterovirus were amplified when the samples were seeded with rotavirus and were assayed with primers specific to rotavirus. This seems to indicate that inhibition of the reaction may be associated with annealing of the primers to the template, as well as with the action of the reaction enzymes. It is possible that using different primers or using more than two primers in a reaction may enhance the detection of viruses.
PCR assays compared with cell culture assays.
We expected that the results of the PCR and cell culture analyses of environmental samples might not correlate well. The minimum level of detection of viruses in any sample when the cell culture method is used is 1 PFU, which may be equivalent to one virion to several virions. Cell culture techniques can detect only viruses which are infectious and culturable. In addition, cell culture techniques, as currently practiced, may be less than ideal for detecting even culturable and infectious viruses. Most protocols (including U.S. EPA Information Collection Rule procedure) call for a 14-day initial passage and a 14-day second passage of a sample on cells, followed by a 7-day confirmation passage of putative positive samples. However, there have been reports that a much longer incubation period can be used to show that samples which have been found to be negative in standard assays are really positive. Finally, since each environmental sample is unique, little is known about what components of a sample may inhibit viral infection of cells in culture.
RT-PCR is potentially a much more sensitive test for the presence of virus since it is possible to detect as little as a single molecule of RNA. However, RT-PCR cannot distinguish infectious viruses from noninfectious viruses or to detect the presence of intact virus particles (it is possible that “naked” RNA or DNA could be detected in an assay). However, based on our sample collection and processing procedures (positively charged filter and elution with 1.5% beef extract), a positive RT-PCR assay most likely indicates that intact virus particles, not naked RNA, are present in the sample (data not shown).
Given its sensitivity and the fact that it detects viral nucleic acid sequences, PCR analysis should result in more positive findings than cell culture analysis. Since either cell culture analysis or PCR can reveal only a “snapshot” of the quality of the groundwater being sampled, PCR seems to be a desirable and rapid initial screening tool since the presence of even noninfectious viruses indicates that a groundwater supply has been contaminated.
While detection of viral RNA does not indicate that there is an infectious level of contamination, the presence of viral RNA does indicate that there is a source of viral contamination and thus a potential health risk. Thus, the most sensitive method of detection is the most desirable method, even if it cannot confirm the infectivity of the virus in a sample.
Despite the success of this study, there are number of issues which still need to be addressed if PCR technology is going to be used to detect pathogens in water samples. Sample processing and treatments to remove inhibitory substances must be customized for a different quality of water. The molecular methods used to detect pathogens is a fast-growing area in microbiology, and recent advances which simplify the procedures should be considered.
ACKNOWLEDGMENTS
This study was funded by the American Water Works Association Research Foundation and by the U.S. EPA. Support was also provided by the American Water Works Service Company, Inc., and the University of Arizona. In addition, we acknowledge the support of a number of water supply utilities.
REFERENCES
- 1.Abbaszadegan M, Huber M S, Gerba C P, Pepper I L. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:1318–1324. doi: 10.1128/aem.59.5.1318-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craun G F. Health aspects of groundwater pollution. In: Bitton G, Gerba C P, editors. Groundwater pollution microbiology. New York, N.Y: John Wiley & Sons, Inc.; 1984. pp. 135–179. [Google Scholar]
- 3.DeLeon R, Shieh C, Baric R S, Sobsey M D. Proceedings of the 1990 American Water Works Association WQTC. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction. Advances in water analysis and treatment; pp. 833–853. [Google Scholar]
- 4.Edzwald J K, Van Benschoten J E. Proceedings of the 4th Gothenburg Symposium on Chemical Water and Wastewater Treatment. 1990. Aluminum coagulation of natural organic matter; pp. 341–359. [Google Scholar]
- 5.Hyypia T, Auvinen P, Maaronen M. Polymerase chain reaction for the human picornaviruses. J Gen Virol. 1989;70:3261–3268. doi: 10.1099/0022-1317-70-12-3261. [DOI] [PubMed] [Google Scholar]
- 6.Keswick B H, Gerba C P, DuPont H L, Rose J B. Detection of enteroviruses in treated drinking water. Appl Environ Microbiol. 1984;47:1290–1294. doi: 10.1128/aem.47.6.1290-1294.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keswick B H, Gerba C P, Secor S L, Cech I. Survival of enteric viruses and indicator bacteria in groundwater. J Environ Sci Health Part A Environ Sci Eng. 1982;17:903–912. [Google Scholar]
- 8.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses. In: Fields B N, editor. Virology. New York, N.Y: Raven Press; 1990. pp. 549–605. [Google Scholar]
- 9.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–351. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 10.Mullis K B, Ferre F, Gibbs R A. The polymerase chain reaction. Boston, Mass: Birkhauser; 1994. p. 166. [Google Scholar]
- 11.Payment P. Isolation of viruses from drinking water at the Pont-Viau Water Treatment Plant. Can J Microbiol. 1981;27:417–420. doi: 10.1139/m81-063. [DOI] [PubMed] [Google Scholar]
- 12.Pillai S D, Josephson K L, Baily R L, Gerba C P, Pepper I L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991;57:2285–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotbart H A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–494. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 15.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]