Abstract
N6-methyladenosine (m6A) is the most common modification in eukaryotic RNAs. Accumulating evidence shows m6A methylation plays vital roles in various biological processes, including muscle and fat differentiation. However, there is a lack of research on lncRNAs’ m6A modification in regulating pig muscle-fiber-type conversion. In this study, we identified novel and differentially expressed lncRNAs in oxidative and glycolytic skeletal muscles through RNA-seq, and further reported the m6A-methylation patterns of lncRNAs via MeRIP-seq. We found that most lncRNAs have one m6A peak, and the m6A peaks were preferentially enriched in the last exon of the lncRNAs. Interestingly, we found that lncRNAs’ m6A levels were positively correlated with their expression homeostasis and levels. Furthermore, we performed conjoint analysis of MeRIP-seq and RNA-seq data and obtained 305 differentially expressed and differentially m6A-modified lncRNAs (dme-lncRNAs). Through QTL enrichment analysis of dme-lncRNAs and PPI analysis for their cis-genes, we finally identified seven key m6A-modified lncRNAs that may play a potential role in muscle-fiber-type conversion. Notably, inhibition of one of the key lncRNAs, MSTRG.14200.1, delayed satellite cell differentiation and stimulated fast-to-slow muscle-fiber conversion. Our study comprehensively analyzed m6A modifications on lncRNAs in oxidative and glycolytic skeletal muscles and provided new targets for the study of pig muscle-fiber-type conversion.
Keywords: lncRNA, m6A methylation, muscle-fiber-type conversion, pig
1. Introduction
Skeletal muscle is a heterogeneous tissue, composed of different types of muscle fibers, with distinct contractile and metabolic characteristics [1]. Myosin heavy chain (MyHC) is the major contractile protein of skeletal muscle cells [2]. According to the electrophoretic analysis results of myosin heavy chain isoforms in adult mammals, muscle fibers are mainly divided into type I (MyHC I) and type II (MyHC IIa, MyHC IIb, and MyHC IIx/d) muscle fibers, and they are distinguished by their function, biochemical characteristics, and morphological characteristics [1,3,4,5]. Type I muscle fibers are rich in myoglobin and mitochondrial and mainly use lipids as an energy source and carry out aerobic respiration, while type IIb muscle fibers have a high content of glycogen and glucose, and mainly depend on glycolysis to obtain energy [6]. The other two types of muscle fibers, IIa and IIx/d, have intermediate biophysical properties [7]. In livestock, the composition of muscle fiber types is closely related to meat quality [8,9]. For example, Kim et al. reported that an increase in the proportion of IIb fiber decreases muscle pH and increases muscle cooking loss and lightness [10]. Different types of muscle fibers can transform into each other, and mutual conversion between MyHC subtypes follows an obligatory order: I ↔ IIx ↔ IIa ↔ IIb [11]. Muscle-fiber-type conversion is regulated by multiple signaling factors, such as myogenic regulation factors (MRFs) [12,13,14,15], nuclear factor of activated T cells (NFAT) [16], insulin-like growth factors (IGFs) [17,18], peroxisome proliferative activated receptor-γ activation of auxiliary factor 1 Alpha (PGC-1α) [19], and other unknown regulatory factors.
Long noncoding RNAs (LncRNAs), a class of RNAs with a length greater than 200 nucleotides that are unable to code proteins, have been found to participate in a variety of biological processes, including myogenic differentiation [20,21]. LncRNAs have also been reported to play an important regulatory role in muscle fiber conversion in livestock. For example, lncRNA MyHC-IIa/X-as promotes MyHC-IIX by sponging miR-130b, maintaining the fast fiber phenotype [22]. LncRNA-FKBP1C regulates muscle-fiber-type switching by affecting the stability of MYH1B [23]. LncRNAs function through diverse mechanisms. In the nucleus, lncRNAs act on regulating the transcription program via chromatin interaction and remodeling and establishing spatial organization of nuclear compartments through scaffolding. In the cytoplasm, lncRNAs mediate signal transduction pathways, posttranscriptional control of gene expression, and translational processes [24].
N6-methyladenosine (m6A) is the most abundant internal modification on mRNAs [25]. It is a reversible RNA modification that prefers to occur on a consensus motif RRACH (in which R represents A or G, and H represents A, C, or U) near mRNA stop codon and in long internal exons [25]. m6A is introduced by a nuclear methyltransferase complex, including METTL3, METTL14, WTAP, and RBM15 (or RBM15B), which have been designated as m6A “writers” [26], and can be removed by m6A demethylases, such as FTO and ALKBH5, which have been designated as m6A “erasers” [26]. Furthermore, multiple proteins, including YTHDF1, YTHDF2, YTHDF3, and YTHDC1, acting as m6A “readers”, can bind to modified sites and regulate a variety of post-transcriptional processes [27]. The functions of m6A in mRNA include nuclear transport, splicing, stability, and translation [28]. Many lncRNAs have also been found to be modified by m6A, and m6A modification affects these lncRNAs’ expression levels and mechanisms of action. For example, MALAT1 can regulate the variable splicing and gene transcription of pre-mRNA. It is found that MALAT1 contains multiple m6A modification sites. m6A can change the binding ability of MALAT1 to protein by changing its RNA structure, and as such, affect the splicing and transcription function of MALAT1 [29]. Xist is also highly methylated by m6A, knocking out m6A methyltransferase METTL3 inhibits Xist-mediated X chromosome silencing [30]. However, the effect of m6A modification on lncRNAs is far from clear. Recently, using methylated-RNA immunoprecipitation sequencing (MeRIP-seq), Xie et al. revealed the temporal expression profile and m6A methylation status of lncRNAs during skeletal myogenesis [31]. However, little is known about the m6A methylation status of lncRNAs and the regulation of m6A on lncRNAs during muscle-fiber-type conversion.
In this study, we identified 5607 novel lncRNAs in oxidative and glycolytic skeletal muscles using RNA-seq, and further disclosed a large number of m6A sites and specific m6A modification patterns in lncRNAs using MeRIP-seq. We highlight the potential role of m6A-modified lncRNAs in muscle fiber conversion by regulating the expression of the muscle-development-related gene in cis. Our results reveal the tissue-specific expression and m6A methylation status of lncRNAs in different types of muscle fibers, which provides a basic reference for further study of the regulation of m6A on lncRNAs.
2. Results
2.1. Difference in Phenotypic Traits and m6A-Related Gene Expression Levels between Oxidative and Glycolytic Skeletal Muscles
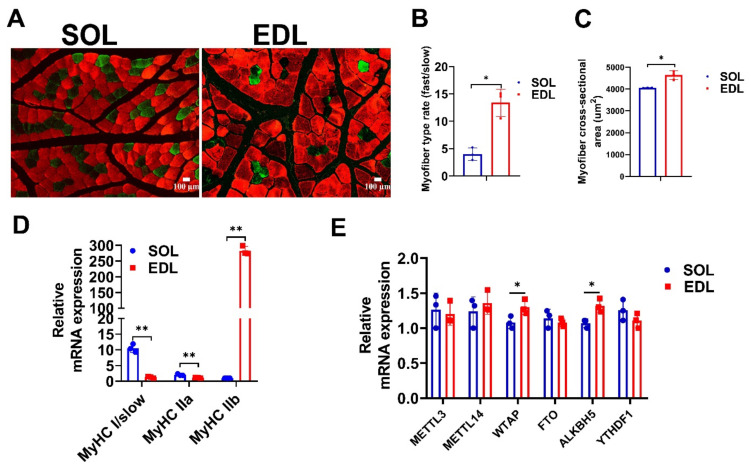
First, we detected the phenotypic traits of soleus (SOL) and extensor digitorum longus (EDL) muscles of a 6-month-old Duroc pig. The immunofluorescence staining results of the glycolytic muscle-fiber marker MyHC IIb and the oxidative muscle-fiber marker MyHC I showed that the composition of glycolytic muscle fiber in EDL was almost three times that of SOL (Figure 1A,B), and the average cross-sectional area of individual myofiber in EDL was larger than that of SOL (Figure 1C). Moreover, the mRNA expression of MyHC IIb was higher in EDL than in SOL, while MyHC I and MyHC IIa mRNA expressions were higher in SOL than in EDL (Figure 1D). The above results indicate that EDL is a typical glycolytic skeletal muscle, while SOL is a typical oxidative skeletal muscle. We also detected the expression of m6A writers (METTL3, METTL14, and WTAP), erasers (FTO and ALKBH5), and read (YTHDF1) genes in SOL and EDL. The qPCR results showed that the mRNA levels of WTAP and ALKBH5 were extraordinarily lower in SOL than in EDL (Figure 1E).
Figure 1.
Detection of expression levels of muscle-fiber-type related genes and m6A-related genes. (A) Representative images of MyHC I (green) and MyHC IIb (red) immunofluorescent staining in SOL and EDL muscle sections. Scale bar: 100 μm. (B) Quantification of the cross-sectional area of SOL and EDL muscle sections in (A). (C) Statistical analysis of fast and slow muscle fibers of SOL and EDL muscles in (A). (D) qPCR experiment detects the expression level of MyHC I, MyHC IIa, and MyHC IIb in SOL and EDL. (E) qPCR experiment detects the expression level of m6A-related genes METTL3, METTL14, WATAP, FTO, ALKBH5, and YTHDF1 in SOL and EDL. The relative RNA expressions were standardized to that of the control gene β-actin. Data represent mean ± SD of three independent biological replicates. * p < 0.05, ** p < 0.01.
2.2. Dynamic Changes of lncRNAs’ Transcriptome in Oxidative and Glycolytic Skeletal Muscles
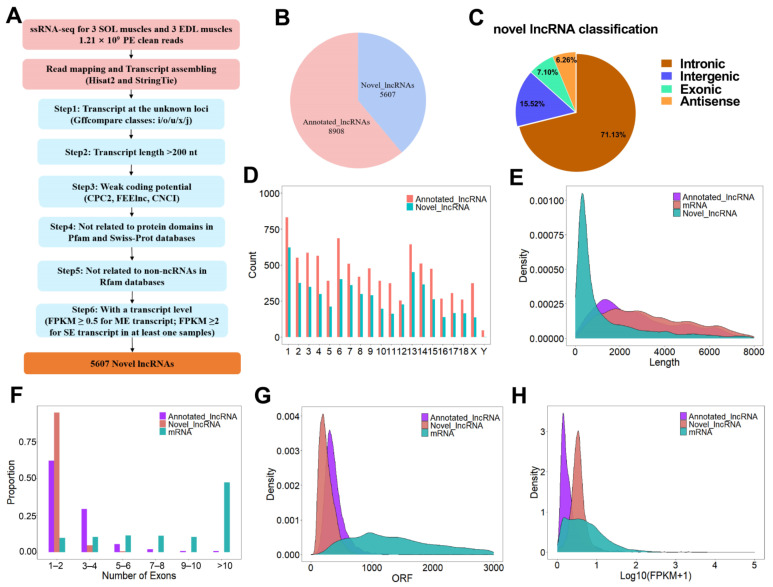
LncRNAs have recently been reported to have important roles in regulating muscle development and muscle-fiber conversion. To understand the genetic basis of lncRNA-regulated skeletal-muscle-fiber development, we performed RNA sequencing (MeRIP-seq input library) on the SOL and EDL muscles of 6-month-old Duroc pigs. A total of 530,385,924 raw reads were generated from three SOL and three EDL muscle samples. After filtering out reads containing adapters and reads of low quality, a total of 445,953,950 properly paired mapped reads were mapped to the pig reference genome (Sscrofa 11.1.94), accounting for 84.08% of the raw reads. After a series of filtering steps, shown in Figure 2A, a total of 5607 novel lncRNAs were obtained in SOL and EDL (Figure 2B and Table S1). It is worth noting that 71.13% (3988), 7.1% (398), 15.52% (870), or 6.26% (351) of the novel lncRNAs were intronic lncRNAs, exonic lncRNAs, lincRNAs (long intergenic non-codingRNAs), or antisense lncRNAs, respectively (Figure 2C). An analysis of genomic location distribution indicated that the lncRNAs were distributed on all chromosomes, without obvious chromosome preference (Figure 2D). To understand the characterization of the identified novel lncRNAs, we made comparisons of the gene structure and expression patterns among novel lncRNAs, annotated lncRNAs, and mRNAs. The results showed that the novel lncRNAs and annotated lncRNAs shared common characteristics, including shorter transcript length, fewer exons, shorter ORF, and lower expression compared with mRNAs (Figure 2E–H).
Figure 2.
Identification and characterization of lncRNAs identified in SOL and EDL. (A) Bioinformatics pipeline for novel lncRNAs identification. nt: nucleotide; ME: multiple exons; SE: single exons. (B) Pie chart showing the proportion of novel lncRNAs identified in our study and previously annotated pig lncRNAs. (C) Classification of novel lncRNAs identified in this study. (D) Chromosome distribution of lncRNAs. (E–G) Distribution of transcript lengths (E), exon number (F), ORF length (G) in lncRNAs and mRNAs. (H) Distribution of expression level (showing log10 (FPKM + 1)) in lncRNAs and mRNAs.
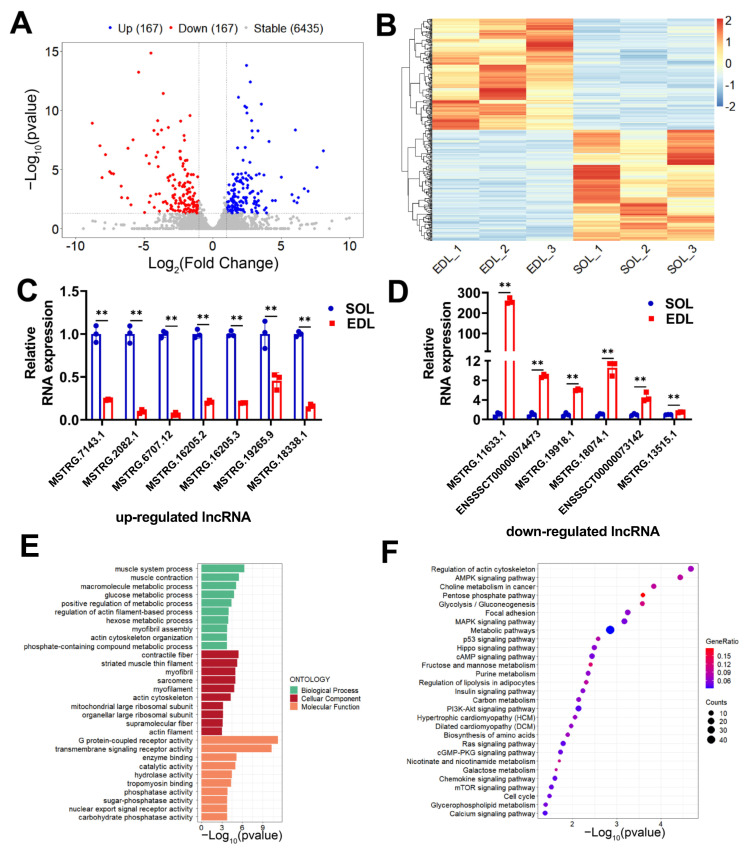
A total of 6769 (including 5607 novel lncRNAs and 1162 annotated lncRNAs) expressed lncRNAs were used for further differential expression analysis (Table S1). We identified 334 differentially expressed lncRNAs (DE lncRNAs) between SOL and EDL (Table S2), among which, 167 were upregulated and 167 were downregulated in SOL (Figure 3A). Hierarchical clustering analysis showed that the expression patterns of DE lncRNAs were, as expected, considerably different between the SOL group and EDL group, while there were small differences among the three biological replicates in each group (Figure 3B). To verify the reliability of our RNA-seq results, 13 DE lncRNAs, including 7 upregulated and 6 downregulated, were randomly selected to validate their expression differences between SOL and EDL by qPCR. The results showed that all detected lncRNAs displayed either higher or lower transcript abundance in SOL or EDL, which was consistent with that of the RNA-seq results (Figure 3C,D). Given that lncRNAs have no encoding potential, their functions are achieved through the regulation of target genes. Hence, we determined the cis target genes to explore the potential function of DE lncRNAs. GO enrichment and KEGG pathway analysis on the cis target genes of DE lncRNAs were conducted. GO analysis results suggested the important role of DE lncRNAs in regulating muscle-fiber switching, as numerous processes related with skeletal-muscle-fiber properties were enriched. For example, muscle system processes, muscle contraction, glucose metabolic process, and myofibril assembly were significantly enriched in the BP term; contractile fiber, myofibril, and mitochondrial large ribosomal subunits were remarkably enriched in the CC term, and G protein-coupled receptor activity, transmembrane signaling receptor activity, enzyme binding, catalytic activity, and phosphatase activity were observably enriched in the MF term (Figure 3E and Table S3). KEGG pathway enrichment analysis showed that pathways related to muscle-fiber properties were enriched, including glycolysis/gluconeogenesis, mTOR-signaling pathways, metabolic pathways, insulin-signaling pathways, AMPK-signaling pathways, and PI3K-Akt-signaling pathways (Figure 3F and Table S3). Taken together, our study identified numerous novel and differentially expressed lncRNAs in different muscle-fiber types and highlighted the potential regulation of lncRNAs in muscle-fiber conversion.
Figure 3.
Screening and functional enrichment analysis of DE lncRNAs in SOL compared with EDL. (A) Volcano plot showing the expression profile of DE lncRNAs between SOL and EDL. Blue dots \\SOL; grey dots represent lncRNAs with stable expression in both tissues. (B) Hierarchical clustering heatmap showing the expression profile of DE lncRNAs between SOL and EDL. (C,D) 7 upregulated (C) and 6 downregulated (D) DE lncRNAs were randomly selected and verified via qPCR. (E) GO enrichment analysis of the nearest target genes of DE lncRNAs. (F) KEGG pathway analysis of the nearest target genes of DE lncRNAs. The relative RNA expressions were standardized to that of the control gene β-actin. Data represent mean ± SD of three independent biological replicates. ** p < 0.01.
2.3. Overall Features of lncRNAs m6A Methylation in Oxidative and Glycolytic Skeletal Muscles
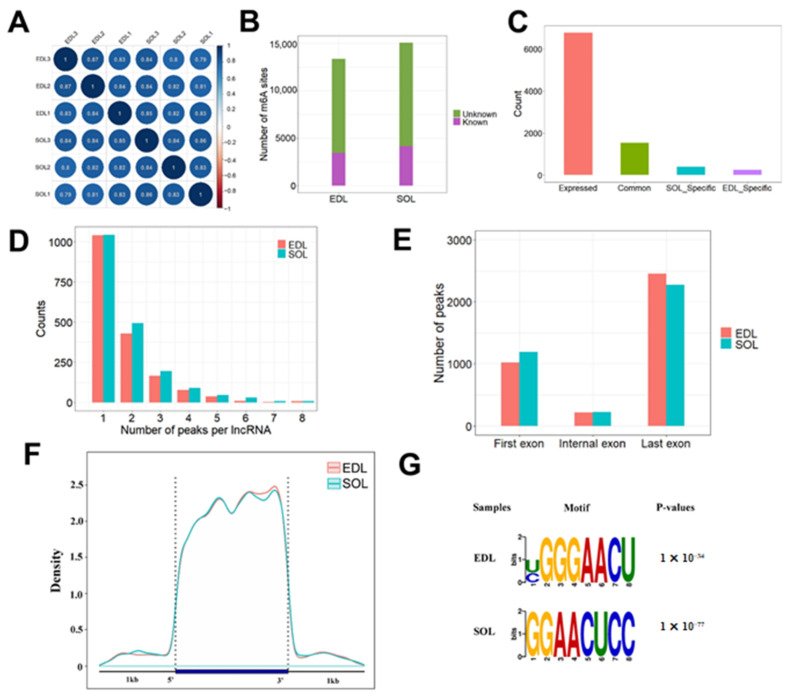
To reveal the m6A methylation profile of lncRNAs expressed in oxidative and glycolytic skeletal muscles, we performed methylated-RNA immunoprecipitation and sequencing (MeRIP-seq) and obtained an average of 80,387,850 properly paired mapped reads per sample, with a greater than 89% mapping rate. Pearson’s correlation analysis showed that the three biological replications of the same samples were clustered well (Figure 4A), suggesting that our sequencing data within the same group were highly consistent. In total, we identified 15,071 and 13,380 m6A peaks (hereafter m6As) in SOL and EDL, respectively. Among these identified peaks, 10,925 m6As in SOL and 9949 m6As in EDL were novel m6As that have not been reported in RMbase (Figure 4B). We further focused on the m6As located in lncRNAs. We identified 2888 m6As in 1925 expressed lncRNAs in SOL and 2527 m6As in 1782 expressed lncRNAs in EDL; among them, 1534 lncRNAs were m6A-modified in two tissue types, and 391 and 248 lncRNAs were only m6A-modified in SOL and EDL, respectively (Figure 4C and Table S4). Among the 391 lncRNAs specifically modified by m6A in SOL, 11 lncRNAs were specifically expressed in SOL and 9 lncRNAs were specifically expressed in EDL. Among the 248 lncRNAs specifically modified by m6A in EDL, 13 lncRNAs were specifically expressed in SOL and 23 lncRNAs were specifically expressed in EDL (Table S4). We checked the number of m6As in each lncRNA and found that most lncRNAs contained 1–2 m6As in SOL and EDL tissues (Figure 4D). We then analyzed the peak distribution for lncRNAs, and found that m6As were preferentially enriched in the last exon of the lncRNAs (Figure 4E). Furthermore, the distribution pattern of m6A on the lncRNA transcripts with a 1 kb flanking region was roughly uniform, but with a slight increase in the 3′ end (Figure 4F), as reported in a previous study [32].
Figure 4.
Overall features of lncRNAs m6A methylation in oxidative and glycolytic skeletal muscles. (A) Pearson correlation analysis of MeRIP-seq data between each pair of samples. The samples were hierarchically clustered. The intensity of the color represented the similarity, and the black outlining was drawn according to the hierarchical cluster. (B) The number of m6As found in each tissue. Any m6A site that did not overlap with any site in RMBase were labelled as ‘unknown’. (C) Histogram showing the distribution of lncRNAs with m6As. Expressed: expressed lncRNAs; Common: lncRNAs with m6As in two tissues; SOL-specific: lncRNAs with m6As in SOL only; EDL-specific: lncRNAs with m6As in EDL only. (D) The distribution of peak number of m6A-modified lncRNAs. (E) Metagene profile of enrichment of m6As in lncRNAs. (F) Histogram showing the distribution of m6As in the three regions of lncRNAs. (G) The enriched consensus motif of m6As in lncRNAs.
To identify the common sequence elements of the m6A peaks on lncRNAs, we used HOMER software to identity a consensus motif with default parameters that will generate motifs ranging from three to eight bases. We found the enriched motif containing the well validated consensus m6A motif RRACH [33,34] in the SOL and EDL groups (Figure 4G), which were similar to the previously identified m6A motif in the lncRNAs [31]. All the above analyses could provide a fundamental reference for the m6A epitranscriptome in skeletal muscle with different types of muscle fiber.
2.4. Association Analysis of m6A with lncRNAs Expression
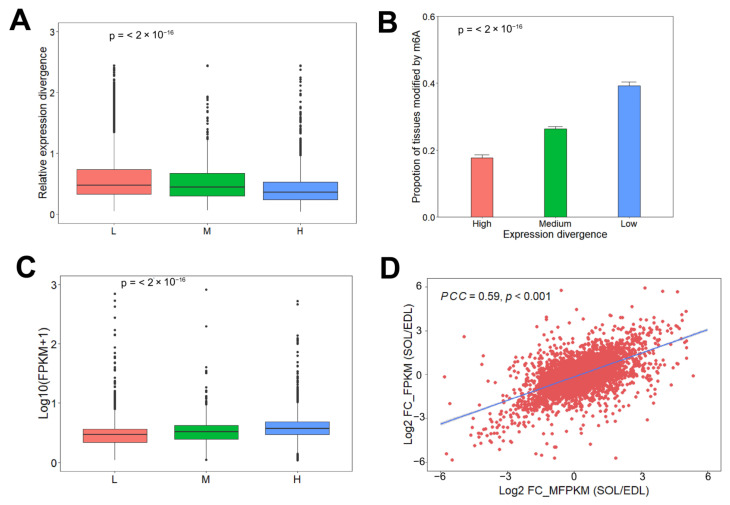
To further examine the role of m6A methylation in lncRNAs’ expression dynamic, we classified the 6769 expressed lncRNAs in SOL and EDL into three sets, according to the proportion of transcripts with m6A peaks in two tissues, L: lncRNAs without m6A peaks in two tissue types, M: lncRNAs with m6A peaks in only one tissue type, and H: lncRNAs with m6A peaks in both tissue types. We found that lncRNAs in the H sets had the most stable expression levels, while lncRNAs in the L sets showed the least stable expression levels (Figure 5A). We next clustered the 6769 lncRNAs into Low, Median, and High groups, according to the quantile of the expression divergence. We found that lncRNAs with more stable expression levels were also more likely to have a higher proportion of transcripts modified by m6A methylation in two tissue types (Figure 5B). These results indicated that m6A methylation was positively correlated with lncRNAs’ expression homeostasis. Furthermore, we explored the association between m6A methylation abundance and lncRNAs’ expression levels. We found that the expression level of m6A-modified lncRNAs was higher than that of non-m6A-modified lncRNAs, and lncRNAs with m6A methylation in both tissue types had the highest expression levels (Figure 5C). We then examined the correlation of lncRNAs’ expression levels with m6A methylation levels; Pearson correlation coefficient analysis results showed that lncRNAs’ m6A levels were positively correlated with their expression levels (R = 0.72, p < 0.001) (Figure 5D). These results demonstrate that m6A may play a role in lncRNAs transcription activation.
Figure 5.
Association analysis of m6A with lncRNAs expression. (A) Relative expression divergence of lncRNAs in the L set (4596 lncRNAs), M set (639 lncRNAs) and H set (1534 lncRNAs). Significance was evaluated by a two-sided Mann–Kendall trend test, p < 2.2 × 10−16. (B) The lncRNAs were clustered into Low (1693 lncRNAs), Median (3383 lncRNAs), and High (1693 lncRNAs) groups according to the quantile of the expression divergence. Significance was evaluated by a two-sided Mann–Kendall trend test, p < 2.2 × 10−16. (C) Expression level in lncRNAs in the L set (4596 lncRNAs), M set (639 lncRNAs) and H set (1534 lncRNAs). Significance was evaluated by a two-sided Mann–Kendall trend test, p < 2.2 × 10−16. (D) Scatter plot showing the positive correlation between m6A levels and expression level of lncRNAs between EDL and SOL. FC: Fold change; PCC: Pearson correlation coefficient.
2.5. Conjoint Analyses of MeRIP-seq and RNA-seq Data
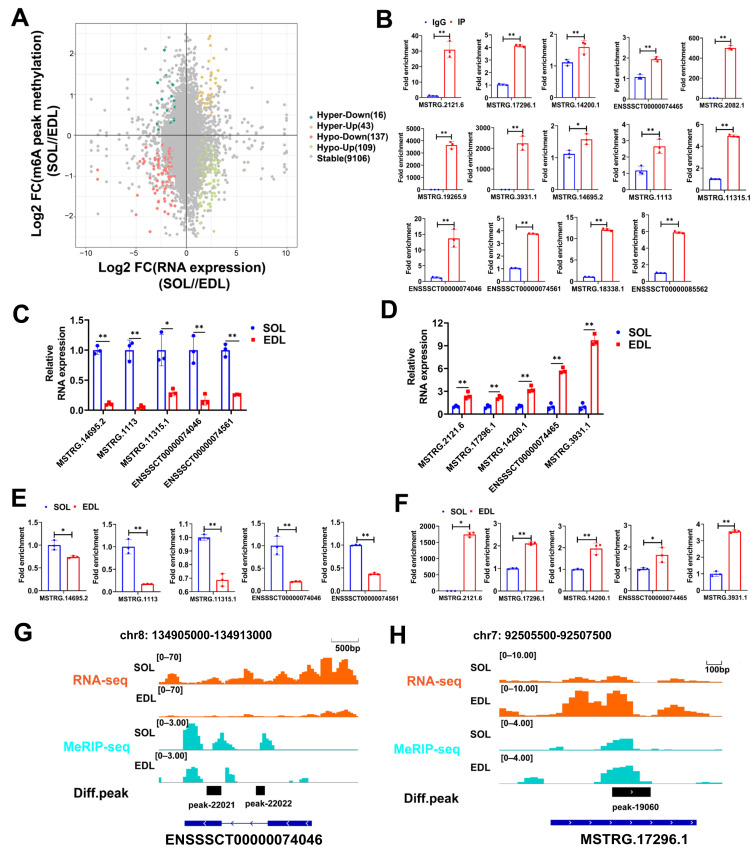
To explore the potential role of m6A on lncRNAs, we performed conjoint analysis of MeRIP-seq and RNA-seq data. Of the 334 DE lncRNAs in Figure 3A, 305 (88.9%) lncRNAs were also shown to be differentially m6A modified, and these lncRNAs were referred to as dme-lncRNAs. A high proportion of DE lncRNAs modified by m6A indicated that m6A might participate in muscle-fiber-type conversion by regulating the transcription of lncRNAs. Among the 305 dme-lncRNAs, 152 upregulated lncRNAs were significantly hyper-methylated (43; Hyper-Up) or hypo-methylated (109; Hypo-Up) in SOL, and153 downregulated lncRNAs were significantly hyper-methylated (16; Hyper-Down) or hypo-methylated (137; Hypo-Down) in SOL (Figure 6A). To confirm the significantly differentially m6A-modified lncRNAs, we randomly selected 14 significantly differentially m6A-modified lncRNAs and performed methylated RNA immunoprecipitation, followed by real-time PCR (MeRIP-qPCR). The results showed that all selected lncRNAs were remarkably enriched in the m6A group, compared to the IgG control (Figure 6B). Moreover, we compared the m6A enrichment and expression differences of the above 14 lncRNAs in SOL and EDL. We finally validated 10 (5 Hyper-Up lncRNAs and 5 Hypo-Down lncRNAs) out of 14 lncRNAs, with the same change in the m6A and expression levels between SOL and EDL (Figure 6C–F), which was consistent with the analysis results, indicating that the analysis results were highly reliable. As shown in the IGV, lncRNA ENSSSCT00000074046 had higher m6As enrichment in SOL, and its expression level was also higher in SOL (Figure 6G), while lncRNA MSTRG. 17296.1 had a higher m6As enrichment in EDL, and its expression level was also higher in EDL (Figure 6H). Taken together, these findings suggest that the m6A enrichment change in lncRNAs may affect their expression levels.
Figure 6.
Conjoint Analyses of MeRIP-Seq and RNA-Seq Data. (A) Four-quadrant graph showing the distribution of lncRNAs with a marked change in both RNA expressions and m6A methylation levels in SOL compared with EDL. Different colors were used to identify representative lncRNAs. (B) MeRIP-qPCR validated 14 m6A-methylated lncRNAs. IgG was used as negative control. (C,D) MeRIP-qPCR results showed that 5 Hyper-Up lncRNAs of the 14 m6A-methylated lncRNAs have higher m6As enrichment in SOL than EDL (C), 5 Hypo-Down lncRNAs of the 14 m6A-methylated lncRNAs have higher m6As enrichment in EDL than SOL (D); data were normalized by IgG. (E) qPCR results showed that the expression of 5 Hyper-Up lncRNAs were increased in SOL compared with EDL. (F) qPCR results showed that the expression of 5 Hypo-Down lncRNAs were decreased in SOL compared with EDL. (G,H) Genome browser tracks showing RNA-seq (orange) and MeRIP-seq (light blue) data at lncRNA ENSSSCT00000074046 (G, scale bar: 500 bp) and MSTRG. 17296.1 (H, scale bar: 100 bp) loci in SOL and EDL. The relative RNA expressions were standardized to that of the control gene β-actin. Data represent mean ± SD of three independent biological replicates. * p < 0.05; ** p < 0.01.
2.6. Identification of Key Muscle-Fiber-Types-Related lncRNAs
Previous studies have shown that different muscle fibers are closely related to animals’ economic traits, such as meat quality and muscle growth [35,36]. To identify m6A-modified lncRNAs that regulate muscle-fiber-type conversion, we conducted an enrichment analysis by mapping dme-lncRNAs to the QTL regions in pigs. The animal QTLdb (PigQTLdb) database has a total of 31,455 pig quantitative trait loci (QTLs), representing 695 different traits [37]. We found 162 dme-lncRNAs were located in 2919 QTLs; among them, “meat and carcass traits”-related QTLs accounted for the largest proportion (68.8%). In addition, QTLs related to “healthy traits”, “production traits”, “reproduction traits”, and “exterior traits” accounted for 14.1%, 10.5%, 3.8%, and 2.8%, respectively. We mainly focused on QTLs associated with meat-associated traits, and we identified 49 dme-lncRNAs that were associated with “meat and carcass traits”; among them, some were closely associated with muscle fiber characteristics, such as meat color, fatty acid content, pH, flavor, and enzyme activity.
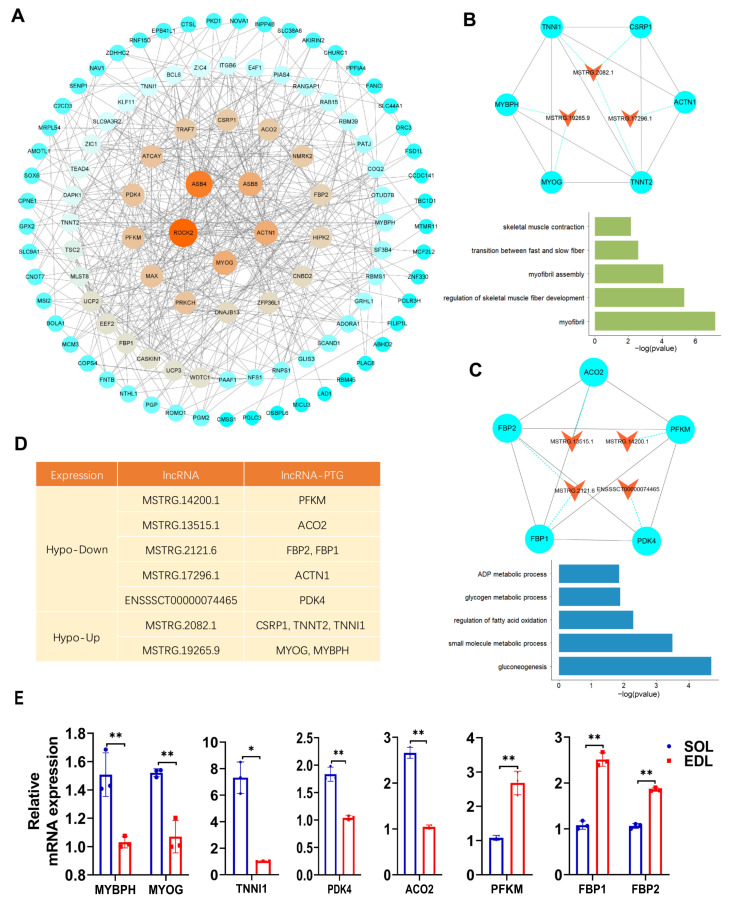
It has been reported that lncRNAs can regulate their neighboring gene transcription in cis [38]. To further explore the potential function of the 49 dme-lncRNAs, we identified their potential cis target genes (PTGs) by searching for expressed protein-coding genes around 100 kb of these lncRNAs. In this way, we found 119 cis PTGs for 49 dme-lncRNAs. Then, we performed protein–protein interaction (PPI) analysis for 119 cis PTGs, and found the muscle-related genes, such as MYOG, ACTN1, that were the key codes in the PPI network (Figure 7A), suggesting these dme-lncRNAs may regulate muscle-related genes to participate in the process of muscle-fiber-type conversion. Furthermore, we screened out the nodes and conducted MCODE analysis to identify highly connected clusters in a large PPI network (Table S5). We selected the genes in the top two clusters for GO enrichment analysis and found that they were significantly enriched in the pathways related to muscle-fiber properties and energy metabolism (Figure 7B,C). We investigated six PTGs involved in muscle-fiber property pathways, corresponding to three dme-lncRNAs, MSTRG.2082.1, MSTRG.19265.9, and MSTRG.17296.1, and five PTGs involved in energy metabolism pathways, corresponding to four dme-lncRNAs, MSTRG.13515.1, MSTRG.14200.1, MSTRG.2121.6, and ENSSSCT00000074465 (Figure 7D and Table S6). The cis PTGs of seven dme-lncRNAs might play an important role in the conversion of muscle-fiber types. In particular, MSTRG.2082.1 was involved in the regulation of TNNI1, which is a marker gene of slow muscle fiber. MSTRG.13515.1 was involved in the regulation of ACO2, which is a part of the citric acid cycle and metabolizes α-ketoglutarate, a product of glutamine oxidation [39]. Furthermore, we detected the mRNA expression of PTGs of these seven dme-lncRNAs in SOL and EDL; the qPCR results showed that the mRNA expression of MYBPH, MYOG, TNNI1, PDK4, and ACO2 was upregulated in SOL, while PFKM, FBP1, and FBP2 mRNA expression was downregulated in SOL (Figure 7E). Notably, the mRNA expression of MYBPH, MYOG, TNNI1, PFKM, FBP1 and FBP2 was consistent with their nearby lncRNAs, MSTRG.19265.9 and MSTRG.2082.1, and MSTRG.14200.1 and MSTRG.2121.6, respectively. In contrast, the mRNA expression of ACO2 and PDK4 was opposite to their nearby lncRNAs, MSTRG.13515.1 and ENSSSCT00000074465, respectively (Figure 7E). In summary, we identified seven key m6A-modified lncRNAs that may affect muscle-fiber-type conversion by positively or negatively regulating their cis target genes.
Figure 7.
QTL analysis and functional enrichment of dme-lncRNAs. (A) Protein–protein interaction (PPI) network of PTGs of lncRNAs visualized by using Cytoscape. The size and color of circles represented the degree of interaction between the genes. (B,C) Above: the top two clusters from PPI network identified by using MCODE. Bottom: GO enrichment analysis of PTGs in corresponding cluster. The orange triangle node represented lncRNAs, the blue cycle node represented PTGs. Each pair of PTGs and lncRNAs was indicated by blue dotted line. The interaction of PTGs was indicated by black solid line. (D) The information of lncRNAs and their adjacent mRNAs from (E). (E) qPCR result showing the adjacent mRNA expressions in SOL and EDL. The relative mRNA expression was standardized to that of the control gene β-actin. Data represent mean ± SD of three independent biological replicates. * p< 0.05; ** p < 0.01.
2.7. Inhibition of MSTRG.14200.1 Reduced PSCs Differentiation and Stimulated Fast-to-Slow Muscle-Fiber Conversion
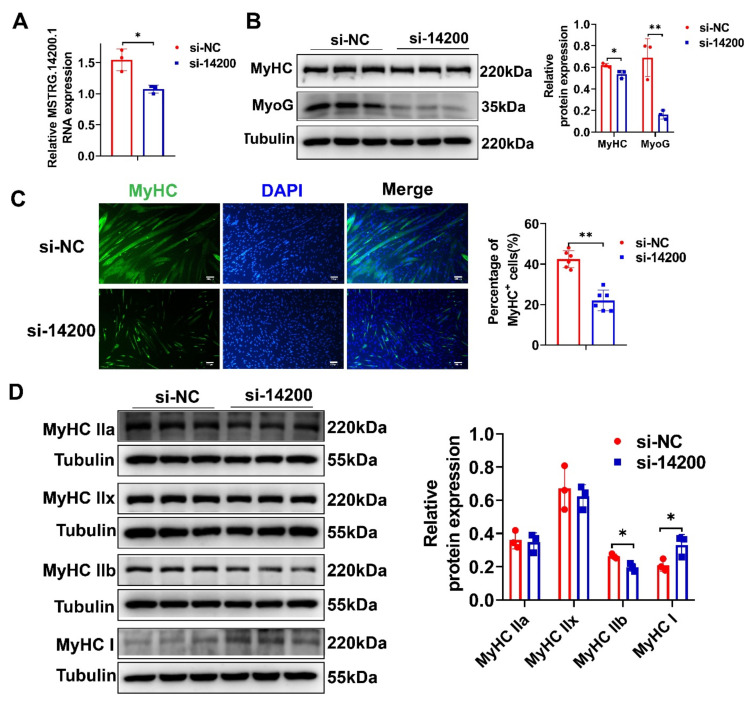
To further explore the role of lncRNAs in pig skeletal muscle satellite cell (PSCs) differentiation and muscle-fiber-type conversion, we chose lncRNA MSTRG.14200.1, which has a higher expression level in EDL than in SOL, for further loss-of-function assays. The expression level of MSTRG.14200.1 was successfully knocked down using siRNA (Figure 8A). By Western blotting and immunofluorescence staining, we found that MSTRG.14200.1 knockdown significantly decreased MyoG and MyHC protein expression (Figure 8B) and MyHC+ cells proportion (Figure 8C), indicating the MSTRG.14200.1 knockdown inhibited PSC differentiation. The MSTRG.14200.1 knockdown also remarkably reduced MyHC IIb protein expression, while increasing MyHC I protein expression (Figure 8D), which showed that the MSTRG.14200.1 knockdown stimulated fast-to-slow muscle-fiber conversion. All the above results demonstrate that MSTRG.14200.1 has an important role in promoting PSC differentiation and inducing slow-to-fast muscle-fiber conversion.
Figure 8.
Inhibition of MSTRG.14200.1 reduced PSC differentiation and stimulated fast-to-slow muscle-fiber conversion. (A) qPCR result showed that MSTRG.14200.1 expression was significantly reduced. (B) Western blotting result showed that the protein expression levels of MyoG and MyHC were remarkably decreased after MSTRG.14200.1 knockdown. (C) Immunofluorescence staining result of MyHC showed that knockdown of MSTRG.14200.1 significantly reduced the proportion of MyHC+ cells. Scale bar: 200 μm. (D) Western blotting result showed that MSTRG.14200.1 knockdown notably inhibited MyHC IIb protein expression and enhanced MyHC I protein expression. The relative RNA and protein expression were standardized to that of the control gene β-actin and Tubulin, respectively. Data represent mean ± SD of three independent biological replicates. * p < 0.05; ** p < 0.01.
3. Discussion
Skeletal muscle fiber formation and the conversion of different types of muscle fibers are complex processes, regulated by many factors. The current research on skeletal muscle-fiber-type conversion is mainly focused on signal transduction pathways, protein-coding gene regulation, and nutritional intervention [40,41,42]. Recent reports have also shown the important roles of lncRNAs in skeletal muscle development and muscle-fiber-type conversion [21,22,23]. A more recent report has revealed the m6A methylation status of lncRNAs during skeletal myogenesis [31]. However, it is unclear whether lncRNAs’ m6A methylation is involved in muscle-fiber-type conversion. In the present study, we hypothesized that lncRNAs might also be regulated by m6A and participate in skeletal muscle-fiber-type conversion. Hence, we comprehensively identified differentially m6A-methylated lncRNAs in oxidative and glycolytic skeletal muscle by MeRIP-seq. To the best of our knowledge, this study is the first systematic evaluation of m6A methylomes profiles of lncRNAs in oxidative and glycolytic skeletal muscle. Our work provides a valuable resource for the future study of the regulation and function of m6A modification on lncRNAs in muscle-fiber-type conversion.
The size of the pig genome is roughly the same as that of the human and mouse genome. However, fewer lncRNAs were annotated in pigs compared with the above two species [43,44,45]; this indicates that a great quantity of pig lncRNAs have not been discovered. In this study, we identified 5607 novel lncRNAs, which broaden the annotation of the pig lncRNAs. We found that the transcript length, number of exons, ORF length, and gene expression abundance of the novel identified lncRNAs are similar to previously annotated lncRNAs, but differ from the mRNAs, in accordance with those of other studies [46,47]. We used three kinds of coding potential software (CNCI, FEElnc, and CPC2) to exclude transcripts with protein-coding potential [48,49,50]. We noted that some lncRNAs also have long putative ORF, equivalent to mRNA ORF, such as those with ORF ≥ 500 nt; these lncRNAs may protein code mRNAs containing unknown protein domains. Furthermore, we identified 334 DE lncRNAs between SOL and EDL through RNA-seq, providing key candidate lncRNAs involved in the regulation of skeletal muscle-fiber-type conversion.
Studies have demonstrated that m6A is a ubiquitous modification in mRNAs and plays a key role in regulating gene expression [34]. m6A modification in lncRNAs has also attracted the attention of researchers. In this study, we identified a large number of m6A peaks in lncRNAs, in both SOL and EDL muscles, through MeRIP-seq, indicating lncRNAs are extensively modified by m6A. We found typical m6A consensus motifs within lncRNAs, as previously identified in lincRNAs [51]. m6A was evenly enriched in the lncRNA body, increased slightly at the 3′ end, and more located in the last exon of lncRNA, which was similar to a previous report [32,52]. These results show that the m6A peaks identified in our study are credible. m6A methylation in mRNA affects nuclear transport, splicing, stability and translation of mRNA [28]. m6A methylation in lncRNAs can also regulate their expression. For example, METTL3-mediated m6A methylation modification, resulting in LINC00958 upregulation by stabilizing its RNA transcript and the high level of LINC00958, led to the poor overall survival of hepatocellular carcinoma patients [53]. Another study reported that the METTL14 knockdown can abolish the m6A level of XIST and enhanced XIST expression, and m6A-methylated XIST can also be recognized by YTHDF2 to mediate the degradation of XIST [54]. In the present study, we systematically analyzed the role of m6A methylation in lncRNA expression dynamics, and found the lncRNAs’ m6A levels were positively correlated with their expression levels, indicating m6A methylation may positively regulate these lncRNAs’ expressions. m6A-reader proteins are responsible for recognizing m6A sites and play a role in regulating RNA stability. The m6A-reader protein YTHDF2 was shown to mediate RNA decay [55,56] and other reader proteins, such as IGF2BP1/2/3, YTHDC1, and YTHDF1/3, and can strengthen RNA stability [30,56,57]. Thus, we speculate that the lncRNAs, whose expression levels are positively regulated by m6A in our study, may be mainly recognized and bound by IGF2BP1/2/3, YTHDC1, or YTHDF1/3 and, thus, enhance their expression. However, further research is needed to confirm this hypothesis, and more efforts should be undertaken to clarify how m6A methylation affects lncRNAs expression.
Certain types of skeletal muscle fibers are known to exert a pivotal function in determining the meat quality of livestock after death, such as meat color, drip loss, and pH [58]. To identify lncRNAs that were specifically involved in muscle-fiber-type conversion, we took advantage of the animal QTLdb (PigQTLdb) database, and performed a correlation analysis by mapping dme-lncRNAs to the QTL regions related to pig-meat-related traits. This analysis allowed us to identify 49 dme-lncRNAs closely associated with muscle-fiber-type characteristics, such as meat color, fatty acid content, PH, flavor, and enzyme activity. A large number of studies have shown that the biological function of lncRNAs can be predicted by evaluating the relevant cis genes [46]. Therefore, we further predicted the function of the 49 dme-lncRNAs by searching for their PTGs, and we found 119 cis PTGs of 49 dme-lncRNAs. These genes were further used for lncRNAs’ functional annotation and enrichment analysis. Finally, we identified seven dme-lncRNAs, whose PTGs were enriched in muscle-fiber properties and energy metabolism (for example, MSTRG.2082.1 paired with CSRP1, TNNT2, and TNNI1). Of note, TNNI1, encoding the slow skeletal muscle isoform, is specifically expressed in slow muscle fibers and has been used as a model gene to study the specific expression mechanism of slow fibers [59,60]. Our qPCR results also found that both MSTRG.2082.1 and TNNI1 expressions were upregulated in SOL compared with in EDL, indicating MSTRG.2082.1 co-located with TNNI1 may positively affect the expression of TNNI1 and, thus, regulate the conversion of skeletal muscle-fiber types. Besides, MSTRG.14200.1 paired with PFKM, as PFKM is a key regulator of glycolysis, encodes a muscle subtype of phosphofructose kinase, called phosphofructose kinase 1 [61]. This is a strong candidate for skeletal muscle gene expression, associated with glycemic traits [62]. Interestingly, we found both MSTRG.14200.1 and PFKM have a higher expression in EDL than in SOL, indicating MSTRG.14200.1 may play a role in regulating glycolytic muscle fiber through enhancing PFKM expression. However, whether the seven dme-lncRNAs play a functional role in skeletal muscle-fiber-type conversion needs further systematic functional research.
In summary, we analyzed the differential expressions and m6A-methylation profiles of lncRNAs in oxidative and glycolytic skeletal muscles, and identified seven m6A-modified lncRNAs that may play an important role in muscle-fiber-type conversion. In addition, we preliminarily verified that lncRNA MSTRG.14200.1 can promote PSC differentiation and slow-to-fast muscle-fiber conversion. Our study provides the resource of m6A-modified lncRNAs profile for future studies on the function and mechanism of m6A-modified lncRNAs, and it opens up a new way for the study of RNA epigenetics in muscle-fiber-type conversion.
4. Materials and Methods
4.1. Sample Preparation
Three 6-month-old Duroc pigs were obtained from the breeding pig farm of Guangdong Wen’s Foodstuffs Group Co., Ltd. (Yunfu, China). Samples of soleus muscles (SOL) and extensor digitorum longus muscles (EDL) were immediately put into liquid nitrogen and then stored at −80 °C for RNA extraction or fixed in 4% paraformaldehyde for histology staining. All animal experiments were conducted based on the National Research Council Guide for the Care and Use of Laboratory Animals and approved the Animal Care and Use Committee of the South China Agricultural University, Guangzhou, China.
4.2. m6A-seq and RNA-seq Library Preparation
Total RNA of three 6-month-old Duroc pigs was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. RNA amount and purity were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and confirmed by agarose gel electrophoresis. Ribosomal RNA was depleted from total RNA using the Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, CA, USA), and the ribosomal-depleted RNA was fragmented using Magnesium RNA Fragmentation Module (Cat.e6150, NEB, Ipswich, MA, USA) at 86 °C for 7 min. Then the cleaved RNA fragments were incubated with m6A-specific antibody (No. 202003, Synaptic Systems, San Jose, CA, Germany) and Dynabeads antibody Coupling Kit (Thermo Fisher Scientific, Waltham, MA, USA) in IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% Igepal CA-630) for 2 h at 4 °C. The IP RNA then underwent a series of processing, including reverse transcription to produce cDNA, synthesis of second-strand DNA, incorporation of dUTP solution into the second strand, addition of A-base at the end of each strand, and finally PCR amplification to form a library with fragment size of 300 ± 50 bp. Both the Input samples without immunoprecipitation and the m6A IP samples libraries were sequenced on an illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) to produce paired-end 150 bp reads.
4.3. Identification of lncRNAs
The clean data were obtained by removing reads containing adapters, reads containing over 10% of poly (N) by fastq from the raw data [63]. Hisat2 was used to map the clean reads to reference genome (Ensembl Sscrofa 11.1.94) under chain-specific parameters: “rna-strandness RF” [64]. Unique mapped reads with a mapping quality greater than 20 using samtools [65]. The mapped paired-reads from each library were assembled with StringTie (v2.1.1) [66] to construct and identify transcripts through a reference-based approach, and calculate fragments per kilo-base of exon per million fragments (FPKM) of lncRNAs and coding genes. The assembled transcripts were compared with known gene models using gffcompare [67], and transcripts with class codes ‘i’, ‘j’, ‘o’, ‘u’, and ‘x’ were selected. We further removed transcripts that were shorter than 200 nt in length. The Coding-Non-Coding Index (CNCI) [48], Flexible Extraction of LncRNAs (FEElnc) [49] and Coding Potential Calculator 2 (CPC2) [50] were used to evaluate the coding potential of filtered transcripts with default parameters. The transcripts without coding potential on the above three software were retained. The remaining transcripts were compared with the Swiss-Prot (https://www.ebi.ac.uk/uniprot, (accessed on 16 July 2021), Pfam protein (http://pfam.xfam.org, (accessed on 16 July 2021) and Rfam (http://rfam.xfam.org, (accessed on 16 July 2021) databases to exclude the potential protein-coding genes and known ncRNAs including tRNA, ribosomal RNA (rRNA), small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA). Finally, the transcripts with FPKM ≥ 0.5 (2 for single-exon transcripts) at least in one sample were considered as “novel lncRNAs” (Figure 2A). Both datasets of known and novel lncRNAs were combined into the final lncRNAs set used in this research.
4.4. Differential Expression Analysis
mRNAs and lncRNAs with ≥0.5 FPKM in at least one library were considered expressed and were used for further analysis. Differential gene expression analysis between two groups was performed using the R package DESeq2 [68]. LncRNAs with adjusted p-value < 0.05 using Benjamini–Hochberg and log2 fold change (log2FC) ≥ |1| were considered differentially expressed lncRNAs.
4.5. Functional Enrichment Analysis
To predict the functions of the lncRNAs, the expressed mRNAs within 100 kb of lncRNAs were defined as cis target genes and submitted to functional enrichment analysis. Gene ontology (GO) analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using PANTHER [69] and KOBAS-i [70], respectively. The GO terms and pathways categories with p value < 0.05 were considered significant. The pig quantitative trait loci (QTLs) database was downloaded from the Animal QTLdb (PigQTLdb) (http://www.animalgenome.org/QTLdb/pig.html, (accessed on 19 October 2021). The command “intersect” in bedtools was used to acquire lncRNAs enriched in QTL traits locus [71].
4.6. Establishment of PPI Network
PPI network was established by the STRING (v10.5) [72], and those experimentally validated interactions with a combined score > 0.4 were selected. The screened networks were visualized by the Cytoscape 3.6.1 [73]. Then, the “Molecular Complex Detection” (MCODE), a clustering algorithm identifying locally densely connected regions in a large PPI network based on node-weighting arithmetic, was performed to establish PPI network modules with parameters (Degree cutoff = 2, Node score cutoff = 0.3, k-core = 2, Max. Depth = 100).
4.7. MeRIP-Seq Analysis
The mapped reads from the IP and Input libraries were fed into the R package exomePeak2 (v1.6.0) for calling peaks under default parameters settings, and identifying differential peaks (adjusted p-value < 0.05) [74]. Identified peaks were annotated by intersecting with gene architecture using bedtools and custom shell script. Peaks located at expressed lncRNAs were selected for downstream analysis. Peaks that did not overlap with any m6A modification site in RMBase [75] were determined to be unknown. The distribution of m6As in lncRNAs was characterized by Guitar R package (v1.16.0) [76]. The DREME tool in the MEME suite (http://meme-suite.org/tools/dreme, (accessed on 10 August 2021) was used to discover relatively short (up to 8 bp) motifs that were enriched within a set of target sequences (m6A peak sequences) [77]. The FPKM of Input and IP samples were calculated by StringTie. The m6A enrichment levels of lncRNAs were represented as MFPKM (MFPKM = FPKM_IP/FPKM_INPUT) averaged in the three biological replicates. The Pearson correlation analysis and principal component analysis (PCA) of sequencing data were performed using DeepTools [78]. Read coverage of regions was visualized via the Integrative Genomics Viewer (IGV) [79].
4.8. MeRIP Assay
The MeRIP experiment procedure was performed as mentioned in ‘m6A-seq library preparation’, except that total RNA was not subjected to ribosomal RNA (rRNA) removal and fragmentation. The Input RNA and immunoprecipitated RNA from SOL and EDL muscle tissues were reverse-transcribed to produce cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA), and analyzed using real-time quantitative PCR (qPCR).
4.9. Real-Time Quantitative PCR (qPCR)
Total RNA of PSCs and tissues was obtained using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed to create cDNA by Evo M-MLV RT Kit (AG, Changsha, China). qPCR was performed on an ABI QuantStudio 7 Flex system (Thermo Fisher Scientific, Waltham, MA, USA) using PowerUpTM SYBRTM Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The Ct (2−ΔΔCt) method was used to analyze relative RNA expression. All primers used for qPCR in this study are listed in Table S7.
4.10. Histology Staining
The fixed tissues were embedded in paraffin and cut into 4 μm thin slices. Then the sections were used for immunofluorescence staining of MyHC I (BF-D5; DSHB; Iowa City, IA, USA) and MyHC IIb (BF-F3; DSHB; Iowa City, IA, USA). Immunofluorescence staining on paraffin muscle sections was performed in accordance with previous reports [80]. The images were captured using a Nikon ECLIPSE Ci microscope (Nikon, Tokyo, Japan) and analyzed using the ImageJ software (NIH, Bethesda, MD, USA).
4.11. Porcine Skeletal Muscle Satellite Cells Isolation and Culture
Porcine skeletal muscle satellite cells (PSCs) were isolated from leg muscles of gilts within a week. The gilt was sacrificed, washed with flowing water, and disinfected with 75% alcohol, then the leg muscles were removed and placed in PBS containing 1% penicillin-streptomycin (Gibco, Carlsbad, CA, USA). The muscles were cut into pieces and digested in 2 mg/mL collagenase I (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 2 or 3 h with shaking, until the mixture became uniform. Digestion was then stopped and the mixture was in turn filtered through 100-, 200-, and 400-mesh sieves. The cells were differential-adhesion cultured in RPMI 1640 medium supplemented with 20% FBS (Gibco, Carlsbad, CA, USA), 1% non-essential amino acids (Gibco, Carlsbad, CA, USA), 1% GlutaMAX (Gibco, Carlsbad, CA, USA), 0.5% chicken embryo extract (Gemini Bio, Sacramento, CA, USA), 4 ng/mL basic fibroblast growth factor (Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin-streptomycin for 2 h to remove fibroblasts. The supernatant containing purified cells was then transferred to a cell culture bottle coated with collagen for proliferating culture at 37 °C and 5% CO2.
For the differentiation of PSCs, when the cells reached 70–80% confluence, the proliferating medium was replaced with RPMI 1640 containing 2% horse serum (Gibco, Carlsbad, CA, USA).
4.12. siRNA Synthesis, and Cell Transfection
siRNA for MSTRG.14200.1 and negative control (NC) were designed and synthesized by GenePharma (Shanghai, China). siRNA oligos sequences were as follows: MSTRG.14200.1 (sense 5′- GCCUACUUAGUGCAGAAACTT’), NC (sense 5′- UUCUCCGAACGUGUCACGUTT-3′). siRNA oligos transfection of PSCs were performed with Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
4.13. Western Blotting
Total protein of PSCs was extracted with RIPA buffer containing 1% PMSF (Beyotime, Shanghai, China), and the protein was then incubated at 95 °C for 5 min for denaturation in 6× protein loading buffer. Western blotting was performed according to previous reports [81]. The primary antibodies used include: MyHC I (sc-53089; Santa Cruz; Delaware, Santa Cruz, CA, USA), MyHC IIb (A15293; ABclonal; Wuhan, China), MyHC IIa (A15292; ABclonal; Wuhan, China), MyHC IIx (A6935; ABclonal; Wuhan, China), MyHC (sc-376157; Santa Cruz; Delaware, Santa Cruz, CA, USA), MyoG (sc-12732; Santa Cruz; Delaware, Santa Cruz, CA, USA), and β-Tubulin (GB11017B; Servicebio; Wuhan, China). The secondary antibodies used include: Goat Anti-Mouse IgG (A0216; Beyotime; Shanghai, China) and Goat Anti-Rabbit IgG (A0208; Beyotime; Shanghai, China). Protein band intensity was quantified using ImageJ.
4.14. Cell Immunofluorescence Staining
PSCs were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 30 min, respectively. After three washes, the cells were blocked in QuickBlock™ Blocking Buffer (Beyotime, Shanghai, China) for 2 h, incubated with a primary antibody overnight and incubated with a secondary antibody for 1 h, in turn. Finally, the nuclei were stained with DAPI reagent. Images of at least three random fields of view were captured using a Nikon ECLIPSE Ti microscope (Nikon, Tokyo, Japan) and quantified using ImageJ. The primary antibody was MyHC (sc-376157; Santa Cruz; Delaware, Santa Cruz, CA, USA). The secondary antibody was FITC-labeled Goat Anti-Mouse IgG (A0568; Beyotime, Shanghai, China).
4.15. Statistical Analysis
Statistical analyses for the results of qPCR were performed by two-tailed, unpaired Student’s t-test using SPSS software (IBM, Armonk, NY, USA) and the histogram plots were generated by GraphPad Prism 8.0.2 software (GraphPad Software, San Diego, CA, USA). Data were presented as means ± standard deviation (SD) in at least triplicate. A p < 0.05 was considered as statistically significant. The significance was marked as * p < 0.05 and ** p < 0.01.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23094600/s1.
Author Contributions
S.W. performed the experiments, interpreted results, and wrote manuscript. B.T. analyzed data and interpreted results. L.X., J.Z. and X.Z. collected samples and helped perform some experiments. L.H., J.Y., G.C. and E.Z. participated in the design of the project and revised the manuscript. T.G. and Z.W. conceived the project and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the China Postdoctoral Science Foundation (Program 2021M691075), the Guangdong Basic and Applied Basic Research Fund: Regional Joint Fund-Youth Fund Project (2020A1515110319).
Institutional Review Board Statement
All animal testing was conducted under a protocol approved by the Ethics Committee of the Laboratory Animal Center of South China Agricultural University (Permit Number 2021F036, Permit Date 2 March 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA810786/, (accessed on 3 March 2022), PRJNA810786.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Frontera W.R., Ochala J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015;96:183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin K.M., Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J. Appl. Physiol. 2001;90:345–357. doi: 10.1152/jappl.2001.90.1.345. [DOI] [PubMed] [Google Scholar]
- 3.Schiaffino S., Reggiani C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 4.Bottinelli R., Reggiani C. Human skeletal muscle fibres: Molecular and functional diversity. Prog. Biophys. Mol. Biol. 2000;73:195–262. doi: 10.1016/S0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 5.Schiaffino S., Reggiani C. Myosin isoforms in mammalian skeletal muscle. J. Appl. Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 6.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 7.Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr. Opin. Genet. Dev. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen L.Y., Luo J., Lei H.G., Jiang Y.Z., Bai L., Li M.Z., Tang G.Q., Li X.W., Zhang S.H., Zhu L. Effects of muscle fiber type on glycolytic potential and meat quality traits in different Tibetan pig muscles and their association with glycolysis-related gene expression. Genet. Mol. Res. 2015;14:14366–14378. doi: 10.4238/2015.November.13.22. [DOI] [PubMed] [Google Scholar]
- 9.Choe J.H., Kim B.C. Association of blood glucose, blood lactate, serum cortisol levels, muscle metabolites, muscle fiber type composition, and pork quality traits. Meat Sci. 2014;97:137–142. doi: 10.1016/j.meatsci.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Kim G.D., Ryu Y.C., Jeong J.Y., Yang H.S., Joo S.T. Relationship between pork quality and characteristics of muscle fibers classified by the distribution of myosin heavy chain isoforms. J. Anim. Sci. 2013;91:5525–5534. doi: 10.2527/jas.2013-6614. [DOI] [PubMed] [Google Scholar]
- 11.Pette D., Staron R.S. Mammalian skeletal muscle fiber type transitions. Int. Rev. Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- 12.Ekmark M., Rana Z.A., Stewart G., Hardie D.G., Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. Pt 2J. Physiol. 2007;584:637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen D.L., Sartorius C.A., Sycuro L.K., Leinwand L.A. Different pathways regulate expression of the skeletal myosin heavy chain genes. J. Biol. Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L.N., Ren Y., Chen J.Q., Wang Y.Z. Effects of myogenin on muscle fiber types and key metabolic enzymes in gene transfer mice and C2C12 myoblasts. Gene. 2013;532:246–252. doi: 10.1016/j.gene.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Moretti I., Ciciliot S., Dyar K.A., Abraham R., Murgia M., Agatea L., Akimoto T., Bicciato S., Forcato M., Pierre P., et al. MRF4 negatively regulates adult skeletal muscle growth by repressing MEF2 activity. Nat. Commun. 2016;7:12397. doi: 10.1038/ncomms12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo P., Wang L., Luo L., Wang L., Yang K., Shu G., Wang S., Zhu X., Gao P., Jiang Q. Ca2+-Calcineurin-NFAT pathway mediates the effect of thymol on oxidative metabolism and fiber-type switch in skeletal muscle. Food Funct. 2019;10:5166–5173. doi: 10.1039/C8FO02248H. [DOI] [PubMed] [Google Scholar]
- 17.Schiaffino S., Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: Insights from genetic models. Skelet. Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao S., Ren H., Li Y., Zhou J., Duan C., Lu L. Differential regulation of IGF-I and IGF-II gene expression in skeletal muscle cells. Mol. Cell Biochem. 2013;373:107–113. doi: 10.1007/s11010-012-1479-4. [DOI] [PubMed] [Google Scholar]
- 19.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 20.Akhade V.S., Pal D., Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv. Exp. Med. Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- 21.Wang S., Jin J., Xu Z., Zuo B. Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells. 2019;8:1107. doi: 10.3390/cells8091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dou M., Yao Y., Ma L., Wang X., Shi X., Yang G., Li X. The long noncoding RNA MyHC IIA/X-AS contributes to skeletal muscle myogenesis and maintains the fast fiber phenotype. J. Biol. Chem. 2020;295:4937–4949. doi: 10.1074/jbc.RA119.010498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J.A., Wang Z., Yang X., Ma M., Li Z., Nie Q. LncRNA-FKBP1C regulates muscle fiber type switching by affecting the stability of MYH1B. Cell Death Discov. 2021;7:73. doi: 10.1038/s41420-021-00463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridges M.C., Daulagala A.C., Kourtidis A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021;220:e202009045. doi: 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Eckert M.A., Harada B.T., Liu S.M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y., et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazi F., Fatica A. Interplay Between N (6)-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 2019;7:116. doi: 10.3389/fcell.2019.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie S.J., Tao S., Diao L.T., Li P.L., Chen W.C., Zhou Z.G., Hu Y.X., Hou Y.R., Lei H., Xu W.Y., et al. Characterization of Long Non-coding RNAs Modified by m6A RNA Methylation in Skeletal Myogenesis. Front. Cell Dev. Biol. 2021;9:762669. doi: 10.3389/fcell.2021.762669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y., Zhang X., Zhang X., Zhao K., Zhang J., Yang C., Chen Y. Comprehensive Analysis of the Transcriptome-Wide m6A Methylome in Pterygium by MeRIP Sequencing. Front. Cell Dev. Biol. 2021;9:670528. doi: 10.3389/fcell.2021.670528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 35.Lee S.H., Joo S.T., Ryu Y.C. Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci. 2010;86:166–170. doi: 10.1016/j.meatsci.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 36.Koohmaraie M., Shackelford S.D., Wheeler T.L., Lonergan S.M., Doumit M.E. A muscle hypertrophy condition in lamb (callipyge): Characterization of effects on muscle growth and meat quality traits. J. Anim. Sci. 1995;73:3596–3607. doi: 10.2527/1995.73123596x. [DOI] [PubMed] [Google Scholar]
- 37.Hu Z.L., Park C.A., Reecy J.M. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 2016;44:D827–D833. doi: 10.1093/nar/gkv1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan P., Luo S., Lu J.Y., Shen X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017;46:170–178. doi: 10.1016/j.gde.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 39.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Zhang M., Xue Y., Liang D., An W., Jia G., Zhao H., Liu G., Huang Z. Effect of dietary L-theanine supplementation on skeletal muscle fiber type transformation in vivo. J. Nutr. Biochem. 2022;99:108859. doi: 10.1016/j.jnutbio.2021.108859. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S., Chen X., Huang Z., Chen D., Yu B., Chen H., He J., Luo J., Zheng P., Yu J., et al. Leucine promotes porcine myofibre type transformation from fast-twitch to slow-twitch through the protein kinase B (Akt)/forkhead box 1 signalling pathway and microRNA-27a. Br. J. Nutr. 2019;121:1–8. doi: 10.1017/S000711451800301X. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Shen T., Randall W.R., Schneider M.F. Signaling pathways in activity-dependent fiber type plasticity in adult skeletal muscle. J. Muscle Res. Cell Motil. 2005;26:13–21. doi: 10.1007/s10974-005-9002-0. [DOI] [PubMed] [Google Scholar]
- 43.Xu J., Bai J., Zhang X., Lv Y., Gong Y., Liu L., Zhao H., Yu F., Ping Y., Zhang G., et al. A comprehensive overview of lncRNA annotation resources. Brief. Bioinform. 2017;18:236–249. doi: 10.1093/bib/bbw015. [DOI] [PubMed] [Google Scholar]
- 44.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I., et al. Gencode 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou C., Li J., Luo W., Li L., Hu A., Fu Y., Hou Y., Li C. Transcriptome analysis reveals long intergenic non-coding RNAs involved in skeletal muscle growth and development in pig. Sci. Rep. 2017;7:8704. doi: 10.1038/s41598-017-07998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong L., Hu Q., Zang X., Xie Y., Zhou C., Zou X., Li Y., Deng M., Guo Y., Liu G., et al. Analysis and Screening of Reproductive Long Non-coding RNAs Through Genome-Wide Analyses of Goat Endometrium During the Pre-attachment Phase. Front. Genet. 2020;11:568017. doi: 10.3389/fgene.2020.568017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen L., Gan M., Tang Q., Tang G., Jiang Y., Li M., Chen L., Bai L., Shuai S., Wang J., et al. Comprehensive Analysis of lncRNAs and circRNAs Reveals the Metabolic Specialization in Oxidative and Glycolytic Skeletal Muscles. Int. J. Mol. Sci. 2019;20:2855. doi: 10.3390/ijms20122855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wucher V., Legeai F., Hédan B., Rizk G., Lagoutte L., Leeb T., Jagannathan V., Cadieu E., David A., Lohi H., et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017;45:e57. doi: 10.1093/nar/gkw1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang Y.-J., Yang D.-C., Kong L., Hou M., Meng Y.-Q., Wei L., Gao G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao S., Cao S., Huang Q., Xia L., Deng M., Yang M., Jia G., Liu X., Shi J., Wang W., et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell Biol. 2019;21:651–661. doi: 10.1038/s41556-019-0315-4. [DOI] [PubMed] [Google Scholar]
- 52.Nie Y., Tian G.G., Zhang L., Lee T., Zhang Z., Li J., Sun T. Identifying cortical specific long noncoding RNAs modified by m6A RNA methylation in mouse brains. Epigenetics. 2021;16:1260–1276. doi: 10.1080/15592294.2020.1861170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., Cao M., Cai J., Wu J., Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X., Zhang S., He C., Xue P., Zhang L., He Z., Zang L., Feng B., Sun J., Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H., Xu Y., Yao B., Sui T., Lai L., Li Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613. doi: 10.1038/s41419-020-02833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S., Wang J.Z., Chen D., He Y.T., Meng N., Chen M., Lu R.X., Chen X.H., Zhang X.L., Yan G.R. An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nat. Commun. 2020;11:1685. doi: 10.1038/s41467-020-15403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryu Y.C., Kim B.C. The relationship between muscle fiber characteristics, postmortem metabolic rate, and meat quality of pig longissimus dorsi muscle. Meat Sci. 2005;71:351–357. doi: 10.1016/j.meatsci.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 59.Corin S.J., Juhasz O., Zhu L., Conley P., Kedes L., Wade R. Structure and expression of the human slow twitch skeletal muscle troponin I gene. J. Biol Chem. 1994;269:10651–10659. doi: 10.1016/S0021-9258(17)34109-1. [DOI] [PubMed] [Google Scholar]
- 60.Polly P., Haddadi L.M., Issa L.L., Subramaniam N., Palmer S.J., Tay E.S., Hardeman E.C. hMusTRD1alpha1 represses MEF2 activation of the troponin I slow enhancer. J. Biol. Chem. 2003;278:36603–36610. doi: 10.1074/jbc.M212814200. [DOI] [PubMed] [Google Scholar]
- 61.Nakajima H., Noguchi T., Yamasaki T., Kono N., Tanaka T., Tarui S. Cloning of human muscle phosphofructokinase cDNA. FEBS Lett. 1987;223:113–116. doi: 10.1016/0014-5793(87)80519-7. [DOI] [PubMed] [Google Scholar]
- 62.Keildson S., Fadista J., Ladenvall C., Hedman A.K., Elgzyri T., Small K.S., Grundberg E., Nica A.C., Glass D., Richards J.B., et al. Expression of phosphofructokinase in skeletal muscle is influenced by genetic variation and associated with insulin sensitivity. Diabetes. 2014;63:1154–1165. doi: 10.2337/db13-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pertea G., Pertea M. GFF Utilities: GffRead and GffCompare. F1000Res. 2020;9:304. doi: 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bu D., Luo H., Huo P., Wang Z., Zhang S., He Z., Wu Y., Zhao L., Liu J., Guo J., et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meng J., Cui X., Rao M.K., Chen Y., Huang Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29:1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W.-J., Li J.-H., Liu S., Wu J., Zhou H., Qu L.-H., Yang J.-H. RMBase: A resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44:D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui X., Wei Z., Zhang L., Liu H., Sun L., Zhang S.-W., Huang Y., Meng J. Guitar: An R/Bioconductor Package for Gene Annotation Guided Transcriptomic Analysis of RNA-Related Genomic Features. Biomed. Res. Int. 2016;2016:8367534. doi: 10.1155/2016/8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bailey T.L. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang S., Xu X., Liu Y., Jin J., Zhu F., Bai W., Guo Y., Zhang J., Zuo H., Xu Z., et al. RIP-Seq of EZH2 Identifies TCONS-00036665 as a Regulator of Myogenesis in Pigs. Front. Cell Dev. Biol. 2020;8:618617. doi: 10.3389/fcell.2020.618617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y., Li W., Zhu M., Li Y., Xu Z., Zuo B. FHL3 differentially regulates the expression of MyHC isoforms through interactions with MyoD and pCREB. Cell Signal. 2016;28:60–73. doi: 10.1016/j.cellsig.2015.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA810786/, (accessed on 3 March 2022), PRJNA810786.