Abstract
In a previous study (S. G. Acinas, F. Rodríguez-Valera, and C. Pedrós-Alió, FEMS Microbiol. Ecol. 24:27–40, 1997), community fingerprinting by 16S rDNA restriction analysis applied to Mediterranean offshore waters showed that the free-living pelagic bacterial community was very different from the bacterial cells aggregated or attached to particles of more than about 8 μm. Here we have studied both assemblages at three depths (5, 50, and 400 m) by cloning and sequencing the 16S rDNA obtained from the same samples, and we have also studied the samples by scanning electron microscopy to detect morphology patterns. As expected, the sequences retrieved from the assemblages were very different. The subsample of attached bacteria contained very little diversity, with close relatives of a well-known species of marine bacteria, Alteromonas macleodii, representing the vast majority of the clones at every depth. On the other hand, the free-living assemblage was highly diverse and varied with depth. At 400 m, close relatives of cultivated γ Proteobacteria predominated, but as shown by other authors, near the surface most clones were related to phylotypes described only by sequence, in which the α Proteobacteria of the SAR11 cluster predominated. The new technique of rDNA internal spacer analysis has been utilized, confirming these results. Clones representative of the A. macleodii cluster have been completely sequenced, producing a picture that fits well with the idea that they could represent a genus with at least two species and with a characteristic depth distribution.
The subject of prokaryotic biodiversity in the sea has received new and substantial attention with the development of molecular techniques to describe and identify the microbial components of natural communities. PCR amplification and the cloning of diagnostic molecules, mostly the 16S rRNA genes, permits extensive studies of the microbial diversity of ecosystems without the bias imposed by pure-culture techniques (or at least with a different one). In any case, molecular techniques represent a new approach to the extremely complex problem of describing microbial diversity. With the proliferation of studies based on cloning and sequencing ribosomal DNA (rDNA) retrieved from ocean samples, it has been recognized that bacterioplankton is dominated by a relatively limited subset of broad phylogenetic groups that are widely distributed and often exhibit clear trends in their vertical distribution in the water column (17, 21, 22, 34, 38).
We previously applied (1) a community-fingerprinting (36, 43) analysis to the water column in the stratified section of a Western Mediterranean station (halfway between Barcelona and the island of Mallorca). Offshore marine waters in tropical and subtropical latitudes often have a typical vertical temperature structure; in the case of temperate waters, this is mostly so during the summer. During this season, temperature and density vary over the first 100 m of depth. Under these conditions there is a characteristic chlorophyll-depth profile with a maximum, frequently sharp, known as the deep chlorophyll maximum (DCM). The DCM corresponds to a layer of maximum primary productivity and phytoplankton concentration. It is located between 40 and 100 m below the surface, where environmental conditions, particularly nutrient concentrations, are apparently optimal for many photosynthetic microorganisms (11, 14, 15, 24, 27). Our previous study by amplified rDNA restriction analysis (ARDRA) community fingerprinting showed that the prokaryotic assemblages at the surface, the DCM, and the deep water mass (400 m) varied (1). Within the bacterial community the main difference found was between the cells that lived in association with large particles (particles over ca. 8 μm, retained by a glass fiber prefilter) and the free-living cells (which passed through the filter).
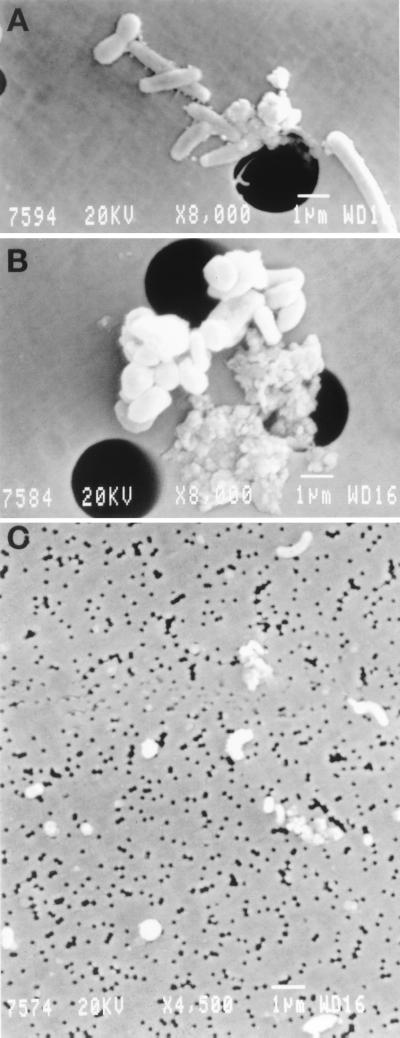
Attached bacteria are often larger, and are present in higher local concentrations, than those found free living in water (10) (Fig. 1). Although they are relatively few in the open ocean (compared to free-living cells), they could have an important role in carbon cycling (9, 25). There is information in the literature about the community present in relatively large aggregates, such as marine snow (4, 13), but smaller, more widespread aggregates of microscopic size have not been investigated.
FIG. 1.
Scanning electron microscopy of attached bacterial communities (A and B) and free-living bacterial communities (C).
Here we have studied in further depth the samples that seemed most promising from our previous ARDRA community fingerprinting (1). We have analyzed both free-living and attached bacterial communities found at three different depths (5 m, DCM, and 400 m) by sequencing 16S rDNA clones. Scanning electron microscopy has been used to examine the morphology of cells collected on each filter. The attached subsample gave very little diversity by the random cloning and sequencing approach used. In an attempt to retrieve more phylotypes from this sample, we have also used a methodology in which the predominant sequences amplified in one environment are characterized by ribosomal internal spacer analysis (RISA) (8).
MATERIALS AND METHODS
Sampling.
Three samples from different depths were selected from station D (about halfway between Barcelona and Mallorca [1], over a bottom of 2,000 m) in the Western Mediterranean. The work was done during the cruise FRONTS95 of the B/O García del Cid from 16 to 25 June 1995 (1). We analyzed free-living and attached assemblages found at three different depths: the surface (5 m); DCM, located at 52 m in this sample; and 400 m. Water samples were collected with a 30-liter double Van Dorn bottle and dispensed into plastic carboys.
The filtering protocol has been described earlier (1). First, Millipore AP20 glass fiber filters were used to remove larger particles and eukaryotes. Free-living prokaryotes were then collected by positive-pressure filtration on 0.22-μm-pore-size filters. The glass fiber filter was then rinsed to remove the attached prokaryotic fraction rather than directly extracting it, in order to avoid contamination from eukaryotes that would interfere due to PCR amplification of chloroplast rDNA by bacterial primers. This was accomplished as follows. The glass fiber filter was placed with the organisms facing down on top of a second AP20 filter. The filtrate from the 0.22-μm-pore-size filter (10 to 20 liters) was circulated at a high flow rate and with positive pressure through this “sandwich.” The bacteria washed out from the system were collected again with a 0.22-μm-pore-size filter. The number of bacterial cells thus retrieved was about 10% of the total number of bacterial cells in the untreated sample (1).
DNA extraction and purification.
DNA extraction and purification followed the protocol of Fuhrman et al. (18), with slight modifications as previously described (1).
PCR amplification of 16S rDNA and RISA.
16S rRNA genes were amplified from total DNA by PCR with two bacterial primers: ANT-1 and S (Table 1). The ribosomal internal transcribed spacers plus a stretch of the 16S rDNA (ca. 500 nucleotides) for RISA were amplified with primers B1055 and 23SOR (Table 1). All primers were subjected to CHECK PROBE SSU Prok (Ribosomal Database Project) (31) to confirm their adequacy. PCRs were performed with a Perkin-Elmer 480 thermal cycler. Reaction mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 9), 1.5 mM MgCl2, 0.1% Triton X-100, 200 mM of each deoxyribonucleotide triphosphate (dATP, dCTP, dGTP, and dTTP) (Pharmacia Biotechnology LKB, 2 U of TaqI DNA polymerase (Promega Corporation, Madison, Wis.), 0.2 mM (each) oligonucleotide primer, and 100 ng of template DNA in a total volume of 50 μl. The reaction mixtures were overlaid with mineral oil (Light White Oil; Sigma). The following conditions were used for amplification: a cycle of 95°C for 5 min; 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; plus an extension step of 10 min at 72°C. Five microliters of PCR product was analyzed in 1% agarose gels (SeaKem; FMC Bioproducts) in 1× Tris-acetic acid-EDTA (TAE) buffer, stained with 0.5 mg of ethidium bromide/μl, and visualized with UV. Products generated from RISAs were separated in 2% Methaphor agarose gels (FMC Bioproducts) in 1× Tris-borate-EDTA (TBE) buffer.
TABLE 1.
Oligonucleotides used in this study
| Name | Use | E. coli numbering | Sequence | Reference |
|---|---|---|---|---|
| ANT-1 | PCR | 8–27 | 5′-AGAGTTTGATCATGGCTCAG-3′ | 32 |
| S | PCR | 1510–1492 | 5′-GGTTACCTTGTTACGACTT-3′ | 32 |
| B1055 | PCR, sequencing | 1055–1074 | 5′-AATGGCTGTCGTCAGCTCGT-3′ | 5 |
| 23SOR | PCR | 21–38 | 5′-TGCCAAGGCATCCACCGT-3′ | 23 |
| Macle F | Sequencing | 338–357 | 5′-AATGGGGGAAACCCTGATGC-3′ | This paper |
| Macle R | Sequencing | 1060–1041 | 5′-ACTTAACCCAACATCTCACG-3′ | This paper |
Purification of RISA products.
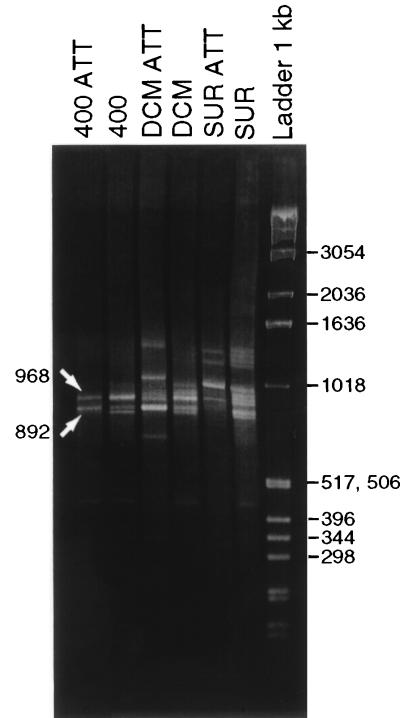
Major bands (Fig. 2) were cut out of the gel and purified with a Sephaglas BandPrep kit (Pharmacia Biotech), following the manufacturer’s instructions. The DNA was recovered in 20 μl of Tris-EDTA (TE) buffer.
FIG. 2.
Methaphor 2% gel fragments from RISA analyses of free-living and attached (ATT) bacterioplankton communities from surface (SUR), DCM, and 400 m (400). The arrows show the bands that were cloned and sequenced.
Clone library construction.
Clone libraries from PCR products were constructed with the TA cloning kit (Invitrogen Corporation, San Diego, Calif.), following the manufacturers’ recommendations.
Recombinant plasmids were extracted by using the QIAprep spin miniprep kit (Qiagen), as described in the manufacturers’ instructions. The purified plasmids were digested with EcoRI to separate the insert, and the product was run in agarose gels to determine the insert size. One hundred and thirty clones were grown in Luria-Bertani medium at 37°C for 18 h and kept at −80°C.
Sequencing.
The nucleotide sequences of plasmid inserts were determined by using the ABI PRISM dye terminator cycle-sequencing ready-reaction kit (Perkin-Elmer) and an ABI PRISM 377 sequencer (Perkin-Elmer), according to the manufacturers’ instructions. The 16S rRNA genes of 17 clones related to Alteromonas macleodii and chosen to represent the three depths sampled were completely sequenced with M13 Forward (−21) and M13 Reverse primers from the TA cloning kit and the internal primers Macle R and Macle F shown in Table 1. The 16S rRNA genes of the other 103 clones were partially sequenced (approximately 350 nucleotides from each end of the gene) with the standard M13 Forward (−21) and M13 Reverse primers.
The 10 clones obtained by RISA were partially sequenced with the B1055 primer (Table 1).
Phylogenetic analysis.
Sequences were evaluated by the program CHECK CHIMERA, provided by the Ribosomal Database Project, to check chimerical gene artifacts.
The sequences were compared to 16S rRNA sequences available in the GenBank and EMBL databases obtained from the National Center for Biotechnology Information database by the BLAST search. Similarity percentages were calculated manually.
The sequences were aligned with the Clustal W program (Genetics Computer Group package). In this alignment we used the sequences determined in this study and small-subunit rDNA sequences of the following bacteria from the γ subdivision of Proteobacteria which were obtained from the National Center for Biotechnology Information database: A. macleodii IAM 1290T (X82145), Shewanella alga “Bry” (X81621), Shewanella putrefaciens ATCC 8071T (X82133), Vibrio alginolyticus ATCC 17749 (X56576), Pseudoalteromonas atlantica IAM 12927T (X82134), Pseudoalteromonas haloplanktis subsp. haloplanktis ATCC 14393T (X67024), Pseudoalteromonas haloplanktis subsp. tetraodonis IAM 14160 (X82139), Pseudoalteromonas peptidysin F12-50-A1 (AF007286), Pseudoalteromonas rubra ATCC 29570T (X82147), and Pseudoalteromonas luteoviolacea NCIMB 1893T (X82144).
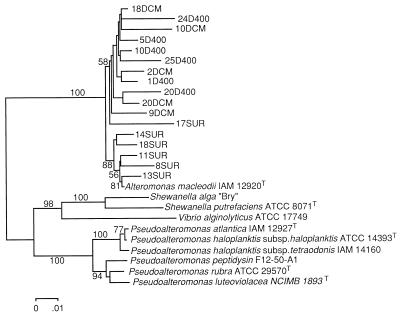
The phylogenetic tree in Fig. 3 was calculated with the neighbor-joining algorithm (40) by using the program MEGA (Molecular Evolutionary Genetics Analysis) version 1.01 obtained from the Institute of Molecular Evolutionary Genetics, the Pennsylvania State University, University Park. Bootstrap analysis of neighbor-joining data (500 resamplings) (16) was used to evaluate the tree topologies recovered for 1,492 positions.
FIG. 3.
Phylogenetic tree based on 1,492 nucleotide positions showing relationships of the surface (SUR), DCM, and 400-m (400) clones related to A. macleodii and representative bacterial 16S rRNA genes within the γ subdivision of Proteobacteria. An unrooted phylogenetic tree was obtained by performing a neighbor-joining analysis. Bootstrap values over 50% are shown below the segments.
The similarity matrices were calculated by the method of Jukes and Cantor (26) in the MEGA program.
Scanning electron microscopy.
Aliquots of 96 ml from each sample were fixed in 1% glutaraldehyde at 4°C overnight. The sample was filtered through a 3-μm-pore-size Millipore filter to recover the attached assemblage. The free-living bacteria that had passed through the 3-μm-pore-size filter were recovered on a 0.22-μm-pore-size filter. The filters were serially dehydrated in 25, 50, 70, and 100% ethanol solutions (three times for 10 min in each stage), critical-point dried, mounted on scanning electron micrograph stubs, sputter coated with gold, and viewed on a JEOL JSM 840 scanning electron microscope. Nucleotide sequence accession numbers. GenBank nucleotide sequence accession numbers for completely sequenced clones are from AF114495 to AF114509. Accession numbers for partial sequences of clones recovered at 400 m in the attached fraction are AF114510 to AF114533 and of those in the free-living fraction are AF114534 to AF114577 and AF114654 to AF114657. At the DCM, accession numbers for the attached fraction are AF114578 to AF114598 and AF114643 to AF114644 and those for the free-living fraction are AF114658 to AF114695. At the surface, accession numbers for the attached fraction are AF114599 to AF114622 and AF114623 to AF114642 and those for the free-living fraction are AF114645 to AF114653.
RESULTS
We sequenced ca. 40 clones from each depth (5 m, DCM, and 400 m). About 20 corresponded to bacterial 16S rDNAs recovered from the particulate fraction, and the other 20 corresponded to those from the free-living population. Each clone contained a nearly complete 16S rRNA gene (8 to 1510 [Escherichia coli numbering]) and was sequenced from both ends, producing an average of ca. 350 nucleotides from each end. Both segments contain hypervariable regions that are often included in environmental studies. The final similarity value was obtained by aligning both ends (ca. 700 nucleotides) to the complete database sequence. The results are shown in Tables 2 to 5, described below.
TABLE 2.
Phylogenetic affiliation of clones obtained from attached and free-living bacteria at 400-m depth
| Best matcha | Similarity (%)b | No. of clonesc | No. of nucleotides compared (minimum–maximum) | Phylogenetic affiliation |
|---|---|---|---|---|
| ATT (no. of clones = 18) | ||||
| A. macleodii IAM 12920T | 94.2–97.7 | 15 | 653–937 | γ Proteobacteria |
| A. macleodii IAM 12920T | 93.8 | 1 | 578 | |
| A. macleodii IAM 12920T | 94.3 | 1 | 837 | |
| Pseudoalteromonas sp. SW29 | 93.5 | 1 | 740 | |
| Free (no. of clones = 24) | ||||
| A. macleodii IAM 12920T | 86.9–97.6 | 12 | 662–895 | γ Proteobacteria |
| A. macleodii IAM 12920T | 92.2 | 1 | 657 | |
| A. macleodii IAM 12920T | 94.2 | 1 | 661 | |
| A. macleodii IAM 12920T | 95.4 | 1 | 850 | |
| P. antarctica CECT 4664 | 95.8 | 1 | 811 | |
| P. antarctica CECT 4664 | 94.4 | 1 | 737 | |
| Klebsiella planticola | 85.5 | 1 | 726 | |
| S. alga ATCC 8073 | 84.8 | 1 | 674 | |
| Unidentified marine bacterium strain E401 | 85.3 | 1 | 605 | |
| Unidentified γ Proteobacteria OM60 | 89.1 | 1 | 691 | |
| Uncultured proteobacterium OCS44 cluster SAR86 | 95.1 | 1 | 630 | |
| Unidentified eubacterium clone SAR324 | 94.3 | 1 | 651 | δ Proteobacteria |
| Unidentified α Proteobacteria cluster SAR11 | 90.1 | 1 | 865 | α Proteobacteria |
Name in boldface given by authors depositing the sequence and corresponding to uncultured bacteria. ATT, attached; Free, free living.
The range of similarities indicates the highest and lowest values of similarity to the best match when more than one clone with over 97% identity was found.
Clones with more than 97% nucleotide identity were considered identical.
TABLE 5.
Distribution by phylogenetic affiliation of 120 16S rDNA clones from different subsamples and depths
| Characteristic or group | No. of clones
|
|||||
|---|---|---|---|---|---|---|
| Surface free living | Surface attached | DCM free living | DCM attached | 400 m free living | 400 m attached | |
| Total | 17 | 19 | 24 | 18 | 24 | 18 |
| γ Proteobacteria | ||||||
| Total | 6 | 18 | 12 | 16 | 22 | 18 |
| A. macleodii IAM 12920T | 0 | 9 | 2 | 16 | 15 | 17 |
| Unidentified marine bacterium E401 (isolate) | 0 | 7 | 0 | 0 | 1 | 0 |
| Cluster SAR86 | 3 | 0 | 7 | 0 | 1 | 0 |
| S. alga | 0 | 1 | 0 | 0 | 1 | 0 |
| Other γ Proteobacteria | 3 | 1 | 3 | 0 | 4 | 1 |
| α Proteobacteria | ||||||
| Total | 9 | 1 | 6 | 2 | 1 | 0 |
| Cluster SAR11 | 6 | 0 | 4 | 2 | 1 | 0 |
| Cluster SAR116 | 1 | 1 | 0 | 0 | 0 | 0 |
| Sulfitobacter pontiacus | 1 | 0 | 1 | 0 | 0 | 0 |
| Other α Proteobacteria | 1 | 0 | 1 | 0 | 0 | 0 |
| Cyanobacteria | ||||||
| Total | 1 | 0 | 2 | 0 | 0 | 0 |
| Clone SAR7 | 1 | 0 | 1 | 0 | 0 | 0 |
| Other Cyanobacteria | 0 | 0 | 1 | 0 | 0 | 0 |
| Cytophagales | ||||||
| Total | 1 | 0 | 2 | 0 | 0 | 0 |
| Gelidibacter algens | 1 | 0 | 1 | 0 | 0 | 0 |
| Other Cytophagales | 0 | 0 | 1 | 0 | 0 | 0 |
| β Proteobacteria ARC33 | 0 | 0 | 1 | 0 | 0 | 0 |
| δ Proteobacteria clone SAR324 | 0 | 0 | 0 | 0 | 1 | 0 |
| High G+C gram positive unidentified firmicute OM1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Average pairwise nucleotide identity (%) | 43.3 | 89.1 | 39.1 | 94.1 | 80.22 | 94.2 |
Sequences recovered at 400 m.
At 400 m the assemblages recovered in the attached and free-living fractions were relatively similar. From the attached assemblage we have sequenced 18 clones for an average length of 800 nucleotides (minimum, 578 nucleotides; maximum, 937 nucleotides [Table 2]). This is the sample showing the least sequence diversity, with an average pairwise nucleotide identity of 94.2%. Of the 18 clones, 17 showed a high similarity to the 16S rDNA of A. macleodii; 7 of the clones showed a similarity of 97% or higher, so they very probably belong to a very close taxon, and the lowest similarity found among this cluster was 93.8% (still closely related). The one remaining clone had the best match with Pseudoalteromonas sp. strain SW29 and was therefore also related to Alteromonas (19).
From the free-living subsample, 24 clones were sequenced. Again, diversity was low, with all but two of the sequences belonging to the γ Proteobacteria. A cluster of 12 clones with within-cluster similarities ranging from 86.9 to 97.6% (Table 2) had A. macleodii as the best match, with over 96% similarity in 5 of them. Another three also had A. macleodii as the best match, and two others had Pseudoalteromonas antarctica CECT 4664 as the best match. The remaining seven clones showed similarity to uncultured organisms or had only low similarities to cultured organisms. One of the γ Proteobacteria clones had 95.1% similarity to the OCS44 sequence, a member of the SAR86 cluster retrieved in coastal Oregon waters. The retrieval of one sequence with a 94.3% similarity to the δ proteobacterium clone SAR324, described as abundant in Atlantic waters below 200 m, (44), is consistent with a deeper-water distribution for this phylotype.
DCM (52 m).
The attached assemblage at the DCM is similar to the 400-m subsample. Sixteen of 18 clones had the best match to A. macleodii (Table 3), most with very high similarities, indicating close relationship; 7 of these had similarities over 97%. The two remaining clones, which could be ascribed to the α Proteobacteria, are related to SAR 407, a SAR11 A2 cluster (17) representative.
TABLE 3.
Phylogenetic affiliation of clones obtained from attached and free-living bacteria at DCM
| Best matcha | Similarity (%)b | No. of clonesc | No. of nucleotides compared (minimum–maximum) | Phylogenetic affiliation |
|---|---|---|---|---|
| ATT (no. of clones = 18) | ||||
| A. macleodii IAM 12920T | 92.5–98.9 | 15 | 564–828 | γ Proteobacteria |
| A. macleodii IAM 12920T | 94.4 | 1 | ||
| Unidentified α Proteobacteria; clone SAR 407; cluster SAR11 | 92.5 | 1 | 796 | α Proteobacteria |
| Unidentified α Proteobacteria; clone SAR 407; cluster SAR11 | 92 | 1 | 684 | |
| Free (no. of clones = 24) | ||||
| Uncultured proteobacterium OCS5; cluster SAR86 | 91.1–99 | 3 | 533–567 | γ Proteobacteria |
| Uncultured proteobacterium OCS5; cluster SAR86 | 93.2 | 1 | 634 | |
| Uncultured proteobacterium OCS5; cluster SAR86 | 92.2 | 1 | 773 | |
| Uncultured proteobacterium OCS44; cluster SAR86 | 93–96.6 | 2 | 641–767 | |
| Proteobacterial SCB11 | 92.7 | 1 | 508 | |
| A. macleodii IAM 12900T | 96 | 1 | 756 | |
| A. macleodii IAM 12900T | 97.1 | 1 | 650 | |
| P. agarici | 92 | 1 | 724 | |
| Aeromonas sp. | 96.5 | 1 | 734 | |
| Unidentified α Proteobacteria; clone SAR 407; cluster SAR11 | 85.7–98 | 2 | 530–617 | α Proteobacteria |
| Unidentified α Proteobacteria; clone SAR 220; cluster SAR11 | 90.8 | 1 | 591 | |
| Uncultured proteobacterium; OCS180; cluster SAR11 | 95 | 1 | 670 | |
| Paracoccus solventivorans | 88.5 | 1 | 609 | |
| Sulfitobacter pontiacus ChLG-10 | 94.8 | 1 | 582 | |
| Microcystis elabens NIES42 | 91.8 | 1 | 723 | Cyanobacteria |
| Unknown marine bacterioplankton; clone SAR7 | 96 | 1 | 565 | |
| Psychroserpens burtonensis ACAM188 | 86.2 | 1 | 764 | Cytophagales |
| Gelidibacter algens C8ST5 | 82.4 | 1 | 579 | |
| Unidentified proteobacterium ARC33 | 93 | 1 | 685 | β Proteobacteria |
| Unidentified firmicute OM1 | 96.6 | 1 | 682 | High G+C gram positive |
Name in boldface given by authors depositing the sequence and corresponding to uncultured bacteria. ATT, attached; Free, free-living.
The range of similarities indicates the highest and lowest values of similarity to the best match when more than one clone with over 97% identity was found.
Clones with more than 97% nucleotide identity were considered identical.
From the free-living subsample, 24 clones were sequenced (Table 3). These clones show much more diversity and a clear predominance of uncultured phylotypes. Here the similarity to cultivated strains is low. Exceptions are one clone with a 96.5% similarity to Aeromonas sp. and two with over 96% similarity to A. macleodii. Other cultivated microorganisms with similarities near 90% were Microcystis elabens, Paracoccus solventivorans, Sulfitobacter pontiacus (94.8%), and Pseudomonas agarici. The majority of the clones similar to putative organisms known only by sequence were related to the SAR86 cluster (seven clones) of uncultivated γ Proteobacteria, specifically to a sequence retrieved from the Oregon coast. Some of them show extremely high matches, in the range expected for strains belonging to the same species. Some were related to the uncultivated α Proteobacteria cluster SAR11 (four clones). One clone gave a good match to a high-G+C gram-positive bacterium (96.6%), and another was a good match to a β proteobacterium from an Arctic lake. Two clones were grouped with similarities over 85% to the order Cytophagales. It is remarkable that only two clones among the free-living bacteria at the DCM belong to the cyanobacteria group.
Surface (5 m).
Nineteen clones were sequenced from the attached subsample (Table 4). The results are very similar to those of the rest of the attached samples, with nine clones highly related to A. macleodii. Seven clones had significant similarities to an unidentified marine isolate, E401, from Tanabe Bay, Japan, which is apparently related to the Aeromonas genus of aquatic γ Proteobacteria. The free-living subsample (17 clones) was dominated by bacteria related to uncultivated putative organisms of the α Proteobacteria (Table 4). Four clones were similar to the SAR11 A-2 cluster known to be more abundant at the surface (17). Remarkably, one clone had a very high similarity (97.9% over 705 nucleotides) to a sequence retrieved from the Oregon coast and belonging to the SAR116 cluster, a widespread phylotype (34). Among the γ Proteobacteria three clones had high similarities to the uncultured Oregon coast sequence OCS44 that belongs to the SAR86 cluster. One clone had 95.6% similarity to uncultured clone SAR7 (cyanobacteria), and another clone (over 83%) belonged to the Cytophagales group.
TABLE 4.
Phylogenetic affiliation of clones obtained from attached and free-living bacteria at surface
| Best matcha | Similarity (%)b | No. of clonesc | No. of nucleotides compared (minimum–maximum) | Phylogenetic affiliation |
|---|---|---|---|---|
| ATT (no. of clones = 19) | ||||
| A. macleodii IAM 12900T | 89.7–98.6 | 8 | 575–810 | γ Proteobacteria |
| A. macleodii IAM 12900T | 96.1 | 1 | 807 | |
| Unidentified marine bacterium strain E401 | 87.4–93.2 | 6 | 617–796 | |
| Unidentified marine bacterium strain E401 | 83.5 | 1 | 501 | |
| Vibrio logei ATCC 15832 | 86.2 | 1 | 539 | |
| S. alga ATCC 8073 | 91.7 | 1 | 612 | |
| Uncultured proteobacterium OCS28; cluster SAR116 | 94 | 1 | 655 | α Proteobacteria |
| Free (no. of clones = 17) | ||||
| Unidentified α proteobacterium; clone SAR 241; cluster SAR 11 | 86.1–88 | 2 | 613–700 | α Proteobacteria |
| Unidentified α proteobacterium; clone SAR 407; cluster SAR 11 | 91.4 | 1 | 784 | |
| Unidentified α proteobacterium; clone SAR 407; cluster SAR 11 | 86 | 1 | 628 | |
| Unidentified α proteobacterium; clone SAR 407; cluster SAR 11 | 89.6 | 1 | 668 | |
| Unidentified α proteobacterium; clone SAR 407; cluster SAR 11 | 95.2 | 1 | 598 | |
| Uncultured proteobacterium OCS126; cluster SAR 116 | 97.9 | 1 | 705 | |
| Prionitis lancedata gall symbiont | 92.4 | 1 | 613 | |
| Sulfitobacter pontiacus | 94.4 | 1 | 524 | |
| Uncultured proteobacterium OCS44; cluster SAR 86 | 95.5–97.1 | 3 | 549–792 | γ Proteobacteria |
| Coxiella burnetii | 90.3 | 1 | 514 | |
| Deleya salina | 86.5 | 1 | 655 | |
| Deleya salina | 88.7 | 1 | 563 | |
| Gelidibacter algens C8ST5 | 82.9 | 1 | 579 | Cytophagales |
| Unknown marine bacterioplankton; clone SAR 7 | 95.6 | 1 | 627 | Cyanobacteria |
Name in boldface given by authors depositing the sequence and corresponding to uncultured bacteria. ATT, attached; Free, free-living.
The range of similarities indicates the highest and lowest values of similarity to the best match when more than one clone with over 97% identity was found.
Clones with more than 97% nucleotide identity were considered identical.
Some clones show extremely low similarity to any known sequence, e.g., 82.4% over 579 nucleotides. However, it is remarkable that in this work the large majority of the retrieved sequences have similarities of over 90% to entries of either isolated strains or uncultured phylotypes.
A. macleodii cluster.
The 16S rRNA genes of 17 clones belonging to the A. macleodii cluster were completely sequenced. The relationships among the 17 sequences are shown in Fig. 3. Two clearly defined clusters were found. All of the sequenced clones appeared with A. macleodii in a cluster separated from other genera of the γ subdivision of Proteobacteria, such as Pseudoalteromonas and Shewanella. The sequences retrieved from the surface were closely related to A. macleodii IAM 12920T (within-cluster similarity, 98.4 to 99.1% [Table 6]), whereas DCM and 400-m clones formed a different subcluster (with similarities from 95.8 to 98.5% [Table 6]).
TABLE 6.
Similarity matrix (based on 1,492 nucleotide positions) among the clones related to A. macleodii and 16S rRNA gene sequences from representatives within the γ subdivision of Proteobacteria
| Organism | % Similarity to organism:
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
| 1. CLONE 1OD4OO | 96.8 | 97.0 | 97.1 | 97.1 | 96.1 | 99.1 | 97.1 | 97.0 | 96.5 | 97.5 | 98.0 | 97.4 | 98.0 | 88.5 | 88.5 | 89.1 | 88.7 | 88.7 | 98.4 | 98.4 | 96.5 | 97.9 | 88.9 | 87.3 | 86.6 | 86.3 | |
| 2. CLONE 10DCM | 95.8 | 94.9 | 95.7 | 94.1 | 97.2 | 95.2 | 94.8 | 95.0 | 95.4 | 96.3 | 95.4 | 96.1 | 86.3 | 86.3 | 86.8 | 86.3 | 87.5 | 96.4 | 96.8 | 94.3 | 95.7 | 86.5 | 85.2 | 84.9 | 84.6 | ||
| 3. CLONE 11SUR | 98.1 | 98.4 | 95.8 | 97.1 | 97.8 | 95.7 | 95.3 | 96.1 | 96.8 | 97.0 | 96.5 | 88.2 | 88.0 | 88.1 | 87.9 | 88.2 | 96.7 | 97.3 | 97.7 | 98.9 | 87.9 | 87.3 | 87.1 | 85.8 | |||
| 4. CLONE 13SUR | 97.7 | 95.5 | 97.2 | 97.4 | 95.7 | 95.0 | 95.8 | 96.3 | 97.0 | 96.6 | 88.2 | 88.1 | 88.7 | 88.6 | 88.2 | 96.7 | 96.9 | 97.3 | 99.1 | 88.2 | 87.8 | 87.6 | 86.6 | ||||
| 5. CLONE 14SUR | 95.2 | 97.5 | 98.2 | 95.8 | 95.6 | 96.5 | 97.1 | 97.0 | 96.7 | 88.4 | 88.2 | 88.3 | 88.2 | 88.4 | 96.8 | 97.6 | 97.3 | 98.6 | 88.2 | 87.3 | 87.2 | 86.0 | |||||
| 6. CLONE 17SUR | 96.3 | 95.1 | 94.7 | 94.0 | 94.4 | 95.0 | 95.2 | 95.3 | 86.4 | 86.5 | 86.8 | 86.3 | 86.8 | 95.8 | 95.7 | 94.7 | 96.2 | 86.7 | 85.4 | 85.0 | 84.0 | ||||||
| 7. CLONE 18DCM | 97.6 | 97.1 | 97.2 | 97.5 | 98.0 | 97.6 | 98.1 | 88.7 | 88.7 | 89.2 | 88.8 | 88.7 | 98.7 | 98.5 | 96.6 | 98.1 | 89.1 | 87.5 | 86.9 | 86.5 | |||||||
| 8. CLONE 18SUR | 96.2 | 95.1 | 96.2 | 96.6 | 96.8 | 97.0 | 88.3 | 88.3 | 88.2 | 88.1 | 88.3 | 97.0 | 97.3 | 97.1 | 98.4 | 88.1 | 87.4 | 86.9 | 85.5 | ||||||||
| 9. CLONE 20D400 | 95.2 | 95.3 | 96.1 | 96.0 | 97.4 | 86.8 | 86.8 | 87.3 | 86.8 | 87.0 | 96.9 | 96.8 | 94.9 | 96.5 | 87.2 | 86.3 | 85.4 | 84.8 | |||||||||
| 10. CLONE 24D400 | 95.3 | 96.1 | 95.1 | 96.0 | 86.3 | 86.2 | 87.1 | 86.7 | 86.4 | 96.4 | 96.7 | 94.3 | 95.7 | 87.1 | 85.1 | 84.7 | 84.7 | ||||||||||
| 11. CLONE 25D400 | 96.9 | 96.1 | 96.4 | 87.3 | 87.3 | 87.8 | 87.3 | 87.5 | 96.9 | 97.1 | 95.6 | 96.8 | 87.5 | 86.1 | 85.6 | 85.1 | |||||||||||
| 12. CLONE 2DCM | 97.1 | 97.3 | 87.5 | 87.5 | 88.0 | 87.5 | 87.7 | 98.2 | 98.0 | 95.7 | 97.3 | 87.7 | 86.4 | 85.9 | 85.6 | ||||||||||||
| 13. CLONE 9DCM | 97.4 | 87.8 | 87.6 | 87.8 | 87.6 | 87.9 | 97.2 | 97.0 | 96.6 | 97.8 | 87.6 | 86.5 | 86.3 | 85.1 | |||||||||||||
| 14. CLONE 20DCM | 88.0 | 88.0 | 88.6 | 88.2 | 88.1 | 97.7 | 97.7 | 96.0 | 97.4 | 88.3 | 87.0 | 86.3 | 85.8 | ||||||||||||||
| 15. P. haloplanktis subsp. haloplanktis | 99.3 | 96.8 | 96.4 | 99.1 | 88.0 | 88.2 | 87.4 | 89.2 | 96.5 | 90.8 | 89.5 | 88.9 | |||||||||||||||
| 16. P. atlantica | 97.3 | 96.7 | 99.7 | 88.0 | 88.2 | 87.3 | 89.1 | 96.9 | 91.1 | 89.7 | 89.3 | ||||||||||||||||
| 17. P. rubra | 98.4 | 97.2 | 88.6 | 88.5 | 87.3 | 89.1 | 98.4 | 90.4 | 88.6 | 89.3 | |||||||||||||||||
| 18. P. luteoviolacea | 96.6 | 88.2 | 88.2 | 87.2 | 89.0 | 97.6 | 90.3 | 89.0 | 89.3 | ||||||||||||||||||
| 19. P. haloplanktis subsp. tetraodonis | 88.2 | 88.3 | 87.4 | 89.2 | 96.7 | 90.8 | 89.5 | 89.3 | |||||||||||||||||||
| 20. CLONE 1D400 | 98.0 | 95.9 | 97.5 | 88.3 | 86.9 | 86.3 | 85.9 | ||||||||||||||||||||
| 21. CLONE 5D400 | 96.3 | 97.7 | 88.2 | 87.2 | 86.6 | 86.0 | |||||||||||||||||||||
| 22. CLONE 8D400 | 98.3 | 87.2 | 86.2 | 86.3 | 85.0 | ||||||||||||||||||||||
| 23. A. macleodii | 89.1 | 88.1 | 87.8 | 86.6 | |||||||||||||||||||||||
| 24. P. peptidysin | 90.1 | 88.5 | 89.2 | ||||||||||||||||||||||||
| 25. S. alga | 95.3 | 90.0 | |||||||||||||||||||||||||
| 26. S. putrefaciens | 90.5 | ||||||||||||||||||||||||||
| 27. Vibrio alginilyticus | |||||||||||||||||||||||||||
Morphology.
To assess the morphological types present in the attached assemblage versus those in the free-living assemblage, we examined by scanning electron microscopy the glass fiber filtrate retrieved on a 0.22-μm-pore-size filter (free living) and a small aliquot of the raw sample collected on a 3-μm-pore-size absolute filter (attached) from the surface sample. The differences were quite obvious (Fig. 1). The free-living assemblage was composed of small cells (with diameters well below 1 μm) with very different morphologies (Fig. 1C). On the other hand, the attached assemblage consisted of much larger cells, with diameters around 1 μm and with much less morphological diversity (Fig. 1A and B). Coccobacillary forms (as A. macleodii appears to be in culture) were abundant in the attached sample, although some elongated rods, spirals, and other shapes were observed as well. Aggregates and clustered cells, often bound to detrital material, were also abundant, as expected.
RISA.
The RISA technique permits separating different types of 16S rDNAs in a mixed-community DNA sample by simply using a primer located at the 5′ end of the 23S rRNA gene so that the spacer between the two genes is amplified together with a section of the 16S gene (8). We have amplified a region spanning from position 1055 (E. coli numbering) in the 16S rDNA to the beginning of the 23S rDNA (position 38), so the expected size range was 600 to 1,600 bp (the 16S rDNA fragment plus the spacer region). The PCR products corresponding to different organisms can be separated by size in an agarose gel, and then different bands can be excised from the gel and the 16S region can be sequenced to identify the organism. Figure 2 shows a Methaphor agarose gel in which the PCR products from the different depths and assemblages are shown. Used in this way, the technique is not very informative as a community-fingerprinting methodology due to the relatively small number of discernible bands. However, in the 400-m attached sample, in which we assume the diversity to be very low, two major PCR products of ca. 890 and 970 bp appeared; we will refer to them as RISA1 and RISA2, respectively. These bands were cloned, and five clones from each one were analyzed. The clones were partially sequenced with the B1055 internal primer (Table 1). The five RISA2 clones were found to belong to the A. macleodii cluster, with similarities between 86.6 and 96.5%. The other five clones sequenced from the band of ca. 890 bp (RISA1 clones) were principally related to the Pseudoalteromonas group of γ Proteobacteria. Two clones were similar to P. antarctica, with 89.9 and 98% similarities over 434 and 414 nucleotides, respectively. One clone had a 95.8% similarity to Pseudoalteromonas espejiana over 263 nucleotides. Finally, the last two clones were more distantly related, one giving a 92.9% identity in 282 nucleotides to an unidentified Cytophaga isolate (S23328 environmental sample) and the other giving a 93% identity in 422 nucleotides to Methylobacter sp. strain BB5. The sequence diversity retrieved with RISA was thus higher than that obtained from the sequencing of 18 random clones.
DISCUSSION
Community structure depth variation.
The sequencing results confirm the initial data obtained by ARDRA (1) showing a marked community structure variation with depth in the superficial stratified waters of the Western Mediterranean. There is also abundant information in the literature supporting the thesis that prokaryotic diversity in the open ocean varies with depth (17, 28, 44). Our results support this view, specifically for the first 100 m. The predominance of clones belonging to the α subclass of Proteobacteria at the surface has been described by several authors and seems to be a widespread occurrence (17, 38). As was found previously (17), the abundance of SAR11 A-1 and A-2 clusters decreases sharply from 0 to 40 m, although SAR11 G1 increases slightly with depth. Our clones also belong to a relatively restricted range of phylotypes, perhaps reflecting a clearly predominant ecotype adapted to live in the relatively warm waters of the upper layer of the ocean. However, just about 50 m below, at the DCM, the community structure changes significantly and a much larger representation of γ Proteobacteria was found. That could be an effect of the peculiar conditions of the DCM, with a much higher abundance of phytoplankton and perhaps a higher availability of organic nutrients, or it could simply reflect the change in physical conditions, mostly water temperature and light intensity. At 400 m the change is even more dramatic.
In terms of the phylogenetic groups retrieved, our results are not very different from others obtained in offshore oligotrophic waters of the Pacific and Atlantic Oceans, and they strengthen the opinion (38) that a relatively few major phylogenetic clusters are widespread and could predominate numerically in marine bacterioplankton, at least in temperate latitudes.
The community living in particulate matter or in aggregates is very different from the pelagic community (see below). However, this difference is much more pronounced in surface waters than in the deep sample, i.e., the pelagic and attached bacterial communities at 400 m seemed more similar, in terms of the phylotypes retrieved (as well as by community fingerprinting [Fig. 2]), than they are at the surface or at the DCM.
Attached versus free living.
Our results, as well as some previous reports (1–3, 7, 13, 29, 30), indicated that the bacterial community in aquatic environments is, in terms of species composition, markedly different for cells associated with particles and those that are free living. The attached community shows amazingly little diversity, with most clones belonging to the γ Proteobacteria and highly similar to the cultivated marine bacterium A. macleodii IAM 12920T, a strain isolated in the 1970s from coastal waters near Oahu, Hawaii (6). The pelagic assemblage is dominated by a more heterogeneous population that varies with depth. Here the best matches correspond to uncultivated entries only known by sequence, as shown in many previous studies. Amorphous aggregates that appear in natural aquatic environments can have various origins, e.g., bacteria attached to zooplankton fecal pellets, bacteria attached to each other by polymers, or bacteria attached to animal debris, such as the cast houses of mucous netfeeders (33). Aggregates may form a microhabitat providing protection from some bacteriovores (12) as well as nutrient abundance when advective flow through porous aggregates occurs (33).
In their work with marine snow (macroscopic detrital aggregates of >0.5-mm diameter) DeLong and coworkers found the majority of clones to be associated with Cytophaga, Planctomyces, or γ Proteobacteria. In our own results the particle-associated cells show much less diversity and belong almost exclusively to the γ Proteobacteria, although one cytophaga-related sequence was retrieved by RISA. Macroaggregates are of a very different nature and contain large amounts of detrital organic matter, which could explain the apparent discrepancy. Probably the most striking result of this work (Table 5) is the large representation of clones highly related specifically to the marine γ proteobacterium A. macleodii. This is the only described species of the genus Alteromonas, and it represents a rather isolated phylogenetic branch, as shown by comparison of its 16S rRNA with that of other marine isolates (19). It is a heterotrophic marine aerobe characterized by a wide range of substrates that can be used as sources of carbon and energy. Our results indicate that it could represent an important genus of marine bacteria specialized for particle (or aggregate)-associated niches. Other authors have already detected significant representation of this marine organism in samples from the Mediterranean (35) or the Atlantic (42). Their well-known capabilities as copiotrophs of relatively large size (0.7 to 1 μm in diameter and 2 to 3 μm long) (19) fit well with the adaptation to a high-nutrient and/or predator-free microenvironment. The differences found at the sequence level, and particularly the depth-dependent distribution of the sequences, are consistent with the existence of different species and/or ecotypes, adapted to different depths, within this cluster. One, including the original strain A. macleodii, would be predominant in surface waters, while the other, with no known cultivated representatives, is found mostly in deeper waters. The A. macleodii cluster is also well represented in the free-living fraction at 400 m. Apparently, that is contrary to the hypothesis formulated above. However, if we assume that the preferred habitat of the A. macleodii cluster is attached to particles, their presence in the free-living fraction at 400 m could simply reflect the fact that sinking particles are one of the main sources of bacterial biomass, including pelagic cells, in deep waters. The scarcity of nutrients would also make the growth associated with particles a good survival strategy at great depths.
We used RISA as an alternative methodology for community fingerprinting to compare the attached and free-living assemblages at different depths and for 16S rDNA sequence retrieval in the case of the 400-m attached subsample. The two techniques (RISA and cloning and sequencing of PCR-amplified 16S rDNA) include a PCR step that can bias their results. However, considering that the RISA primers are different, the similarity in the conclusions reached by both techniques is noteworthy. Although with the specific methodology used here RISA gave very poor fingerprinting results, probably due to the low resolution of the agarose gels, it is remarkable that, at least for the analyzed sample, it allows the recovery of more diversity than the random cloning and sequencing of 18 clones of PCR-amplified 16S rDNA.
Cultured versus uncultured.
Our results can shed some additional light on the classical discrepancy between culture- and PCR-based methods to describe biodiversity. On one hand, there is evidence indicating that most bacterial cells living in the ocean are not cultivated on standard marine media and belong to taxa distantly related to those well known from pure-culture studies (41). On the other hand, there are a number of studies in which it is shown that cultured strains of marine bacteria can represent significant fractions of the bacterial biomass in sea water (37, 39). The existence of two largely different assemblages sharing the habitat would be an important factor to consider in this kind of comparison. If a seawater sample is directly plated on an agar medium, the assemblage of large cells, belonging mostly to easily cultivated γ Proteobacteria attached to particles, will rapidly grow and override any other microbial group in the “culturable” harvest. The other assemblage, composed of smaller cells that live swimming in the medium, would be very difficult to retrieve if they had to compete. Both groups are probably of similar relevance in terms of biomass (or rRNA present in the sample) since, although in terms of cell numbers the free-living bacteria could be orders of magnitude more abundant, the attached fraction contains much larger cells. Therefore, depending on the technique used to collect biomass and/or detect the presence of one group or another, very different conclusions could be reached. For example, if total biomass is collected and hybridized to a probe for the attached (cultured) representatives (39), a good proportion of the hybridization signal could be accounted for by this fraction, although numerically they represent a very minor component of the community.
ACKNOWLEDGMENTS
This work was supported by European Commission Grant ENV4-CT96-0218, ELOISE project 077.
We are grateful to K. Hernández for secretarial assistance and to S. Ingham for graphics.
REFERENCES
- 1.Acinas S G, Rodríguez-Valera F, Pedrós-Alió C. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rDNA. FEMS Microbiol Ecol. 1997;24:27–40. [Google Scholar]
- 2.Albright L J, MacCrae S K, May B E. Attached and free-floating bacterioplankton in Howe Sound, British Columbia, a coastal marine fjord embayment. Appl Environ Microbiol. 1986;51:614–621. doi: 10.1128/aem.51.3.614-621.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfreider A, Pernthaler J, Amman R, Sattler B, Glöckner F O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alldredge A L, Cole J J, Caron D A. Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol Oceanogr. 1986;31:68–78. [Google Scholar]
- 5.Amman R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann L, Baumann P, Mandel M, Allen R D. Taxonomy of aerobic marine eubacteria. J Bacteriol. 1972;110:402–429. doi: 10.1128/jb.110.1.402-429.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidle K D, Fletcher M. Comparison of free-living and particle-associated bacteria communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl Environ Microbiol. 1995;61:944–952. doi: 10.1128/aem.61.3.944-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cammen L M, Walker J A. Distribution and activity of attached and free-living suspended bacteria in the Bay of Fundy. Can J Fish Aquat Sci. 1982;39:1655–1663. [Google Scholar]
- 10.Caron D A, Davis P G, Madin L P, Sieburth J M. Heterotrophic bacteria and bacteriovorous protozoa in oceanic macroaggregates. Science. 1982;218:795–797. doi: 10.1126/science.218.4574.795. [DOI] [PubMed] [Google Scholar]
- 11.Cullen J J. The deep chlorophyll maximum: comparing vertical profiles of chlorophyll a Can. J Fish Aquat Sci. 1982;39:791–803. [Google Scholar]
- 12.Curds C R. The ecology and role of protozoa in aerobic sewage treatment processing. Annu Rev Microbiol. 1982;36:27–46. doi: 10.1146/annurev.mi.36.100182.000331. [DOI] [PubMed] [Google Scholar]
- 13.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacteria assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 14.Eppley R W, Swift E, Redalje D G, Landry M R, Hass L W. Subsurface chlorophyll maximum in August-September 1985 in the CLIMAX area of the North Pacific. Mar Ecol Prog Ser. 1988;42:289–301. [Google Scholar]
- 15.Estrada M, Marrasé C, Latasa M, Berdalet E, Delgado M, Riera T. Variability of deep chlorophyll maximum characteristics in the Northwestern Mediterranean. Mar Ecol Prog Ser. 1993;92:289–300. [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evaluation. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuhrman J A, Comeau D E, Hagström A, Chan A M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier G, Gauthier M, Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 20.Giovannoni S J, Cary S C. Probing marine systems with ribosomal RNAs. Oceanography. 1993;6:95–104. [Google Scholar]
- 21.Giovannoni S J, Rappé M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 24.Herbland A, Le Borgne R, Le Bouteiller A, Voituriez B. Structure hydrologique et production primaire dans l’Atlantique tropical oriental. Oceanogr Trop. 1983;17:15–25. [Google Scholar]
- 25.Irriberry J, Unanue M, Ayo B, Barcina I, Egea L. Bacterial production and growth rate estimation from [3H]thymidine incorporation for attached and free-living bacteria in aquatic systems. Appl Environ Microbiol. 1990;56:483–487. doi: 10.1128/aem.56.2.483-487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 27.Kimor B, Berman T, Schneller A. Phytoplankton assemblages in the deep chlorophyll maximum layers off the Mediterranean coast of Israel. J Plankton Res. 1987;9:433–443. [Google Scholar]
- 28.Lee S, Fuhrman J A. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- 29.Lochte K, Turley C M. Bacteria and cyanobacteria associated with phytodetritus in the deep sea. Nature. 1988;333:67–69. [Google Scholar]
- 30.Logan B E, Hunt J R. Advantages to microbes of growth in permeable aggregates in marine systems. Limnol Oceanogr. 1987;32:1034–1048. [Google Scholar]
- 31.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Foge K, Blandy J, Woese C R. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Murcia A J, Acinas S G, Rodríguez-Valera F. Evaluation and prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;17:247–256. [Google Scholar]
- 33.Morita R Y. Bacteria in oligotrophic environments. New York, N.Y: Chapman & Hall; 1997. [Google Scholar]
- 34.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 35.Muyzer, G. Personal communication.
- 36.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinhassi J, Zweifel U L, Hagström Å. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA gene clones from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 39.Rehnstam A S, Bäckman S, Smith D C, Azam F, Hagström Å. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol. 1993;102:161–166. [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Maarel. Personal communication.
- 43.Weidner S, Arnold W, Pühler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright T D, Vergin K L, Boyd P W, Giovannoni S J. A novel δ-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]