Abstract
The concept of pan-genome, which is the collection of all genomes from a population, has shown a great potential in genomics study, especially for crop sciences. The rice pan-genome constructed from the second-generation sequencing (SGS) data is about 270 Mb larger than Nipponbare, the rice reference genome (NipRG), but it is still disadvantaged by incompleteness and loss of genomic contexts. The third-generation sequencing (TGS) with long reads can help to construct better pan-genomes. In this paper, we report a high-quality rice pan-genome construction method by introducing a series of new steps to deal with the long-read data, including unmapped sequence block filtering, redundancy removing, and sequence block elongating. Compared to NipRG, the long-read sequencing-based pan-genome constructed from 105 rice accessions, which contains 604 Mb novel sequences, is much more comprehensive than the one constructed from ∼3000 rice genomes sequenced with short reads. The repetitive sequences are the main components of novel sequences, which partially explain the differences between the pan-genomes based on TGS and SGS. Adding six wild rice accessions, there are about 879 Mb novel sequences and 19,000 novel genes in the rice pan-genome in total. In addition, we have created high-quality reference genomes for all representative rice populations, including five gapless reference genomes. This study has made significant progress in our understanding of the rice pan-genome, and this pan-genome construction method for long-read data can be applied to accelerate a broad range of genomics studies.
Since its first establishment in bacteria research (Tettelin et al. 2005), the concept of pan-genome refers to a notion that an individual only contains a portion of genes of its species. Thus, pan-genome construction and determination of gene presence-absence variations (PAVs) have been an important subject in prokaryotic and eukaryotic genome research, particularly with the rapid progress of sequencing technologies. Using the second-generation sequencing (SGS) technology, especially the Illumina sequencing platform (sequencing by synthesis) with reads shorter than 200 bp, pan-genomes of major crops have been constructed, including rice (Wang et al. 2018; Zhao et al. 2018), maize (Haberer et al. 2020), and soybean (Li et al. 2014). Significant amounts of gene PAVs in major crop species have been revealed, and some of them are important in crop improvement (Della Coletta et al. 2021). However, pan-genomes constructed from SGS data are disadvantaged by incomplete genome coverage and inaccurate gene prediction. These problems can be largely resolved by long-read sequencing (LRS, also known as the third-generation sequencing [TGS]) technology, which has been applied for constructing pan-genomes in rapeseed (Song et al. 2020), soybean (Liu et al. 2020), tomato (Alonge et al. 2020), barley (Jayakodi et al. 2020), apple (Sun et al. 2020), and rice (Qin et al. 2021). However, the quality of a species pan-genome is determined by the sample representativeness and population size as well as the construction methods. Due to their importance, previously reported pan-genomes of major crop species constructed from SGS data and from small/or less representative samples remain to be improved and validated.
Here, we report an effort to construct high-quality pan-genomes for Asian cultivated rice, Oryza sativa L. (OS) using 105 OS accessions representing all of its major populations plus six accessions of its wild relatives, Oryza rufipogon (OR), using both TGS and SGS. In addition to the gapless high-quality reference genomes for five of the major OS populations, the constructed rice pan-genomes validated and greatly expanded the current rice pan-genome constructed from the 3010 rice genomes (3K-RG) using SGS.
Results
High-quality sequencing and assembly of genomes for major OS populations
We selected 75 diverse rice accessions (Methods; Supplemental Table S1) and sequenced them with both Oxford Nanopore Technologies long-read (with a mean depth of 68.71×) (Supplemental Table S2) and Illumina short-read (with a mean depth of 69.04×) (Supplemental Table S3) platforms. With a total of 3 TB of raw data, the 75 rice genomes were first de novo assembled and polished with both long reads and short reads, with a mean N50 of 22.21 Mb for contigs. The polished contigs were corrected and assembled into chromosome-level scaffolds with a mean N50 of 33.08 Mb and a mean Benchmarking Universal Single-Copy Orthologs (BUSCO) score of 98.25% for these 75 genomes.
Another 13 assembled genomes from other batches (Supplemental Table S4) in addition to 25 TGS OS genomes from public databases (Supplemental Table S5) were also included in this study.
Assembly of gapless and high-quality genomes for representative rice populations
A total of nine genomes with fewest gaps for representative rice populations (Supplemental Table S6) were selected, including the rice reference genome NipRG. For NipRG, most (43, 58.9%) of its 73 gaps were filled with corrected reads and polished contigs from 15 Geng/japonica (GJ) genomes (Methods), and its genome was increased from 373 to 395 Mb (Supplemental Fig. S1A; Supplemental Table S6). For representative rice populations, we applied similar methods to create five gapless and three high-quality genomes. The five gapless genomes were for populations Aus (cA, NATELBORO, 386 Mb), Xian (indica)-1B (XI-1B, PR106, 391 Mb), Xian-2 (XI-2, LARHAMUGAD, 391 Mb), Xian-3 (XI-3, LIMA, 393 Mb), and tropical Geng (GJ-trp, KETANNANGKA, 389 Mb). The three high-quality genomes were for Basmati (cB, ARC10497, 387 Mb), subtropical Geng (GJ-sbtrp, CHAOMEO, 379 Mb), and temperate Geng (GJ-tmp, Qiutianxiaoting, 388 Mb) with 4, 3, and 54 gaps, respectively (Supplemental Fig. S1B–I; Supplemental Table S6).
Construction of the rice pan-genome
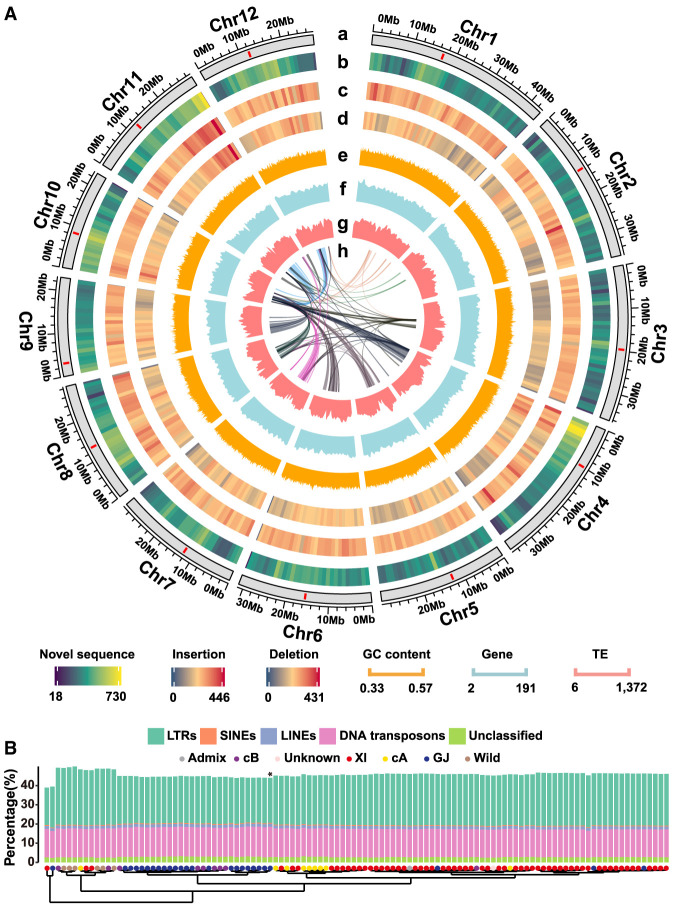
We constructed a high-quality rice pan-genome from 111 rice genomes (Supplemental Table S1; Supplemental Fig. S2A–D). A series of new steps were adopted (Supplemental Fig. S3) to deal with long reads. The rice pan-genome had a total of 879 Mb nonredundant novel sequences when considering homologies of more than 500 bases with sequence identity of at least 90%. The size of novel sequences was still huge (>500 Mb) even if we set the sequence identity threshold to 50% (Supplemental Fig. S4A). Transposable elements (TEs) comprised more than half of novel sequences, including retroelements (52.71%) and DNA transposons (16.05%). Gypsy, a long terminal repeat (LTR) retroelement, accounted for 47.83% of novel sequences. The novel sequences were distributed widely but not evenly across each chromosome (Fig. 1A). Chr 1 contained the highest number of novel sequences, and Chr 11 had the highest length of novel sequences, as compared to other chromosomes (Supplemental Fig. S4B,C). Genomic regions containing high densities of novel sequences tended to locate near centromeres (P < 0.001, Wilcoxon rank-sum test [WRST]), except two peaks near the telomeres of Chr 4 and Chr 11 (Supplemental Fig. S4D–F). Different distribution patterns were observed for structural variations (SVs), although high densities of novel sequences tended to have high densities of deletions and translocations (Fig. 1A; Supplemental Figs. S4G, S5A–I). Among all the rice genomes, the OR genomes contained significantly more LTRs than OS genomes did (P = 5.4 × 10−5, WRST) (Fig. 1B), whereas genomes of HR12 and Suijing18 had far fewer LTRs, consistent with previous studies (Mahesh et al. 2016; Nie et al. 2017).
Figure 1.
Genomic features of the rice pan-genome derived from 111 rice accessions. (A) A Circos display of the rice pan-genome. From outer to inner circles, they stand for (a) chromosomes (centromeres are in red), (b) novel sequences distributions, (c) insertion and (d) deletion distributions (≥50 bp), (e) GC content distributions (orange), (f) gene distributions (blue), (g) transposable element distributions (light red), and (h) frequent translocation regions (≥20 translocation events in 1 Mb). (B) Composition of transposable elements in terms of the percentages of the genome size in the 111 rice accessions. (*) NipRG.
A total of 19,319 novel protein-coding genes (2132 novel gene families) absent in the MSU7 were predicted in the rice pan-genome. Of these novel genes, 89.5% genes contained at least one functional domain, including 66.3% genes annotated by Pfam (Supplemental Fig. S6A). Using 122 RNA-seq data sets from roots and leaves of 61 various rice accessions (Kawakatsu et al. 2021), we found 19.9% of novel genes and 42.6% of MSU7 genes had expression evidence in at least one sample (Supplemental Fig. S6B).
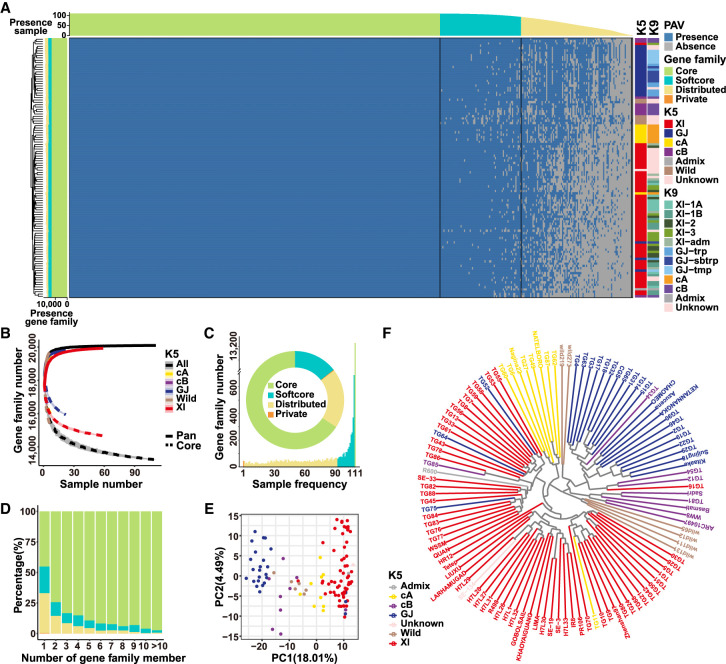
With the rice pan-genome, we used the “map-to-pan” strategy (Wang et al. 2018) to reveal gene (or gene family) PAVs (Fig. 2A). We first showed that different long-read sequencing platforms and sequencing depths had very limited impact on the gene family PAV assessment (Supplemental Fig. S7A). We then estimated that the total number of gene families could reach 20,000 when the sample size was approaching 60, although the number of the core gene families was expected to be lower if more samples were included (Fig. 2B; Supplemental Fig. S7B). In the rice pan-genome, 65.7% (13,227) were core gene families (presented in all samples), 14.4% (2890) were softcore or candidate core gene families (presented in more than 90% of samples), 19.6% (3,938) were distributed or dispensable gene families (present in <90% but more than one of the samples), and only 0.2% (45) were private or unique gene families (present only in one single sample) (Fig. 2C). All private gene families had only one member per gene family (Fig. 2D). When single genes instead of gene families were used as the units, the rice pan-genome consisted of 75,305 (55,986 MSU7 + 19,319 novel) genes with 36.5% (27,460) core genes, 15.0% (11,325) softcore genes, and 48.5% (36,504) distributed genes. Gene families with more members tended to be core, and the opposite was also true for the softcore or distributed gene families (Fig. 2D). The core or softcore genes were enriched in GO terms like regulation of flower development, negative regulation of transcription, and DNA-templated and trichome morphogenesis (Supplemental Fig. S7C), whereas distributed or private genes were enriched in GO terms like plant-type hypersensitive response, cellular water homeostasis, and oligosaccharide metabolic process (Supplemental Fig. S7D).
Figure 2.
Gene family PAVs in 111 rice accessions. (A) Heat map of gene family PAVs of 111 rice accessions. (B) The pan-genome size estimation using 111 rice accessions for all rice accessions and subpopulations (All, Wild, XI, cA, GJ, cB). (C) The sample numbers and percentages of core, softcore, distributed, and private gene families. (D) The percentage of core, softcore, distributed, and private members in gene families. (E) The PCA analysis of 111 rice accessions using gene family PAVs. (F) The clustering of 111 rice accessions using gene family PAVs.
Different populations/subpopulations varied in terms of pan-genome sizes and the proportions of their core and distributed gene families/genes. XI, GJ, cA, cB, and OR had the pan-genomes of 19,947/72,799, 19,848/70,943, 19,528/68,615, 19,674/69,400, and 19,679/70,775 gene families/genes, respectively, and 73.5%/46.5%, 76%/50.7%, 81.3%/59.2%, 78.2%/54.5%, and 78.3%/55.0% of their gene families/genes were core (Fig. 2B). Gene family PAV-based classification of the 105 OS accessions was largely consistent with previous classification of 3K-RG using single nucleotide polymorphisms (Fig. 2E,F; Supplemental Fig. S7E,F; Wang et al. 2018).
Comparison of PAVs derived from SGS and TGS
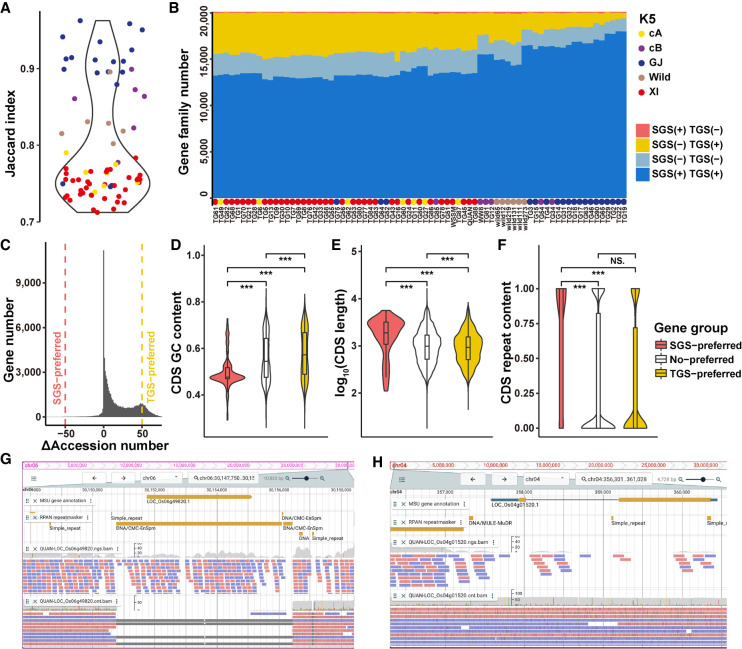
In order to understand the impact of the sequencing technologies on pan-genome analysis, we compared the gene PAVs derived from SGS and TGS data separately for 75 rice accessions. Jaccard Indices (JIs) of gene family PAVs detected by SGS and TGS were 0.7–1.0. In particular, population GJ was the most consistent group with the highest JIs, whereas XI was the most inconsistent group (Fig. 3A). This inconsistency occurred primarily in the TGS-detected regions but not in the SGS-detected regions (Fig. 3B,C). Population XI had a higher percentage of gene families/genes detected by TGS, but not SGS, than that of GJ (Fig. 3B; Supplemental Fig. S8A,B). We divided genes into three groups: TGS-preferred, SGS-preferred, and no-preferred. GO enrichment analysis indicated that TGS-preferred genes were enriched in GO terms like sulfur amino acid metabolic process, response to glucose, and response to sucrose (Supplemental Fig. S8C), whereas SGS-preferred genes were enriched in the GO terms like protein N-linked glycosylation (Supplemental Fig. S8D). We compared the gene features preferred in the two different sequencing technologies. TGS-preferred genes had higher GC contents and shorter CDS lengths than SGS-preferred ones (Fig. 3D,E). Most SGS-preferred genes had all CDS regions overlapping with repeat elements (Fig. 3F), which were related to DNA transposons and LTRs (Supplemental Fig. S8E,H). However, genes with a higher percentage of LINEs (P = 3.2 × 10−11, one sided WRST, FDR = 9.3 × 10−10) and RC/Helitron (P = 5.9 × 10−9, one sided WRST, FDR = 8.5 × 10−8) were more frequently detected in TGS than in SGS (Supplemental Fig. S8E–J). These results suggest that SGS data tend to have a higher false-positive rate in detecting gene PAVs, especially for genes containing repetitive sequences. A few examples of gene absence (Fig. 3G; Supplemental Fig. S8K) and gene presence (Fig. 3H; Supplemental Fig. S8L) identified by TGS, but missed by SGS, are shown. Because the read alignment depths of the mapped regions in these examples were approximately the same as that for TGS data (∼60×), we conclude that gene PAVs or SVs supported by TGS data are real.
Figure 3.
Comparison of gene family PAVs derived from the SGS and TGS data. (A) Jaccard Indices of gene family PAVs derived from SGS and TGS. (B) The number of gene families in each rice accession detected from SGS or TGS. In most rice accessions, especially in population XI, many more gene families were detected from the TGS data than the SGS data. (C) The number of genes versus their ΔAccession numbers. For each gene, its ΔAccession number = number of accessions with this gene detected by TGS − number of accessions with this gene detected by SGS; >50: TGS-preferred genes (n = 9321); <−50: SGS-preferred genes (114); and −50 ∼ 50: No-preferred genes (65,870). (D–F) CDS feature comparison for genes preferred by SGS or TGS. (D) GC contents, (E) lengths, (F) repeat contents. A two-sided Wilcoxon rank-sum test was used to measure the significance of differences. (***) P < 0.001. (G) The read alignment of gene LOC_Os06g49820 in rice accession QUAN indicates that a deletion near this gene is ignored from SGS. (H) The read alignment of another gene LOC_Os04g01520 indicates insufficient reads from SGS are mapped to this gene region, whereas reads from TGS cover it.
Comparison of the pan-genomes of 111 rice accessions and 3K-RG
In order to compare the completeness of TGS based pan-genomes from 111 rice accessions and the previously published 3K-RG pan-genome derived from SGS data (Wang et al. 2018), we constructed two pan-genomes SGS (63-SGSRG) and TGS (63-TGSRG), from the same 63 rice accessions used in both projects, and then mapped the repeat-masked novel sequences to different pan-genomes at the genome sequence level. Twelve pairwise comparisons were made between novel pan-genome sequences from the following data sets: 111-TGSRG (192 Mb/879 Mb nonrepeat sequences), 3K-RG (174 Mb/268 Mb nonrepeat sequences), 63-SGSRG (65 Mb/120 Mb nonrepeat sequences), and 63-TGSRG (84 Mb/339 Mb nonrepeat sequences) (Supplemental Fig. S9A–L). At the genomic sequence level, 92.4% (60 Mb/65 Mb) of the 63-SGSRG novel sequences could be mapped to the 63-TGSRG, and 81.8% (69 Mb/84 Mb) of the 63-TGSRG novel sequences (length ≥ 100 bps, identity ≥ 90%, E-value ≤ 1 × 10−5) could be mapped to the 63-SGSRG (Supplemental Fig. S9I–L). We identified few differences presented in the total lengths of the mapped regions when the identity percentage cutoff was set lower than 90% (Supplemental Fig. S9A–L). Two conclusions were reached from these comparisons. First, with the same sequencing technology but different numbers of accessions, more novel sequences could be detected from more accessions (Supplemental Fig. S9C,E,H,J). Second, with the same accessions but different sequencing technologies, more novel sequences could be detected from TGS than from SGS (Supplemental Fig. S9I,L).
We also compared the novel genes discovered from 111-TGSRG, 63-TGSRG, 3K-RG, and 66-RG (53 OS and 13 OR representative accessions) (Zhao et al. 2018). With the same methods to drop incomplete or short transcripts and select the protein sequence of the longest transcript for each gene from MSU7 and 66-RG, a total of 22,250 novel genes were obtained. Similar to the genomic sequences results, we observed a high mapping rate of the 3K-RG to the 63-TGSRG and 111-TGSRG at the gene sequence level, indicating that the pan-genomes constructed from TGS were more complete. A total of 10,844 (87.0%) novel genes discovered from 3K-RG could be mapped to the 111-TGSRG and 9760 (78.3%) novel genes could be mapped to the 63-TGSRG. Similarly, 17,997 (80.9%) novel genes from 66-RG could be mapped to the 111-TGSRG. However, only 10,754 (55.7%) of 111-TGSRG novel genes and 5397 (58.4%) of 63-TGSRG novel genes could be mapped to the 3K-RG (Supplemental Table S7). Overall, the gene level mapping rates were lower than genomic sequence level mapping rates for pan-genome comparison (Supplemental Table S8). Similar results were obtained at the protein level comparison of the pan-genomes at the global identity cutoffs of 95% and 50% (Supplemental Tables S9, S10). Thus, the TGS data in this study significantly increased the total number of gene families/genes in the rice pan-genome. Based on these comparisons and considering the presence of distributed and private gene families, our results suggest that the current OS pan-genome consisting of 75,305 (20,122) protein-coding genes (families) might be an underestimate of the real numbers of genes and gene families in the world collection of the OS germplasm.
Comparison of OS and OR pan-genomes
To understand the difference between the OS and OR pan-genomes, we compared the pan-genome constructed from 105 OS rice accessions (105OS-TGSRG) and that from six OR rice accessions (6OR-TGSRG). In contrast to the size of 604 Mb (133 Mb nonrepeat) novel sequences in the 105OS-TGSRG pan-genome, a total of 363 Mb (90 Mb nonrepeat) novel sequences were identified in the 6OR-TGSRG pan-genome. With repeat sequences masked, 73.7% (98 Mb/133 Mb) of novel sequences from 105OS-TGSRG could be mapped to 6OR-TGSRG, and 80% (72 Mb/90 Mb) of novel sequences from 6OR-TGSRG can be mapped to 105OS-TGSRG.
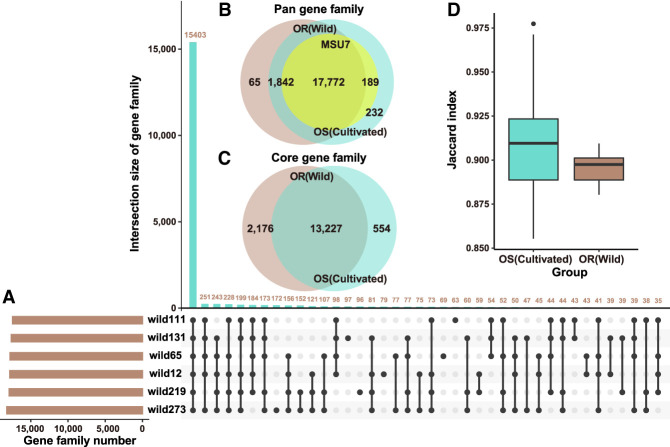
Compared with the total 17,961 gene families in MSU7, 105OS-TGSRG and 6OR-TGSRG contain 20,035 and 19,679 gene families, and 98.9% (17,772/17,961) gene families in MSU7 could be found in 6OR-TGSRG, 97.9% (19,614) 105OS-TGSRG gene families were present in the 6OR-TGSRG, and 99.7% (19,614) 6OR-TGSRG gene families were detectable in the 105OS-TGSRG (Fig. 4A,B). Overall, 68.5%, 11.8%, 19.1%, and 0.2% of the gene families in the 105OS-TGSRG were core, softcore, distributed, and private, respectively. Most (13,227) of the core gene families were shared among 105OS-TGSRG and 6OR-TGSRG (Fig. 4C). The similarity among 6OR-TGSRG was lower than that among 105OS-TGSRG (Fig. 4D). The 105-TGSRG pan-genome contained a total of 17,961 gene families each with at least one MSU7 gene and 2074 gene families with no MSU7 gene. When including the novel sequences and 65 novel gene families from the 6OR-TGSRG, we obtained a total of 275 Mb (879 Mb minus 604 Mb) newly discovered novel sequences and 2139 novel gene families. The GO enrichment analysis indicated that these OR-specific genes were enriched in cellular water homeostasis, oligosaccharide metabolic process, oligopeptide transport, plant-type hypersensitive response, iron-sulfur cluster assembly, glucosinolate biosynthetic process, regulation of defense response, plant ovule development, cellular response to nitrogen starvation, flavonoid biosynthetic process, and unidimensional cell growth.
Figure 4.
Overlap between genes and gene families in the OS (Cultivated) and OR (Wild) rice pan-genomes. (A) Intersection of gene family PAVs from six OR rice accessions. (B) Intersections of pan gene families from six OR rice accessions, MSU7, and 105 OS rice accessions. (C) Intersections of core gene families from six OR rice accessions and 105 OS rice accessions. (D) Similarity in cultivated and wild rice subpopulations.
A well-known gene, GW6 (grain width 6, LOC_Os06g15620) encoding a GA-regulated GAST family protein and positively regulating grain width and weight (Shi et al. 2020), is only present in seven OS accessions (Suijing18, Kitaake, JADO, MUKKALA BAZAL, MAEKJO, Qingjinzaosheng, and 91–382) and two OR ones (wild65 and wild111), whereas a thaumatin-like gene, PR5 (LOC_Os12g43430), is present in 6OR-TGSRG but absent in 105OS-TGSRG.
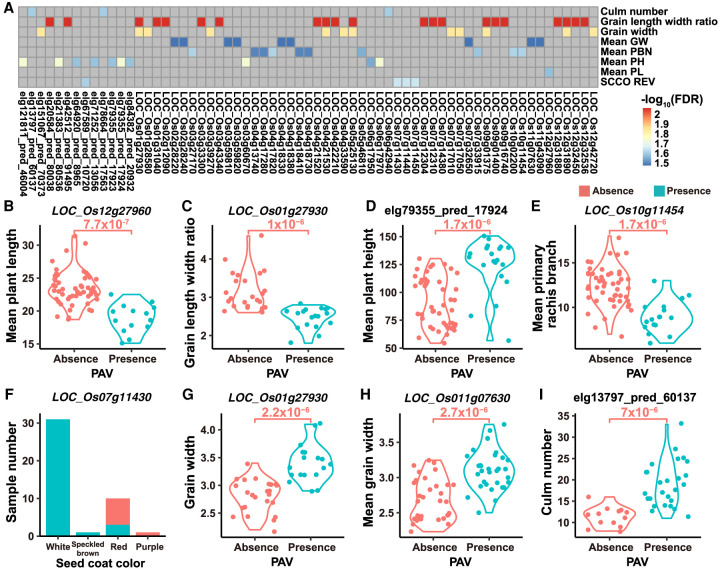
Associations between gene PAVs and phenotypes
To demonstrate how the gene PAVs contribute to rice phenotypic variations, we performed genome-wide association analysis (GWAS) for gene PAVs in 105OS-TGSRG with phenotypes and detected 14,471 significant gene PAV-phenotype associations (P < 0.05, FDR < 0.05). These associations included 8130 genes (5604 MSU7 genes and 2526 novel genes) significantly associated with nine phenotypes (Supplemental Fig. S10A; Supplemental Table S11). Because of missing phenotypic data and limited sample size, these gene PAV-phenotype associations with less significance (low P-values but high FDRs) may also be valuable (Fig. 5A; Supplemental Fig. S10B–E). Figure 5, B–I, shows several associations between gene PAVs and important agronomic traits. For example, the absence of LOC_Os01g27930 (a retrotransposon protein) is associated with an increased grain length-width ratio (Fig. 5C), whereas the presence of LOC_Os01g27930 is associated with increased grain width (Fig. 5G). The absence of the well-known Green Revolution gene, SD-1 (LOC_Os01g66100), is associated with significantly reduced plant height (P = 7 × 10−4, FDR = 0.07). These results indicate that gene PAV is an important contributor to the phenotypic variation in rice populations.
Figure 5.
The associations between phenotypes and gene PAVs. (A) The association heat map of gene PAVs and phenotypes (only associations with P < 1 × 10−5 and FDR < 5 × 10−2 are displayed). (Mean GW) mean grain width, (Mean PBN) mean primary rachis branch, (Mean PH) mean plant height, (Mean PL) mean plant length, and (SCCO REV) seed coat color. The phenotypes started with “mean” are measured in this research. (B–I) Some examples of gene PAVs significantly associated with phenotypes: (B) Mean plant length and LOC_Os12g27960 (P = 7.7 × 10−7, FDR = 2.5 × 10−2); (C) Grain length width ratio and LOC_Os01g27930 (P = 1.0 × 10−6, FDR = 8.2 × 10−3); (D) Mean plant height and a predicted gene elg79355_pred_17924 (P = 1.7 × 10−6, FDR = 1.7 × 10−2); (E) Mean primary rachis branch and LOC_Os10g11454 (P = 1.7 × 10−6, FDR = 2.3 × 10−2); (F) Seed coat color and LOC_Os07g11430 (P = 2 × 10−6, FDR = 2.1 × 10−2); (G) Grain width and LOC_Os01g27930 (P = 2.2 × 10−6, FDR = 1.3 × 10−2); (H) Mean grain width and LOC_Os11g07630 (P = 2.7 × 10−6, FDR = 3.2 × 10−2); (I) Culm number and a predicted gene elg13797_pred_60137 (P = 7.0 × 10−6, FDR = 2.3 × 10−2).
Discussion
One of the primary objectives in this study was to construct a high-quality rice pan-genome. To achieve this, we carefully selected 105 OS accessions, most from the core collections of 3K-RG, representing different rice populations of diverse geographic origins and used different sequencing platforms. In addition to the 105 OS accessions, six wild relatives were also included to make the rice pan-genome complete. The constructed reference genomes and pan-genomes for major OS populations allowed us to understand some technical elements regarding how to efficiently construct them using both TGS and SGS technologies and to uncover several properties of the rice pan-genomes.
We adopted the strategy of “map-to-pan” to construct the rice pan-genome and obtain gene PAVs (Sun et al. 2017; Wang et al. 2018; Song et al. 2020; Li et al. 2021). Because this strategy depends on the quality of the reference genome, we chose Nipponbare as the reference genome (NipRG). We further introduced the concept of unmapped sequence blocks to reduce the sizes of highly similar sequences in pan-genomes derived from TGS data, applied a new algorithm to the acceleration of the clustering of representative sequences, removed potential contaminants using the NT sequence database, and verified novel sequences with mapped reads and elongated these sequences to keep complete gene structures in gene prediction. There are still some drawbacks in our approach. The size of novel sequences may be underestimated because dropping short unmapped sequence blocks, clustering the sequences, and choosing the representative sequences is a process of losing sequences. Even though a method of sequence elongation was introduced to make their structures complete for genes overlapping with the boundaries of novel sequences, the number of novel predicted genes is underestimated. The constructed rice pan-genome contains a total of 70,624 protein-coding genes clustered (with 95% identity) in 20,122 gene families, including 2132 novel gene families missing in MSU7 and 1153 novel gene families missing in 3K-RG.
One of the primary goals in constructing a species pan-genome is to reveal the gene PAVs, an important but largely uncharacterized component of genomic variations in its populations. Our classification of the sequenced OS accessions based on the obtained gene PAVs was consistent with known rice population structure (Wang et al. 2018), indicating gene PAVs are a major contributor to the rice population differentiation. However, we noted significant differences between TGS and SGS in detecting gene PAVs. For example, SGS data often fail to detect genomic sequence variations found by TGS data, especially deletions containing or near repeat elements with high similarity. Finally, the large numbers of gene PAV-phenotype associations identified by GWAS clearly demonstrated that gene PAVs are an important contributor to the phenotypic variation in rice populations.
Considerable efforts have been taken to construct high-quality gap-free reference genomes in many crop plants, but high-quality gap-free reference genomes have been only constructed in rice (Song et al. 2021), because this can be a very expensive and challenging task for species of large genome sizes, even with the TGS technologies. In this study, we used an efficient strategy to construct high-quality reference genomes of rice by filling gaps with closely related genomes from the same population, resulting in nine high-quality reference genomes for all major rice populations, including five gap-free ones. It should be pointed out that each of these reference genomes might be slightly shorter than the real one, as we used the shortest sequences of the related accessions to fill the gaps.
The high-quality rice genomes and pan-genomes resources, and the gene PAVs obtained in this study will facilitate the global efforts of rice functional genomics and improvement in the future.
Methods
Sample collection
A total of 111 rice accessions with 113 samples were used in this research (Supplemental Table S1), including 69 + 13 OS genomes and six OR genomes from three batches obtained in this study plus publicly available TGS data of 25 OS genomes in addition to the NipRG sequences (Supplemental Fig. S2; Supplemental Tables S1, S5).
In the first batch, nine OS accessions (H7L1 [Huanghuazhan], H7L26 [CDR22], H7L27 [PsBRC28], H7L28 [PsBRC66], H7L29 [IR64], H7L30 [Teqing], H7L31 [IR50], H7L32 [OM1723], and H7L33 [Phalguna]) from the breeding program at the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences were sequenced by the Pacific Biosciences (PacBio) and Illumina platforms.
In the second batch, four OS accessions (SE-3 [BR 24], SE-19 [Zhong 413], SE-33 [BG 300], and SE-134 [Haonnong]) were sequenced by the Nanopore and Illumina platforms.
In the third batch, 67 were selected from 3K-RG to represent nine subpopulations (XI-3, XI-2, XI-1B, XI-1A, GJ-trp, GJ-tmp, GJ-sbtrp, cB, cA) each with 6–11 samples of different geographic origins (Wang et al. 2018). Two additional high-yield rice cultivars (QUAN and WSSM) and six wild rice accessions from different regions were chosen as well. These 75 rice accessions were sequenced by the Nanopore and Illumina platforms.
Another 25 OS samples were downloaded from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra), including CHAO MEO::IRGC 80273-1, Azucena, KETAN NANGKA::IRGC 19961-2, ARC 10497::IRGC 12485-1, IR 64, PR 106::IRGC 53418-1, LIMA::IRGC 81487-1, KHAO YAI GUANG::IRGC 65972-1, GOBOL SAIL (BALAM)::IRGC 26624-2, LIU XU::IRGC 109232-1, LARHA MUGAD::IRGC 52339-1, NATEL BORO::IRGC 34749-1 (Zhou et al. 2020), R600 (PRJNA564911), Tetep (PRJNA482013), Suijing18 (Nie et al. 2017), IR64, Sadri (Choi et al. 2020), Basmati (Choi et al. 2020), HR-12 (Mahesh et al. 2016), Nagina 22 (PRJNA315689), Shuhui498/R498 (Du et al. 2017), Carolina Gold Select (PRJNA503892), Zhenshan 97 (PRJNA302542), IR8 (PRJNA353946), and Kitaake (Jain et al. 2019). Three different sequence data sets of IR64 (one newly sequenced sample and two public samples), a well-known rice cultivar widely grown in Southeast and South Asia, were included as samples to compare the differences in sequencing platforms and sequencing depth.
De novo assembly, polishing, scaffolding and evaluation
For 75 newly sequenced OS accessions, each genome size was estimated using KmerGenie v1.7051 (Chikhi and Medvedev 2014) with short reads. The raw Nanopore long reads were checked by NanoPlot v1.0.0 and trimmed (≥Q7, ≥1000 bp) by NanoFilt v2.6.0 (De Coster et al. 2018). The trimmed long reads were corrected and assembled using NextDenovo v2.2.0 (https://github.com/Nextomics/NextDenovo). After genome assembling, the contigs were polished with both long and short reads. First, contigs were polished using Racon v1.4.11 (Vaser et al. 2017) and Medaka v0.11.5 (https://github.com/nanoporetech/medaka) with long reads. Next, all short reads were quality-controlled using FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and trimmed using Trimmomatic v0.39 (Bolger et al. 2014). The contigs were mapped with short reads using Bowtie 2 v2.3.5.1 (Langmead and Salzberg 2012) and polished one round using Pilon v1.23 (Walker et al. 2014).
For nine accessions sequenced by PacBio and Illumina platforms, the long reads were assembled using FALCON v1.8.7 (Chin et al. 2016). The assembled contigs were then polished using smrtlink v4.0 (https://www.pacb.com/support/software-downloads) with long reads and using Pilon with short reads.
Four of the 13 accessions were sequenced by Nanopore and Illumina platforms (SE-3 [BR 24], SE-19 [Zhong 413], SE-33 [BG 300], and SE-134 [Haonnong]).The long reads were first corrected using NextDenovo and then assembled using smartdenovo (https://github.com/ruanjue/smartdenovo). The assembled contigs were polished using Pilon three times with short reads.
For other rice accessions, we directly used assemblies and long reads published in the NCBI SRA database in downstream analysis. With the NipRG's guide, contigs misassembled were corrected and chromosome-level scaffolds were achieved using RaGOO RagTag v1.0.0 (Alonge et al. 2019), which invoked MUMmer v3.9.4 (Kurtz et al. 2004) at the mapping step and minimap2 v2.17 (Li 2018) at the checking step. The quality of each genome assembly was evaluated by mapping to the NipRG with at least 90% as the threshold using QUAST v5.0.2 (Mikheenko et al. 2018). The completeness of each genome assembly was evaluated using BUSCO v5.1.2 (Seppey et al. 2019) with the database embryophyta_odb10 (eukaryota, 2020-09-10).
Structural variation calling, merging, and filtering
The long reads were mapped to the NipRG using minimap2 v2.17 (Li 2018) and sorted using SAMtools v1.9 (Li et al. 2009). Structural variations were detected with Sniffles v1.0.11 (Sedlazeck et al. 2018) as recommended (Zhou et al. 2019). The samples were filtered (allele frequency ≥ 0.05, support reads ≥ 10, SV size ≥ 50 bp), and all filtered SVs were merged (distance < 1000 bp between breakpoints, taking SV type and strand into account) into 316,611 nonredundant SV records using SURVIVOR v1.0.7 (Jeffares et al. 2017).
Gap-filling in constructing OS reference genomes
In constructing reference genomes for different OS populations, gap-filling was performed using TGS-GapCloser v1.1.1 (Xu et al. 2020) with both corrected reads and polished contigs. For NipRG (http://rice.plantbiology.msu.edu/annotation_pseudo_current.shtml), corrected Nanopore long reads from 15 accessions of GJ were used separately to fill gaps with the recommended parameters of “‐‐tgstype ont ‐‐min_idy 0.9 ‐‐min_match 300 ‐‐ne”. Polished contigs from 15 GJ accessions were further used separately to fill gaps with the parameters of “‐‐tgstype ont ‐‐min_idy 0.9 ‐‐min_ match 1000 ‐‐ne”. The gaps are filled with the reads or contigs according to the highest QS (Quality Score) defined in TGS-GapCloser, which considered the alignment length and the alignment identity percentage between a candidate sequence (a read or contig) and the flanking sequence next to the gap.
When multiple sequences from different accessions could fill a gap, the sequence (a corrected read or polished contig) of minimum length was selected.
Similar methods were performed for gap-filling to construct high-quality reference genomes of all K9 subpopulations except XI-1A, with specific samples in Supplemental Table S6.
Pan-genome construction
The system diagram of our new pan-genome construction method is shown in Supplemental Figure S3. From the QUAST outputs of the genome assemblies, the unmapped regions of sequences were defined as unmapped sequence blocks. The unmapped sequence blocks longer than 500 bp were retrieved from both fully and partially unaligned sequences. These blocks were mapped to NipRG (including mitochondrion and plastid) again using minimap2 v2.17 (Li 2018), and the sequences mapped with ≥90% identity and 80% coverage were removed. The remaining sequences were clustered into nonredundant sequences with identity cutoff of 90% using Gclust v1.0.0 (Li et al. 2019) and EUPAN v0.44 (Hu et al. 2017) blastCluster. After that, the remaining sequences were mapped to NT database (June 18, 2020) using BLAST+ v2.10.1 (Camacho et al. 2009) BLASTN. The sequences with hits not from Viridiplantae were dropped and the rest of the sequences were defined as candidate novel sequences.
A coverage-based method was used to check misassemblies in candidate novel sequences. Reads were mapped to NipRG added candidate novel sequences and sorted using SAMtools v1.9 (Li et al. 2009). We calculated the candidate novel sequence coverage from either short or long reads using BEDTools v2.29.2 (Quinlan and Hall 2010). Finally, the candidate novel sequences with coverage ≥ 90% in at least one sample were considered as verified novel sequences.
We tried to elongate sequences with 5000 bp to retain the whole gene body in the gene annotation step and shortened sequences with no novel genes in elongated regions after a similar gene-removing step. The novel representative genes were kept. The final pan-genome was generated by combining NipRG and novel sequences, with MSU7 and novel genes. The genomic features of the pan-genome were plotted using RCircos v1.2.1 (Zhang et al. 2013) in R v4.0.2 (R Core Team 2020).
Transposable element annotations
For the chromosome-level scaffolds and novel sequences of OS accessions, transposable elements were identified using RepeatMasker v4.1.0 (http://www.repeatmasker.org) with the parameter “-no_is -nolow”. The library is the manually curated rice TE database (rice6.9.5.libin) downloaded from GitHub (https://github.com/oushujun/EDTA/blob/master/database) (Ou et al. 2019).
Gene prediction of novel sequences
All protein-coding genes in the obtained novel sequences were predicted with MAKER2 v2.31.10 (Holt and Yandell 2011), a gene prediction system combining ab initio predictions, transcripts, and protein homologies. Oryza genus proteins’ sequences from the NCBI Protein database (date: Dec. 3, 2020, species from plants, source from PDB, RefSeq, UniProtKB/Swiss-Prot) and rice EST mRNAs v163a were downloaded from PlantGDB (Duvick et al. 2008). The low complex repeat sequences were soft-masked with RepeatMasker v4.1.0 (search engine: NCBI/RMBLAST v2.10.0, database: Dfam_3.1 [Storer et al. 2021], Repbase-20181026 [Bao et al. 2015]) first before running MAKER2. The evidence such as ab initio predictions from AUGUSTUS v3.3.3 (Stanke et al. 2008), ESTs and proteins were collected to evaluate the quality of gene prediction models. Exonerate v2.4.0 (https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate) polished BLAST hits, and each of the realigned sequences identified by BLAST around splice sites were retained. A total of 30,883 transcripts were dropped because of incomplete coding regions (lack of start/stop codons). Finally, 96,425 transcripts each with a complete coding region were predicted. Of these, 28,529 transcripts overlapped with novel sequences that do not locate in the elongated sequences.
Novel genes and gene families
For the gene level analysis, the longest transcript isoforms of both the 55,986 reference (MSU7) genes and predicted genes in the novel sequences were translated into protein sequences and clustered with 95% identity using CD-HIT v4.8.1 (Fu et al. 2012). In total, 70,624 clusters were identified. Of these, 19,319 novel genes were identified.
For the gene family-level analysis, the protein sequences of the longest gene transcript isoforms were clustered using Markov Chain-based algorithm OrthoMCL v2.0.9 (Li et al. 2003) with the parameter “percentMatchCutoff = 50, evalueExponentCutoff = −5”, which invokes BLAST+ v2.10.1 (Camacho et al. 2009) BLASTP at the step of sequence comparison. In the end, 20,122 gene families were identified (9778 families with at least two genes, 10,344 families each with one single gene), including 17,990 gene families with at least one reference gene and 2132 gene families with only novel predicted genes. The proteins each with the longest protein coding length in the gene families were treated as representative ones. The novel representative genes in the gene families were kept, including (1) novel gene families (the novel representative genes with no reference gene member in the same gene family), and (2) existing gene families with novel representative genes (the novel representative genes with at least one reference gene member in the same gene family). Finally, 2132 novel gene families and 3260 existing gene families with novel representative genes were obtained.
RNA-seq validation, functional domains, and GO annotations of the predicted novel genes
RNA-seq data from 122 public samples (61 rice accessions of two tissues) were collected to validate the expressions of genes. All raw reads were quality-controlled using FastQC v0.11.8 and trimmed using Trimmomatic v0.39. The trimmed reads were mapped to all transcripts (including 55,986 MSU7 genes and 19,319 novel genes). Only reads mapped in a proper pair were considered using SAMtools v1.9. The coverage of transcripts was computed using BEDTools v2.29.2. The protein sequences of the predicted novel genes were extracted and input to InterProScan v5.45-80.0 (Jones et al. 2014) to predict domains and important sites of their proteins. The GO terms of proteins were annotated as described in a previous study (Wang et al. 2018). Finally, 75.9% (14,658/19,319) of the predicted novel genes were annotated with at least one GO term. GO enrichment analysis was performed using the package clusterProfiler v3.16.1 (Yu et al. 2012) in R v4.0.2. The GO terms with adjust P < 0.05 using the Benjamini–Hochberg (BH) method were retained.
Gene PAVs analysis
The trimmed short and long reads were mapped to the rice pan-genome with Bowtie 2 v2.3.5.1 (Langmead and Salzberg 2012) and minimap2 v2.17 (Li 2018). Then, the alignment results were sorted with SAMtools v1.9 (Li et al. 2009). The coverage of each position in the all OS genomes was computed using BEDTools v2.29.2 (Quinlan and Hall 2010) with the parameter “genomecov -bga -split”. A gene was considered as present when its gene body and CDS coverages were over 85% and 95%, respectively, according to its mapped short reads or long reads. Otherwise, it was considered as absent. A gene family was considered as present when at least one of its gene members was present. The Jaccard Index (also called Jaccard similarity coefficient) was used to quantify genetic similarity between two genomes. For gene (or gene family) sets A and B of two samples, it was computed as
The higher the Jaccard Index is, the more similar two samples are. All predicted genes or gene families were divided into the core, softcore, distributed, and private predicted genes or gene families in a specified population. The classification of genes/gene families as softcore and distributed ones are based on exact binomial tests (P ≥ 0.05). The pan-core gene family curve was computed with PAVs of subsamples randomly selected from different subpopulations 100 times. The outliers in sample clustering according to TEs and PAVs were not considered in the subsampling (TG52, TG64, TG75, TG16, TG11, TG34, TG54, TG34, TG12, and TG85).
Comparisons between different rice pan-genomes derived from 111/63 rice accessions (111-TGSRG/63-TGSRG/63-SGSRG) and 3010 rice accessions (3K-RG)
Sixty-three OS accessions from 3K-RG were selected to build OS pan-genomes as reported previously (Wang et al. 2018) in three steps of getting unaligned contigs (≥500 bp), removing redundant sequences, and dropping contaminants.
The repeat-masked novel sequences in 111-TGSRG/3K-RG/63-TGSRG/63-SGSRG were mapped to 111-TGSRG/3K-RG/63-TGSRG/63-SGSRG. A query sequence with various identity and length was considered according to a different cutoff.
At the gene level, we mapped the transcripts and proteins of novel genes to the constructed pan-genomes, transcripts, and proteins. We mapped the transcripts of novel genes to construct the sequences from different rice pan-genomes using BLASTN. The identities ≥95% were considered as mapped regions, and transcripts with ≥95% of their regions covered were considered as mapped ones. We also mapped the transcripts and proteins of novel genes to each other from different rice pan-genomes using cd-hit-est-2d and cd-hit-2d in CD-HIT v4.8.1.
Detection of PAV-phenotype associations
The phenotypic data for agronomic traits of the 105 OS accessions were downloaded from the RFGB (http://www.rmbreeding.cn/phenotype) (Wang et al. 2020) and Rice SNP-Seek Database (https://snp-seek.irri.org) (Mansueto et al. 2017). Fisher's exact test was used to detect gene PAV-discrete phenotype associations, and the Wilcoxon rank-sum test was used to detect gene PAV-continuous phenotype associations in R v4.0.2. P-values were adjusted using the FDR method, and a threshold of FDR < 0.05 was used to claim a significant gene PAV-phenotype association.
Data access
The raw sequence data generated in this study have been submitted to the Genome Sequence Archive in China National Center for Bioinformation (https://ngdc.cncb.ac.cn/bioproject/) under accession numbers PRJCA005926, PRJCA007821, and PRJCA007822. The source codes used in this study are available at GitHub (https://github.com/SJTU-CGM/TGSRICEPAN) and as Supplemental Code. The results data are available at https://cgm.sjtu.edu.cn/TGSrice/index.html.
Supplementary Material
Acknowledgments
We thank the High Performance Computing Center (HPCC) at Shanghai Jiao Tong University for the computation services. This work has been supported by grants from the Natural Science Foundation of Shanghai (20ZR1428200), the National Natural Science Foundation of China (32170643, 31971928, and U21A20214), the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (320LH044), the Hainan Yazhou Bay Seed Lab Project (B21HJ0215, B21HJ0223, and B21HJ0508), the Agricultural Science and Technology Innovation Program and the Cooperation and Innovation Mission (CAAS-ZDXT202001), the SJTU JiRLMDS Joint Research Fund (MDS-JF-2019A07), the CAAS Innovative Team Award, and the National High-level Personnel of Special Support Program. The funding agencies had no role in the experimental design, data collection and analyses, decision to publish, or preparation of the manuscript.
Author contributions: Z.K.L., W.S.W., J.L.X., and C.C.W. conceived the project; F.Z., W.S.W., M.L., and X.M.Z. sequenced the rice genomes; H.Z.X. performed pan-genome analysis; H.Z.X., X.R.D., C.C.W., W.S.W., F.Z., and Z.K.L. interpreted results and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276015.121.
Competing interest statement
The authors declare no competing interests.
References
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20: 224. 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182: 145–161.e123. 10.1016/j.cell.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11. 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R, Medvedev P. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30: 31–37. 10.1093/bioinformatics/btt310 [DOI] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13: 1050–1054. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Lye ZN, Groen SC, Dai X, Rughani P, Zaaijer S, Harrington ED, Juul S, Purugganan MD. 2020. Nanopore sequencing-based genome assembly and evolutionary genomics of circum-basmati rice. Genome Biol 21: 21. 10.1186/s13059-020-1938-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34: 2666–2669. 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Coletta R, Qiu Y, Ou S, Hufford MB, Hirsch CN. 2021. How the pan-genome is changing crop genomics and improvement. Genome Biol 22: 3. 10.1186/s13059-020-02224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Yu Y, Ma Y, Gao Q, Cao Y, Chen Z, Ma B, Qi M, Li Y, Zhao X, et al. 2017. Sequencing and de novo assembly of a near complete indica rice genome. Nat Commun 8: 15324. 10.1038/ncomms15324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. 2008. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res 36: D959–D965. 10.1093/nar/gkm1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, et al. 2020. European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet 52: 950–957. 10.1038/s41588-020-0671-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Sun C, Lu KC, Chu X, Zhao Y, Lu J, Shi J, Wei C. 2017. EUPAN enables pan-genome studies of a large number of eukaryotic genomes. Bioinformatics 33: 2408–2409. 10.1093/bioinformatics/btx170 [DOI] [PubMed] [Google Scholar]
- Jain R, Jenkins J, Shu S, Chern M, Martin JA, Copetti D, Duong PQ, Pham NT, Kudrna DA, Talag J, et al. 2019. Genome sequence of the model rice variety KitaakeX. BMC Genomics 20: 905. 10.1186/s12864-019-6262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakodi M, Padmarasu S, Haberer G, Bonthala VS, Gundlach H, Monat C, Lux T, Kamal N, Lang D, Himmelbach A, et al. 2020. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588: 284–289. 10.1038/s41586-020-2947-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bahler J, Sedlazeck FJ. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8: 14061. 10.1038/ncomms14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Teramoto S, Takayasu S, Maruyama N, Nishijima R, Kitomi Y, Uga Y. 2021. The transcriptomic landscapes of rice cultivars with diverse root system architectures grown in upland field conditions. Plant J 106: 1177–1190. 10.1111/tpj.15226 [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Zhou G, Ma J, Jiang W, Jin LG, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, et al. 2014. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol 32: 1045–1052. 10.1038/nbt.2979 [DOI] [PubMed] [Google Scholar]
- Li R, He X, Dai C, Zhu H, Lang X, Chen W, Li X, Zhao D, Zhang Y, Han X, et al. 2019. Gclust: a parallel clustering tool for microbial genomic data. Genomics Proteomics Bioinformatics 17: 496–502. 10.1016/j.gpb.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yuan D, Wang P, Wang Q, Sun M, Liu Z, Si H, Xu Z, Ma Y, Zhang B, et al. 2021. Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol 22: 119. 10.1186/s13059-021-02351-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou GA, Zhang H, Liu Z, Shi M, et al. 2020. Pan-genome of wild and cultivated soybeans. Cell 182: 162–176.e113. 10.1016/j.cell.2020.05.023 [DOI] [PubMed] [Google Scholar]
- Mahesh HB, Shirke MD, Singh S, Rajamani A, Hittalmani S, Wang GL, Gowda M. 2016. Indica rice genome assembly, annotation and mining of blast disease resistance genes. BMC Genomics 17: 242. 10.1186/s12864-016-2523-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, Sanciangco M, Palis K, Copetti D, Poliakov A, et al. 2017. Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res 45: D1075–D1081. 10.1093/nar/gkw1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34: i142–i150. 10.1093/bioinformatics/bty266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie SJ, Liu YQ, Wang CC, Gao SW, Xu TT, Liu Q, Chang HL, Chen YB, Yan PC, Peng W, et al. 2017. Assembly of an early-matured japonica (Geng) rice genome, Suijing18, based on PacBio and Illumina sequencing. Sci Data 4: 170195. 10.1038/sdata.2017.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol 20: 275. 10.1186/s13059-019-1905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, He Q, Ou S, Zhang H, Li X, et al. 2021. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184: 3542–3558.e3516. 10.1016/j.cell.2021.04.046 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15: 461–468. 10.1038/s41592-018-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962: 227–245. 10.1007/978-1-4939-9173-0_14 [DOI] [PubMed] [Google Scholar]
- Shi CL, Dong NQ, Guo T, Ye WW, Shan JX, Lin HX. 2020. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J 103: 1174–1188. 10.1111/tpj.14793 [DOI] [PubMed] [Google Scholar]
- Song JM, Guan Z, Hu J, Guo C, Yang Z, Wang S, Liu D, Wang B, Lu S, Zhou R, et al. 2020. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6: 34–45. 10.1038/s41477-019-0577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JM, Xie WZ, Wang S, Guo YX, Koo DH, Kudrna D, Gong C, Huang Y, Feng JW, Zhang W, et al. 2021. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol Plant 14: 1757–1767. 10.1016/j.molp.2021.06.018 [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. 10.1093/bioinformatics/btn013 [DOI] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA 12: 2. 10.1186/s13100-020-00230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hu Z, Zheng T, Lu K, Zhao Y, Wang W, Shi J, Wang C, Lu J, Zhang D, et al. 2017. RPAN: rice pan-genome browser for ∼3000 rice genomes. Nucleic Acids Res 45: 597–605. 10.1093/nar/gkw958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jiao C, Schwaninger H, Chao CT, Ma Y, Duan N, Khan A, Ban S, Xu K, Cheng L, et al. 2020. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat Genet 52: 1423–1432. 10.1038/s41588-020-00723-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci 102: 13950–13955. 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27: 737–746. 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49. 10.1038/s41586-018-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Yu H, Huang J, Wang WS, Faruquee M, Zhang F, Zhao XQ, Fu BY, Chen K, Zhang HL, et al. 2020. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol J 18: 14–16. 10.1111/pbi.13215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Guo L, Gu S, Wang O, Zhang R, Peters BA, Fan G, Liu X, Xu X, Deng L, et al. 2020. TGS-GapCloser: a fast and accurate gap closer for large genomes with low coverage of error-prone long reads. Gigascience 9: giaa094. 10.1093/gigascience/giaa094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Meltzer P, Davis S. 2013. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 14: 244. 10.1186/1471-2105-14-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. 2018. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50: 278–284. 10.1038/s41588-018-0041-z [DOI] [PubMed] [Google Scholar]
- Zhou A, Lin T, Xing J. 2019. Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol 20: 237. 10.1186/s13059-019-1858-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Chebotarov D, Kudrna D, Llaca V, Lee S, Rajasekar S, Mohammed N, Al-Bader N, Sobel-Sorenson C, Parakkal P, et al. 2020. A platinum standard pan-genome resource that represents the population structure of Asian rice. Sci Data 7: 113. 10.1038/s41597-020-0438-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.