Abstract
When detected at single-base-pair resolution, the genome-wide location, occupancy level, and structural organization of DNA-binding proteins provide mechanistic insights into genome regulation. Here we use ChIP-exo to provide a near-base-pair resolution view of the epigenomic organization of the Escherichia coli transcription machinery and nucleoid structural proteins at the time when cells are growing exponentially and upon rapid reprogramming (acute heat shock). We examined the site specificity of three sigma factors (RpoD/σ70, RpoH/σ32, and RpoN/σ54), RNA polymerase (RNAP or RpoA, -B, -C), and two nucleoid proteins (Fis and IHF). We suggest that DNA shape at the flanks of cognate motifs helps drive site specificity. We find that although RNAP and sigma factors occupy active cognate promoters, RpoH and RpoN can occupy quiescent promoters without the presence of RNAP. Thus, promoter-bound sigma factors can be triggered to recruit RNAP by a mechanism that is distinct from an obligatory cycle of free sigma binding RNAP followed by promoter binding. These findings add new dimensions to how sigma factors achieve promoter specificity through DNA sequence and shape, and further define mechanistic steps in regulated genome-wide assembly of RNAP at promoters in E. coli.
The genome-wide positional organization of the Escherichia coli transcription machinery and related structural proteins has been studied in vivo but at moderate resolution (Wade et al. 2006; Davis et al. 2011; Myers et al. 2015). This has allowed promoter regions that are enriched with these factors to be defined. Yet, the structural organization of proteins at promoters has yet to be defined on a genomic scale at single-base-pair resolution.
E. coli has over 4000 genes, with many transcribed into a single polycistronic mRNA (Gama-Castro et al. 2016). Consequently, there are fewer than a thousand promoters. Promoter count is confounded by the potential for multiple distinct promoters to initiate polycistronic messages that cover essentially the same genes, and some promoters are used only in certain environments. Furthermore, additional promoters may exist within polycistronic units, including some that may drive noncoding transcription (Conway et al. 2014; Thomason et al. 2015).
E. coli promoters have been annotated in large part through computational consensus motif searches (Gama-Castro et al. 2016; He et al. 2018) and transcription start site mapping (Nonaka et al. 2006; Wade et al. 2006; Conway et al. 2014; Thomason et al. 2015). Unlike eukaryotes, bacterial promoters tend to have well-defined consensus sequences that can help define where promoters reside (Browning and Busby 2004, 2016). However, it remains uncertain whether all genome-wide instances of a consensus suffice to recruit the transcription machinery. E. coli promoters typically include a −10 element (core and extended), a −35 element, and a discriminator region that define where transcription starts (Browning and Busby 2004, 2016). The −35 and −10 elements (or equivalent) are recognized by sigma (σ) factors that provide promoter specificity for RNA polymerase (RNAP). In many cases, they function in concert with sequence-specific transcription factors (Travers and Burgess 1969; Martínez-Antonio and Collado-Vides 2003; Feklístov et al. 2014; Browning and Busby 2016). RNAP has five subunits: two alpha subunits (RpoA), one beta subunit (RpoB), one beta prime subunit (RpoC), and one omega subunit (Ebright 2000). RpoA interacts with A/T-rich upstream promoter (UP) elements to the extent that they are present (Ross et al. 2001).
E. coli has seven sigma factors that direct large programs of gene expression in response to environmental signaling (Burgess 2001; Browning and Busby 2016). By one model, they compete to bind RNAP, thereby directing RNAP to specific sets of genes (Maeda et al. 2000; Gruber and Gross 2003; Browning and Busby 2004; Mauri and Klumpp 2014). RpoD (σ70) is required to transcribe most housekeeping genes. RpoH (σ32) is specific for heat shock genes (Straus et al. 1987). RpoN (σ54) is unique as it has a distinct evolutionary history compared with other sigma factors (Studholme and Buck 2000; Browning and Busby 2004). RpoN binding is determined by −24 and −12 elements rather than at −35 and −10 (Wigneshweraraj et al. 2008; Browning and Busby 2016). Unlike other sigma factors, RpoN-directed initiation involves ATP-dependent activators to initiate transcription (Wigneshweraraj et al. 2008; Browning and Busby 2016).
Biochemical and structural studies have led to the general model that sigma factors engage with RNAP before promoter binding (McClure 1985; Kulbachinskiy et al. 1999; Young et al. 2001). A complex of sigma and RNAP then selects the correct promoter based on DNA sequence recognition. Once bound to DNA to form a closed complex, it proceeds through isomerization steps that produce an open complex in which promoter DNA has melted and been placed in the RNAP active site (Ruff et al. 2015). For RpoN, this requires distal enhancer proteins that are brought into close proximity to the promoter, in some cases by IHF (Rappas et al. 2007; Wigneshweraraj et al. 2008). It remains to be established whether an RpoN/RNAP closed promoter complex is constitutively present genome-wide that is awaiting activation. In contrast, RpoH is thought to be rate-limited in promoter binding by its concentration (Straus et al. 1987). Once transcription initiates, RNAP clears the promoter and sigma either dissociates (Travers and Burgess 1969; Hansen and McClure 1980; Straney and Crothers 1987; Krummel and Chamberlin 1989) or, in some studies, travels with or reassociates with RNAP (Ring et al. 1996; Kapanidis et al. 2005; Harden et al. 2016). Although mechanisms for how sigma factors direct RNAP to promoters and initiate transcription have been well established through biochemical assays, supporting evidence for the applicability of such models on a genomic scale in vivo is limited, owing in part to the low resolution of genome-wide assays.
Genome-wide protein–DNA interactions in vivo can be mapped and studied through the chromatin immunoprecipitation and sequencing (ChIP-seq) assay. We have developed ChIP-exo, a version of ChIP-seq having ultrahigh spatial resolution along chromosomal DNA (Rhee and Pugh 2011). ChIP-exo uses formaldehyde to rapidly fix protein–DNA interactions in vivo. A 5′-3′ exonuclease is used to digest DNA up to the cross-link. This allows precise mapping of protein–DNA interactions. The ability of ChIP-exo to map genome-wide protein–DNA interactions with a greater than 100-fold increase in positional resolution compared with ChIP-seq has been shown in eukaryotes but not in prokaryotes like E. coli (Rhee and Pugh 2011).
As part of genomic regulation, the bacterial chromosome is compacted into a nucleoid structure (Browning and Busby 2016). The nucleoid is formed as a result of DNA supercoiling, molecular crowding, and nucleoid-associated proteins (NAPs) (Dillon and Dorman 2010). Even though some NAPs may function similarly to eukaryotic histone proteins, they are a diverse group of proteins, with some NAPs recognizing specific DNA sequences and regulating transcription (Martínez-Antonio and Collado-Vides 2003). E. coli has at least 12 NAPs including factor for inversion stimulation (Fis) and integration host factor encoded by the ihfB (also known as himD) locus (Dillon and Dorman 2010; Browning and Busby 2016).
This study examines at single-base-pair resolution the genome-wide binding and organization of key subsets of sigma factors, RNAP, and NAPs that regulate transcription and nucleoid structure of the bacterial genome. Although mapping genome-wide protein–DNA interactions in exponentially growing cells is highly informative about steady-state interactions, additional mechanistic insights may be achieved by observing any redistribution of interactions when cells are rapidly reprogrammed by environmental signals. We therefore chose to map the positional organization of the selected factors under conditions of exponential growth at 30°C and upon acute heat shock. Previous western blot analysis shows that RpoH concentration peaks at 6 min of heat shock at 42°C (Straus et al. 1987) and, so, is the condition that we used. We looked for changes in the precise genome-wide binding locations with the goal of testing predictions of in vitro models of σ/RNAP assembly at promoters. We examine the DNA sequence elements associated with factor binding, assessing potential contributions other than direct sequence readout to achieve genome-wide site recognition.
Results
We used ChIP-exo to map the genome-wide binding locations of various targets: RNAP subunits (RpoA, -B, -C), sigma factors (RpoD/σ70, RpoH/σ32, and RpoN/σ54), and NAPs (Fis and IhfB [also known as IHF-β]) in TAP-tagged DY330 E. coli. We initially chose D-galactose/methyl-galactoside ABC transporter periplasmic binding protein (MglB [also known as GBP]) as a negative control because it has no annotations linking it to DNA binding. If correct, its DNA interaction status should be negative and appear as background. With all ChIP-exo data, patterns of genome-wide binding (x-axis positions of peak locations) can be compared among different targets. However, peak intensity levels (y-axis occupancy) are not comparable across different targeted factors due in part to protein-specific differences in cross-linking efficiency with DNA.
In addition to examining factor binding in rich media (LB) at normal 30°C, we measured any reorganization upon acute heat stress by instantaneously shifting log-phase cells for 6 min to 42°C before covalently trapping interactions with formaldehyde (see Methods). Cells were lysed and their chromosomal DNA fragmented. Specific targets were immunoprecipitated using antibodies directed against a TAP epitope tag fused to the target's C-terminus. DNA was then trimmed 5′-3′ to the site of cross-linking with lambda exonuclease and subsequently deeply sequenced. Sequencing also confirmed the correct target fusion in the data sets. Two biological replicates were merged after confirming data reproducibility.
We generated a reference list consisting of 638 transcription units (TUs), as defined in RegulonDB, a comprehensive genomic database for E. coli (Gama-Castro et al. 2016), as well as the published literature (Supplemental Table S1; Salgado et al. 2000; Cho et al. 2009; Mao et al. 2015). The actual number of TUs may vary with growth condition and is subject to some uncertainty, but our number is within a previously predicted range of 630–700 (Salgado et al. 2000). We did not examine TUs identified in related strains having distinct reference genomes (Conway et al. 2014; Thomason et al. 2015). We first looked at target occupancy in and around promoter regions to better understand their relationship with TUs, and then we focused on cognate DNA motifs genome-wide to better understand mechanisms of site specificity and initiation. We start with RpoD, using other targets for comparison and for controls. Then we used the same type of analysis to examine RpoH, RpoN, and NAPs.
RpoD and RNAP accumulate at promoters
As expected, ChIP-exo showed RpoD enrichment at promoter regions (TU ATG starts) (Fig. 1A, upper), with ∼70% of 638 TU promoters having a RpoD signal above background (Fig. 1B). The distribution of RpoD among TUs was essentially unchanged upon 6 min of heat shock (Supplemental Fig. S1), indicating that the heat shock response involves little or no reprogramming of RpoD-bound promoters. Across all TUs, RNAP (RpoA, -B, -C) occupancy correlated with RpoD occupancy, whether at 30°C or upon 42°C heat shock (Fig. 1C). Little or no correlation was observed between RpoD/RNAP and other sigma factors and NAPs, thereby showing promoter specificity of the assay and the targeted factors. Promoter-proximal RpoH, RpoN, Fis, and IhfB were well correlated with themselves at 30°C versus 42°C, but not with each other. Thus, we confirm that RpoD/RNAP represents the predominant transcription initiation complex under normal growth and under acute heat shock. It has no predominant positional ties along the genome to RpoH, RpoN, Fis, and IHF.
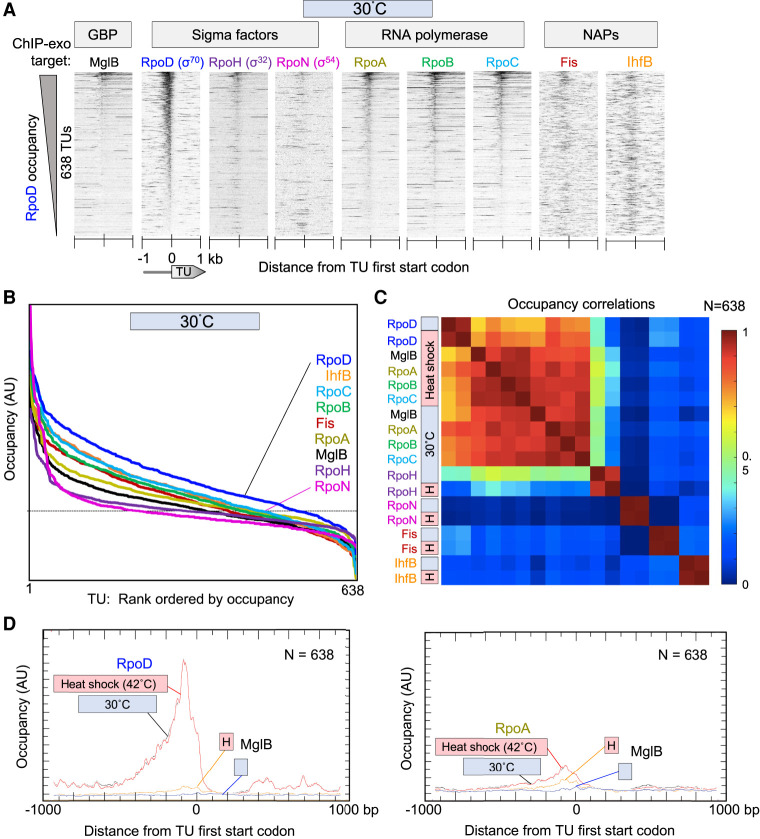
Figure 1.
Sigma factors and RNAP binding at RpoD promoter regions. (A) Heatmap of occupancy distribution of the indicated protein targets as measured by ChIP-exo. Transcription units (TUs; rows, N = 638 TUs) are aligned by their ATG start codons (5′-3′, left to right) and sorted by RpoD occupancy (summed −80 to +80 bp from TU ATG start) at 30°C. ATG starts were used instead of TSSs because TSSs have more experimental error, condition-specific biological variability, and variance among strains compared with DNA elements. Sense and antisense tags were shifted in the 3′ direction by 6 bp (to adjust for the headroom of lambda exonuclease) and merged. Data files have an x-axis bin size of 2 bp. (B) Relative occupancy level in arbitrary units (AUs) on a linear scale for individual target proteins, rank ordered by their occupancy level at 638 TUs from panel A. The dashed line represents the approximate midpoint observed with targets that are expected to be background. (C) Heatmap matrix of correlation coefficients for co-occupancy of targets from panel A (from −500 to +100 of a TU ATG start). Heat shock, or H, denotes instantaneous change from 30°C to 42°C for 6 min. (D) Composite (averaged) plots of RpoD, RpoA, and MglB occupancy from panel A (30°C) and from Supplemental Figure S1 (6 min, 42°C). AU denotes arbitrary linear units. Data files have an x-axis bin size of 2 bp. The MglB data are the same in both panels. To compare y-axis magnitudes between same-target samples from 30°C and 42°C, gene-averaged y-axis values were empirically scaled (i.e., one scaling factor per data set) to achieve similar y-axis minima values in the x-axis window (i.e., to achieve similar average local background). This assumes that the minima are background.
MglB as a negative control was expected to produce low background signals, which it did (Fig. 1A,B). However, its signal across promoter regions was well correlated with RpoD and RNAP occupancy at 30°C and 42°C, but not with RpoH, RpoN, Fis, and IhfB (Fig. 1C) This indicates that MglB is not behaving as background; otherwise, it would have been uncorrelated with all tested sigma factors and NAPs. One potential exception is where the genome-wide background is highly reproducible but far from uniform (i.e., nonrandom structure to the background). In such a case, the promoter background for all data sets, including RpoH, RpoN, Fis, and IhfB, would have correlated with MglB, as they comprise mostly background binding in the regions defined by RpoD/RNAP binding. As this was not the case, the RpoD/RNAP-specific correlation of MglB indicates that MglB colocalizes with RpoD/RNAP across the genome, albeit with low cross-linking efficiency, and not with other sigma factors.
On average, RpoD and RNAP occupancies were most enriched <400 bp upstream of TU ATG start codons with their peak locations being coincident and ∼30 bp upstream (Fig. 1D). Except for a small number of highly expressed TUs, RNAP was lowly or not detectable in gene bodies (Fig. 1A,D). This is consistent with other reports (Reppas et al. 2006). Although it is possible that promoter-bound and elongating RNAP differ in their cross-linking efficiencies, if we assume them to be similar, then RNAP forms a positionally more stable promoter complex relative to any other distance downstream. This is consistent with slow steps in initiation and/or promoter clearance compared with elongation (Ruff et al. 2015; Winkelman and Gourse 2017). Our simplest interpretation of the data supports existing models that the transition from transcription initiation to elongation is a rate-limiting step on DNA in RpoD-directed transcription, as previously suggested genome-wide (Reppas et al. 2006) and suggested from biochemical experiments (McClure 1985).
Mechanism of RpoD promoter binding and selectivity
We searched for motifs associated with RpoD binding by first identifying the top 500 RpoD-occupied regions and then applying MEME for de novo motif discovery (Supplemental Table S2; Bailey et al. 2009). The top-scoring motif is shown in Figure 2A and includes the well-known −10 and −35 region of RpoD promoters (Hawley and McClure 1983; Lisser and Margalit 1993; He et al. 2018). There was an almost identical match with the extended consensus defined computationally by He et al. (2018; Fig. 2A, cf. middle and bottom motifs). This included a C/G-rich sequence immediately downstream (3′) from the −10 consensus sequence. However, we further detected an A/T-rich sequence just upstream of the −35 motif (from −44 to −40), which is likely the UP element (Ross et al. 1993). The concordance with He et al. (2018) is noteworthy given that their RpoD motif discovery was based on computational algorithms that examined position-specific trinucleotide propensity and electron–ion interaction pseudopotentials of nucleotides, whereas our identifications are based on experimentally measured binding of RpoD.
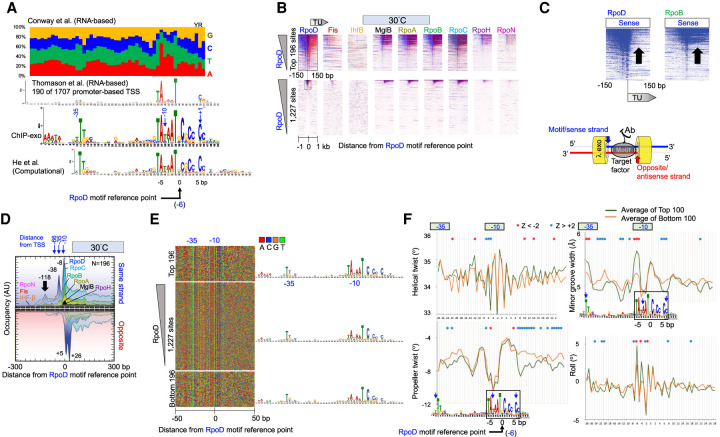
Figure 2.
Properties of genome-wide RpoD–DNA interactions. (A) MEME motif of RpoD-bound locations. All panels are aligned to UP, −35, and −10 regions. Blue numbers refer to approximate base-pair distances from transcript start sites (Zuo and Steitz 2015). Negative values reflect distances in the 5′ direction on the sense/motif strand. Black numbers (−5, 0, 5) refer to base-pair distances from an arbitrary reference point within the motif (x = 0) and is the reference point used for all other panels in this figure. (Top) Nucleotide frequencies reported in Supplemental Table S3 of Conway et al. (2014). (First middle) MEME logo as reported in Supplemental Figure S7A of Thomason et al. (2015). (Second middle) MEME logo as defined by the top 500 RpoD-bound locations determined by ChIP-exo in this study. (Bottom) MEME logo as shown in Figure 2 of He et al. (2018). (B) Heatmap of occupancy distribution of the indicated protein targets as measured by ChIP-exo at 30°C. Rows correspond to RpoD motif instances (N = 1227; bottom panel) aligned by the reference point indicated in panel A. Blue indicates exonuclease stop sites on the sense/motif strand (sequence tag Read_1 5′ ends; see illustration at lower right); red reflects data on the antisense/opposite strand. Top panels show a zoom-in of the relevant section of the bottom panels. All data are sorted based on RpoD occupancy ±50 bp from the motif reference point. (C) Heatmaps from panel B, showing only the sense strand data from RpoD and RpoB. Arrows point out the TU region, in which there is an occupancy differential between RpoD and RpoB. (D) Composite plots of panel B (top), where the antisense/opposite strand data are inverted (N = 196). y-axis scales are linear, but with arbitrary units of occupancy (tag counts), and are thus not comparable between targets. Data files have an x-axis bin size of 2 bp and are smoothed with a 5-bp moving average. Peak locations are indicated. (E) Four-color plot of nucleotide sequences covering ±50 from the reference point in panel A. Corresponding MEME motifs are shown to the right. (F) Composite plots of DNA shape parameters in the vicinity of the RpoD motif reference point, comparing the top 100 RpoD-bound motifs (green) to the bottom 100 bound motifs (yellow). Positions exceeding a statistical Z-score threshold of two are indicated with asterisks.
In the UP, −35, and −10 regions, our motif agreed well with mRNA-based start site mapping (Conway et al. 2014; Thomason et al. 2015). However, there appeared to be some discrepancy at the transcription start site (+1). Although it is generally understood that the −1/+1 nucleotides are enriched with YR nucleotides (IUPAC nomenclature), this determination was based on only 88 promoters (Hawley and McClure 1983). Genome-wide studies based on mapping RNA 5′ ends have found the YR motif to be enriched, with 79% of the +1 nucleotide being an R (Kim et al. 2012), whereas another study had R at 56% (Mendoza-Vargas et al. 2009). Thomason et al. (2015) report YR enrichment, but this was observed only when analyzing 190 out of 1707 promoter transcription start sites (Fig. 2A). Our ChIP-exo data report dispersed YR enrichment at −1/+1 based on distance from the consensus −10 element, rather than RNA 5′ end mapping. Our data find C being enriched at +1, rather than YR, which is in accord with that of He et al. (2018). This occurred whether RpoD occupancy was high or low (see Fig. 2E), indicating that transcription initiation efficiency as defined by the +1 nucleotide was not accounting for differences in RpoD cross-linking. Furthermore, because RNAP can initiate transcription at variable distances downstream from the −10 element (Jeong and Kang 1994; Liu and Turnbough 1994; Lewis and Adhya 2004), a canonical start site would not be particularly evident in a MEME logo that is based on fixed distances. Thus, if YR is the predominant site of initiation, then it exists at variable distances from the −10 element. A model whereby RNAP “scrunches” the downstream DNA during initiation might allow for a YR search at variable distances. However, if transcription initiation predominates at a fixed distance downstream from the −10 element, then YR is not a predominant site of initiation.
We next searched for all instances of the RpoD motif (e-value threshold = 10−4) across the genome, whether occupied by RpoD or not. We found 1227 instances of the RpoD motif. Exonuclease stop sites (ChIP-exo tag 5′ ends) for RpoD and other factor targets were aligned to a fixed reference point within each motif instance (Fig. 2B). Here the data are plotted at single-base-pair resolution with the exonuclease stop sites shown separately on the motif strand (blue) and its opposite strand (red). The vast majority of the 1227 RpoD motif instances either were not bound by RpoD at 30°C or 42°C or had low occupancy (Supplemental Fig. S2A).
When comparing RpoD (σ70) ChIP-exo signal on the sense strand to that of RpoB (RNAP), RpoD was generally depleted in gene bodies (Fig. 2C, arrows), although enrichment was observed across a small number of specific genes (blue horizontal “lines” across certain TUs). This is consistent with other studies showing that at certain genes RpoD dissociates upon or soon after promoter clearance, whereas at other genes, RpoD continues to associate or reassociate with RNAP during elongation (Ring et al. 1996; Bar-Nahum and Nudler 2001; Mukhopadhyay et al. 2001; Kapanidis et al. 2005; Mooney et al. 2005; Perdue and Roberts 2011; Harden et al. 2016). Our ChIP-exo data suggest that RpoD generally dissociates as RNAP starts across gene bodies, except in specific cases. Whether RpoD travels with RNAP or reassociates cannot be determined from these experiments.
Three major exonuclease stop sites were detected on the sense strand, encompassing the −35 and −10 regions (averaged in Fig. 2D, individual genes in Fig. 2B), whereas two major stop sites were detected on the antisense strand encompassing the transcription start site. Lambda exonuclease stop sites are typically ∼6 bp more 5′ from where cross-linking occurs, as expected of its ∼6-bp head room between its active sites and the leading edge of the exonuclease. Because multiple adjacent stop sites were observed on each DNA strand in a population average, not all potential sites of cross-linking had achieved cross-linking; otherwise, only the most 5′ stop sites on each strand for a given binding site would have been detectable. One interpretation of incomplete cross-linking is that there is diminished local accessibility to formaldehyde or diminished local proximity of the cross-linkable protein–DNA residues to each other.
Additional RpoD cross-linking was detected up to ∼150 bp upstream of the TSS (based on the −35/−10 motif) (Fig. 2D, black arrow). Because in vitro structures of site-bound RpoD would not predict interactions this far upstream, this may reflect indirect cross-linking through other proteins bound upstream. This upstream pattern did not change upon heat shock (Supplemental Fig. S2B). No significant motifs were picked up by MEME in this upstream region, indicating that there may not be a predominant site-specific protein present. Taken together, these results indicate that RpoD binds to small fraction of potential RpoD sequence motifs in the genome. At every bound site, RpoD makes the same type of interaction with its core motif (i.e., canonical points of cross-linking relative to the motif). This does not change upon heat shock (6 min at 42°C).
We next investigated why as many as 80% of the RpoD consensus motifs were not bound by RpoD. We compared the DNA sequence of the top 196 bound motifs to the bottom 196 unbound motifs and found only slight differences in their composition (Fig. 2E). The primary differences occurred at the TATAAT portion of the −10 region, which is where the primary contact of RpoD with promoters occurs. This region was more degenerate at unbound sites (albeit A/T-rich), which confirms that sequence variation at the −10 region contributes to RpoD occupancy levels. Additionally, high-occupancy motif instances were three times more likely to be located within 500 bp of an annotated TU ATG than unbound sites, indicating that a substantial fraction of unbound sites are not likely to be biologically relevant RpoD sites.
Part of this study was to examine the contributions of DNA shape to site-specific binding. Four aspects of DNA shape readouts (helical twist, propeller twist, minor groove width, and roll) are computational predictions based on Monte Carlo simulations (Rohs et al. 2009; Li et al. 2017). We first examined DNA shape parameters that distinguished bound versus unbound RpoD in the −10 region (Fig. 2F). The −10 region is where both promoter recognition and DNA melting occurs (Browning and Busby 2004). Significant shape differences between bound and unbound motifs were located at the first TA of TATAAT, particularly in helical twist, roll, and minor groove width. Because the TA composes part of the canonical motif sequences, the discrimination between bound and unbound likely includes both direct base-specific contacts and DNA shape readout that is imparted by at least T versus A at positions −12 and −13.
Distinct shape readouts for bound versus unbound were most evident outside of the canonical motif sequences (Fig. 2F), with the predominant differentials of binding being a reduction in both propeller twist and minor groove width at the transcription start site, as well as reduction in propeller twist further downstream. Similar differentials were observed just downstream from the −35 region. Taken together, these results suggest that DNA shape readout may contribute to RpoD site recognition (or stability) particularly just downstream from the −35 and −10 regions. An ability to initiate transcription site-specifically through interactions at the −35, −10, and other upstream regions may further involve proper DNA shape of the general initiation region.
Rate-limiting recruitment of RNAP to a refined set of 46 RpoH promoters
We next focused on a small number (N = 120) (Supplemental Table S3) of annotated heat shock–induced genes, as previously defined (Nonaka et al. 2006; Wade et al. 2006; Gama-Castro et al. 2016; Keseler et al. 2017), with the goal of confirming or more accurately specifying the set of RpoH-regulated (σ32) TUs. The existing set was largely defined through increased mRNA expression upon ectopic RpoH overexpression (Nonaka et al. 2006). We took an orthogonal approach of examining transcription complex assembly before and after heat shock in the presence of endogenous RpoH levels. We first examined the immediate upstream region of the 120 annotated heat shock–induced genes, including those contained within larger polycistronic TUs. As can been seen in Figure 3A and Supplemental Figure S3A, only a small fraction of these intergenic regions contained RpoH, which is consistent with many of them being contained within approximately 70 polycistronic TUs, as previously determined (Nonaka et al. 2006).
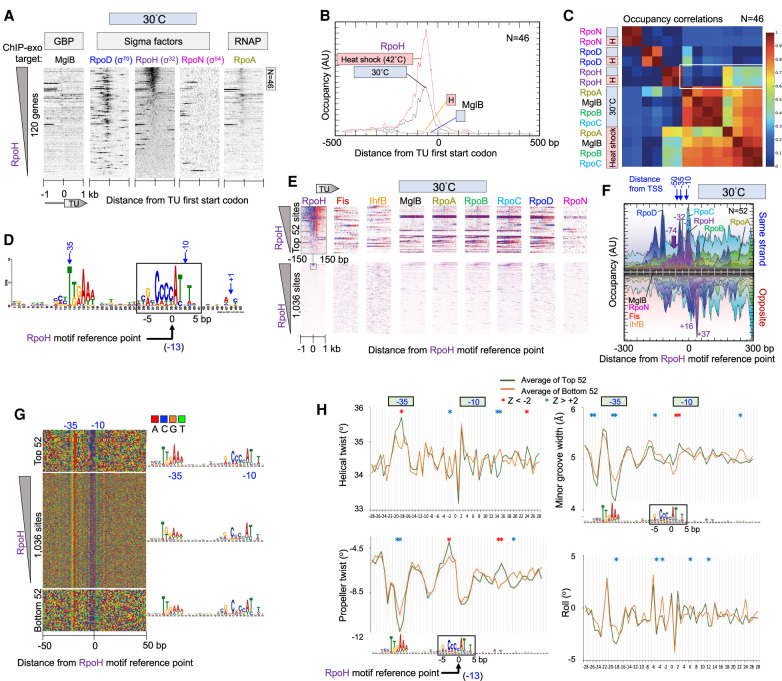
Figure 3.
Genome-wide interactions of RpoH at RpoH promoters. (A) Heatmap of occupancy distribution of the indicated targets around TU ATG start codons of 120 previously defined heat shock genes (Nonaka et al. 2006; Gama-Castro et al. 2016). Data are sorted based on RpoH occupancy ±120 bp from the ATG start of each gene. The top 46 genes are indicated. For additional figure details, see Figure 1A. (B) Composite plots of RpoD and MglB occupancy of the top-most RpoH-bound TUs from panel A (N = 46). Plots also include heat shock data from Supplemental Figure S3. For details, see Figure 1D. (C) Heatmap matrix of correlation coefficients for target occupancy co-occurrence at 46 promoters (from −500 to +100 of a TU ATG start) defined in panel A. Heat shock, or H, denotes instantaneous change from 30°C to 42°C for 6 min. A white box is drawn around key correlation relationships. (D) MEME motif derived from the top 500 RpoH-occupied regions as described in Figure 2A. (E) Heatmap of occupancy for the indicated targets at 30°C distributed around the RpoH motif reference point. Data are sorted based on RpoH occupancy ±50 bp from the reference point. For additional plotting details, see Figure 2B. (F–H) Distribution around 52 RpoH motifs of RpoH and other target occupancies at 30°C (F), nucleotide sequence (G), and DNA shape (H). For plotting details, see Figure 2, D through F, for plotting details.
Based on current annotations, we further whittled this existing list to 54 nonoverlapping TUs, of which 40 contained a previously reported RpoH promoter and another 14 did not (Supplemental Table S3; Nonaka et al. 2006). We applied multiple criteria to validate this existing set: RpoH ChIP-exo occupancy, detection of an RpoH motif in our MEME analysis, and/or the presence of an RpoH promoter defined more recently by EcoCyc (Keseler et al. 2013, 2017). We confirmed 39 of the 40 TUs that had a previously reported RpoH promoter but only seven of the 14 where an RpoH promoter was not previously reported. We found RpoH occupancy to be weak in many of the latter. In total, we confirmed 46 annotated TUs (Fig. 3A; Supplemental Table S3). Those TUs reportedly having an internal RpoH promoter (e.g., yhgH [also known as gntX] and yrfG) were confirmed (Nonaka et al. 2006). However, of the three reported to have multiple RpoH promoters, one (htpG) was found to have only a single predominant site of RpoH binding, another (mutL) had no binding, and the third (dnaK) was confirmed (Supplemental Table S3). We identified 23 additional RpoH-bound cognate motifs that could not be assigned to a promoter region (i.e., >500 bp from an annotated TU promoter) (Supplemental Table S3). These may represent novel RpoH promoters. Typically, they were located inside gene bodies.
In total, based on ChIP-exo and prior studies (Nonaka et al. 2006; Wade et al. 2006), we estimate that there are about 70 RpoH control regions of which about 46 are directly tied to annotated heat shock promoters. Other heat shock–inducible genes may be under the control of the alternative heat shock sigma factor RpoE, as previously reported (Erickson et al. 1987; Wang and Kaguni 1989; Rouvière et al. 1995; Ades et al. 2003) but not studied here.
Upon heat shock, we observed increased RpoH occupancy at RpoH-bound promoters regions (Fig. 3B; Supplemental Fig. S3B). RpoN was essentially absent from RpoH promoter regions, whereas many of the 120 annotated heat shock genes that lacked RpoH nevertheless contain RpoD, as previously observed (Wade et al. 2006). All 46 RpoH promoters were constitutively occupied by RpoH at substantial levels before heat shock, with the level of increase upon heat shock being rather moderate. This includes RpoH promoters with high and low pre-existing RpoH levels (Supplemental Fig. S3A). Similarly, RpoA occupancy increased in step with increased occupancy of RpoH upon heat shock, with RpoH and RNAP (RpoA, -B, -C) having correlated occupancy at 42°C (Fig. 3C). The same was true for MglB. These heat shock RpoH–RNAP correlations were specific to the 46 RpoH promoters, as such correlations did not exist when all TUs were examined (Fig. 1C). RpoH and RNAP occupancy did not correlate before heat shock. This indicated that RpoH occupied RpoH promoters even when RNAP was absent and that RpoH was largely inactive for RNAP recruitment at 30°C. Nevertheless, upon heat shock (42°C), there was substantial recruitment of RNAP, and therefore, activation of RpoH occurred that is distinct from DNA binding. This is not inconsistent with seemingly alternative in vitro models of sigma factor recruitment solely through a pre-existing complex with RNAP, in that both mechanisms could occur.
MEME identified the canonical RpoH motif as its top-scoring motif among the top-scoring 500 RpoH-occupied peaks (Fig. 3D). We found 1036 instances of this motif across the genome, of which only 52 instances had detectable binding of RpoH (Fig. 3E; Supplemental Fig. S3C). ChIP-exo peak patterning detected multiple exonuclease stop sites on each strand, each at about the same position relative to each other and its cognate motif, as was seen with RpoD to its cognate motif (cf. Figs. 2D and 3F; also Supplemental Fig. S3D). However, their relative positions were not identical to the base pair, indicating some distinctiveness in the way RpoD and RpoH interact with their cognate sites in vivo. These results suggests that RpoH and RpoD contact their cognate motifs in similar, but not identical, ways across the genome, as expected of their related structures.
An additional difference was that RpoD had upstream cross-links up to about −150 (Fig. 2D), whereas RpoH had upstream cross-links up to about −100 (Fig. 3F). These cross-linking locations are further upstream than what would be expected of these sigma factors alone and, so, may represent indirect cross-linking arising from sigma interactions with upstream regulatory proteins. If so, then promoter occupancy by RpoH in the absence of RNAP might include these upstream interactions. RpoH also cross-linked at about +30 (exonuclease stop site at +37 in Fig. 3F). This would seem to be beyond where RpoH alone would cross-link and thus could reflect additional downstream indirect interactions with other factors.
We next investigated motif discrimination. There was remarkably little sequence distinction between RpoH-bound and unbound RpoH motif instances (Fig. 3G). This suggests that RpoH does not achieve specificity solely through direct DNA sequence readout of its cognate motif. When DNA shape properties were examined (Fig. 3H), there were shape distinctions between bound and unbound RpoH sites, but not as strong as seen for RpoD (although the relative weakness may be partially owing to only about half of the set of “top-bound” sites actually having detectable RpoH). Maximal differences were observed in propeller twist in regions that lie between the −10 and −35 region. These were the same relative regions identified as discriminatory in RpoD site selection (cf. Figs. 2F and 3H). Taken together, RpoH also appears to use DNA shape to help discriminate sites but perhaps to a lesser extent than RpoD.
RNAP-independent binding of RpoN
We detected binding of RpoN just upstream of 50 annotated TUs (Fig. 4A), similar to what was observed previously (Reitzer and Schneider 2001; Bonocora et al. 2015). Many of the highly occupied sites were enriched for operons that regulate nitrogen stress response under nitrogen-limiting conditions. Binding to these promoters changed little upon heat shock, as expected (Supplemental Fig. S4A).
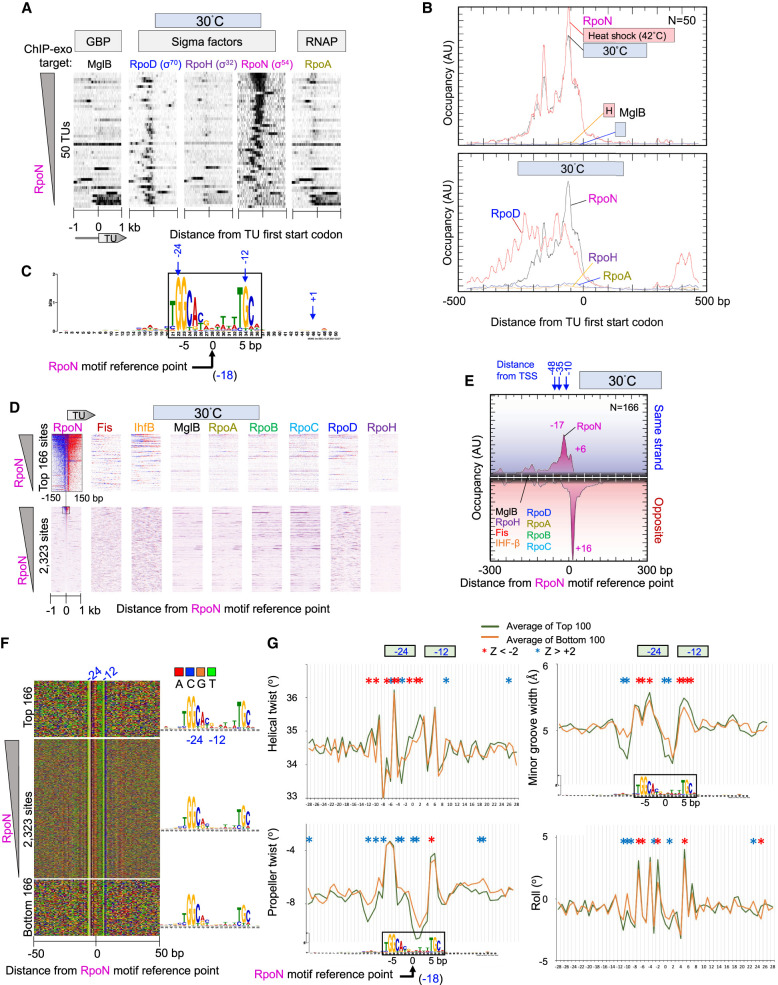
Figure 4.
Genome-wide interactions of RpoN at RpoN promoters. (A) Distribution of RpoN around the ATG start codons of 50 RpoN-bound TU. Data are sorted based on RpoN occupancy ±500 bp from the ATG start of each gene. For additional figure details, see Figure 1A. (B) Composite plots of sigma factor, RpoA, and MglB occupancy at RpoN-bound TUs from panel A. Plots also include heat shock data from Supplemental Figure S4. For details, see Figure 1D. (C) MEME motif derived from the top 500 RpoN-bound locations as described in Figure 2A. (D) Heatmap occupancy of indicated protein targets at 30°C distributed around RpoN motif reference points. For plotting details, see Figure 2B. (E–G) Distribution around RpoN motifs of RpoN and other target occupancies at 30°C (E), nucleotide sequence (F), and DNA shape (G). For plotting details, see Figure 2, D through F.
RpoN-bound promoter regions contained detectable levels of RpoD and were generally absent of RpoH (Fig. 4A). Thus, like RpoH promoters, these RpoN promoters generally reside among RpoD promoters. RNAP was generally absent from RpoN promoter regions, except where it aligned with RpoD (Fig. 4A,B, lower panel). Thus, RpoN occupies its target promoters in the absence of RNAP. Except for what can be accounted for by RpoD, these promoters are not necessarily expected to be active despite RpoN binding, as they are in nitrogen-replete media (Merrick 1993; Zafar et al. 2014). In this regard, they are much like RpoH promoters, which are less active in the absence of heat shock despite having RpoH bound. The absence of RNAP from RpoN-bound sites does not support a model in which RNAP is constitutively bound to RpoN-bound sites, awaiting activation into an open promoter complex (Wigneshweraraj et al. 2008; Browning and Busby 2016).
RpoN is distinct from most other sigma factors with no sequence similarity to RpoD. It binds to the −24/−12 promoter region with the consensus sequence TGGC-N9-GC (Browning and Busby 2004, 2016; Wigneshweraraj et al. 2008), although another report has a longer consensus: TGGCACG-N4-TTGCT (Lloyd et al. 2017). From the top 500 RpoN-bound locations, MEME identified the cognate motif for RpoN: the −24 GG element and the −12 GC element, having the consensus sequence TGGCAYRNWWNWTGC (Fig. 4C). We found 2323 instances of this RpoN motif across the genome. In contrast to RpoD and RpoH, about half of these sites had detectable RpoN occupancy (Fig. 4D). This binding was site specific as the ChIP-exo peaks aligned precisely (to the base pair) to their sites like the high-occupancy RpoN motif sites (Fig. 4D, lower panel, RpoN vertical stripe; also Fig. 4E; Supplemental Fig. S4B,C).
We did not detect a general enrichment of IHF (or Fis) upstream of RpoN-bound sites (Fig. 4D,E; Supplemental Fig. S4B,C), as would be expected if these proteins were facilitating an RpoN enhancer looping mechanism (Wigneshweraraj et al. 2008). However, there are many instances in which IHF is enriched at highly bound RpoN promoters (Fig. 4D). Thus, involvement of IHF may be selective to a limited number of cases and not be general to most RpoN-regulated promoters. Although it is possible that IHF may be recruited to other RpoN promoter regions during gene activation, our detailed study of IHF binding below finds that nearly all IHF motifs are constitutively bound by IHF. This would leave few sites for inducible binding.
The vast majority of RpoN sites were intragenic, which confirms a prior report (Bonocora et al. 2015). However, ChIP-exo detected many more bound sites. What these large number of binding events represent is unclear but may correspond to undefined TUs that are not active under conditions in which operons were defined (Bonocora et al. 2015). Previous ChIP-seq of RpoN in Gram-negative Pseudomonas aeruginosa detected binding of RpoN at about 1000 sites, with only 10% of these sites in annotated promoter regions (Shao et al. 2018).
Unlike RpoD and RpoH, RpoN had only a single predominant exonuclease stop site on the antisense strand but with multiple minor stop sites on the sense strand (Fig. 4E; Supplemental Fig. S4B). This suggests that RpoN is relatively well-anchored to the downstream portion of its site and is either more loosely anchored on its upstream site or has less efficient cross-linking there. Its contact with genomic DNA (ChIP-exo patterning) is quite distinct compared with RpoD and RpoH, as the distinctiveness of their cognate motifs would suggest.
When we compared the DNA sequence of the top 166 RpoN-bound sites to 166 unbound sites, nearly the same consensus motif was achieved (Fig. 4F). Thus, as with the other sigma factors, a specific sequence is insufficient to define RpoN site specificity in vivo. We compared averaged DNA shape profiles at these bound and unbound sites. Despite the identical consensus sequence, all four computationally predicted shape parameters (roll, minor groove width, helical twist, and propeller twist) showed statistically significant differences between highly and lowly occupied sites (Fig. 4G). Like RpoD and RpoH, the sequences adjacent to the consensus motif sequences contributed significantly to shape differences between highly and lowly bound sites, suggesting that DNA shape contributes to RpoN site discrimination. RpoN (and RpoH) differed from RpoD shape differentials (bound vs. unbound) in lacking an additional contribution where transcription starts. However, significant shape contributions were evident with all three sigma factors in the region located between the core −35/−24 and −10/−12 motifs.
DNA sequence as a predominant factor driving Fis and IHF site affinity
The bacterial chromosome is condensed by supercoiling and interactions with NAPs, including Fis, IHF, and Hns. Previous studies have implicated these proteins in influencing transcription both positively and negatively (Browning et al. 2000; McLeod and Johnson 2001). Our analysis showed that Fis and IHF are enriched <400 bp upstream of annotated TUs (Fig. 5A,B). However, their occupancies were not correlated with RpoD or RNAP (Fig. 1C). Their occupancy in these regions may avoid conflicts with transcription in gene bodies. There, it could act in the structural organization of the bacterial nucleoid, insulate neighboring genes, and/or regulate transcription initiation both positively and negatively (McLeod and Johnson 2001; Gerstel et al. 2003; Chintakayala et al. 2013). At the time of their discovery, NAPs were thought to compact DNA by seemingly nonspecific binding and bending (Finkel and Johnson 1992; Feldman-Cohen et al. 2006). However, it is now known that both Fis and IHF bind somewhat degenerate, but sequence-specific sites (Rice et al. 1996; Cho et al. 2008; Stella et al. 2010; Kahramanoglou et al. 2011; Prieto et al. 2012).
Figure 5.
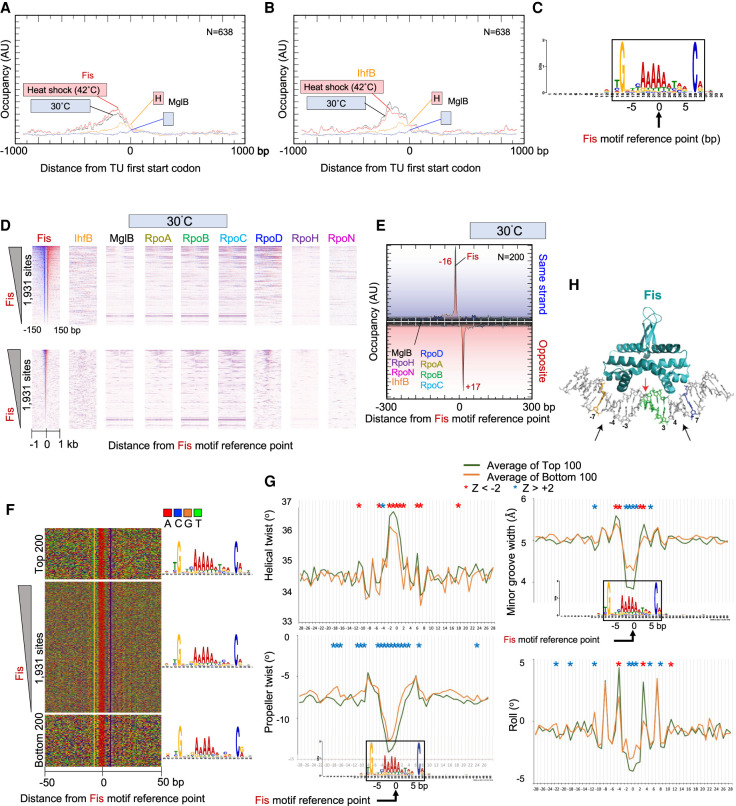
Genome-wide interactions of Fis. (A,B) Composite plots of Fis (A) or IHF (B) and MglB occupancy around the ATG start codons of 638 TUs. Plots also include heat shock data from Supplemental Figure S5. (C) MEME motif derived from the top 500 Fis-bound locations as described in Figure 2A. (D) Heatmap occupancy of indicated protein targets at 30°C distributed around the Fis motif reference point. For plotting details, see Figure 2B. (E–G) Distribution around Fis motifs of Fis and other target occupancies at 30°C (E), nucleotide sequence (F), and DNA shape (G). For plotting details, see Figure 2, D through F. (H) Crystal structure of Fis–DNA interactions (Stella et al. 2010). Black arrows indicate sites of contact. G and C nucleotides at −7 and +7 are colored (gold and blue, respectively), as is the poly(dA:dT) tract (green).
From the 500 most-occupied Fis locations, MEME extracted the expected cognate motif (Fig. 5C; Cho et al. 2008; Kahramanoglou et al. 2011). We identified 1931 Fis motifs genome-wide. Unlike the sigma factors, Fis bound to essentially all consensus motifs throughout the genome, both at 30°C and upon acute heat shock at 42°C (Fig. 5D; Supplemental Fig. S5A,B). Detected occupancy levels varied over an almost 10-fold range at these sites. RpoD/RNAP occupancy was highest in regions where Fis binding was highest, despite a general lack of correlation. This suggests that Fis may be linked to high levels of gene expression in specific cases, rather than being generally true.
Exonuclease stop sites on each strand were located 16–17 bp from the motif midpoint (i.e., ∼33 bp apart) (Fig. 5E). This distance indicates that there is one major site of cross-linking on the edge of each side of the complex. We did not detect opposite-strand peaks located ∼12 bp apart in the 3′ direction, which would have occurred if cross-linking was incomplete (as is seen with sigma factors and other proteins). This suggests that the variance in site occupancy across the genome by Fis was not owing to variances in cross-linking efficiency at different genomic sites. Moreover, nonspecific background in the −1- to +1-kb region surrounding the Fis motif did not vary with Fis site occupancy (Fig. 5D, bottom panels), indicating that other technical aspects like DNA extraction efficiency were not accounting for variance in site occupancy.
Highly occupied sites displayed the same motif consensus as lowly bound sites (Fig. 5F), indicating that other factors were contributing to Fis occupancy levels in addition to direct nucleotide sequence recognition. Fis binding sites were of particular interest because they only have one nondegenerate G-C bp per Fis monomer (at position 7 relative to the motif center), which is expected to lack sufficient sequence-based specificity to define site-specific Fis binding.
When DNA shape was examined, each metric showed significant differences in shape between the high and low occupancy sites (Fig. 5G). Compared with lowly bound sites, high occupancy sites had a narrower minor groove at the motif center, with a wider minor groove at both flanks (±3–5 bp from center). They also showed significant negative roll deviations at the center, along with positive deviations 4 bp from the center, which correspond to major groove interfaces with the Fis reading heads.
A crystal structure of Fis at its motif shows that Fis-bound DNA has an overall curvature of ∼65° (Fig. 5H), and the close spacing between the helix–turn–helix domain is accommodated by severe compression of the central minor groove (Fig. 5H, red arrow; Stella et al. 2010). Although the central 5 bp of the motif is not directly in contact with Fis, the narrowing of the minor groove within this region is essential for insertion of the alpha helices within adjacent major grooves (Stella et al. 2010). This led to a model that Fis selects sites with an intrinsically small central minor groove (Stella et al. 2010), which our data support on a genome-wide scale in vivo. Thus, the intrinsic shape of DNA may be a substantial contributor by which Fis binds site-specifically with different levels of affinity.
IHF binds DNA as a heterodimer of IhfA and IhfB subunits in a sequence-specific manner. Upon binding, IHF maintains sharp bending of the DNA, which allows interaction of other proteins with the DNA. This aspect has been implicated in regulating transcription, replication, and recombination (Dillon and Dorman 2010). From the 500 most-occupied IHF locations, we extracted the expected cognate motif (Fig. 6A; Hales et al. 1994). A genome-wide search of this motif found 2293 instances. As was the case with Fis binding to its cognate site, nearly all IHF motifs were bound by IHF (Fig. 6B; Supplemental Fig. S6A). IHF displayed a major and minor exonuclease stop site on each strand, revealing multiple (at least four) points of cross-linking (Fig. 6C; Supplemental Fig. S6B).
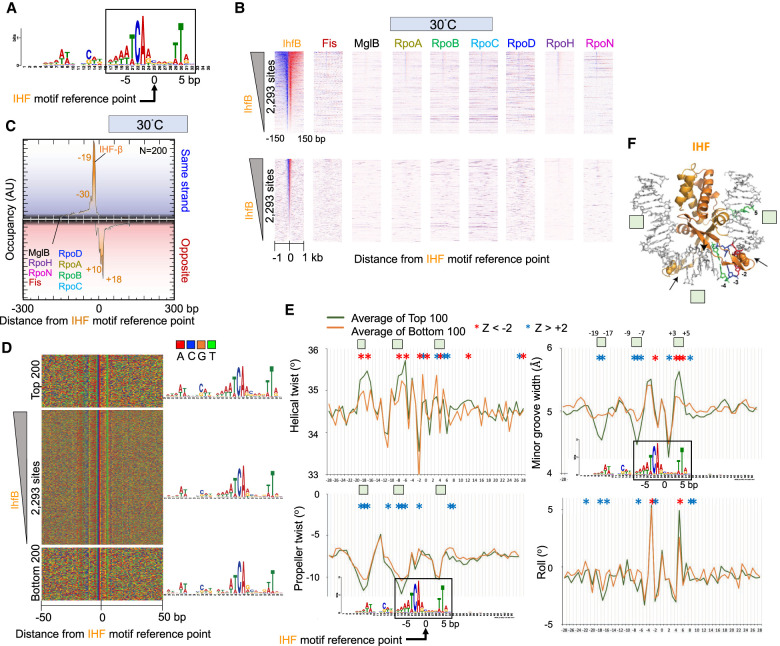
Figure 6.
Genome-wide interactions of IHF. (A) MEME motif derived from the top 500 IHF-bound locations as described in Figure 2A. (B) Heatmap occupancy of indicated protein targets at 30°C distributed around the IHF motif reference point. For plotting details, see Figure 2B. (C–E) Distribution of IHF and other target occupancies at 30°C (C), nucleotide sequence (D), and DNA shape around IHF motifs (E). For plotting details, see Figure 2, D through F. Green boxes correspond to regions shown in panel F. (F) Crystal structure of IHF (Rice et al. 1996). Select motif base coloring is as defined in panel D.
As with Fis, the highest and least occupied IHF bound motifs had essentially the same consensus motif (Fig. 6D), indicating that high versus low occupancy is not entirely defined for IHF through direct sequence readout. The shape differences between the high IHF occupancy sites and the low IHF occupancy sites were most evident at positions −19 to −17, −9 to −7, and +3 to +5 relative to its motif reference point (Fig. 6E). IHF is a minor groove binding protein. A narrow minor groove has been implicated in the stability of the IHF–DNA complex (Rice et al. 1996). Upon DNA binding, IHF induces a bend in the DNA by >160°, causing it to wrap around the protein. Bending occurs largely at two kinks, causing a disruption of the base stacking by intercalation of a proline residue (Fig. 6F, space filled residues; Rice et al. 1996). IHF appears to exclusively contact the phosphodiester backbone and minor groove of DNA (Fig. 6F, black arrows). Thus, it likely relies substantially on indirect non-sequence-specific features rather than canonical major groove sequence-specific contacts, typical of other proteins, to recognize its binding site.
In accordance with this, we observed that the more highly occupied IHF sites tend to have a larger helical twist, smaller minor groove width, and smaller propeller twist specifically around the −19- to −17-bp and −9- to −7-bp position of the motif, which corresponds to two distinct A/T-rich regions of the motif. Positions −19 to −17 and +3 to +5 have similar placements on the IHF structure, well beyond where DNA kinks are made (Fig. 6F). However, unlike −19 to −17, the +3 to +5 region has less reliance on propeller twist. Thus, certain locally placed shape features of DNA may add to IHF site-specific recognition beyond what sequence alone offers.
Discussion
Sigma factors and RNAP find their cognate binding sites at the start of TUs among a backdrop of a much larger number of unrecognized potential sites throughout the bacterial genome. These potential sites have essentially all the proper sequence motifs in the proper positions (minimally −35/−24 or −10/−12 elements) to be recognized by their cognate sigma factors, but this is not enough. How they find their cognate binding sites among a vast excess of seemingly identical but unbound sites remains an open question. Certainly, the primary mode of specificity is a base-specific readout. Additionally, we found that bound and unbound motifs have distinct preferences for intrinsic DNA shape features that emanate from the core motifs and can account for differential site recognition. The same is true for NAPs.
Intrinsic DNA shape features include DNA helical twist and minor groove width, along with base propeller twist and roll. These features are definable by combinations of adjacent nucleotides and, so, are not readily evident in PWMs reported by MEME or equivalent software. Multiple combinations of sequences place these shape parameters into favorable or unfavorable ranges that can augment or inhibit DNA binding. As shown here and elsewhere, DNA shape contributes to site specificity for sigma factors and for RNAP (Lara-Gonzalez et al. 2020). These shape features may properly apposition the binding sites and flanking DNA in a manner that accommodates sigma factor binding. Beyond DNA sequence and shape readout, local site-specific DNA-binding proteins may regulate sigma factors and RNAP occupancy at promoters. This aspect was not investigated here. When compared with sigma factors, site specificity appears different for NAPs like Fis and IHF, where essentially all instances of their sites (at the same motif e-value threshold) appear to be bound. With NAPs, shape differentials appear to quantitatively affect binding site affinity rather than bound versus unbound seen with sigma factors.
A general view of RNAP recruitment is that sigma factors and RNAP first assemble into a holoenzyme complex that is subsequently recruited to promoters. In doing so, sigma factors may compete with each other for RNAP (Malik et al. 1987; Kulbachinskiy et al. 1999; Maeda et al. 2000; Young et al. 2001; Browning and Busby 2004; Mauri and Klumpp 2014). Using ChIP-exo, we find that at least RpoH and RpoN can occupy their cognate sites within promoters in the absence of RNAP, with their steady-state levels being defined in part by DNA sequence/shape readout along with other factors. The low levels of cross-linking observed upstream of and downstream from the sigma factors, beyond what can be accounted for by known sigma/DNA contacts, could reflect additional interactions with factors that we have not discerned in this study.
The absence of RNAP is strongest at RpoN promoters under conditions in which such promoters are not expected to be active. There, RpoN occupancy is robust. At RpoH promoters, RNAP is absent under non–heat shock conditions (30°C) but present upon acute heat shock (42°C). RpoH occupancy is robust under both conditions, with additional binding upon heat shock. Increased occupancy is in accord with heat shock activation being administered in part through increased concentrations of RpoH (Straus et al. 1987; Yura 2019). Our results with RpoH and RpoN suggest that at least two types of sigma factors can be maintained at promoters independent of RNAP and that RNAP is not recruited to these sigma-bound promoters unless triggered to do so. This suggests that RNAP recruitment to sigma-bound promoters is triggered by activating signals. Thus, promoter-bound sigma factors provide a mechanism to recruit RNAP. However, free RpoD is unable to recognize the −10 region unless RNAP first drives a conformational change (Kulbachinskiy et al. 1999). Consequently, there might be multiple mechanisms to recruit RNAP to promoters. For example, an initial round of recruitment might occur through a preformed sigma/RNAP complex, and there may be a second round of recruitment in which RNAP associates with promoter-bound sigma before sigma has had a chance to dissociate. This would allow stress-specific sigma factors to engage in bursting, where multiple initiation events accompany a single sigma binding event (Engl et al. 2020). Whether sigma factors are slow to dissociate from DNA in vivo relative to reassociation of RNAP is not known and thus speculative. In vitro, RpoD rapidly dissociates upon transcription initiation (Travers and Burgess 1969; Hansen and McClure 1980; Straney and Crothers 1987; Krummel and Chamberlin 1989). In vivo, sequence-specific transcription factors could offer stability to promoter-bound sigma factors, thereby slowing their dissociation. Thus, an open question is whether transcription factors stabilize sigma factors in the absence of RNAP.
Our work confirms many previous studies on the location of the targets studied here. The high-resolution view afforded here increases the confidence of location detection and provides structural information on binding. Consequently, we observe that most coding TUs have detectable RpoD occupancy and that a small subset of about 46 TUs binds RpoH, and another small subset of about 50 TUs binds RpoN. However, we observed widespread binding of RpoN at cognate sites that are not annotated TU promoters, as others have reported (Bonocora et al. 2015). RpoN binds to these sites no differently than at other sites, and thus, they are not incidental sites of contact or background. Whether they point to condition-specific TUs remains to be determined.
At 30°C, RpoD promoters are active, whereas RpoH and RpoN promoters are largely inactive. Upon acute heat shock (6 min at 42°C), we find that RpoD promoters remain active and essentially unchanged, RpoN promoters remain inactive and unchanged, and RpoH promoters switch from inactive to active. Although all three sigma factors are bound at 30°C, increased occupancy is observed only with RpoH upon heat shock. However, this quantitative increase is insufficient to explain the qualitative off-to-on change in RNAP binding, which might therefore involve switching the promoter-bound RpoH from an inactive to an active state. The data do not exactly align with a model whereby free sigma factors compete for RNAP interactions in vivo, even locally imposed, because we are not observing a loss of RpoD at promoters when RpoH is increasing. Moreover, there are about 10 times more RpoD promoters (some very highly active) than RpoH promoters, making the impact of any competition rather limited. Nonetheless, we envision that promoter-bound RpoH and RpoN become activated by sigma-specific environmental signals and recruit RNAP. Our results do not exclude free sigma factors from also binding RNAP and then occupying promoters. The key discovery here is that at least RpoH and RpoN can occupy cognate promoters without RNAP.
Methods
Cell growth
DY330 E. coli TAP-tagged strains were purchased from GE Healthcare (OEC4630). Strains grew at similar rates, indicating that C-terminal tagging was not substantially impacting the functions of the tag factors. Cell cultures were grown in 50 mL of LB media at 30°C in a shaking incubator with a starting OD600 = 0.1 and then were treated when the final OD600 = 0.7–0.8. For inducing heat shock, 25 mL of the media at 54°C was added to 25 mL of culture at OD600 = 0.7–0.8 to achieve the final temperature of 42°C. Twenty-five milliliters of room temperature media was added to the control sample. The heat shock sample was placed for 6 min in a 42°C shaker, whereas the control sample was placed for 6 min in a 25°C shaker . To cross-link the protein–DNA interactions, 25 mL of cold formaldehyde (Thermo Fisher Scientific) was added to the heat shock sample and 25 mL of room temperature formaldehyde to the control sample so that the final formaldehyde concentration was 1%. Note that formaldehyde cross-linking is nearly instantaneous and most efficient at 25°C (Poorey et al. 2013). Both 42°C heat shock and 30°C samples were placed for 15 min in a 25°C shaker. Excess formaldehyde was quenched by adding 2.5 M glycine to the final concentration of 125 mM glycine for 5 min. Cells were harvested by centrifugation at 4000g for 5 min at 4°C. Cell pellets were washed with ST buffer (10 mM Tris-HCl at pH 7.5 and 50 mM NaCl) with protease inhibitors, flash-frozen, and stored at −80°C until used. At least two independent biological replicates were performed for each experiment.
Cell lysis
Harvested cells were lysed in 1 mL of lysis buffer (10 mM Tris-Cl at pH 8.0, 1% sucrose, 10 mM EDTA, 50 mM NaCl) supplemented with complete protease inhibitor (CPI, Roche). One hundred microliters of lysozyme (10 mg/mL) was added per gram of the cell pellet. Samples were incubated for 30 min at room temperature with rotation and centrifuged for 10 min at 4000 rpm and room temperature. The supernatant was removed, and the pellets were resuspended in 250 μL of sonication buffer (50 mM HEPES, 150 mM NaCl, 2 mM EDTA, 0.1% sodium deoxycholate, 1% Triton X-100, 0.2% sodium dodecyl sulfate) and transferred to a 1.5-mL polystyrene tube containing ∼75 μL of 0.1-mm zirconium beads. The samples were then sonicated in a Bioruptor (Diagenode) for four cycles with 30-sec on/off intervals to obtain DNA fragments 100 to 500 bp in size.
ChIP-exo
A 10–15 mL culture equivalent at OD600 = 0.8 of E. coli chromatin was incubated overnight at 4°C with 10 μL bed volume of IgG-Dynabeads in 200 μL final volume. ChIP-exo 5.0 was performed exactly as previously described (Rossi et al. 2018). Essentially this involves chromatin fragmentation to <500 bp through sonication, followed by chromatin immunoprecipitation of the target protein. Although the DNA is still on the resin, library construction is initiated along with resection of each DNA strand in the 5′-3′ direction using lambda exonuclease until digestion is impeded by a protein–DNA cross-link. From this final DNA. libraries are constructed and sequenced.
DNA sequencing
DNA sequencing was performed using the paired end mode on a NextSeq 500 or 550 to produce 2 × 40 bp reads. Sequenced reads were aligned to the ec2 genome using BWA-MEM (v0.7.9a) (Li 2013). PCR duplicates were removed from the aligned reads.
Data analysis
See Data Access section for links to data files and detailed bioinformatic methods. Scripts may be found under Supplemental Code. In summary, base calls for the paired-end reads were performed using Bcl2fq version 2.16 (Illumina). ChIP-exo reads were aligned to the E. coli W3110 reference genome (ec2, the reference genome for DY330) using BWA version 0.7.9a with default parameters. NCIS normalization (500-bp windows, 0.75 minimum fraction) was performed to control for differences in sequencing depth of different data sets, using MglB at their respective temperatures as negative controls (Liang and Keleş 2012). As the results of this study indicated that MglB may not be an appropriate negative control, we emphasize that the y-axis values (z-axis in heatmaps) represent arbitrary units (AUs) on a linear scale and are not directly comparable across different targets and temperature conditions. Additionally, different targets are expected to have different cross-linking efficiencies and thus preclude direct comparison of y-axis magnitudes (z-axis for heatmaps) between targets. However, x-axis peak locations are comparable between targets. To compare y-axis magnitudes between same-target samples from 30°C and 42°C, gene-averaged y-axis values within their respective windows were empirically scaled (i.e., one scaling factor per data set) to achieve similar y-axis minima (i.e., to achieve similar local background levels). This assumes that regions of minima are predominantly background).
For sequence-specific factors, peak calling was performed using GeneTrack (s5, e20, F1) and ChExMix algorithm for the 30°C and 42°C data sets (Albert et al. 2008; Yamada et al. 2020). Unique peaks from both GeneTrack and ChExMix were combined for each sequence-specific factor from the 30°C and 42°C shock data sets. The top 500 peaks were used for de novo motif discovery using MEME to determine the presence of known DNA sequence motifs ±50 bp from midpoint of the paired peaks (Supplemental Table S2; Bailey et al. 2009). For motif searching, the PWM was generated and used to identify nonoverlapping motif sites across the whole E. coli genome (ec2) using default parameters in FIMO (Grant et al. 2011). Motif sites were aligned to the cognate motif reference point and sorted by protein occupancy at 30°C condition. The cutoff value for sigma factor motif sites was determined using summation of tags in the sorting window and setting the threshold value to be 200 tags for RpoD and RpoH and 100 tags for RpoN, respectively. Four color plots, composite plots, and heatmaps were then oriented around these motif sites unless specified otherwise. DNA shape analysis at highly and lowly bound locations was performed using the DNAshape webserver (Zhou et al. 2013).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE189206. Coordinate files, detailed methods, and plot values used here can be found at GitHub (https://github.com/CEGRcode/2022_EColi_John-Jabbar). GUI ScriptManager v.012 (RRID:SCR_021797) was used for analysis of all data and is available for download at GitHub (https://github.com/CEGRcode/scriptmanager).
Supplementary Material
Acknowledgments
We thank Jeff Roberts (Cornell) for valuable feedback on the manuscript. This work was supported by National Institutes of Health grant ES013768 to B.F.P. The authors used ScriptManager (RRID:SCR_021797), PEGR (Platform for Epigenomic and Genomic Research, RRID:SCR_021861), and Galaxy (RRID:SCR_006281) for data analysis and FAIR data retention compliance.
Author contributions: J. John designed and conducted experiments, performed initial data analysis, and cowrote the manuscript. J. Jabbar conducted the final analysis of the data. N.B. conducted initial data analysis, manuscript writing, and project oversight. M.J.R. provided bioinformatic support on DNA shape analysis. W.K.M.L. provided bioinformatic support and oversight. B.F.P. conceptualized the project and conclusions and cowrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276544.121.
Competing interest statement
B.F.P. is an owner of and has a financial interest in Peconic, LLC, which uses the ChIP-exo technology (US patent 20100323361A1) implemented in this study and could potentially benefit from the outcomes of this research.
References
- Ades SE, Grigorova IL, Gross CA. 2003. Regulation of the alternative σ factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J Bacteriol 185: 2512–2519. 10.1128/JB.185.8.2512-2519.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF. 2008. GeneTrack—a genomic data processing and visualization framework. Bioinformatics 24: 1305–1306. 10.1093/bioinformatics/btn119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Nudler E. 2001. Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell 106: 443–451. 10.1016/S0092-8674(01)00461-5 [DOI] [PubMed] [Google Scholar]
- Bonocora RP, Smith C, Lapierre P, Wade JT. 2015. Genome-scale mapping of Escherichia coli σ54 reveals widespread, conserved intragenic binding. PLoS Genet 11: e1005552. 10.1371/journal.pgen.1005552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65. 10.1038/nrmicro787 [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. 2016. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol 14: 638–650. 10.1038/nrmicro.2016.103 [DOI] [PubMed] [Google Scholar]
- Browning DF, Cole JA, Busby SJ. 2000. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol Microbiol 37: 1258–1269. 10.1046/j.1365-2958.2000.02087.x [DOI] [PubMed] [Google Scholar]
- Burgess RR. 2001. Sigma factors. In Encyclopedia of genetics (ed. Brenner S, Miller JH), pp. 1831–1834. Academic Press, New York. [Google Scholar]
- Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC. 2013. E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet 9: e1003152. 10.1371/journal.pgen.1003152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B-K, Knight EM, Barrett CL, Palsson BØ. 2008. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res 18: 900–910. 10.1101/gr.070276.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 27: 1043–1049. 10.1038/nbt.1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T, Creecy JP, Maddox SM, Grissom JE, Conkle TL, Shadid TM, Teramoto J, San Miguel P, Shimada T, Ishihama A, et al. 2014. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio 5: e01442-14. 10.1128/mBio.01442-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SE, Mooney RA, Kanin EI, Grass J, Landick R, Ansari AZ. 2011. Mapping E. coli RNA polymerase and associated transcription factors and identifying promoters genome-wide. Methods Enzymol 498: 449–471. 10.1016/B978-0-12-385120-8.00020-6 [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8: 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- Ebright RH. 2000. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol 304: 687–698. 10.1006/jmbi.2000.4309 [DOI] [PubMed] [Google Scholar]
- Engl C, Jovanovic G, Brackston RD, Kotta-Loizou I, Buck M. 2020. The route to transcription initiation determines the mode of transcriptional bursting in E. coli. Nat Commun 11: 2422. 10.1038/s41467-020-16367-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JW, Vaughn V, Walter WA, Neidhardt FC, Gross CA. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev 1: 419–432. 10.1101/gad.1.5.419 [DOI] [PubMed] [Google Scholar]
- Fekliístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial σ factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68: 357–376. 10.1146/annurev-micro-092412-155737 [DOI] [PubMed] [Google Scholar]
- Feldman-Cohen LS, Shao Y, Meinhold D, Miller C, Colón W, Osuna R. 2006. Common and variable contributions of Fis residues to high-affinity binding at different DNA sequences. J Bacteriol 188: 2081–2095. 10.1128/JB.188.6.2081-2095.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE, Johnson RC. 1992. The Fis protein: It's not just for DNA inversion anymore. Mol Microbiol 6: 3257–3265. 10.1111/j.1365-2958.1992.tb02193.x [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS, Alquicira-Hernández K, Martínez-Flores I, Pannier L, Castro-Mondragón JA, et al. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 44: D133–D143. 10.1093/nar/gkv1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U, Park C, Römling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol 49: 639–654. 10.1046/j.1365-2958.2003.03594.x [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. 2003. Multiple σ subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57: 441–466. 10.1146/annurev.micro.57.030502.090913 [DOI] [PubMed] [Google Scholar]
- Hales LM, Gumport RI, Gardner JF. 1994. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J Bacteriol 176: 2999–3006. 10.1128/jb.176.10.2999-3006.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen UM, McClure WR. 1980. Role of the σ subunit of Escherichia coli RNA polymerase in initiation. II. Release of σ from ternary complexes. J Biol Chem 255: 9564–9570. 10.1016/S0021-9258(18)43429-1 [DOI] [PubMed] [Google Scholar]
- Harden TT, Wells CD, Friedman LJ, Landick R, Hochschild A, Kondev J, Gelles J. 2016. Bacterial RNA polymerase can retain σ70 throughout transcription. Proc Natl Acad Sci 113: 602–607. 10.1073/pnas.1513899113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11: 2237–2255. 10.1093/nar/11.8.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Jia C, Duan Y, Zou Q. 2018. 70ProPred: a predictor for discovering σ70 promoters based on combining multiple features. BMC Syst Biol 12: 44. 10.1186/s12918-018-0570-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W, Kang C. 1994. Start site selection at lac UV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res 22: 4667–4672. 10.1093/nar/22.22.4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res 39: 2073–2091. 10.1093/nar/gkq934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S. 2005. Retention of transcription initiation factor σ70 in transcription elongation: single-molecule analysis. Mol Cell 20: 347–356. 10.1016/j.molcel.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, et al. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41: D605–D612. 10.1093/nar/gks1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, et al. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45: D543–D550. 10.1093/nar/gkw1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hong JS, Qiu Y, Nagarajan H, Seo JH, Cho BK, Tsai SF, Palsson BO. 2012. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet 8: e1002867. 10.1371/journal.pgen.1002867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel B, Chamberlin MJ. 1989. RNA chain initiation by Escherichia coli RNA polymerase: structural transitions of the enzyme in early ternary complexes. Biochemistry 28: 7829–7842. 10.1021/bi00445a045 [DOI] [PubMed] [Google Scholar]
- Kulbachinskiy A, Mustaev A, Goldfarb A, Nikiforov V. 1999. Interaction with free β′ subunit unmasks DNA-binding domain of RNA polymerase σ subunit. FEBS Lett 454: 71–74. 10.1016/S0014-5793(99)00778-4 [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez S, Dantas Machado AC, Rao S, Napoli AA, Birktoft J, Di Felice R, Rohs R, Lawson CL. 2020. The RNA polymerase α subunit recognizes the DNA shape of the upstream promoter element. Biochemistry 59: 4523–4532. 10.1021/acs.biochem.0c00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DE, Adhya S. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol Microbiol 54: 692–701. 10.1111/j.1365-2958.2004.04318.x [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Li J, Sagendorf JM, Chiu TP, Pasi M, Perez A, Rohs R. 2017. Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res 45: 12877–12887. 10.1093/nar/gkx1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Keleş S. 2012. Normalization of ChIP-seq data with control. BMC Bioinformatics 13: 199. 10.1186/1471-2105-13-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser S, Margalit H. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res 21: 1507–1516. 10.1093/nar/21.7.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Turnbough CL Jr. 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol 176: 2938–2945. 10.1128/jb.176.10.2938-2945.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd MG, Lundgren BR, Hall CW, Gagnon LB, Mah TF, Moffat JF, Nomura CT. 2017. Targeting the alternative σ factor RpoN to combat virulence in Pseudomonas aeruginosa. Sci Rep 7: 12615. 10.1038/s41598-017-12667-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Fujita N, Ishihama A. 2000. Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 28: 3497–3503. 10.1093/nar/28.18.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Zalenskaya K, Goldfarb A. 1987. Competition between σ factors for core RNA polymerase. Nucleic Acids Res 15: 8521–8530. 10.1093/nar/15.20.8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Ma Q, Liu B, Chen X, Zhang H, Xu Y. 2015. Revisiting operons: an analysis of the landscape of transcriptional units in E. coli. BMC Bioinformatics 16: 356. 10.1186/s12859-015-0805-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martı´nez-Antonio A, Collado-Vides J. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol 6: 482–489. 10.1016/j.mib.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Mauri M, Klumpp S. 2014. A model for σ factor competition in bacterial cells. PLoS Comput Biol 10: e1003845. 10.1371/journal.pcbi.1003845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure WR. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54: 171–204. 10.1146/annurev.bi.54.070185.001131 [DOI] [PubMed] [Google Scholar]
- McLeod SM, Johnson RC. 2001. Control of transcription by nucleoid proteins. Curr Opin Microbiol 4: 152–159. 10.1016/S1369-5274(00)00181-8 [DOI] [PubMed] [Google Scholar]
- Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juárez K, Contreras-Moreira B, et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4: e7526. 10.1371/journal.pone.0007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ. 1993. In a class of its own: the RNA polymerase σ factor σ54 (σN). Mol Microbiol 10: 903–909. 10.1111/j.1365-2958.1993.tb00961.x [DOI] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R. 2005. σ and RNA polymerase: an on-again, off-again relationship? Mol Cell 20: 335–345. 10.1016/j.molcel.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. 2001. Translocation of σ70 with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106: 453–463. 10.1016/S0092-8674(01)00464-0 [DOI] [PubMed] [Google Scholar]
- Myers KS, Park DM, Beauchene NA, Kiley PJ. 2015. Defining bacterial regulons using ChIP-seq. Methods 86: 80–88. 10.1016/j.ymeth.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev 20: 1776–1789. 10.1101/gad.1428206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue SA, Roberts JW. 2011. σ70-dependent transcription pausing in Escherichia coli. J Mol Biol 412: 782–792. 10.1016/j.jmb.2011.02.011 [DOI] [PubMed] [Google Scholar]
- Poorey K, Viswanathan R, Carver MN, Karpova TS, Cirimotich SM, McNally JG, Bekiranov S, Auble DT. 2013. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science 342: 369–372. 10.1126/science.1242369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM. 2012. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res 40: 3524–3537. 10.1093/nar/gkr1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappas M, Bose D, Zhang X. 2007. Bacterial enhancer-binding proteins: unlocking σ54-dependent gene transcription. Curr Opin Struct Biol 17: 110–116. 10.1016/j.sbi.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of ς54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65: 422–444. 10.1128/MMBR.65.3.422-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K. 2006. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell 24: 747–757. 10.1016/j.molcel.2006.10.030 [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419. 10.1016/j.cell.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87: 1295–1306. 10.1016/S0092-8674(00)81824-3 [DOI] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW. 1996. Function of E. coli RNA polymerase σ factor σ70 in promoter-proximal pausing. Cell 86: 485–493. 10.1016/S0092-8674(00)80121-X [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. 2009. The role of DNA shape in protein–DNA recognition. Nature 461: 1248–1253. 10.1038/nature08473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262: 1407–1413. 10.1126/science.8248780 [DOI] [PubMed] [Google Scholar]
- Ross W, Ernst A, Gourse RL. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove. Genes Dev 15: 491–506. 10.1101/gad.870001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MJ, Lai WKM, Pugh BF. 2018. Simplified ChIP-exo assays. Nat Commun 9: 2842. 10.1038/s41467-018-05265-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière PE, De Las Peñas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. 1995. rpoE, the gene encoding the second heat-shock σ factor, σE, in Escherichia coli. EMBO J 14: 1032–1042. 10.1002/j.1460-2075.1995.tb07084.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EF, Record MT Jr, Artsimovitch I. 2015. Initial events in bacterial transcription initiation. Biomolecules 5: 1035–1062. 10.3390/biom5021035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. 2000. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci 97: 6652–6657. 10.1073/pnas.110147297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Zhang X, Zhang Y, Zhu M, Yang P, Yuan J, Xie Y, Zhou T, Wang W, Chen S, et al. 2018. RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J Bacteriol 200: e00205-18. 10.1128/JB.00205-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Cascio D, Johnson RC. 2010. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev 24: 814–826. 10.1101/gad.1900610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straney DC, Crothers DM. 1987. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J Mol Biol 193: 267–278. 10.1016/0022-2836(87)90218-X [DOI] [PubMed] [Google Scholar]
- Straus DB, Walter WA, Gross CA. 1987. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 329: 348–351. 10.1038/329348a0 [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Buck M. 2000. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol Lett 186: 1–9. 10.1111/j.1574-6968.2000.tb09074.x [DOI] [PubMed] [Google Scholar]
- Thomason MK, Bischler T, Eisenbart SK, Förrstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197: 18–28. 10.1128/JB.02096-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers AA, Burgess RR. 1969. Cyclic re-use of the RNA polymerase σ factor. Nature 222: 537–540. 10.1038/222537a0 [DOI] [PubMed] [Google Scholar]
- Wade JT, Castro Roa D, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. 2006. Extensive functional overlap between σ factors in Escherichia coli. Nat Struct Mol Biol 13: 806–814. 10.1038/nsmb1130 [DOI] [PubMed] [Google Scholar]
- Wang QP, Kaguni JM. 1989. A novel σ factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol 171: 4248–4253. 10.1128/jb.171.8.4248-4253.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, et al. 2008. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol Microbiol 68: 538–546. 10.1111/j.1365-2958.2008.06181.x [DOI] [PubMed] [Google Scholar]
- Winkelman JT, Gourse RL. 2017. Open complex DNA scrunching: a key to transcription start site selection and promoter escape. Bioessays 39: 1600193. 10.1002/bies.201600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Kuntala PK, Pugh BF, Mahony S. 2020. ChExMix: a method for identifying and classifying protein–DNA interaction subtypes. J Comput Biol 27: 429–435. 10.1089/cmb.2019.0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BA, Anthony LC, Gruber TM, Arthur TM, Heyduk E, Lu CZ, Sharp MM, Heyduk T, Burgess RR, Gross CA. 2001. A coiled-coil from the RNA polymerase β′ subunit allosterically induces selective nontemplate strand binding by σ70. Cell 105: 935–944. 10.1016/S0092-8674(01)00398-1 [DOI] [PubMed] [Google Scholar]
- Yura T. 2019. Regulation of the heat shock response in Escherichia coli: history and perspectives. Genes Genet Syst 94: 103–108. 10.1266/ggs.19-00005 [DOI] [PubMed] [Google Scholar]
- Zafar MA, Carabetta VJ, Mandel MJ, Silhavy TJ. 2014. Transcriptional occlusion caused by overlapping promoters. Proc Natl Acad Sci 111: 1557–1561. 10.1073/pnas.1323413111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R. 2013. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res 41: W56–W62. 10.1093/nar/gkt437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Steitz TA. 2015. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell 58: 534–540. 10.1016/j.molcel.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.