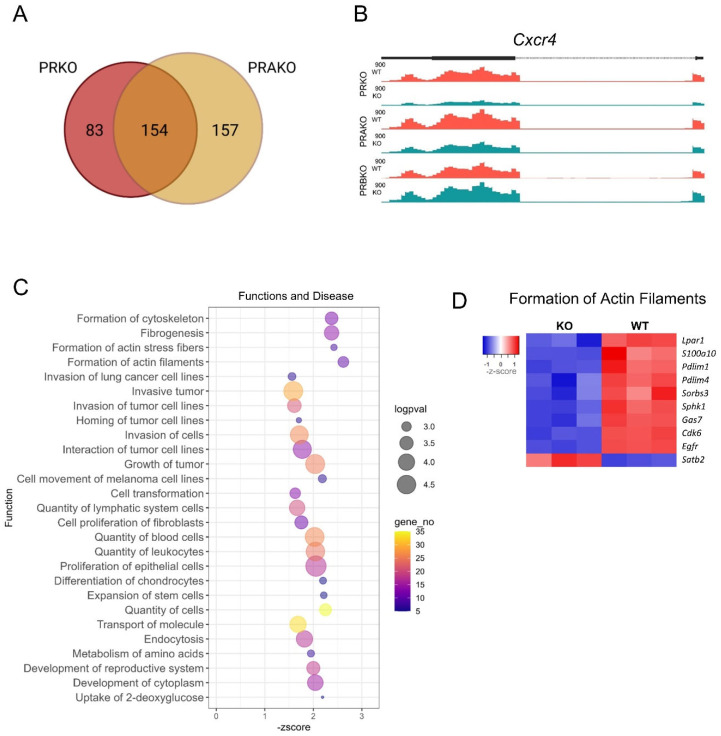
Figure 6.
RNA-seq of PRKO, PRAKO and PRBKO granulosa cells identifies PGR-regulated genes and pathways. RNA-seq was performed on WT and KO granulosa cells of the PRKO, PRAKO and PRBKO strains. (A) RNA-seq analysis identified 236 DEGs in PRKO GCs and 310 for PRAKO GCs with 154 common DEGs (logFC ≥ 1, padj ≤ 0.01). (B) Example of mapped RNA-seq reads for PRKO, PRAKO and PRBKO depicting genome site for Cxcr4. (C) The 154 DEGs common to PRKO and PRAKO were analyzed in IPA and most highly downregulated function and diseases identified, depicted with log p-value (size), the number of DEGs associated with the function (color) and z-score indicating degree of downregulation (X-axis). Functions are grouped based on similarities. (D) Heatmap depicting the DEGs associated with “formation of actin filaments” function. RNA-seq reads per samples (log-counts per million, LCPM) are shown for PRKO (left) and PRWT (right) where blue is low and red is high expression.