Abstract
Escherichia coli isolates from rectal swabs from 62 chickens and stools from 42 children living in close contact with chickens on the same farms in Kiambu district, Kenya, were compared for their genetic relatedness. Antibiotic susceptibility profiles broadly categorized isolates from the children and from the chickens into two separate clusters: the majority (144; 85.5%) of the E. coli isolates from children were multidrug resistant, while the majority (216; 87.1%) of the E. coli isolates from chickens were either fully susceptible or resistant only to tetracycline. Sixty- and 100- to 110-MDA plasmids were found to encode the transferable resistance to co-trimoxazole and tetracycline. HindIII restriction endonuclease digestion of the 60- and 100- to 110-MDA plasmids produced four distinct patterns for isolates from children and three distinct patterns for isolates from chickens. XbaI digestion of genomic DNA followed by pulsed-field gel electrophoresis (PFGE) analysis produced 14 distinct clusters. There were six distinct PFGE clusters among the isolates from children, while among the isolates from chickens there were seven distinct clusters. Only one PFGE cluster contained isolates from both children and chickens, with the isolates displaying an approximately 60% coefficient of similarity. This study showed that although several different genotypes of E. coli were isolated from children and chickens from the same farms, the E. coli strains from these two sources were distinct.
There has been controversy over the natural ecology of Escherichia coli and its infectious plasmids. Whether E. coli isolates and their plasmids are derived from humans and are transmitted to animals or vice versa is still unclear. It is thought by some that animals may serve as reservoirs for E. coli strains found in humans (6) and that the frequency of transmission to humans of E. coli strains containing antibiotic resistance plasmids could be as high as 78% within 2 days of exposure. In contrast, other studies (10) did not observe any relatedness between plasmids from E. coli isolated from humans handling carcasses of chickens and those from the carcasses. However, restriction endonuclease (RE) digests of plasmids from some chicken isolates from the same flocks were found to be similar. As the stability of plasmid DNA characteristics in bacterial strains over time is not clearly understood, Krause et al. (5) suggested that this typing method be used in conjunction with other methods, such as pulsed-field gel electrophoresis (PFGE) of endonuclease-digested chromosomal DNA, to investigate epidemiological relationships. Due to the importance of E. coli as a carrier for infectious plasmids and the possibility of their zoonotic transmission to humans, we studied genotypic relationships between several human and animal isolates.
In one study, for example, PFGE was successfully used to demonstrate differences between epidemiologically unrelated E. coli strains of identical phage type (1). In another study by Krause et al. (5), all but a few strains of E. coli O157:H7 from outbreaks in Scotland, which had identical restriction fragment length polymorphism profiles, were of the same phage types. However, the observation that strains belonging to the same phage type had coefficients of similarity as low as 40% indicated clearly that phage type and genotypic characteristics have evolved along separate lines. The aim of the present study was to employ genotypic methods of characterizing bacteria to determine whether E. coli isolates found in the stools of children were the same as those isolated from chickens living in close contact with them on 12 small-scale farms in the Kiambu district of Kenya.
MATERIALS AND METHODS
Study sites and cases.
Children between 6 months and 2 years of age presenting with diarrhea at the outpatient departments and pediatric wards of two rural hospitals in Thika and Kiambu, Kenya (serving a population of 200,000 from Kiambu district and situated 20 and 40 km, respectively, from Nairobi, Kenya) were enrolled. The children came from a small-scale-farming region of Kiambu district (plots of 1 to 5 acres), where food crops such as maize, beans, and coffee are grown. In addition, each household keeps chickens for commercial egg production. The chickens run around the compound during the daytime and roost in a chicken shed at night. For farmers with over 50 chickens, the birds are fed and housed in chicken sheds with a modified deep-litter system. Children have easy access to the sheds together with their parents to feed the chickens in the morning and to collect eggs in the afternoon.
Specimen collection.
Although most households have treated tap water for domestic use, fecal contamination of water by either animals or humans was investigated by using the most-probable-number (MPN) method. Briefly, 100 ml of tap water was collected from each of the farms in sterile plastic containers. Water sampling was done twice, at the beginning and at the end of the study. Each water sample was divided into five portions, each further divided into 10-, 1-, and 0.1-ml samples, and then each portion was inoculated into test tubes containing 10 ml of MacConkey broth, with inverted Durham tubes for gas collection. Samples were incubated at 37°C for 18 h. Positive reactions (lactose fermentation/gas production) were evaluated for each of the five tubes per test portion of water. The MPN of coliforms per 100-ml water sample was then determined with standard probability tables (16). Water samples with no coliforms isolated were regarded as excellent. Water samples with a mean count of 1 to 10 coliforms/100 ml were regarded as acceptable. Water giving counts greater than 10 coliforms/100 ml were considered fecally contaminated.
Stool specimens were collected within a 3-month period, July to September 1994. A stool specimen was obtained from each child with diarrhea in the outpatient departments and the pediatric wards and was examined before commencing treatment. The index patients were followed to their homes at the addresses provided by consenting parents, where, using saline-moistened sterile cotton-tipped wooden applicators, rectal swabs from chickens were collected into Stuart’s transport medium (Oxoid Ltd., Basingstoke, United Kingdom) in universal containers. An average of one specimen per 10 chickens was collected from each of the 12 farms visited during the study. The specimens were delivered to the laboratory within 2 h of collection.
Isolation and identification of bacteria.
The specimens were processed by standard techniques (3). Briefly, each of the specimens was cultured initially in selenite-F broth (Oxoid) at 37°C for 18 h. An aliquot (1 μl) from the broth culture was then streaked onto MacConkey agar (Oxoid) and incubated at 37°C for 18 h. Four colonies resembling E. coli were randomly picked from the MacConkey agar plates and confirmed by biochemical tests on API 20E strips (bioMerieux, Basingstoke, United Kingdom). Isolates were stored at −70°C on protect beads (Technical Services Consultants Ltd., Heywood, United Kingdom) until they were analyzed.
Antibiotic susceptibility testing.
Escherichia coli isolates were tested for susceptibility to antibiotics by the MIC method, according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (9). The following antibiotics (all from Mast Laboratories, Liverpool, United Kingdom) were used: ampicillin, tetracycline, trimethoprim, sulfamethoxazole, chloramphenicol, streptomycin, gentamicin, co-amoxiclav, ciprofloxacin, and nalidixic acid. The MICs of the antibiotics were determined by using doubling dilutions of the antibiotics in diagnostic sensitivity test agar. Adjusted bacterial inocula (106 CFU/ml) were delivered onto the plates with a multipoint inoculator. E. coli ATCC 25922, for which the MICs of the antibiotics are known was included as a control for antibiotic potency. Inoculated agar plates were incubated at 37°C for 18 h. The MICs were interpreted by using guidelines from the NCCLS (9).
Plasmid studies.
Plasmid DNA was isolated from E. coli strains as described by Kado and Liu (4). DNA bands were visualized with a UV transilluminator (UVP Inc., San Gabriel, Calif.) and photographed with an MP-3 camera (Polaroid, Cambridge, Mass.). Plasmid molecular masses were determined by electrophoresis with plasmids of known molecular mass from E. coli V517 (7) and 39R861 (11). In vitro conjugation was attempted to determine mobility for antibiotic-resistant isolates by using E. coli K-12 (nalidixic acid resistant) as a recipient, as previously described by Walia et al. (15). Transconjugants were then selected on MacConkey agar (Oxoid) supplemented with nalidixic acid (32 mg/liter each) and ampicillin or chloramphenicol (32 mg/liter each). To determine the transferable antibiotic resistance, transconjugants were tested for susceptibility to the battery of antibiotics previously used for the donor E. coli. In order to verify transferable resistance plasmids against the original plasmid sizes and RE digest patterns, plasmid DNA from transconjugants was isolated and digested with HindIII (Life Technologies Ltd., Paisley, United Kingdom) according to the manufacturer’s instructions. RE digest fragments were separated by electrophoresis on 1% agarose gels at 100 V for 1 h.
Preparation of chromosomal DNA.
Chromosomal DNA was prepared in agarose plugs as described by Thong et al. (14) with modifications. Briefly, an overnight bacterial culture in Luria broth was harvested by centrifugation and resuspended in cell suspension buffer (10 mM Tris [pH 7.2], 20 mM NaCl, 50 mM EDTA). Equal volumes of the bacterial cell culture and 2% CleanCut agarose (Bio-Rad Laboratories, Richmond, Calif.) were mixed in a mold to form plugs. Two plugs were prepared for each isolate. The agarose plugs were incubated overnight at 37°C in lysis solution (25 mg of lysozyme/ml in 10 mM Tris [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% Sarkosyl). The plugs were then deproteinated by incubating overnight at 50°C in proteinase K solution (25 mg of proteinase K/ml in 100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% Sarkosyl). Cell debris and any excess proteinase K were removed by washing once with 1.7% phenylmethylsulfonyl fluoride in isopropanol and twice with a Tris-EDTA buffer (20 mM Tris [pH 8.0], 50 mM EDTA) for 1 h each at room temperature.
RE digestion and PFGE.
Agarose plugs were first equilibrated for 1 h in 0.5 ml of REACT2 buffer (Life Technologies). The plugs were then incubated overnight at 37°C in fresh buffer (300 μl) containing 25 U of XbaI. PFGE of agarose plug inserts was then performed on a CHEF-DR II system (Bio-Rad Laboratories) in a 1% agarose gel in 0.5× TBE (0.1 M Tris [pH 8.0], 0.1 M boric acid, 0.2 M EDTA) buffer for 22 h at 120 V, with a pulse time of 1 to 40 s at 14°C. To eliminate the possibility of the RE digestion of the large plasmids from E. coli influencing the PFGE patterns, plasmids previously isolated were digested with XbaI. Large plasmids were shown to produce fragments of less than 20 kb, which were not scored during the analysis. To determine the reproducibility of the PFGE fragment patterns obtained, a second set of agarose plugs was digested again with XbaI by the same procedure.
Lambda DNA digests consisting of a ladder (ca. 22 fragments) of increasing size from 48.5 to approximately 1,000 kb were included as a DNA size standard (Bio-Rad). The gel was stained with ethidium bromide and photographed on an UV transilluminator (UVP Inc.). The RE digest patterns were interpreted by considering the migration distances and intensities of all visible bands, as described previously (12). Genetic similarity was calculated by the Dice coefficient (13) and clustered by the unweighted-pair group arithmetic averaging method generated by the molecular fingerprinting program (Molecular Analyst version 1.4.1; Bio-Rad).
RESULTS
Descriptive epidemiology. (i) Children.
In order to establish the likely role of antibiotic usage in the isolation of resistant E. coli from children, we investigated the use of antibiotics for the treatment of diarrhea. Of the 42 children from the present study, 32 who had moderate-to-severe diarrhea were given either ampicillin or co-trimoxazole as first-line treatment. These children were discharged on the same day to continue treatment at home. We established a poor compliance in completing treatment at home, as most mothers discarded the medicines as soon as the child’s diarrhea resolved. However, 10 very ill children were admitted for 5 to 7 days and given gentamicin injections in addition to rehydration therapy.
(ii) Farm studies.
Twelve farms were visited in the course of the 3-month study. For the purposes of the present study, the investigated farms were arbitrarily assigned identification numbers from 1 to 12 according to the order in which the investigators first visited them. In order to assess the impact of chicken-farming practices on the isolation of antibiotic-resistant E. coli, we investigated the farmers’ use of antibiotics for growth promotion. The most widely used antibiotic was tetracycline. Each farmer used tetracycline as a powder, which was added to drinking water for the birds in widely varying dosages of 0.5 to 5 g/liter. Tetracycline was also present in commercially available feed supplements that were used in chicken rearing on all of the farms.
Bacteria.
The MPN test for water from 4 of the 12 farms gave a mean count of 1 to 3 coliforms/100 ml of water. Tap water from the other eight farms did not give any coliforms. Initially, 168 E. coli isolates were obtained from the children (4 isolates per child); 128 isolates came from 32 children seen at the outpatient departments and 40 isolates came from 10 children from the pediatric wards at the two hospitals in a 3-month period. In addition, 248 E. coli isolates were obtained from rectal swabs of chickens (4 isolates per swab) from the corresponding 12 farms.
Antibiotic susceptibility.
Of the E. coli isolates from the children, 144 (85.7%) were multidrug resistant, commonly to ampicillin and co-trimoxazole or ampicillin and tetracycline. Only eight (4.8%) isolates were fully susceptible to all antibiotics tested. In contrast only 32 of 248 (12.9%) isolates from chickens were multidrug resistant. Ninety-two (37.1%) of the isolates from chickens were fully susceptible. A large proportion of isolates from the children and the chickens (118 [70.2%] and 148 [59.7%], respectively) were resistant to tetracycline, with a MIC at which 50% of the isolates are inhibited of >64 mg/liter. The antibiotic susceptibility patterns of all four isolates of E. coli from each of the specimens from either the chickens or the children were the same.
Plasmid studies.
Plasmids from each of the four individual E. coli isolates from the same specimen from either chickens or children produced the same plasmid profiles. Therefore, a single isolate was randomly selected from each specimen for further plasmid analysis, giving a total of 42 isolates from children and 62 isolates from chickens. The most common plasmid profiles were of 100- to 110-MDa plasmids, alone or with 24-MDa plasmids. These were found in 28 (66.7%) E. coli isolates from children and 22 (35.5%) isolates from chickens. Ten (23.8%) isolates from children and four isolates from chickens contained 60- and 10-MDa plasmids. In addition, 2 isolates from children and 13 (21%) isolates from chickens contained plasmids of 10 to 25 MDa. Two antibiotic-susceptible E. coli isolates from children and 23 (37.1%) isolates from chickens contained only small plasmids of 2 to 10 MDa. Of the 42 E. coli isolates from children, 40 ampicillin-resistant isolates were examined for transferable antibiotic resistance. A total of 36 of 40 (90%) transferred resistance to one or more antibiotics to E. coli K12 (Table 1). For E. coli from chickens, conjugational transfer of resistance was attempted for 37 tetracycline-resistant isolates. A total of 24 of 37 (64.9%) transferred resistance to tetracycline alone or together with resistance to co-trimoxazole. In isolates from chickens or children, resistance was carried on the 100- to 110- and 60-MDa plasmids. HindIII RE digestion of the 60- and the 100- to 110-MDa plasmids produced seven combinations of RE digest patterns of 4, 6, 10, and 12 fragments (Table 1). Four RE digest patterns were distinct for isolates from children and three patterns were distinct for isolates from chickens.
TABLE 1.
Antibiotic resistance transfer profiles and RE digest patterns for resistance plasmids from E. coli isolates
| Resistance pattern | No. of isolates | Pheno-type trans-ferreda | Plasmid (MDa) | No. of isolates with RE digest profileb:
|
|||
|---|---|---|---|---|---|---|---|
| 4 | 6 | 10 | 12 | ||||
| Isolates from children (n = 36) | |||||||
| Am Tm Su Te | 14 | Am Tm Su | 100 | 6 | 3 | 4 | 1 |
| Am Tm Su Te | 2 | Am Te | 110 | 2 | 0 | 0 | 0 |
| Am Cm Te | 8 | Am Cm | 60 | 3 | 3 | 2 | 0 |
| Am Te | 12 | Te | 100 | 8 | 4 | 0 | 0 |
| Isolates from chickens (n = 24) | |||||||
| Am Tm Su Te | 2 | Am Tm Su | 60 | 1 | 1 | 0 | 0 |
| Am Te | 3 | Te | 100 | 0 | 0 | 3 | 0 |
| Te | 19 | Te | 100 | 0 | 19 | 0 | 0 |
Am, ampicillin; Cm, chloramphenicol; Su, sulphamethoxazole; Te, tetracycline; Tm, trimethoprim.
Profile designations indicate number of fragments.
Genotypes.
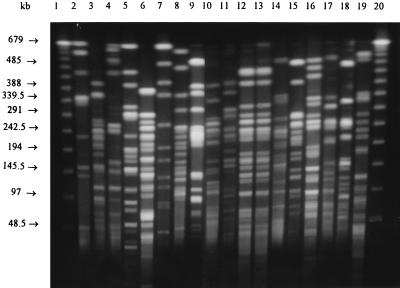
To determine the reproducibility of the PFGE digest patterns, a repeat PFGE procedure was carried out on E. coli isolates from five children (20 isolates) and five chickens (20 isolates) randomly selected from the study group. In each case, the same PFGE pattern was obtained on both occasions, thus indicating that XbaI-digested fragment patterns from E. coli from the present study were reproducible. Four E. coli isolates from each child and from each chicken gave the same PFGE pattern, indicating that each group of subjects had been colonized by the same E. coli strain. Therefore, further genotype analysis by PFGE was done with a single isolate from each specimen. As XbaI digestion of the 100- to 110-MDa plasmids produced fragments of less than 20 kb (data not shown), bands of this size were excluded from the analysis of PFGE fragment patterns. Figure 1 is a representative gel showing XbaI-digested DNA from E. coli from children and chickens from the various farms.
FIG. 1.
RE fragment patterns of XbaI-digested genomic DNA from representative E. coli isolates from children and chickens from eight farms, illustrating the diversity of E. coli strain types. Lanes 1 and 20, 48.5-kb DNA molecular size standard; lanes 2 and 4, Ch9695 and Hu8819 from farm 7; lanes 3 and 5, Ch9761 and Hu8823 from farm 10; lanes 6 and 8, Ch9762 and Hu9649 from farm 8; lanes 7 and 9, Ch9771 and Hu9687 from farm 12; lanes 10 and 12, Ch9794 and Hu50 from farm 2; lanes 11 and 13, Ch9765 and Hu51 from farm 2; lanes 14 and 16, Ch9697 and Hu8538 from farm 3; lanes 15 and 17, Ch9725 and Hu8529 from farm 1; lanes 18 and 19, Ch9728 and Hu72 from farm 9. E. coli isolates of type Ch were from chickens, and isolates of type Hu were from children.
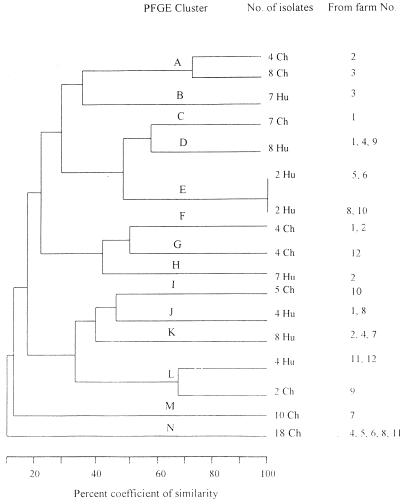
Analysis of the genetic relatedness of E. coli isolates by the Dice coefficient method and by physical examination of PFGE gel pictures demonstrated that isolates that produced RE digest patterns showing a greater-than-60% coefficient of similarity were possibly closely related (fewer than four bands difference) as defined by Tenover et al. (12). PFGE patterns showing less than a 60% coefficient of similarity appeared sufficiently different to represent unrelated isolates. Fourteen clusters were produced from PFGE analysis of the 42 E. coli isolates from children and the 62 isolates from chickens. The number of fragments produced by XbaI digestion ranged from 11 to 20, with sizes of approximately 22 to 680 kb. E. coli isolates from children fell into six clusters, and the isolates from chickens fell into seven different clusters. There was only one PFGE cluster which contained isolates from chickens (two from farm 9) as well as isolates from children (four from farms 11 and 12). This was PFGE cluster L, within which isolates had an approximately 60% coefficient of similarity (Fig. 2).
FIG. 2.
Dendrogram showing estimated genetic relationships of E. coli isolates from 62 chickens (Ch) and 42 children (Hu) from various farms. The patterns of PFGE fragments showing a 100% coefficient of similarity were indistinguishable; these were likely to be subtypes of the same strain. Isolates with PFGE patterns showing a >60% coefficient of similarity were likely to be related, while those showing a <60% coefficient of similarity were unrelated. Genetic similarity was calculated by the Dice coefficient and clustered by the unweighted-pair group arithmetic averaging method. Genotypes and farm identifications were arbitrarily defined.
Four of the seven E. coli PFGE clusters that were found among chicken isolates were unique to particular farms: C, G, I, and M were unique to farms 1, 12, 10, and 7, respectively (Fig. 2). Similarly, as demonstrated in Table 2, chickens from 10 of the 12 farms studied were found to excrete only one genogroup of E. coli. However, 18 E. coli isolates from chickens from five different farms (no. 4, 5, 6, 8, and 11) situated at least 2 to 5 km from each other were of the same genotype, N, and thus shared significant genetic relatedness. Although the PFGE results suggested close relatedness of the chicken isolates, there was no significant relatedness of these E. coli isolates with those obtained from children from the linked households.
TABLE 2.
Distribution of PFGE clusters from E. coli from children and chickens from various farms
| Farm no. | No. of isolates/PFGE cluster froma:
|
|
|---|---|---|
| Children | Chickens | |
| 1 | 2/D | 7/C |
| 3/J | 2/F | |
| 2 | 2/K | 4/A |
| 7/H | 2/F | |
| 3 | 7/B | 8/A |
| 4 | 3/D | 3/N |
| 3/K | ||
| 5 | 1/E | 4/N |
| 6 | 1/E | 4/N |
| 7 | 3/K | 10/M |
| 8 | 1/E | 3/N |
| 1/J | ||
| 9 | 3/D | 2/L |
| 10 | 1/E | 5/I |
| 11 | 1/L | 4/N |
| 12 | 3/L | 4/G |
PFGE clusters were arbitrarily assigned.
Only two (PFGE clusters B and H) of the six PFGE clusters found among E. coli from children were unique (to farms 3 and 2, respectively). In addition, as demonstrated in Table 2, children from 8 of the 12 farms were found to excrete only one genogroup of E. coli. In contrast, two PFGE clusters (D and K) of E. coli were found in children from three different farms. This may indicate interactions among children within this farming community. From the genetic-similarity data there was more diversity among E. coli isolates from children than among isolates from chickens from the same farms (P < 0.01). Four E. coli isolates that were from four children in the same ward in the Thika hospital but from four different households formed a PFGE cluster (E). These isolates, which also had 100-MDa plasmids with indistinguishable RE digest patterns, were likely to have been acquired nosocomially. The genetic relationships of these and other E. coli isolates as analyzed by the use of dendrograms are shown in Fig. 2.
DISCUSSION
Antibiotic susceptibility data from the present study demonstrated that E. coli isolates from chickens and from children from the same geographical area were different. The majority (96.8%) of isolates from chickens were either fully susceptible or were resistant only to tetracycline, while the majority (85.7%) of isolates from children were multidrug resistant. Indeed, tetracycline was the only antibiotic to which both the isolates from chickens and those from children were highly resistant. It is probable that the large quantities of tetracycline used in raising chickens provided the selective pressure for the emergence and spread of resistance to it among chicken isolates. What was surprising was that the high usage of tetracycline has not coselected for other resistances in the E. coli isolates from chickens. As tetracyclines were rarely used for treatment of the children, we can only attribute the high level of resistance to their use in the adult population. On the other hand, the use of co-trimoxazole and ampicillin as first-line agents for the treatment of childhood diarrhea in the hospitals in the study may be responsible for the high levels of resistance to these antibiotics.
HindIII digestion of the 60- and 100- to 110-MDa plasmids that encoded the transferable antibiotic resistance in E. coli from both chickens and children further demonstrated that these isolates were different. Plasmids from chickens produced three distinct RE digest patterns, while those from children fell into four distinct patterns. Thus, RE digest patterns indicated that different populations of resistance plasmids were present among the isolates from chickens and children. Although some studies (6) have previously reported transmission of plasmids between E. coli from chickens and humans, this was not demonstrated in the present study, as indicated by the diversity of the plasmid RE digest patterns. Other workers (10) have made similar observations, in which plasmids from E. coli from humans who worked in a poultry-processing plant were highly diverse and were unrelated to those from E. coli isolated from the carcasses.
In their study, Meng et al. (8) recommended the use of PFGE in addition to other typing methods in order to characterize epidemiologically unrelated bacterial strains. In the present study, PFGE of XbaI-digested DNA from isolates from children and from chickens from 12 farms produced a total of 14 different clusters. For E. coli isolates from either children or chickens, coefficients of similarity ranged from 20 to 100%, revealing the large diversity in E. coli strain types that existed within the study population. For E. coli isolates form either children or chickens from the same farms, XbaI RE digestion followed by PFGE was able to further differentiate between isolates within the same plasmid RE profile. PFGE clusters were generally unique for isolates from either children or chickens from the same farms. In only one of the clusters (PFGE cluster L) were isolates likely to be related within a 60% coefficient of similarity.
Recently, Banatvala et al. (1) also successfully applied PFGE to characterize an outbreak of E. coli O157:H7 infection associated with food contamination in retail supermarkets in parts of Connecticut. In their study, restriction fragment length polymorphism of PFGE profiles was used to distinguish between outbreak-related strains and sporadic cases of E. coli O157:H7 infection. In agreement with observations made in the present study, Barrett et al. (2) also reported that XbaI provided the best discrimination, with the most easily interpreted restriction fragments for studies of characterization of E. coli.
Four E. coli isolates obtained from children from four different farms admitted to the pediatric ward in Thika Hospital had indistinguishable PFGE patterns, and HindIII digestion of their 100-MDa resistance plasmids produced similar patterns. These infections were therefore likely to have been acquired nosocomially. For three-quarters of the farms, only one type of PFGE cluster existed within each farm, indicating that a single or a few closely related subtypes of E. coli were likely to be commonly found within a flock of chickens or among children from the same household. However, E. coli isolates from children showed greater genetic diversity than isolates from chickens from the same farms. This may be an indication of a wider variation in E. coli strain types circulating within the human population than among chickens. In the case of the children, the possibility of E. coli infections exists, as during the daytime, children from a number of families on the same or neighboring farms are left in the care of one housegirl while the adults work on the farm. Food or drink contamination is therefore a possible route of transmission of E. coli infections among the children.
From our findings it would appear that periods of feeding chickens and collecting eggs were not sufficient to allow colonization of the children with E. coli from chickens or that E. coli isolates from chickens were unable to colonize the children. As the results of MPN tests of tap water indicate, fecal contamination was not important in the transmission of coliforms between children and chickens. Levy et al. (6) have hypothesized that humans may become colonized with E. coli from chickens by breathing in contaminated dust from chicken sheds. The findings of the present study do not confirm this hypothesis. On the other hand, in chicken rearing the use of open basins for providing water and feed for chickens raises the possibility of fecal contamination and subsequent transmission of the same E. coli strains within a flock.
There was significant relatedness in the PFGE patterns of 18 E. coli isolates (PFGE cluster N) from chickens from five different farms. It is probable that there was movement of chickens between farms, which may have contributed to a cross-farm transmission of closely related strains of E. coli. The PFGE patterns of E. coli isolates from children from the corresponding farms, however, were entirely different and thus not likely to have been acquired from common sources.
In conclusion, despite the activities in chicken rearing that may have exposed children to E. coli strains from chickens, the present study has shown that clusters of E. coli which were prevalent among the chicken flocks were distinct from those in children. Within the E. coli population from either the chickens or the children, further diversity exists, with little overlap between human and animal strain types. Indeed, antibiotic susceptibility profiles and HindIII digest patterns of resistance plasmids further confirmed that E. coli isolates in children from the present study were unlikely to have been acquired from chickens from the same farms.
ACKNOWLEDGMENTS
S.K. was sponsored by the BSAC and the Shell Fellowship.
Many thanks to technologists from the Centre for Microbiology Research, KEMRI, for their support. We thank the Director, KEMRI, for permission to publish this work.
REFERENCES
- 1.Banatvala N, Magnano A R, Cartter M L, Barrett T J, Bibb W F, Vasile L L, Mshar P P, Lambert-Fair M A, Green J H, Bean N H, Tauxe R V. Meat grinders and molecular epidemiology: two supermarket outbreaks of Escherichia coli O157:H7 infection. J Infect Dis. 1996;173:480–483. doi: 10.1093/infdis/173.2.480. [DOI] [PubMed] [Google Scholar]
- 2.Barrettt T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewing W H. Edward’s and Ewing’s identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Publishing Co., Inc.; 1986. [Google Scholar]
- 4.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause U, Thomson-Carter F M, Pennington T H. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J Clin Microbiol. 1996;34:959–961. doi: 10.1128/jcm.34.4.959-961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy S B, FitzGerald G B, Macone A N. Spread of antibiotic resistant plasmids from chicken to chicken and from chicken to man. Nature. 1976;260:40–42. doi: 10.1038/260040a0. [DOI] [PubMed] [Google Scholar]
- 7.Macrina F L, Kopecko D J, Jones K R, Ayers D J, McCowen S M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 8.Meng J, Zhao S, Zhao T, Doyle M P. Molecular characterisation of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis and plasmid DNA analysis. J Med Microbiol. 1995;42:258–263. doi: 10.1099/00222615-42-4-258. [DOI] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 10.O’Brien T F, DiGiorgio J, Parsonnet K C, Kass E H, Hopkins J D. Plasmid diversity in Escherichia coli isolated from processed poultry and poultry processors. Vet Microbiol. 1993;35:243–255. doi: 10.1016/0378-1135(93)90149-2. [DOI] [PubMed] [Google Scholar]
- 11.Rochelle P A, Fry J C, Day M J, Bale M J. An accurate method for estimating sizes of small and large plasmids and fragments by gel electrophoresis. J Gen Microbiol. 1986;132:53–59. doi: 10.1099/00221287-132-1-53. [DOI] [PubMed] [Google Scholar]
- 12.Tenover F C, Arbeit R D, Goering R V the Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Contr Hosp Epidemiol. 1997;18:426–439. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 13.Thong K L, Cheong Y M, Puthucheary S, Koh C L, Pang T. Epidemiological analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135–1141. doi: 10.1128/jcm.32.5.1135-1141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thong K L, Ngeow Y F, Altwegg P N, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walia S K, Madhavan T, Chagh T D, Sharma K B. Characterization of self transmissible plasmids determining lactose fermentation and multiple antibiotic resistance in clinical strains of Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 1987;7:279–284. doi: 10.1016/0147-619x(87)90003-5. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. Drinking water and sanitation 1981–1990. A way to health, 1981. Geneva, Switzerland: World Health Organization; 1981. [Google Scholar]