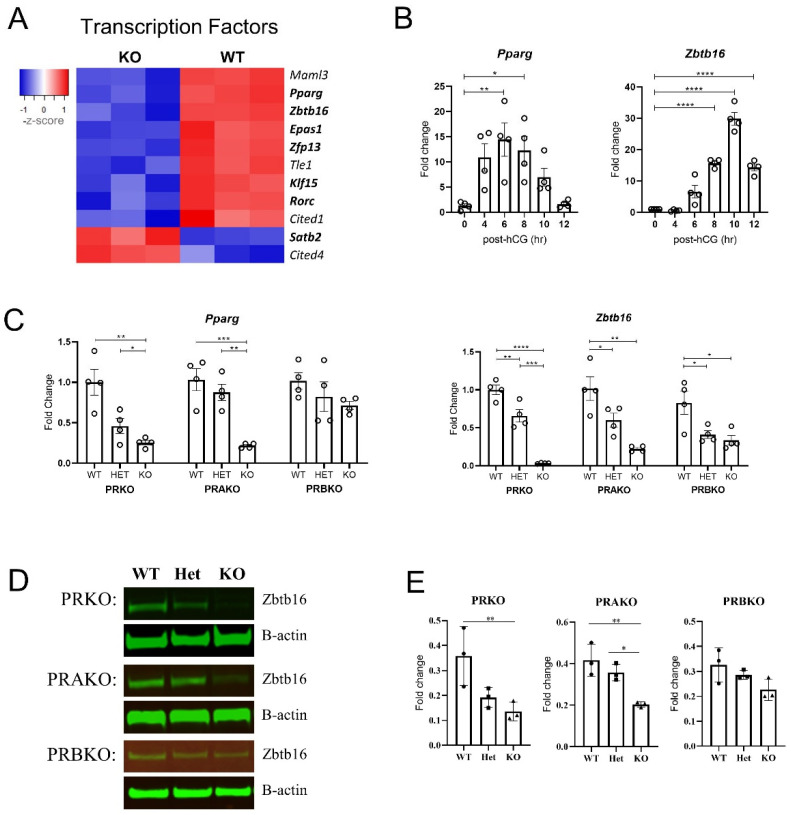
Figure 7.
PGR regulates a network of transcription factors in granulosa cells including Pparg and Zbtb16. (A) Heatmap showing expression level of PGR-regulated transcription factors, including DNA-binding transcription factors (bold) and transcription cofactors (not bold). Shown are PRKO (left) and PRWT (right) RNA-seq reads (LCPM) with blue representing low expression levels and red increased expression levels. (B) Expression of Pparg and Zbtb16 in wildtype granulosa cells over a preovulatory time course from 0–12 h post-hCG using CBA-F1 mice. (C) Expression of Pparg and Zbtb16 in granulosa cells of PRKO (left) PRAKO (center) and PRBKO (right) mice at 8 h post-hCG qPCR data normalized to reference gene Rpl19 and expressed as fold change relative to 0 h or WT control. N = 4 biological replicates of cells pooled from ovaries of 3 females. (D) Western blot for ZBTB16 protein in granulosa cells of PRKO, PRAKO and PRBKO mice at 10 h post-hCG. (E) Quantification of Western blot ZBTB16 expression relative to B-actin loading control. N = 3 pools of cells from 3 females. Mean ± SEM, statistical significance determined by one-way ANOVA. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.