Abstract
The particular interdisciplinary nature of human microbiome research makes organization and reporting of results spanning epidemiology, biology, bioinformatics, translational medicine, and statistics a challenge. Commonly used reporting guidelines for observational or genetic epidemiology studies lack key features specific to microbiome studies. Therefore a multidisciplinary group of microbiome epidemiology researchers adapted guidelines for observational and genetic studies to culture-independent human microbiome studies, also developing new reporting elements for laboratory, bioinformatic, and statistical analyses tailored to microbiome studies. The resulting tool, named Strengthening The Organization and Reporting of Microbiome Studies (STORMS), is composed of a 17-item checklist organized into six sections corresponding to the typical sections of a scientific publication, presented as an editable table for inclusion in supplementary materials. STORMS provides guidance for concise and complete reporting of microbiome studies that will facilitate manuscript preparation, peer review, reader comprehension of publications, and comparative analysis of published results.
Introduction
Changes in the human microbiome have been associated with many disease and health states1. However, reporting the results of human microbiome research is challenging as it often involves approaches from microbiology, genomics, biomedicine, bioinformatics, statistics, epidemiology, and other fields, resulting in a lack of consistent recommendations for reporting of methods and results. Inconsistent reporting can have consequences for the field by affecting the reproducibility of study results2. While researchers have called for better reporting standards,3 such as the Genomic Standards Consortium’s MIxS checklist4, to provide a means for reporting sampling, processing and data generation; no comprehensive standardized guidelines spanning laboratory and epidemiological reporting have been proposed.
Standard reporting guidelines promote research consistency and, as a consequence, encourage reproducibility and improved study design. Editorial adoption of the CONsolidated Standards Of Reporting Trials (CONSORT) guidelines, for example, has been associated with an increase in quality of trial reporting5,6. Other epidemiological reporting guidelines have seen broad adoption, such as Strengthening the Reporting of OBservational studies in Epidemiology (STROBE)7 and STrengthening the REporting of Genetic Association Studies (STREGA)8. STROBE-metagenomics9 proposes an extension to the STROBE checklist for metagenomics studies. Subsequent to the Minimum Information About a Microarray Experiment (MIAME)9, Minimum Information about a MARKer gene Sequence (MIMARKS) and Minimum Information about any (x) Sequence (MIxS) checklists provide detailed guidance on reporting of sequencing studies in general. These are focused on the technical aspects of data generation, however, as are projects such as the Microbiome Quality Control Project (MBQC)10 and International Human Microbiome Standards (IHMS)11,12. Together, these serve as useful foundations, but do not span the full range of reporting of human microbiome studies, include items intended for other types of studies, and provide limited guidance on manuscript preparation.
Studies of the human microbiome share many features with other types of molecular epidemiology, but also require unique considerations with their own methodological best practices and reporting standards. In addition to standard elements of epidemiological study design, culture-independent microbiome studies involve collection, handling and preservation of biological specimens, evolving approaches to laboratory processing with elevated potential for batch effects, bioinformatic processing, statistical analysis of sparse, unusually-distributed, high-dimensional data, and reporting of results on potentially thousands of microbial features13-15. Because there is no agreed-upon gold-standard method for microbiome research and the field has not reached consensus on many of these aspects, inconsistencies in reporting inhibit reproducibility and hamper efforts to draw conclusions across similar studies.
For these reasons, we convened a multi-disciplinary working group to develop guidelines tailored to microbiome study reporting. Members of this group include epidemiologists, biostatisticians, bioinformaticians, physician-scientists, genomicists, and microbiologists. The checklist is designed to balance completeness with burden of use, and is applicable to a broad range of human microbiome study designs and analysis. The “Strengthening The Organizing and Reporting of Microbiome Studies (STORMS)” checklist (Table S1) draws relevant items from related guidelines and adds new tailored guidelines to serve as a tool to organize study planning and manuscript preparation, to improve the clarity of manuscripts, and to facilitate reviewers and readers in assessing these studies.
Methodology
Origin and development
The origins of these guidelines are rooted in a project to create a standardized database of published literature reporting relationships between the microbiome and disease (bugsigdb.org, in preparation). The goal of that project is to create a publicly available, standardized database of microbiome study findings indexed by condition of interest (e.g. disease, health status, diet, environmental factor) microbiome site (e.g. gut, mouth, skin), and microbial taxonomy to aid comparative analysis. As of August 2021, 31 curators (Table S2) have extracted findings from 513 unique published studies (Table S3). Included studies must have examined the relationship between the microbiome and a condition of interest and included findings on a taxonomic level (even if all findings were null).
This review revealed substantial reporting heterogeneity, particularly for epidemiology, such as study design, confounding factors, and sources of bias. It also revealed microbiome-specific issues, including statistical analysis of compositional relative abundance data and handling ‘batch’ effects.16 This heterogeneity highlighted the need for standardized reporting guidelines, similar to those used in other fields of study. The curators determined that standardized reporting guidelines would streamline the review process, but would more importantly help researchers throughout the field of microbiome research communicate their findings effectively.
The resulting multidisciplinary group of bioinformaticians, epidemiologists, biostatisticians, and microbiologists was thus convened to discuss microbiome reporting standards. The group began by reviewing existing reporting standards including STROBE17, STROBE-ME18, STREGA8, MICRO19, MIMARKS4, and STROGAR20. The group also reviewed existing articles containing recommendations for microbiome reporting21,22. The STROBE and STREGA guidelines were used as a starting point for the STORMS checklist, although aspects were incorporated from the other reporting standards.
Following the reporting standards development guidelines recommended by EQUATOR, a comprehensive list of potential guideline items was created. From this list, group members added, modified, and removed items based on their expertise. After the first round of edits, the checklist was then applied to a recent microbiome study23 by group members. Comments, removals, and additions were harmonized after each round. Based on this process, additional changes, simplifications, and clarifications were made. This process was repeated until there was a group consensus that the checklist was ready for use.
In addition to the core working group, outside subject matter experts identified by working group members were then invited to review the guidelines and provide feedback as members of the STORMS Consortium. Substantive feedback (i.e. not grammar, spelling, or other small changes) from 46 authors was organized by topic and compiled into a feedback document (Supplement 1), and responded to as in a response to reviewers letter. After this round of revisions, consortium members were once again invited to review the checklist prior to submission for publication.
Elaboration and explanation of checklist items
Checklist
The latest version of the checklist at time of publication is presented in Table S1 and a summary of items is presented in Figure 1. Of the items in the latest version in the STORMS checklist, nine items or sub-items were unchanged from STROBE, three were modified from STROBE, one was modified from STREGA, and 57 new guidelines were developed. Nine Items with an overlap with MIxS are specified. Rationale for new and modified items are presented below. Documentation of items unmodified from STROBE and STREGA were presented in the publications of those checklists.
Figure 1.
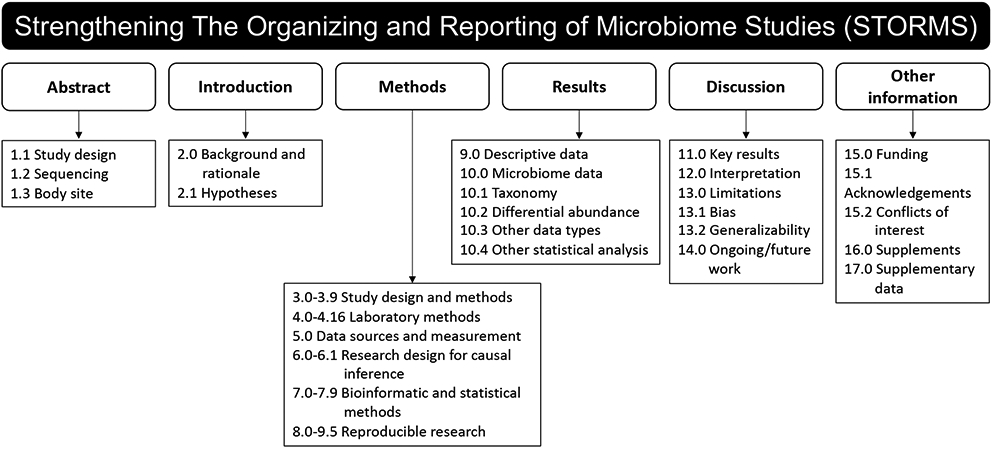
STORMS checklist items by major manuscript heading. For detailed descriptions of each item and additional guidance, see the STORMS checklist (Table S1).
Abstract (1.0-1.3)
Along with commonly included abstract materials such as a basic description of the participants and results, authors should report the study design24--such as cross-sectional, case-control, cohort, or randomized controlled trial--in the abstract of their article (item 1.1), as required by other reporting guidelines. Communicating study design in the abstract allows readers to quickly categorize the type of evidence provided. As part of this basic description, sequencing methods should be mentioned (item 1.2). Body site(s) sampled should also be included (item 1.3).
Introduction (2.0-2.1)
The introduction should clearly describe the underlying background, evidence, or theory that motivated the current study (item 2.0). Among other possibilities, this could include pilot study data, previous findings from a similar study or topic, or a biologically plausible mechanism that has been proposed. This clarifies for the reader the motivations for the present study. If the study is exploratory in nature, it should be explained what motivated the current exploration and the goals of the exploratory study. The hypothesis developed based on the background should be included. If the study was exploratory and did not define a hypothesis, pre-specified study objectives should be included (item 2.1).
Methods (3.0-8.5)
Participants (3.0-3.9)
The methods section should contain sufficient information for study replicability. Because study design is essential to understanding the study, it should be stated in the methods (item 3.0). When describing the participants in the study, the population of interest should be described and how participants were sampled from the source population (item 3.1). Because participant characteristics such as environment25, lifestyle behaviors, diet, biomedical interventions, demographics26, and geography27 (item 3.2) can correspond with substantial differences in the microbiome, it is essential to include this description. Temporal context can be quite important as well so start and end dates for recruitment, follow-up, and data collection should be stated (item 3.3).
Specific criteria used to assess potential participants for eligibility in the study should also be reported, detailing both inclusion and exclusion criteria (item 3.4). Inclusion and exclusion criteria are pre-established characteristics used for selection of participants into the study, and describing these criteria is essential in understanding the study’s target population28. This is expanded from STROBE, which requires eligibility criteria, but does not specify that both inclusion and exclusion criteria should be reported in detail. Any information collected about antibiotics or other treatments that could affect the microbiome should be described (item 3.5) as well as if any exclusion criteria included recent antibiotic or other medication use.
The final analytic sample sizes and read numbers should be stated as well as the reason for any exclusion of participants at any step of the recruitment, follow-up, or laboratory processes (item 3.6). STROBE suggests using a flow diagram to show when and why participants were removed from the study. A template for such flow diagrams is presented in Figure 2 and available in an editable spreadsheet (Table S1), and a public domain version for re-use at https://stormsmicrobiome.org. If participants were lost to follow-up or did not complete all assessments in a longitudinal study, details on how follow-ups were conducted should be stated and time-point specific sample sizes should also be reported (item 3.7). Additionally, studies that matched cases to controls should describe what variables were used in matching (item 3.4).
Figure 2. Examples of flowcharts for item 3.6.
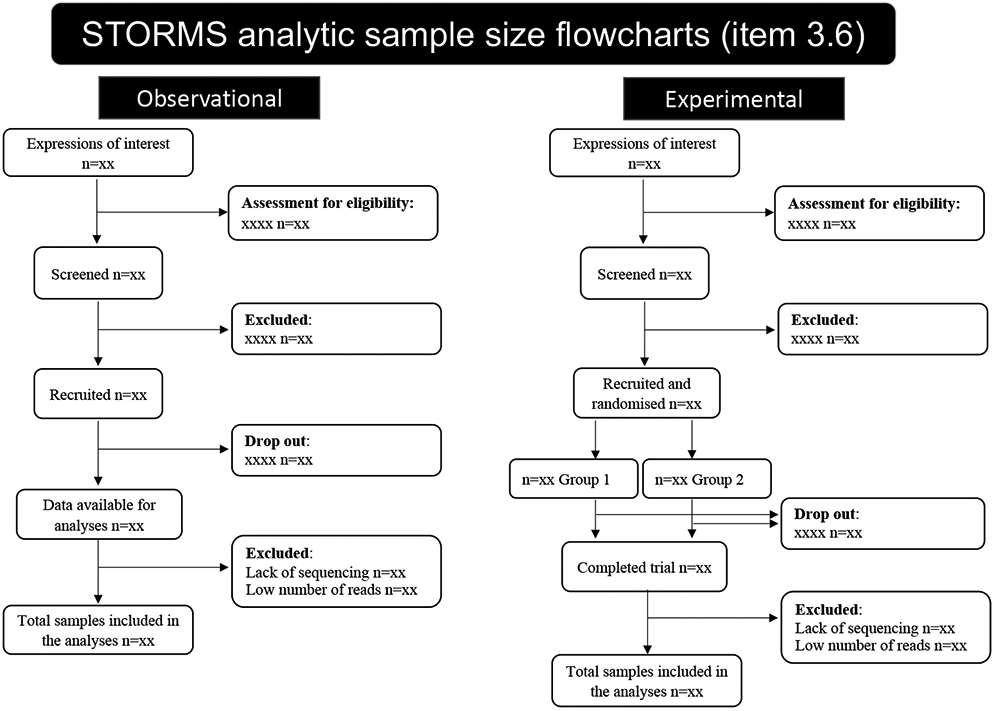
Though not required by STORMS, flowcharts can help visualize how the final analytic sample was calculated.
Laboratory methods (4.0-4.17)
Since STROBE does not cover laboratory methods, new items were developed for STORMS. Describe laboratory methods in sufficient detail to allow replication. The handling of lab samples should be described, including procedures for sample collection (items 4.1), shipping (item 4.2), and storage (item 4.3).
Because DNA extraction can be a major source of technical differences across studies,10 DNA extraction methods should be described (item 4.4). Human DNA removal and microbial DNA enrichment, if performed, should also be included (item 4.5). Likewise, if positive controls (item 4.7), negative controls (item 4.8), or contaminant mitigation methods (item 4.9) were used, they should be identified and described.
Sequencing-related methods should be reported. This includes primer selection and DNA amplification (including the variable region of the 16S rRNA gene if applicable) (item 4.6). Major divisions of sequencing strategy, such as shotgun or amplicon sequencing, should be identified (item 4.11). Finally, the methods used to determine relative abundances should be explained (item 4.12) and the read numbers that serve as denominators recorded.
Batch effects should be discussed as a potential source of confounding, including steps taken to ensure batch effects do not overlap with exposures or outcomes of interest (item 4.13)29. If conducting metatranscriptomics, metaproteomics, or metabolomics, provide details of those methods (items 4.14 to 4.16).
Data sources/measurement (5.0)
For non-microbiome data (e.g. health outcomes, participant socioeconomic, behavioral, dietary, biomedical characteristics, including disease location and activity, and environmental variables), the measurement and definition of each variable should be described (item 5.0). For instance, participant sex and age could be obtained from electronic medical records or from a questionnaire distributed to participants; this data source should be described. Limitations of measurement may also be discussed including potential bias due to misclassification or missing data, as well as any attempts made to address these measurement issues.
Research design considerations for causal inference (6.0-6.1)
Observational data is often used to test associations that aim towards causal inference in situations where the hypothesized causal relationship is not directly observed. Methods include, for example, the use of multivariable analysis or matching to adjust for confounding variables between a hypothesized exposure (such as abundance of a microbial taxon) and the disease or condition under study30. Confounders can be thought of as common causes of the exposure and the outcome under study that can induce a spurious association between the exposure and the outcome31,32. For instance, age could be a common confounder due to its significant influence on the microbiome and on risk of most health outcomes33. Laboratory batch effects could also confound relationships between the microbiome and a condition of interest if steps are not take to avoid imbalance of the condition across batches34. A common method for attempting to control for confounding is to adjust for or stratify on the confounder31. Justification should be provided for variables included or excluded in regression models for causal inference (item 6.0), as adjusting for or stratifying on a non-confounding variable can introduce bias35. As part of this theoretical justification, consider including a directed acyclic graph showing the hypothesized causal relationships of interest36,37.
In addition to considering the theoretical motivations for the present study, discuss the potential for selection or survival bias which can distort the observed relationship between the microbiome and variable of interest (item 6.1). For example, such bias may occur due to loss-to-follow-up (in longitudinal studies) or due to participants not being included in the study due to the condition itself (e.g. participants who have died of aggressive forms of colorectal cancer have not survived to be in a hypothetical study of colorectal cancer microbiomes)38. Other items elsewhere in the checklist may be directly relevant to questions of causal inference including hypotheses (item 2.1), study design (item 3.0), matching (item 3.8), bias (item 13.1) and generalizability (item 13.2). Authors investigating causal questions are encouraged to consider their reporting on these items in the context of causal inference as well.
Bioinformatics and Statistical Methods (7.0-7.9)
Adequate description of bioinformatic and statistical methods is essential to producing a rigorous and reproducible research report. Data transformations (such as normalization, rarefaction, and percentages) should be described (item 7.0). Quality control methods should be fully disclosed including criteria for filtering or removing reads or samples (item 7.1). All statistical methods used to analyze the data should be stated, (item 7.3) including how results of interest were selected (e.g. using a p-value, q-value, or other threshold) (item 7.8). Taxonomic, functional profiling, or other sequence analysis methods should be described in detail (item 7.2) In the interest of reproducibility, all software, packages, databases, and libraries used for the pre-processing and analysis of the data should be described and cited including version numbers (item 7.9).
Reproducible Research (8.0-8.5)
Reproducible research practices serve as quality checks in the process of publication and further transparency and knowledge sharing, as detailed in the rubric proposed by Schloss39. Journals are increasingly implementing reproducible research standards that include the publishing of data and code, and those guidelines should be followed when possible40,41. STORMS itemizes the accessibility of data, methods, and code (items 8.0 through 8.5). If possible, raw (item 8.1) and processed (item 8.2) data should be deposited into independently-maintained public repositories that provide long-term availability, such those maintained by NCBI or EMBL-EBI. Repositories like Zenodo (https://zenodo.org/) or Publisso (https://www.publisso.de/en/) can be used to provide a DOI to processed datasets. If data or code are not or cannot be made publicly available, even in a repository that provides restricted-access options, a description of how interested readers can access the data should be provided. As stated in item 8.0, any protected information should be described along with how such data can be accessed.
Results (9.0-10.4)
Descriptive Data (9.0)
Descriptive statistics about the study population should be reported (item 9.0). At a minimum, age and sex of all participants should be reported, but other important participant characteristics should be reported when possible including medication use or lifestyle factors such as diet. Consider reporting these data in a descriptive statistics table. Packages such as the table1 package in R make creating such a table straightforward42.
Outcome Data (10.0-10.4)
The main outcomes of the study should be detailed including descriptive information, findings of interest, and the results of any additional analyses. Descriptive microbiome analysis (for instance, dimension reduction such as Principal Coordinates Analysis, measures of diversity, gross taxonomic composition) should be reported for each group and each time point (item 10.0). This contextualizes the results of differential abundance analysis for readers. When reporting differential abundance test results, the magnitude and direction of differential abundance should be clearly stated (item 10.2) for each identifiable standardized taxonomic unit (item 10.1). Results from other types of analyses such as metabolic function, functional potential, MAG assembly, and RNAseq should be described in the results as well (items 10.3 and 10.4). Additional results (e.g. non-significant results or full differential abundance results) can be included in supplements, but should not be excluded entirely. Although the problem has been known for decades40, journals across many fields are recognizing the issue of publication bias and therefore the issue of non-reporting of null results43. Including such results in publications will help to reduce the severity of this bias and improve future systematic reviews and meta-analyses.
Discussion (11.0-14.0)
Most recommendations for the Discussion section are similar to STROBE including a discussion of the limitations of the present study and associated methods (item 13.0). One additional recommendation is made: discuss the potential for biases and how they would influence the study findings (item 13.1). Many forms of bias such as residual/unmeasured confounding, bias related to compositional analysis44, measurement bias, or selection bias45 could affect the interpretation of the results of the study and it is important to acknowledge potential sources of bias when discussing the results46. As described in STROBE, authors should also consider the generalizability of their findings and if these findings could be applicable to the target population or other populations (item 13.2). If different forms of bias were not assessed or assumed to be negligible, this should be stated. Finally, authors should discuss potential future or ongoing research based on findings of the present study (item 14.0).
Other Information (15.0-17.0)
In addition to a statement of funding (item 15.0), authors should also include acknowledgements and conflicts of interest statements (items 15.1 and 15.2, respectively). Conflicts of interest statements should be written according to the criteria established by the journal. Finally, the paper should state where supplementary materials and data can be accessed (items 16.0 and 17.0).
Implementation
The STORMS checklist can streamline peer review by providing both a checklist for assessing for completeness and a roadmap with pointers to the manuscript. Prior to submission, we recommend that authors use the “Comments” field to provide explanations where warranted, and to refer to relevant sections of the manuscript, to make the work of peer reviewers more straightforward and more accurate. We provide two examples of pre-publication use of STORMS: i) a multi-site study of associations between essential hypertension and gut microbial metabolic pathways47, and ii) an observational study of the stool microbiota of multiple host species48. Additional post-publication examples are also available49,50, which highlight that references to line numbers either must be updated during production or will continue to refer to the pre-print version.
Discussion
The STORMS checklist for reporting on human microbiome studies was developed with the following priorities. The checklist should 1) be easy to understand and use by researchers from various fields, through straightforward use of language and pruning of items rarely relevant to the current literature, 2) be organized in the outline of a manuscript, so it can serve as a tool for authors and for peer reviewers, particularly when included in manuscript submission as a supplemental table with comments, 3) assist in the complete and organized reporting of a study, not in enforcing any particular methods, 4) reuse or modify items from related checklists where relevant, and 5) represent consensus across a broad cross-section of the human microbiome research community. The checklist facilitates manuscript authors in providing a complete, concise, and organized description of their study and its findings. Included as a supplemental table to a manuscript, it also supports efficient peer review and post-publication interpretation.
While other efforts for extending STROBE for microbiome and metagenomic have been proposed9 and laboratory-focused reporting checklists have been released4,51, to our knowledge STORMS is the first comprehensive reporting checklist for human microbiome research. We aim for the STORMS guidelines to improve the quality and transparency of microbiome epidemiology studies by introducing a shared grammar of study reporting in a structured checklist format. Reporting checklists introduced in other disciplines have been shown to improve the quality of journal articles5,6.
A major strength of STORMS is the rigor and transparency of its development by a diverse, multidisciplinary consortium of subject matter experts. All feedback received from consortium members, and responses, have been included as Supplement 1. The development of STORMS is an ongoing process, and new versions of the checklist will be released to reflect evolving standards and technological processes. A version control system with changelog has been implemented, and annual reviews of the checklist are planned. Additionally, the working group plans to evaluate STORMS impact on microbiome reporting by examining how many articles are fulfilling checklist items before and after its release. We invite interested readers to join the STORMS Consortium by contacting the corresponding author or by visiting the consortium website for more information (https://www.stormsmicrobiome.org/). We also encourage journals to include the STORMS checklist in their instructions to authors and advise peer reviewers to consult the checklist when reviewing submissions.
There are some limitations to the STORMS checklist. The checklist was not created to assess study or methodological rigor. It is meant to aid authors’ organization and ease the process of reader assessment of how studies are conducted and analyzed. Conclusions about the quality of studies should not be made based on their adherence to STORMS guidelines, although we expect the reporting guidelines to help readers review studies critically. It does not encourage, discourage, or assume the use of null hypothesis significance testing52 or methods of compositional data analysis53, topics of some controversy in the field. In general the checklist avoids reference to or guidance on specific statistical methodological decisions.
Through the efforts of the STORMS Consortium working in an iterative, transparent, and collaborative process, the STORMS checklist provides a roadmap for researchers in reporting the results of a human microbiome study. The STORMS Consortium believes that the checklist is sufficiently flexible and user-friendly to support widespread adoption and contribution to microbiome study standards. Its adoption will ideally encourage thoughtful study design, reproducibility, collaboration, and open knowledge sharing between research groups as they explore the human microbiome.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number 5R01CA230551. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additionally, JBD was supported by the Leverhulme Trust.
Consortia Author List
Genomic Standards Consortium
Robert D. Finn23, Lynn Schriml55, Ramona Walls64, Christopher Hunter86, Susanna-Assunta Sansone80
23European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK, UK, 55University of Maryland School of Medicine, Institute for Genome Sciences, Baltimore, MD, USA, USA, 64Bio5 Institute, University of Arizona, USA, 86GigaScience Database, BGI-Hong Kong, Hong Kong, 80Oxford e-Research Centre, Department of Engineering Science, University of Oxford, UK, UK
Massive Analysis and Quality Control Society
Cesare Furlanello27, Benjamin Haibe-Kains30, Wenjun Bao70, Aedin Culhane71, Viswanath Devanarayan72, Joaquin Dopazo73, Xiaohui Fan74, Matthias Fischer75, Wendell Jones76, Rebecca Kusko77, Christopher Mason78, Timothy Mercer79, Andreas Scherer81, Leming Shi82, Shraddha Thakkar83, Weida Tong84, Russ Wolfinger85, Susanna-Assunta Sansone80
27HK3 Lab, Rovereto (TN), Italy, Italy, 30Princess Margaret Cancer Centre, Toronto, Canada, Canada, 70JMP Life Sciences, SAS Institute, CARY, NC, USA, USA, 71Dept of Data Sciences, Dana-Farber Cancer Institute, Boston, MA, USA, USA, 72Eisai, Inc., USA, 73Clinical Bioinformatics Area, Hospital Virgen del Rocio, Sevilla, Spain, Spain, 74College of Pharmaceutical Sciences, Zhejiang University, China, China, 75Experimental Pediatric Oncology, University Children's Hospital, and Center for Molecular Medicine Cologne (CMMC), Medical Faculty, University of Cologne, Germany, Germany, 76Q2 Solutions-EA Genomics, USA, USA, 77Immuneering Corporation, USA, USA, 78Weill Cornell Medicine, New York, USA, USA, 79Garvan Institute of Medical Research, University of New South Wales, Australia, Australia, 81Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland, Finland, 82State Key Laboratory of Genetic Engineering, Human Phenome Institute, School of Life Sciences and Shanghai Cancer Center, Fudan University, Shanghai, China, China, 83Office of Computational Science, OTS, CDER, USA, USA, 84Division of Bioinformatics and Biostatistics, NCTR/FDA, USA, USA, 85Scientific Discovery and Genomics, SAS Institute, Cary, NC, USA, USA, 80Oxford e-Research Centre, Department of Engineering Science, University of Oxford, UK, UK
Footnotes
Competing Interests Disclosure
J.S.E. is employed by a research center that belongs to a food company (Grupo Empresarial Nutresa). T.J.S. is a shareholder for Resilient Biotics. HS received unrestricted study grants from Biocodex; board membership, consultancy, or lecture fees from Carenity, Abbvie, Astellas, Danone, Ferring, Mayoly Spindler, M.S.D., Novartis, Roche, Tillots, Enterome, Maat, BiomX, Biose, Novartis, and Takeda; and is a co-founder of Exeliom bioscience. L.B.T. is an employee and shareholder of BiomCare. P.W. is listed as an inventor on patents PCT/EP2012/065178 and PCT/EP2013/052134, and is a member of the scientific steering committee for a clinical trial by 4D Pharma plc. T.Y. is a Founder and Board Member of Metabologenomics Inc. and Metagen Therapeutics Inc. L.Z. is a co-founder of the Notitia Biotechnologies Company. All other authors declare no competing interests
Data Availability Statement
The STORMS checklist as a versioned spreadsheet, and the analytical sample size flow diagram as editable PPTX and PDF files, are available for adaptation and inclusion in manuscripts at https://www.stormsmicrobiome.org/.
References
- 1.Wirbel J et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med 25, 679–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simoneau J, Dumontier S, Gosselin R & Scott MS Current RNA-seq methodology reporting limits reproducibility. Brief. Bioinform doi: 10.1093/bib/bbz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ten Hoopen P et al. The metagenomic data life-cycle: standards and best practices. GigaScience 6, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yilmaz P et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol 29, 415–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moher D, Jones A, Lepage L & Group, for the C. Use of the CONSORT Statement and Quality of Reports of Randomized Trials: A Comparative Before-and-After Evaluation. JAMA 285, 1992–1995 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Plint AC et al. Does the CONSORT checklist improve the quality of reports of randomised controlled trials? A systematic review. 185, 5 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Vandenbroucke JP et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and Elaboration. PLOS Med. 4, e297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement - Little - 2009 - Genetic Epidemiology - Wiley Online Library. https://onlinelibrary.wiley.com/doi/abs/10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
- 9.Bharucha T et al. STROBE-metagenomics: a STROBE extension statement to guide the reporting of metagenomics studies. Lancet Infect. Dis 20, e251–e260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha R et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol 35, 1077–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costea PI et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol 35, 1069–1076 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Santiago A et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 14, 112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner P et al. Microbiology Investigation Criteria for Reporting Objectively (MICRO): a framework for the reporting and interpretation of clinical microbiology data. BMC Med. 17, 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.STROGAR – STrengthening the Reporting Of Genetic Association studies in Radiogenomics - ScienceDirect. https://www.sciencedirect.com/science/article/pii/S0167814013003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha R, Abnet CC, White O, Knight R & Huttenhower C The microbiome quality control project: baseline study design and future directions. Genome Biol. 16, 276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badal VD et al. Challenges in the construction of knowledge bases for human microbiome-disease associations. Microbiome 7, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Elm E et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J. Clin. Epidemiol 61, 344–349 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Gallo V et al. STrengthening the Reporting of OBservational studies in Epidemiology - Molecular Epidemiology (STROBE-ME): an extension of the STROBE statement. Eur. J. Clin. Invest 42, 1–16 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Microbiology Investigation Criteria for Reporting Objectively (MICRO): a framework for the reporting and interpretation of clinical microbiology data ∣ BMC Medicine ∣ Full Text. https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerns SL et al. STROGAR – STrengthening the Reporting Of Genetic Association studies in Radiogenomics. Radiother. Oncol 110, 182–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha R et al. Next steps in studying the human microbiome and health in prospective studies, Bethesda, MD, May 16–17, 2017. Microbiome 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornung BVH, Zwittink RD & Kuijper EJ Issues and current standards of controls in microbiome research. FEMS Microbiol. Ecol 95, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min Y et al. Sex-specific association between gut microbiome and fat distribution. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce N Classification of epidemiological study designs. Int. J. Epidemiol 41, 393–397 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Jin Y, Wu S, Zeng Z & Fu Z Effects of environmental pollutants on gut microbiota. Environ. Pollut 222, 1–9 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Zhang Y-H, Huang T & Cai Y-D Gene expression profiling gut microbiota in different races of humans. Sci. Rep 6, 23075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patino CM, Ferreira JC, Patino CM & Ferreira JC Inclusion and exclusion criteria in research studies: definitions and why they matter. J. Bras. Pneumol 44, 84–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan L et al. OSAT: a tool for sample-to-batch allocations in genomics experiments. BMC Genomics 13, 689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VanderWeele TJ Principles of confounder selection. Eur. J. Epidemiol 34, 211–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernán MA, Hernández-Díaz S, Werler MM & Mitchell AA Causal Knowledge as a Prerequisite for Confounding Evaluation: An Application to Birth Defects Epidemiology. Am. J. Epidemiol 155, 176–184 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Lv B-M, Quan Y & Zhang H-Y Causal Inference in Microbiome Medicine: Principles and Applications. Trends Microbiol. (2021) doi: 10.1016/j.tim.2021.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh TS, Das M, Jeffery IB & O’Toole PW Adjusting for age improves identification of gut microbiome alterations in multiple diseases. eLife 9, e50240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soneson C, Gerster S & Delorenzi M Batch Effect Confounding Leads to Strong Bias in Performance Estimates Obtained by Cross-Validation. PLOS ONE 9, e100335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole SR et al. Illustrating bias due to conditioning on a collider. Int. J. Epidemiol 39, 417–420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Textor J, van der Zander B, Gilthorpe MS, Liśkiewicz M & Ellison GTH Robust causal inference using directed acyclic graphs: the R package ‘dagitty’. Int. J. Epidemiol dyw341 (2017) doi: 10.1093/ije/dyw341. [DOI] [PubMed] [Google Scholar]
- 37.Loughman A et al. Gut microbiota composition during infancy and subsequent behavioural outcomes. EBioMedicine 52, 102640 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schooling CM Selection bias in population-representative studies? A commentary on Deaton and Cartwright. Soc. Sci. Med. 1982 210, 70 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Schloss PD Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal R The file drawer problem and tolerance for null results. [Google Scholar]
- 41.Munafò M & Neill J Null is beautiful: On the importance of publishing null results. J. Psychopharmacol. (Oxf.) 30, 585–585 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Rich B benjaminrich/table1. [Google Scholar]
- 43.Null is beautiful: On the importance of publishing null results - Munafò Marcus, Neill Jo, 2016. https://journals.sagepub.com/doi/full/10.1177/0269881116638813. [DOI] [PubMed] [Google Scholar]
- 44.McLaren MR, Willis AD & Callahan BJ Consistent and correctable bias in metagenomic sequencing experiments. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernán M & Robins JM. Causal Inference: What If. (Chapman & Hall/CRC, 2020). [Google Scholar]
- 46.Writing a discussion section: how to integrate substantive and statistical expertise ∣ BMC Medical Research Methodology ∣ Full Text. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-018-0490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakai M et al. Essential Hypertension Is Associated With Changes in Gut Microbial Metabolic Pathways: A Multisite Analysis of Ambulatory Blood Pressure. Hypertension 78, 804–815 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Thingholm Louise B. et al. Ecology impacts the decrease of Spirochaetes and Prevotella in the fecal gut microbiota of urban humans. Res. Sq (2021) doi: 10.21203/rs.3.rs-147555/v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q et al. Linking gut microbiome to bone mineral density: a shotgun metagenomic dataset from 361 elderly women. Gigabyte 2021, 1–7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C et al. The female urinary microbiota in relation to the reproductive tract microbiota. Gigabyte 2020, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raes J, Foerstner KU & Bork P Get the most out of your metagenome: computational analysis of environmental sequence data. Curr. Opin. Microbiol 10, 490–498 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Szucs D & Ioannidis JPA When Null Hypothesis Significance Testing Is Unsuitable for Research: A Reassessment. Front. Hum. Neurosci 11, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gloor GB, Macklaim JM, Pawlowsky-Glahn V & Egozcue JJ Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol 8, (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The STORMS checklist as a versioned spreadsheet, and the analytical sample size flow diagram as editable PPTX and PDF files, are available for adaptation and inclusion in manuscripts at https://www.stormsmicrobiome.org/.


