Abstract
A genomic library of Pseudomonas fluorescens DSM 50106 in a λRESIII phage vector was screened in Escherichia coli K-12 for esterase activity by using α-naphthyl acetate and Fast Blue RR. A 3.2-kb DNA fragment was subcloned from an esterase-positive clone and completely sequenced. Esterase EstF1 was encoded by a 999-bp open reading frame (ORF) and exhibited significant amino acid sequence identity with members of the serine hydrolase family. The deduced amino acid sequences of two other C-terminal truncated ORFs exhibited homology to a cyclohexanone monooxygenase and an alkane hydroxylase. However, esterase activity was not induced by growing of P. fluorescens DSM 50106 in the presence of several cyclic ketones. The esterase gene was fused to a His tag and expressed in E. coli. The gene product was purified by zinc ion affinity chromatography and characterized. Detergents had to be added for purification, indicating that the enzyme was membrane bound or membrane associated. The optimum pH of the purified enzyme was 7.5, and the optimum temperature was 43°C. The showed highest purified enzyme activities towards lactones. The activity increased from γ-butyrolactone (18.1 U/mg) to ɛ-caprolactone (21.8 U/mg) to δ-valerolactone (36.5 U/mg). The activities towards the aliphatic esters were significantly lower; the only exception was the activity toward ethyl caprylate, which was the preferred substrate.
Esterases belong to the group of hydrolases (carboxylester hydrolases; E.C. 3.1.1.1) which catalyze the formation or cleavage of ester bonds of water-soluble substrates. The hydrolytic mechanism of most of the known esterases resembles the hydrolytic mechanism of lipases and serine proteases. All of these enzymes contain a catalytic triad that usually consists of a serine, a histidine, and an aspartic acid. The serine is embedded in the consensus sequence G-X-S-X-G at the active site, and ester hydrolysis is mediated by a nucleophilic attack of the active serine on the carbonyl of the substrate in a charge-relay system with the two other amino acid residues (29).
The physiological functions of many esterases are not clear. Some of these enzymes are known to be involved in metabolic pathways that provide access to carbon sources; such enzymes include the acetyl- and cinnamoyl esterases that are involved in degradation of hemicellulose (13, 16). In some plant-pathogenic bacterial and fungal strains these cell wall-degrading esterase activities are believed to be pathogenic factors (25). Detoxification of biocides may be another important role. Insecticide resistance often results from amplification of genes for esterases that hydrolyze the insecticides (3). Some insecticides and neurotoxins inhibit the acetylcholine esterase, which is essential in neurotransmittance. The fusidic acid resistance of Streptomyces lividans is due to a specific esterase which inactivates the antibiotic (39), and a Bacillus subtilis esterase that hydrolyzes the phytotoxin brefeldin A has been described (40).
The fact that different enantiomers interact differently in an organism and may even have hazardous effects, such as the teratogenic activity of the racemic drug thalidomide, has led to a growing demand for enantiomerically pure compounds (37). Lipases and esterases have been used successfully in organic synthesis of optically pure substances. For instance, an esterase from Arthrobacter globiformis was used in the resolution of ethyl chrysanthemate derivatives (27, 28), which are key compounds during the synthesis of pyrethrin insecticides. A heroin-specific esterase has been described, and this esterase selectively converts heroin into morphine; this is followed by further degradation to morphinone by a morphine dehydrogenase (35). A Bacillus carboxyl esterase has been used for stereospecific resolution of R,S-naproxen esters to S-naproxen (34), which is an important anti-inflammatory drug, and a p-nitrobenzyl esterase was genetically engineered in order to synthesize cephalosporin-derived antibiotics (26).
Lactones are widely distributed in nature, and these compounds contribute to the flavor and fragrance of fruits, including raspberry, peaches, and coconuts. Several examples of synthesis of optically pure lactones by lipase-catalyzed hydrolysis have been described previously (15, 19). Furthermore, lactones have also been synthesized from the corresponding ω-hydroxy acids, and consequently, there is a need for specific lipases and esterases.
In this paper, we describe the screening, cloning, sequencing, and biochemical characterization of a new Pseudomonas fluorescens esterase which exhibits the highest activity towards lactones.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli JM109 (41) was used as the host for transformation of plasmid DNA, and E. coli HB101 F′lac[::Tn1739tnpR] (1) was used for infection with λRES phages. These strains were grown in Luria-Bertani (LB) liquid medium or on LB agar plates at 37°C (36). The media were supplemented with 100 μg of ampicillin per ml or 50 μg of kanamycin per ml for selection of plasmids. The vector pIC20H was used for cloning and DNA sequencing experiments (24), and the vector pJOE2775 with a rhamnose-inducible rhaBAD promoter (5) was used for expression of estF1 in E. coli JM109. P. fluorescens DSM 50106 was grown in M9 medium (36) or LB liquid medium supplemented with either 10 mM glucose, 10 mM cyclopentanone, 10 mM cyclohexanone, or 10 mM cycloheptanone.
DNA manipulation techniques.
Restriction enzymes and DNA-modifying enzymes were obtained from Boehringer Mannheim. Standard procedures were used for restriction enzyme analysis and cloning experiments, as described by Sambrook et al. (36). Plasmid DNA was isolated by using a modified protocol described by Kieser (20). E. coli was transformed with plasmid DNA as described by Chung et al. (11). PCR were performed in 100-μl reaction mixtures containing 1 ng of plasmid DNA, 30 pmol of primer, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 10% dimethyl sulfoxide, 2.5 U of Pwo polymerase, and 1× reaction buffer provided by the supplier (Boehringer Mannheim). The DNA was first heated to 100°C for 2 min and then amplified with a Minicycler (Biozym Diagnostic GmbH) by using 30 cycles consisting of 1 min of denaturation at 94°C, 1.5 min of annealing at 5°C below the melting temperature of the primer, and 1.5 min of extension at 72°C.
Screening a genomic library of P. fluorescens in λRESIII for esterase activity.
Construction of a genomic library from P. fluorescens DSM 50106 has been described previously (8). E. coli HB101 F′lac[::Tn1739tnpR] (1) was grown overnight in L broth supplemented with 0.2% maltose and 10 mM MgCl2. A 0.1-ml portion of the overnight culture was infected with 103 to 104 recombinant phages for 10 min at room temperature, and the preparation was incubated at 37°C for 45 min in 2 ml of LB medium supplemented with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and then plated onto LB agar plates containing 50 μg of kanamycin per ml. After growth and replica plating, the plates were overlaid with 5 ml of soft agar (0.5% agar in H2O) containing 80 μl of α-naphthyl acetate (20 mg/ml in N,N′-dimethyl formamide) and 80 μl of Fast Blue RR (80 mg/ml in dimethyl sulfoxide). Esterase-positive colonies developed a brown color in less than 2 min.
DNA sequence analysis.
DNA sequencing of the 3.2-kb MluI-BamHI fragment in pIC20H (pFIS5) was performed by using the chain termination method with double-stranded plasmid DNA as the template. Two strategies were employed. Fragments generated with NaeI and MscI were subcloned in pIC20H. Plasmid pFIS5 and the deletion derivatives were sequenced by using Cy5-labelled M13 universal and reverse primer with an ALFexpress AutoRead sequencing kit (Amersham Pharmacia Biotech). Primer walking was performed by using oligonucleotides obtained from MWG Biotech, Ebersberg, Germany, and a Cy5-dATP labelling mixture along with the ALFexpress AutoRead sequencing kit. The reaction products were separated on a 5.5% Hydrolink Long Ranger gel matrix with an ALFexpress DNA sequencer for 12 h at 55°C and 800 V in the presence of 0.5× TBE buffer. The nucleotide sequence was analyzed with the Genetics Computer Group program package (14), version 8.01. Database searches were performed with the programs BLASTX, BLASTP, and BLASTN by using the electronic mail server of the National Center for Biotechnology Information, Bethesda, Md. (2).
Construction of plasmids pFIS31 and pFIS32.
For expression of estF1 in E. coli the gene was amplified by PCR performed with the primers S1361 (5′-AAAA CAT ATG GCT GTG CAA TGG TT-3′) and S1345 (5′-AAAA GGA TCC TTT GTT CGC CAA GGC AAA-3′). The PCR fragment was cleaved with NdeI and BamHI and inserted into the vector pJOE2775, which was cut with the same enzymes to give pFIS32. For amplification of the gene at the second possible ATG start codon, the DNA was amplified with primers S1328 (5′-AAAA CAT ATG ACA CGG CGG ATT GAA G-3′) and S1345 and inserted into the vector pJOE2775 to give pFIS31. Esterase activity was observed with pFIS32 but not with pFIS31.
Expression of estF1 in E. coli and purification of the esterase.
E. coli JM109 harboring the estF1 gene under control of the rhamnose-inducible rhaBAD promoter on plasmid pFIS32 was grown at 37°C in 250 ml of LB medium supplemented with ampicillin (100 μg/ml) until the early exponential phase (optical density at 600 nm, 0.5 to 0.6). Esterase production was induced when rhamnose (final concentration, 0.2%) was added, and cultivation was continued for 5 h. Cells were collected by centrifugation (15,000 rpm, 155-mm rotor [Jovan, Unterhaching, Germany], 10 min, 4°C) and washed twice with 50 mM sodium phosphate buffer (pH 7.5) at 4°C. Then the detergent Emulgen 913 (Kao Chemicals, Tokyo, Japan) was added to a concentration of 0.5% (vol/vol), and the cells were disrupted by sonification. Cell debris was removed by centrifugation (10,000 rpm, 155-mm rotor [Jovan], 10 min, 4°C). Purification was performed by immobilized metal ion affinity chromatography with a fast protein liquid chromatography system (Pharmacia, Uppsala, Sweden) by taking advantage of the His tag attached to the mature EstF1. A 10-ml portion of the crude cell extract was diluted 1:5 with buffer A (0.5 M betaine, 50 mM sodium phosphate buffer [pH 7.5]), loaded onto a preequilibrated Sepharose fast-flow zinc column (diameter, 1.8 cm; height, 9.8 cm; volume, 25 ml; flow rate, 3.5 ml/min; Pharmacia) with buffer A, and eluted with buffer B (0.3 M imidazole, 0.5 M betaine, 50 mM sodium phosphate buffer [pH 7.5]; flow rate, 3.0 ml/min). Fractions were assayed with the p-nitrophenyl acetate (pNPA) assay as described below. The protein content was determined by using a bicinchoninic acid kit (Pierce, Rockford, Ill.) and bovine serum albumin as the protein standard.
Electrophoresis.
Proteins from the crude extract and from various purification steps were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins in a low-molecular-weight standard mixture obtained from Pharmacia were used as reference proteins. After electrophoresis either the gel was stained with Coomassie brilliant blue or activity staining (zymogram) was performed after renaturion in 50 mM Tris-HCl buffer (pH 7.5) containing α-naphthyl acetate and Fast Red (Sigma), which revealed esterase activity by the formation of a red complex (4).
Esterase activity.
The esterase activity during cultivation, the esterase activity of the crude cell extract, and the esterase activity of the lyophilized enzyme were determined photometrically in a 50 mM sodium phosphate buffer (pH 7.5) solution by using pNPA (10 mM dissolved in dimethyl sulfoxide) at 25°C and 410 nm (ɛ = 15 × 103 M−1 cm−1) or by a pH stat assay performed with 5% (vol/vol) ethyl acetate (equivalent to 500 mM ethyl acetate) at 37°C as described previously (23). One unit of esterase activity was defined as the amount of enzyme that released 1 μmol of p-nitrophenol per min or 1 μmol of acetic acid per min under assay conditions. The values determined for purified EstF1 were corrected for the presence of imidazole, which causes apparently 2.8-fold-higher activity. This phenomenon has been described previously and is related to imidazolyl group-mediated hydrolysis of p-nitrophenol esters (7, 12).
Temperature and pH profiles and the activity of EstF1 towards other esters (but not lactones [see below]) were determined by a pH stat assay in a similar manner by using each substrate at a concentration of 5% (vol/vol), which ensured that there was a large excess of substrate. Temperature stability was determined after incubation of the esterase at a given temperature for 16 h, followed by the pH stat assay performed with ethyl acetate at 37°C and pH 7.5. In each experiment, 1 mg of crude EstF1 or 0.1 mg of purified EstF1 was used. All values were determined in triplicate and were corrected for autohydrolysis of the substrates. The deviations for all data were between 0.2 and 6.8%.
Activity of EstF1 towards lactones.
Lactonase activity was measured with the pH stat apparatus. To 20 ml of 50 mM sodium phosphate buffer (pH 7.5) 100 mM lactone and 5 ml of toluene were added at 37°C. The reaction was started by adding a known amount of esterase. After the level of conversion had reached about 40%, the reaction mixture was transferred to a separating funnel to separate the organic and aqueous phases. The aqueous phase was extracted three times with diethyl ether, and the combined ether phases and the toluene phase were dried separately over anhydrous sodium sulfate. An etheric solution of diazomethane was added to both organic phases until the solutions remained yellow, which indicated that the ω-hydroxy acid formed by lactone hydrolysis had been quantitatively converted to the corresponding methyl ester. After evaporation of the excess solvent in vacuo, the reaction components were analyzed by gas chromatography (Mega series; Fisons Instruments, Mainz, Germany) by using a flame ionization detector and a polar column (Optima 5; film thickness, 0.25 μm; 25 m by 0.25 mm [inside diameter]; Machery & Nagel, Düren, Germany). The analysis was carried out with temperature programming. For γ-butyrolactone the initial temperature was 60°C, the temperature was increased at a rate of 10°C/min to 200°C, and the retention times were as follows: γ-butyrolactone, 2.16 min; and 4-hydroxybutanoic acid methyl ester, 6.27 min. For δ-valerolactone the initial temperature was 60°C, the temperature was increased at a rate of 10°C/min to 160°C, and the retention times were as follows: δ-valerolactone, 3.36 min; and 5-hydroxypentanoic acid methyl ester, 2.89 min. For ɛ-caprolactone the initial temperature was kept at 50°C for 2 min, the temperature was increased at a rate of 5°C/min to 150°C, and the retention times were as follows: ɛ-caprolactone, 7.55 min; and 6-hydroxyhexanoic acid methyl ester, 4.13 min.
Vmax and Km values were calculated from Lineweaver-Burk and Eadie-Hofstee plots derived from the initial measurements of the rates of hydrolysis of the three lactones and ethyl acetate when 0.05 mg of purified EstF1 was used. Lactones were used at concentrations between 5 and 75 mM, and ethyl acetate was used at concentrations ranging from 25 to 100 mM. Hydrolysis was performed essentially as described above; however, the reactions were conducted in the absence of organic solvent. All experiments were performed in triplicate, and the deviations for the data are shown below.
Nucleotide sequence accession number.
The nucleotide sequence of the 3,223-bp MunI-BamHI fragment encoding esterase EstF1 has been deposited in the GenBank database under accession no. AF090329.
RESULTS
Cloning of an esterese gene from P. fluorescens DSM 50106.
Total DNA of P. fluorescens DSM 50106 was partially digested with Sau3AI and inserted between the λ arms of the vector λRESIII as described previously (8). The recombinant phages were converted into autonomous replicating plasmids by infection of E. coli HB101 F′lac[::Tn1739tnpR] (1), and the kanamycin-resistant colonies were screened for esterase activity by overlaying the colonies with soft agar containing α-naphthyl acetate and Fast Blue RR. From the approximately 5,000 colonies screened, we isolated 6 colonies which exhibited esterase activity. The plasmids were isolated from the six colonies and analyzed by restriction enzyme analysis. According to the restriction patterns (and according to the DNA hybridization results [data not shown]) at least two different genes encoding esterase activity were isolated in this way. One of the plasmids, pJOE2967, was investigated further. The plasmid DNA of pJOE2967 was digested with various restriction enzymes, and fragments were inserted into pIC20H and transformed into E. coli JM109. In this way a 3.2-kb BamHI-MunI fragment that encoded the esterase activity was identified. The corresponding plasmid was designated pFIS5. The esterase activity was independent of the orientation of the fragment in the pIC vector and could not be induced by IPTG (data not shown), which indicated that the gene was transcribed from its own promoter.
Nucleotide sequence of the 3.2-kb MluI-BamHI fragment encoding the esterase.
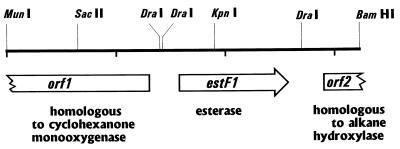
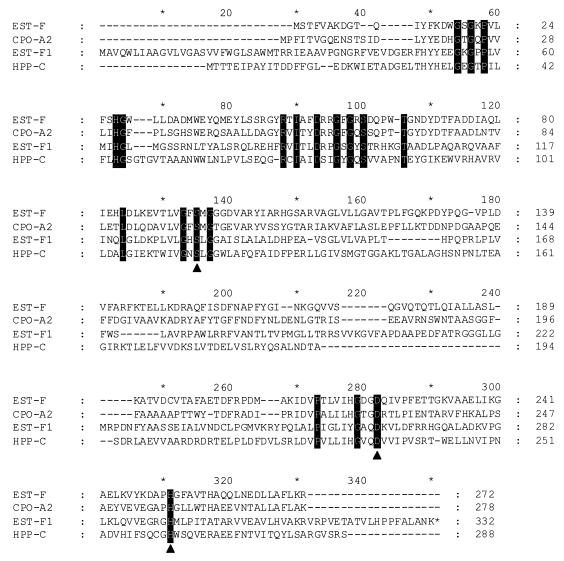
The sequences of both strands of the 3.2-kb MluI-BamHI fragment obtained from P. fluorescens were determined by subcloning various restriction fragments from pFIS5 into pIC20H and by the primer walking method performed with newly constructed oligonucleotides. DNA analysis of the 3,223-bp MluI-BamHI fragment revealed that there were three open reading frames (ORFs) (Fig. 1). The deduced amino acid sequence derived from one of these ORFs exhibited significant levels of similarity to the amino acid sequences of an esterase from P. fluorescens SIK WI (10), bacterial nonheme haloperoxidases, and a dienoate hydrolase (levels of amino acid sequence identity, 23 to 25%) (Fig. 2 shows a multiple alignment); these enzymes belong to subfamilies of the serine hydrolase family. The new enzyme was designated EstF1, and the corresponding gene was designated estF1. The characteristic properties of serine hydrolases include a tertiary structure called the α/β-hydrolase fold and a catalytic triad consisting of serine, histidine, and aspartic acid residues. Alignment of the EstF1 sequence with the EstF, CpoA2, and HppC sequences clearly revealed the presence of the three amino acids of the catalytic triad and the consensus sequence around the active serine (Gly-X-Ser-X-Gly) in EstF1 (Fig. 2). There were two potential ATG start codons. The first potential ATG start codon extended the N-terminal end of the enzyme by 16 to 25 amino acids compared to the related enzymes (Fig. 2). A DNA sequence corresponding to the consensus sequence of a ribosomal binding site was located 7 bp upstream from the first ATG. The second methionine followed at a distance of 25 amino acids in a position similar to the position of the N-terminal ends of the related enzymes, but the corresponding ATG codon was not preceded by a ribosomal binding site. Amplification of the gene by PCR and expression in E. coli resulted in only active enzyme when the first start codon was used (see below).
FIG. 1.
Physical map of the 3,223-bp MunI-BamHI fragment encoding esterase EstF1.
FIG. 2.
Multiple alignment of the esterases EstF from P. fluorescens SIK WI (10) and EstF1 from P. fluorescens DSM 50106 (this study), the nonheme chloroperoxidase CpoA2 from Streptomyces aureofaciens ATCC 10762 (33), and the 2-hydroxy-6-ketonona-2,4-dienoate hydrolase HppC from Rhodococcus globerulus (accession no. U89712). The solid triangles indicate the amino acids of the catalytic triad.
The two other ORFs found on the 3,223-bp fragment were truncated at their C-terminal ends. The deduced amino acid sequence derived from orf1, which was located upstream from estf1 and in an orientation different from that of estf1, exhibited homology (28.6% amino acid sequence identity) to the amino acid sequence of a cyclohexanone monooxygenase of Acinetobacter sp. strain NCIB9871 (9). The deduced amino acid sequence derived from orf2, which was downstream from estF1 and was transcribed in the same direction as estF1, exhibited homology (32% identity) to the amino acid sequence of an alkane hydroxylase (AlkB) of Pseudomonas oleovorans (22).
Production, isolation, and purification of EstF1.
To express estF1 in E. coli and to determine the exact N-terminal end, the gene had to be amplified by PCR. The oligonucleotides used for the two possible start codons introduced a NdeI site exactly at the ATG start codon; this site fit into the cloning site of the rhamnose-inducible expression vector pJOE2775 at the right distance from the t7gene10 ribosomal binding site present in the vector. The C-terminal end of the gene was amplified without the stop codon, and a BamHI site was added by the oligonucleotide in order to obtain a fusion with six His codons in the vector for purification of the protein by affinity chromatography (for construction of the plasmids see Materials and Methods). The resulting plasmids, pFIS31 and pFIS32, were transformed into E. coli JM109. Only plasmid pFIS32, which contained estF1 beginning at the first ATG codon, gave esterase-positive colonies.
Before induction of E. coli JM109(pFIS32) no esterase activity was detected. The highest activity (about 8.6 U/ml in 250 ml of culture broth, as determined by the pNPA assay) was observed 5 h after induction with rhamnose. Surprisingly, the esterase activity could be nearly quantitatively removed from the crude cell extract by centrifugation for 5 min in an Eppendorf centrifuge at 14,000 rpm, and the activity was recovered in the pellet fraction. This finding could not be explained by inclusion body formation since proteins produced as inclusion bodies are usually denatured and enzymatically inactive; rather, it indicated that either aggregation of active enzymes or binding to the cell membrane occurred. In order to facilitate isolation of the esterase, several detergents were added before cell disruption at a concentration of 0.5%. The best results were obtained with Emulgen 913, with which twofold-higher activity occurred in the cell extract compared to nontreated cell extract. Other detergents, such as n-octanoylglucose and cholic acid, had little influence, and Triton X-100 even reduced activity (the activity with Triton X-100 was only one-half the activity without detergent). Additional optimization experiments performed with different detergent concentrations revealed that the highest activity (10 U of lyophilized EstF1 per mg, as determined by the pNPA assay) was obtained in the presence of 1% Emulgen 913.
A total wet cell weight of 1.8 g was obtained from the 250-ml E. coli culture after centrifugation. Crude EstF1 was purified in a single step by immobilized zinc ion affinity chromatography, which yielded an almost homogeneous esterase with a specific activity of 128 U/mg (as determined by the pNPA assay). This corresponded to a purification factor of 12.8. The purity and activity of the purified enzyme were checked by SDS-PAGE, and the molecular weight was estimated to be 44,000 (data not shown). This value differs considerably from the calculated molecular weight (35,915). Activity staining with α-naphthyl acetate and Fast Red (data not shown) resulted in only one red band in the zymogram, which confirmed that no other hydrolytic enzymes were present in the purified esterase sample. Crude EstF1 and pure EstF1 were active at a wide pH range, the optimum pH was 7.5. Whereas crude EstF1 exhibited some activity at pH 4.5, the pure enzyme was active only at pH values greater than 5.5. A pH stat assay performed at different temperatures revealed that the optimal temperatures for crude EstF1 (47°C) and purified EstF1 (43°C) differed slightly, and both enzyme preparations significantly lost activity at temperatures less than 30°C or greater than 45°C. In contrast, the enzyme was only weakly stable after exposure to temperatures greater than 20°C for 16 h; for instance, after incubation at 30°C only 75% the activity of pure EstF1 remained. Lyophilized pure EstF1 even lost activity when it was stored at −20°C. Due to this observation, the eluate obtained by purification was used immediately for further studies.
Substrate spectra.
The substrate specificity of EstF1 was studied by using various carboxylic acid esters and triglycerides. In the case of acyclic aliphatic esters, the highest activities were observed with ethyl caprylate (13.1 U/mg), ethyl butyrate (7.7 U/mg), and ethyl acetate (10.9 U/mg). Changing the alcohol portion resulted in no clear pattern; however, hydrolysis of esters of longer fatty acids (fatty acids with more than eight carbon atoms) resulted in significantly lower activities (Table 1). Triglycerides were hydrolyzed at much lower rates, and triolein was not used as a substrate, as expected for an esterase.
TABLE 1.
Substrate profile of purified EstF1
| Substrate | Relative activity (%) |
|---|---|
| Ethyl acetate | 30 |
| Butyl acetate | 8 |
| Methyl butyrate | 11 |
| Ethyl butyrate | 21 |
| Propyl butyrate | 9 |
| Methyl caprylate | 20 |
| Ethyl caprylate | 36 |
| Methyl decanoate | 7 |
| Ethyl decanoate | 10 |
| Methyl laurate | 4 |
| Methyl myristate | 4 |
| Triacetin | 2 |
| Tributyrin | 3 |
| Triolein | 0.2 |
| γ-Butyrolactone | 50 |
| δ-Valerolactone | 100 |
| ɛ-Caprolactone | 60 |
The activity towards δ-valerolactone (36.5 U/mg) was defined as 100%.
Hydrolysis of lactones.
The presence of a gene homologous to a cyclohexanone monooxygenase gene near estF1 in the genomic DNA prompted us to test the lactone-hydrolyzing activity of EstF1. Crude EstF1 and purified EstF1 exhibited rather high activities towards δ-valerolactone, the product of monooxygenase when cyclopentanone was the substrate, compared to simple aliphatic esters. The specific lactonase activity of the crude enzyme was determined to be 3.1 U/mg for δ-valerolactone, and the activity of the purified enzyme was approximately 12-fold higher (36.5 U/mg). The activity of the purified enzyme towards δ-valerolactone was approximately threefold higher than the activity towards ethyl acetate or ethyl caprylate, although in the photometric assay performed with pNPA the activity was 12.8-fold higher, as a result of a better leaving group (p-nitrophenol instead of ethanol). The specific activities towards γ-butyrolactone and ɛ-caprolactone were 18.1 and 21.8 U/mg, respectively. The specificity of EstF1 towards lactones was confirmed by determining kinetic data (Table 2). The highest Vmax/Km and lowest Km values were obtained for δ-valerolactone, γ-butyrolactone, and ɛ-caprolactone, and ethyl acetate was the worst substrate.
TABLE 2.
Kinetic data determined from hydrolysis of various lactones and ethyl acetate with purified EstF1
| Substrate | Vmax (U/mg) | Km (mM) | Vmax/Km (10−3 min−1 · mg−1) |
|---|---|---|---|
| δ-Valerolactone | 39 (± 2.2) | 7.5 | 5.1 (± 0.34) |
| γ-Butyrolactone | 25 (± 2.4) | 12.6 | 2.0 (± 0.20) |
| ɛ-Caprolactone | 22 (± 2.1) | 23.7 | 0.9 (± 0.09) |
| Ethyl acetate | 17 (± 0.7) | 45.2 | 0.4 (± 0.03) |
DISCUSSION
There are at least four kinds of esterases in P. fluorescens strains; these enzymes differ in substrate specificity, cellular location, and other properties (10). The esterase isolated in this work is clearly related to a group of enzymes consisting of esterases like P. fluorescens SIK WI EstF and a highly homologous esterase of P. putida (31), as well as to bacterial nonheme chloroperoxidases like Streptomyces aureofaciens CpoA2. The physiological roles of these esterases and haloperoxidases are not known. The haloperoxidases catalyze halogenation reactions in vitro, but no in vivo halogenation reaction, such as chlorination of an intermediate to chlortetracycline, has been found. It has been suggested that in vivo the haloperoxidases actually have esterase or other hydrolytic activities with unknown substrates. All of the enzymes belonging to this group contain a catalytic triad, and the α/β-hydrolase fold typical of enzymes belonging to the serine hydrolase family has been identified in CpoA2 by crystallization (18). The EstF1 protein clearly contains the catalytic triad, as shown by the multiple alignment. This enzyme differs from the other enzymes by having an N-terminal extension consisting of about 25 amino acids. According to a Kyte-Dolittle plot (data not shown) this N-terminal region is highly hydrophobic and may anchor the enzyme in the cell membrane. This would explain why detergents were necessary to keep the enzyme soluble for purification. The hydrophobic N-terminal end and a very basic pI (pI 10.9, as calculated from the amino acid composition) may explain the difference between the molecular weight calculated from the amino acid composition and the molecular weight determined by SDS-PAGE; similar differences have also been described for other highly basic proteins (6). The discovery of an ORF encoding a putative cyclohexanone monooxygenase next to estF1 and the high activities towards δ-valerolactone, ɛ-caprolactone, and γ-butyrolactone indicate that EstF1 may play a role in degradation of cyclic ketones. For instance, a cyclohexanone monooxygenase from Acinetobacter sp. was found to selectively catalyze the formation of ɛ-caprolactone from cyclohexanone in a Baeyer-Villiger oxidation (38), and a cyclopentanone oxygenase from Pseudomonas sp. strain NCIB9872 catalyzes the formation of δ-valerolactone from cyclopentanone (17). Further degradation of these lactones by hydrolytic enzymes leads to the corresponding ω-hydroxy acids. Recently, the substrate specificities of the lactonases from Acinetobacter sp. strain NCIMB 9871 and Pseudomonas sp. strain NCIMB 9872 were determined by using crude extracts of the strains (30). None of the lactonase-encoding genes has been sequenced; in addition, the nucleotide sequence of a cyclopentanone monooxygenase gene has never been determined.
To see if EstF1 is involved in degradation of cyclic ketones, P. fluorescens DSM 50106 was incubated in M9 minimal medium supplemented with either glucose, cyclopentanone, cyclohexanone, or cycloheptanone. In contrast to the growth in M9 medium containing glucose, no growth was observed when the ketones were the sole carbon sources. The experiment was repeated with LB medium. Addition of most cyclic ketones to LB medium had no influence on the growth rate of P. fluorescens; the only exception was cycloheptanone, which reduced the growth rate significantly. When the esterase activities of the cells were determined after 7 and 24 h of incubation, no increase in esterase activity was observed with the ketones compared to the activity obtained with glucose. The overall esterase activity (average, about 0.004 U/mg under all conditions) was very low (data not shown). Therefore, despite high activity towards δ-valerolactone EstF1 is not part of a cyclopentanone degradation pathway in P. fluorescens DSM 50106, and the natural substrate of EstF1 remains unknown. Nevertheless, EstF1 provides a useful alternative biocatalyst for synthesis of ω-hydroxy acids, as well as for specific hydrolysis of lactones.
ACKNOWLEDGMENTS
We acknowledge financial support provided by the Deutsche Forschungsgemeinschaft (Bonn, Germany) and supported provided by the German Academic Exchange Service (Bonn, Germany) to V.K.
The technical support provided by U. Schwaneberg and N. Krebsfänger (Institute for Technical Biochemistry, Stuttgart, Germany) is gratefully acknowledged.
REFERENCES
- 1.Altenbuchner J. A new λRES vector with a built-in Tn1721-encoded excision system. Gene. 1993;123:63–68. doi: 10.1016/0378-1119(93)90540-j. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Blackman R L, Spence J M, Field L M, Devonshire A L. Chromosomal location of the amplified esterase genes conferring resistance to insecticides in Mycus persicae (Homoptera: Aphidae) Heredity. 1995;75:297–302. [Google Scholar]
- 4.Bornscheuer U, Reif O W, Lausch R, Freitag R, Scheper T, Kolisis F N, Menge U. Lipase of Pseudomonas cepacia for biotechnological purposes: purification, crystallization and characterization. Biochim Biophys Acta. 1994;1201:55–60. doi: 10.1016/0304-4165(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 5.Bornscheuer U T, Altenbuchner J, Meyer H H. Directed evolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones. Biotechnol Bioeng. 1998;58:554–559. doi: 10.1002/(sici)1097-0290(19980605)58:5<554::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Bourguignon J, Macherel D, Neuburger M, Douce R. Isolation, characterization, and sequence analysis of a cDNA clone encoding L-protein, the dihydrolipoamide dehydrogenase component of the glycine cleavage system from pea-leaf mitochondria. J Biochem. 1992;204:865–873. doi: 10.1111/j.1432-1033.1992.tb16706.x. [DOI] [PubMed] [Google Scholar]
- 7.Breslow R, Singh S. Phosphate ester cleavage catalyzed by bifunctional zinc complexes: comments on the “p-nitrophenyl ester syndrome.”. Bioorg Chem. 1988;16:408–417. [Google Scholar]
- 8.Brünker P, Altenbuchner J, Mattes R. Structure and function of the genes involved in mannitol, arabitol and glucitol utilization from Pseudomonas fluorescens DSM50106. Gene. 1998;206:117–126. doi: 10.1016/s0378-1119(97)00574-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y-C J, Peoples O, Walsh C T. Acinetobacter cyclohexanone monooxygenase: gene cloning and sequence determination. J Bacteriol. 1988;170:781–789. doi: 10.1128/jb.170.2.781-789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi K D, Jeohn G H, Rhee J S, Yoo O J. Cloning and nucleotide sequence of an esterase gene from Pseudomonas fluorescens and expression of the gene in Escherichia coli. Agric Biol Chem. 1990;54:2039–2045. [PubMed] [Google Scholar]
- 11.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleij M C, Drenth W, Nolte R J M. Enantioselective cleavage of esters by histidine-containing tripeptides in micellar solutions of various hexdecyltrialkylammonium bromide surfactants. Recl Trav Chim Pays-Bas. 1993;112:1–6. [Google Scholar]
- 13.Dalrymple B P, Swadling Y, Cybinski D H, Xue G P. Cloning of a gene encoding cinnamoyl ester hydrolase from the ruminal bacterium Butyrivibrio fibrisolvens E14 by a novel method. FEMS Lett. 1996;143:115–120. doi: 10.1111/j.1574-6968.1996.tb08469.x. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enzelberger M M, Bornscheuer U T, Gatfield I, Schmid R D. Lipase-catalyzed resolution of γ- and δ-lactones. J Biotechnol. 1997;56:129–133. doi: 10.1016/s0168-1656(97)00107-7. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira L M A, Wood T M, Williamson G, Faulds C, Hazlewood G P, Black G W, Gilbert H J. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem J. 1993;294:349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin M, Trudgill P W. Purification and properties of cyclopentanone oxygenase of Pseudomonas NCIB 9872. Eur J Biochem. 1976;63:199–209. doi: 10.1111/j.1432-1033.1976.tb10222.x. [DOI] [PubMed] [Google Scholar]
- 18.Hecht H J, Sobek H, Haag T, Pfeifer O, van Pée K H. The metal-ion-free oxidoreductase from Streptomyces aureofaciens has an α/β hydrolase fold. Nat Struct Biol. 1994;1:532–537. doi: 10.1038/nsb0894-532. [DOI] [PubMed] [Google Scholar]
- 19.Kazlauskas R J, Bornscheuer U T. Biotransformations with lipases. In: Rehm H J, Reed G, Pühler A, Stadler P J W, Kelly D R, editors. Biotechnology series. 8a. Weinheim, Germany: Wiley-VCH; 1998. pp. 37–191. [Google Scholar]
- 20.Kieser T. Factors affecting the isolation of ccc DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 21.Kodera Y, Furukawa M, Yokoi M, Kuno H, Matsushita H, Inada Y. Lactone synthesis from 16-hydroxyhexadecanoic acid ethyl ester in organic solvents catalyzed with polyethylene glycol-modified lipase. J Biotechnol. 1993;31:219–224. [Google Scholar]
- 22.Kok M, Oldenhuis R, von der Linden M P G, Raatjes P, Kingma J, van Lelyveld P H, Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene. J Biol Chem. 1989;264:5435–5441. [PubMed] [Google Scholar]
- 23.Krebsfänger N, Zocher F, Altenbuchner J, Bornscheuer U T. Characterization and enantioselectivity of a recombinant esterase from Pseudomonas fluorescens. Enzyme Microb Technol. 1998;22:641–646. [Google Scholar]
- 24.Marsh J L, Erfle M, Weykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 25.McQueen D A R, Schottel J L. Purification and characterization of a novel extracellular esterase from pathogenic Streptomyces scabies that is induced by zinc. J Bacteriol. 1987;169:1967–1971. doi: 10.1128/jb.169.5.1967-1971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore J C, Arnold F H. Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa M, Gomi H, Kishimoto F. Purification and some properties of carboxylesterase from Arthobacter globiformis; stereoselective hydrolysis of ethyl chrysanthemate. Biosci Biotechnol Biochem. 1993;57:594–598. [Google Scholar]
- 28.Nishizawa M, Shimizu M, Ohkawa H, Kanaoka M. Stereoselective production of (+)-trans-chrysanthemic acid by a microbial esterase: cloning, nucleotide sequence, and overexpression of the esterase gene of Arthrobacter globiformis in Escherichia coli. Appl Environ Microbiol. 1995;61:3208–3215. doi: 10.1128/aem.61.9.3208-3215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S, Harel M, Remington S J, Silman I. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 30.Onakunle O A, Knowles C J, Bunch A W. The formation and substrate specificity of bacterial lactonases capable of enantioselective resolution of racemic lactones. Enzyme Microb Technol. 1997;21:245–251. [Google Scholar]
- 31.Ozaki E, Sakimae A, Numazawa R. Nucleotide sequence of the gene for a thermostable esterase from Pseudomonas putida MR-2068. Biosci Biotechnol Biochem. 1995;59:1204–1207. doi: 10.1271/bbb.59.1204. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier I, Altenbuchner J. A bacterial esterase is homologous with non-haem haloperoxidases and displays brominating activity. Microbiology. 1995;141:459–468. doi: 10.1099/13500872-141-2-459. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer O, Pelletier I, Altenbuchner J, van Pée K H. Molecular cloning and sequencing of a non-haem bromoperoxidase gene from Streptomyces aureofaciens ATCC 10762. J Gen Microbiol. 1992;138:1123–1131. doi: 10.1099/00221287-138-6-1123. [DOI] [PubMed] [Google Scholar]
- 34.Quax W J, Broekhuizen C P. Development of a new Bacillus carboxyl esterase for use in the resolution of chiral drugs. Appl Microbiol Biotechnol. 1994;41:425–431. doi: 10.1007/BF00939031. [DOI] [PubMed] [Google Scholar]
- 35.Rathbone D A, Holt P J, Lowe C R, Bruce N C. Molecular analysis of the Rhodococcus sp. strain H1 her gene and characterization of its product, a heroin esterase, expressed in Escherichia coli. Appl Environ Microbiol. 1997;63:2062–2066. doi: 10.1128/aem.63.5.2062-2066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schoffers E, Golebiowski A, Johnson C R. Enantioselective synthesis through enzymatic asymmetrization. Tetrahedron. 1996;52:3769–3826. [Google Scholar]
- 38.Taschner M J, Black D J. The enzymatic Baeyer-Villiger oxidation: enantioselective synthesis of lactones from mesomeric cyclohexanones. J Am Chem Soc. 1988;110:6892–6893. [Google Scholar]
- 39.von den Haar B, Walter S, Schwäpenheuer S, Schrempf H. A novel fusidic acid resistance gene from Streptomyces lividans 66 encodes a highly specific esterase. Microbiology. 1997;143:867–874. doi: 10.1099/00221287-143-3-867. [DOI] [PubMed] [Google Scholar]
- 40.Wie Y, Swenson L, Kneusel R E, Matern U, Derewenda Z S. Crystallization of a novel esterase which inactivates the macrolide toxin brefeldin A. Acta Crystallogr Sect D. 1996;52:1194–1195. doi: 10.1107/S0907444996008414. [DOI] [PubMed] [Google Scholar]
- 41.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]