Abstract
It was shown recently that recombinant Escherichia coli, defective in the β-oxidation cycle and harboring a medium-chain-length (MCL) poly(3-hydroxyalkanoate) (PHA) polymerase-encoding gene of Pseudomonas, is able to produce MCL PHA from fatty acids but not from sugars or gluconate (S. Langenbach, B. H. A. Rehm, and A. Steinbüchel, FEMS Microbiol. Lett. 150:303–309, 1997; Q. Ren, Ph.D. thesis, ETH Zürich, Zürich, Switzerland, 1997). In this study, we report the formation of MCL PHA from gluconate by recombinant E. coli. By introduction of genes coding for an MCL PHA polymerase and the cytosolic thioesterase I (′thioesterase I) into E. coli JMU193, we were able to engineer a pathway for the synthesis of MCL PHA from gluconate. We used two expression systems, i.e., the bad promoter and alk promoter, for the ′thioesterase I- and PHA polymerase-encoding genes, respectively, which enabled us to modulate their expression independently over a range of inducer concentrations, which resulted in a maximum MCL PHA accumulation of 2.3% of cell dry weight from gluconate. We found that the amount of PHA and the ′thioesterase I activity are directly correlated. Moreover, the polymer accumulated in the recombinant E. coli consisted mainly of 3-hydroxyoctanoate monomers. On the basis of our data, we propose an MCL PHA biosynthesis pathway scheme for recombinant E. coli JMU193, harboring PHA polymerase and ′thioesterase I, when grown on gluconate, which involves both de novo fatty acid synthesis and β-oxidation.
Polyhydroxyalkanoates (PHAs) are polyesters of 3-hydroxyacids produced as intracellular granules by a large variety of bacteria (10, 23, 37). Because of their potential use as biodegradable thermoplastics and as biopolymers being produced from renewable resources, PHAs have been the focus of extensive research of groups from academia and industry (2, 5, 36). Pseudomonads synthesize mainly medium-chain-length (MCL) PHAs, formed of monomers of 6 to 14 carbons (9, 21, 24). Although PHAs have been commercially developed and marketed (18), their widespread use has been hindered by the high cost of production (1, 27). Therefore, alternative strategies for PHA production are being investigated. PHA production costs can be reduced by several means including the use of cheap substrates, especially carbohydrates such as sugars or molasses (17, 28, 41), or enhancement of the product yield, e.g., by using recombinant Escherichia coli (27). E. coli holds promise as a source of economical PHA production because of its high productivity, the easy purification of PHA, and the lack of a depolymerase system degrading the synthesized polymer (13, 15, 27). Moreover, transgenic plants are potential candidates for large-scale production at relatively low prices, if PHAs can amount to 20 to 40% of the dry weight (29, 30, 39).
Since all wild-type E. coli strains and wild-type plants are unable to synthesize PHA, these organisms have to be equipped with at least the PHA polymerase-encoding gene, which is the key enzyme for PHA accumulation, since it connects the 3-hydroxyacyl coenzyme A (CoA) units to the polymer. From studies on Pseudomonas oleovorans, it is known that two PHA polymerases, PhaC1 and PhaC2, exist (22). Recently it was found that recombinant E. coli deficient in β-oxidation and harbouring an MCL PHA polymerase can produce MCL PHAs when grown on related substrates such as fatty acids whereas it produced no PHA on carbohydrate substrates such as glucose (25, 32). From studies on Pseudomonas putida KT2442 (3), it is known that three main pathways are involved in the synthesis of the 3-hydroxyalkanoate precursors: β-oxidation, de novo fatty acid biosynthesis, and elongation of 3-hydroxyalkanoates by acetyl-CoA molecules. During growth on fatty acids the β-oxidation pathway is the most active, whereas during growth on carbohydrate or carbohydrate-derived substrates such as sugars or gluconate, the fatty acid synthesis pathway provides the PHA precursors (20). It has been shown that both the β-oxidation and de novo fatty acid biosynthesis routes can function simultaneously in the synthesis of PHA (19). Much less is known about the link between the metabolites of the fatty acid metabolic pathways and the PHA precursor. It has been shown that Pseudomonas contains a 3-hydroxyacyl acyl carrier protein (ACP)→CoA transferase (31). However, it is not known whether additional proteins like a 2-trans-enoyl (ACP→CoA) transferase or a thioesterase also link the fatty acid synthesis pathway with PHA precursor formation in Pseudomonas. In E. coli, a well-characterized protein which could play a role as link is the ′thioesterase I, whereas 3-hydroxyacyl (ACP→CoA) transferase has not been detected.
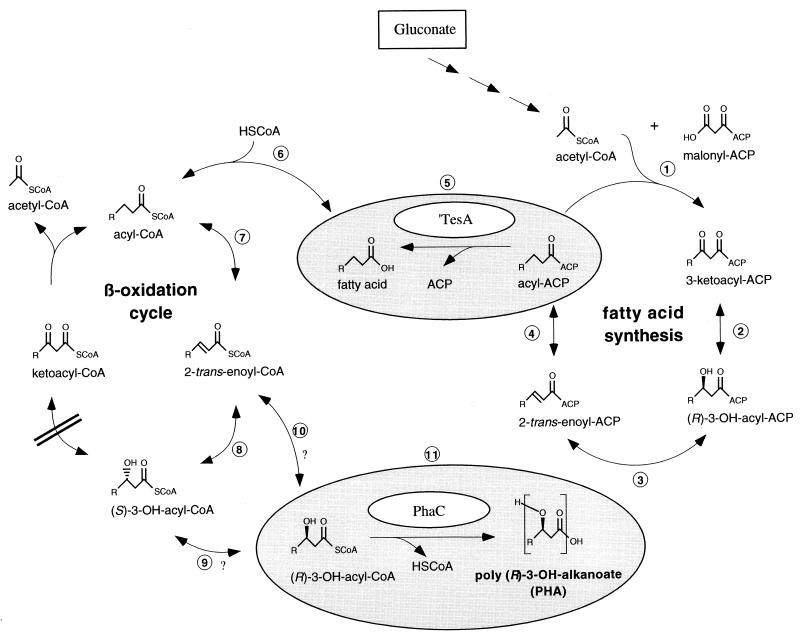
In this study, we equipped E. coli JMU193 (33), deficient in a functional β-oxidation, with a PHA polymerase from P. oleovorans and the thioesterase I from E. coli, which was modified by deletion of its leader sequence, trapping the periplasmic protein in the cytosol (called ′thioesterase I) (6). This recombinant was able to accumulate MCL PHA from gluconate, suggesting a PHA biosynthesis pathway which links de novo fatty acid synthesis and β-oxidation (see Fig. 4).
FIG. 4.
Hypothetical pathway of MCL PHA biosynthesis of PHA polymerase- and ′thioesterase I-containing E. coli JMU193 grown on gluconate. Gluconate is degraded via the central carbohydrate metabolism (indicated by triple arrows), leading to acetyl-CoA. Via four conversions of the fatty acid synthesis pathway, acetyl-CoA is metabolized to acyl-ACP; acetyl-ACP, transferred from acyl-CoA, and malonyl-ACP are coupled to give 3-ketoacyl-ACP by the β-ketoacyl-ACP synthase (step 1). 3-ketoacyl-ACP is converted to (R)-3-hydroxyacyl-ACP by the β-ketoacyl-ACP reductase (step 2). The β-hydroxyacyl-ACP dehydrase (step 3) yields 2-trans-enoyl-ACP, which is metabolized by enoyl-ACP reductase (step 4) to acyl-ACP. Acyl-ACP is hydrolyzed by the ′thioesterase I (step 5), resulting in the corresponding fatty acid, which is activated by acyl-CoA synthase (step 6) to the corresponding acyl-CoA. Acyl-CoA is degraded in the β-oxidation cycle, resulting in 2-trans-enoyl-CoA by the acyl-CoA dehydrogenase (step 7) and yielding (S)-3-hydroxyacyl-CoA by the action of the enoyl-CoA hydratase (step 8). Because of the fadB mutation of E. coli JMU193 (indicated by double sticks), these latter intermediates accumulate and can be transferred by either an isomerase (step 9) or (R)-specific hydratase (step 10) into (R)-3-hydroxyacyl-CoA, which is converted by the PHA polymerase (step 11) into PHA. Reactions enclosed in ellipsoids are carried out by genes which were introduced into E. coli JMU193 encoding the ′thioesterase I and PhaC1 and PhaC2 proteins. Question marks indicate uncertainties about the actual pathway according to data from Pseudomonas.
MATERIALS AND METHODS
Media and growth conditions.
E. coli strains were grown at 37°C in complex Luria-Bertani medium (34) or in minimal medium E2 (24) supplemented with 1% (wt/vol) gluconate. The E2 cultures were inoculated from exponentially growing Luria-Bertani medium precultures. Cells were cultivated in Erlenmeyer flasks and incubated with shaking at 225 rpm. Antibiotics were added as needed: 50 μg of kanamycin per ml, 100 μg of ampicillin per ml, 50 μg of streptomycin per ml, 30 μg of chloramphenicol per ml. Media were solidified with 1.5% (wt/vol) agar for plate experiments. Cell densities were measured spectrophotometrically at 450 nm (40). Cultures were harvested by centrifugation and washed with 10 mM MgSO4 to remove unmetabolized substrate. For determination of the amount of PHA, the cell pellet was lyophilized. For induction of the bad promoter, arabinose at concentrations ranging from 0 to 2% (wt/vol) was added. To induce the alk promoter, dicyclopropylketone (DCPK) was added during the early exponential phase, at an optical density at 450 nm of 0.4, in the range of 0 to 0.05% (vol/vol). Incubation continued overnight, and samples were obtained as described in Results. To induce the alk promoter, pCK01-alkS, which contains the alkS regulatory gene, was cotransformed with the alk promoter expression plasmid pET702. The strains and plasmids used are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JMU193 | fadR::Tn10 fadB64 | 33 |
| P. putida KT2442 | hsdR1 hsdM+, Rfr | 3 |
| Plasmids | ||
| pGEc404 | Kmr, Smr, RSF1010 ori Mob+phaC2 pJRD215 | 22 |
| pBAD22 | Apr, pBR322 ori badp | 14 |
| pHC122 | badp-′tesA, pBAD22 | 6 |
| pET702 | Smr/Spr, RSF1010 ori Mob+alkp– phaC1–VSVG-tag, pVLT35 | 32 |
| pCK01-alkS | lacZp-alkS, Cmr, pSC101 ori V | S. Panke (un-published) |
| pCY322 | Kmr, Apr, pBR322 ori badp | 6 |
| pCY323 | badp-′tesA, pCY322 | 6 |
PHA determination.
For a qualitative analysis of PHA accumulation, cells were observed by light microscopy after being stained with Sudan black (12, 35). Heat-fixed stained samples of cell material were observed with a Leitz DMR phase-contrast light microscope (Leica) at a magnification of ×100. PHA accumulation and composition were analyzed essentially as described by Lageveen et al. (24). Lyophilized cells (5 mg) were subjected to methanolysis (2.5 h at 100°C) in an equal volume of chloroform containing methylbenzoate as an internal standard (2 ml) and a mixture of 15% sulfuric acid and 85% methanol (2 ml). After phase separation and two washes with water, the organic phase was dried with Na2SO4 and analyzed by gas chromatography (GC). The methyl esters were analyzed on a Fisons HRGC Mega2 gas chromatograph equipped with a 30-m-by-0.32-mm Optima-1-0.25-μm column (Machery-Nagel) operating in split mode (split ratio, 25:1) with temperature programming (60°C for 2 min, increments of 5°C/min up to 200°C and increments of 40°C/min up to 280°C, 5 min at 280°C). For peak identification, a PHA standard mixture from P. putida KT2442 was used. The average results of two independent experiments are shown. Moreover, GC-mass spectrometry (MS) spectra were obtained with a Fisons HRGC Mega2 gas chromatograph equipped with a 30-m-by-0.32-mm Optima-1-0.25-μm column attached by a modified assay to a Fisons MD800 mass spectrometer. Spectra were obtained as electron impact spectra (70 eV). For GC-MS analysis, the trimethylsilyl (TMS) derivatives of the 3-hydroxyalkanoic acids were analyzed. TMS derivatization of 3-hydroxyalkanoate methyl esters was accomplished by the addition of 50 μl of N,O-bis(trimethylsilyl)acetamide to a mixture of 200 μl of methanolized sample and 800 μl of chloroform and heating for 15 min at 80°C. An aliquot was analyzed by GC-MS (26).
′Thioesterase I assay.
′Thioesterase I was measured spectrophotometrically in crude extracts by a modification of the assay of Barnes and Wakil (4) by monitoring the increase in absorbance at 412 nm, assuming a molar extinction coefficient of reduced 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) of 13,600 M−1 cm−1 (11). The assay mixture for ′thioesterase I contained 50 mM potassium phosphate buffer (pH 7.4), 0.1 mg of bovine serum albumin per ml, 0.1 mM DTNB, 0.07 mM palmitoyl-CoA, and crude extract containing approximately 1 mg of total protein. The total volume of the reaction mixture was 1 ml, and all the experiments were performed at 25°C. One enzyme unit was defined as the amount of protein necessary for the hydrolysis of 1 μmol of palmitoyl-CoA per min under these conditions, and activity was expressed as milliunits per milligram of protein. The reaction was started by addition of the substrate. Duplicate samples from two independent experiments were measured for determination of enzymatic activity. The ′thioesteraseI-specific substrate hydrolysis in crude extract was determined by total substrate hydrolysis activity in ′tesA-expressing strains corrected for substrate hydrolysis of non-′tesA-expressing strains (background hydrolysis activity caused by chromosomally encoded ′thioesterase I and other thioesterases). The latter typically amounted to 15% of the maximum activity of ′tesA-expressing strains. Protein concentrations were measured by using the Bradford assay (Bio-Rad Laboratories).
RESULTS
Formation of MCL PHA from gluconate by E. coli.
Initial analysis of MCL PHA production from gluconate was carried out with E. coli JMU193, a fadB mutant that is defective in β-oxidation. E. coli JMU193 was equipped with plasmids pGEc404 and pHC122, harboring the PHA polymerase-encoding gene phaC2 from P. oleovorans and the ′thioesterase I-encoding ′tesA gene from E. coli, respectively. The phaC2 gene was constitutively expressed, whereas ′tesA expression was regulated by the addition of arabinose. To achieve this, we used a construct containing the bad promoter of the arabinose operon and its regulatory gene, araC. The AraC protein is both a positive and a negative regulator. In the presence of arabinose, transcription from the bad promoter is turned on. The strain was grown in E2 minimal medium containing gluconate as the sole carbon source, appropriate antibiotics, and 0.01% (wt/vol) arabinose for induction of the bad promoter. After monitoring the culture for PHA accumulation by Sudan black staining, the cells were harvested and assayed for polymer content 45 h after they reached the stationary phase. In the control strain, E. coli JMU193(pGEc404, pBAD), lacking the ′tesA gene, PHA was detected only in trace amounts (<0.1%). In contrast, E. coli JMU193(pGEc404, pHC122) accumulated PHA to 0.6% of cell dry weight (cdw) (Table 2).
TABLE 2.
PHA accumulation from gluconate in recombinant E. coli JMU193a harboring PHA polymerase and ′thioesterase I
| Recombinantb | Plasmid | Genec | Inducer | Amt of PHAs (%, wt/wt) at:
|
|
|---|---|---|---|---|---|
| 25 h | 45 h | ||||
| 1 | pGEc404, pHC122 | phaC2, ′tesA | Arabinose | NDd | 0.6 |
| 2 | pGEc404, pBAD22 | phaC2 | Arabinose | ND | <0.1 |
| 3 | pET702, pCK01-alkS, pCY323 | phaC1-VSVG, alkS, ′tesA | DCPK, arabinose | 1.2 | 2.3 |
| 4 | pET702, pCK01-alkS, pCY322 | phaC1-VSVG, alkS | DCPK, arabinose | 0e | <0.1 |
| 5 | pCY323 | ′tesA | Arabinose | 0 | 0 |
Cells were cultivated in E2 minimal medium with 1% gluconate, antibiotics, and inducers (0.01% arabinose, 0.01% DCPK). At 25 and 45 h after the cells reached the stationary phase, samples were obtained and analyzed by GC as indicated in Materials and Methods.
recombinant E. coli JMU193.
The amount of polymer was calculated as the percentage of the cdw.
ND, not determined
below GC detection limit.
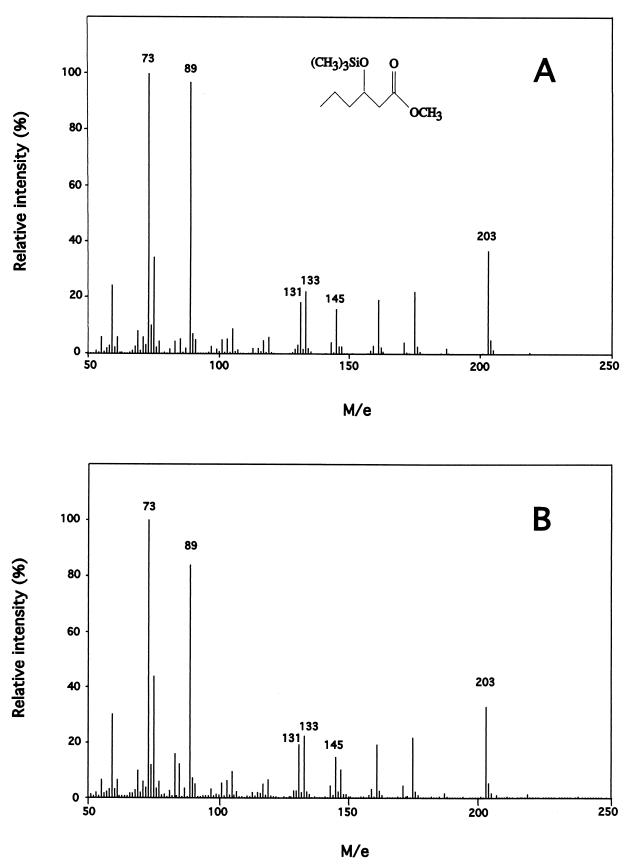
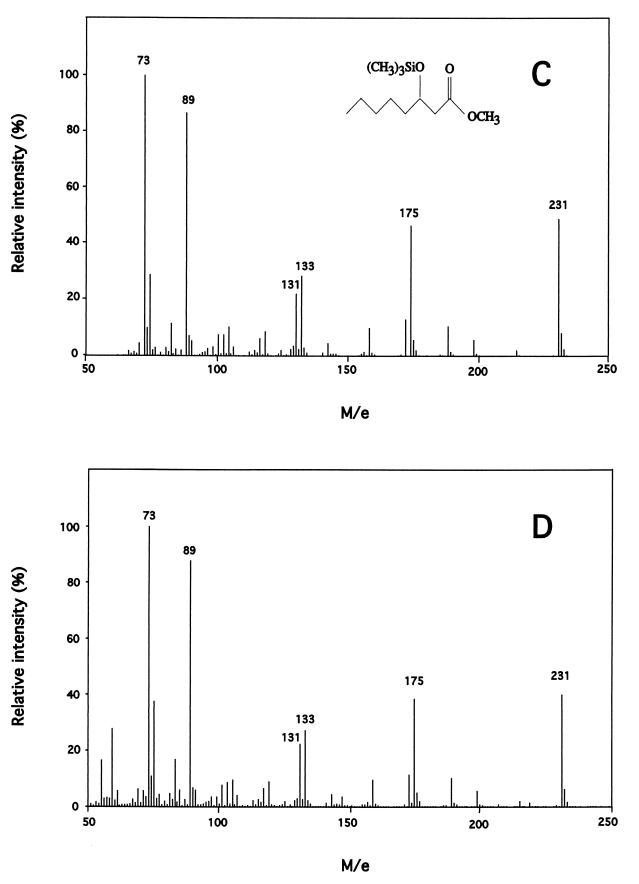
To investigate PHA polymerase PhaC1 of P. oleovorans and because of the instability of the above-described system (data not shown), we tested other constructs for MCL PHA formation in E. coli. We equipped JMU193 with pCY323 harboring the ′tesA gene and carrying a kanamycin resistance gene cassette, pET702 harboring the PHA polymerase phaC1 gene expressed through the inducible alk promoter and containing a sequence encoding the C terminus of the vesicular stomatitis virus glycoprotein (VSVG-tag), and pCK01-alkS encoding the AlkS regulatory protein of the alk promoter. Strains were grown as described above, except that 0.01% (vol/vol) DCPK was added to induce phaC1–VSVG-tag expression through the alk promoter. PHA accumulated to a maximum of 2.3% of cdw 45 h after the cells reached the stationary phase, which is 15% of the amount of MCL PHA accumulated by P. putida KT2442 when grown on gluconate, which served as a positive control (data not shown). Only trace amounts of PHA (<0.1% of cdw) accumulated in control strain JMU193(pET702, pCK01-alkS, pCY322), lacking ′tesA. In control strain JMU193(pCY323), harboring ′tesA and lacking phaC1, no PHA was produced (Table 2). To identify PHA monomers unequivocally from gluconate-grown E. coli JMU193 recombinants, PHA from lyophilized cells was subjected to methanolysis, GC, and GC-MS analysis. Monomers derived from the E. coli polymer were compared to monomers derived from a PHA standard, obtained from P. putida KT2442 grown on gluconate. Figure 1 shows that E. coli JMU193(pET702, pCK01-alkS, pCY323), harboring ′thioesterase I and PHA polymerase, produced PHA with 3-hydroxyhexanoate, 3-hydroxyoctanoate, and 3-hydroxydecanoate monomers.
FIG. 1.
Electron ionization mass spectra of the TMS derivatives of standard and E. coli-derived 3-hydroxyalkanoate methylesters. (A and B) TMS methyl esters of 3-hydroxyhexanoate (Mw 218) derived from a PHA standard from P. putida KT2442 (A) and from E. coli JMU193 (pET702, pCK01-alkS, pCY323) (B) grown on gluconate. Characteristic peaks: basis peak: m/e 203, M−15; m/e 145, M−73, m/e 133, (C5H13SiO2)+, m/e 131, (C5H11SiO2)+, m/e 89, (CH3)3SiO+, m/e 73, (CH3)3Si+. (C and D) TMS methyl esters of 3-hydroxyoctanoate (Mw 246) derived from a PHA standard from P. putida KT2442 (C) and from E. coli JMU193 (pET702, pCK01-alkS, pCY323) (D) grown on gluconate. Characteristic peaks: basis peak: m/e 231, M−15; m/e 175, (CH3)3SiO+⩵CHCH2CO2CH3; m/e 133, (C5H13SiO2)+; m/e 131, (C5H11SiO2)+; m/e 89, (CH3)3SiO+; m/e 73 (CH3)3Si+. (E and F) TMS methyl esters of 3-hydroxydecanoate (Mw 274) derived from a PHA standard from P. putida KT2442 (E) and from E. coli JMU193 (pET702, pCK01-alkS, pCY323) (F) grown on gluconate. Characteristic peaks: m/e 259, M−15; m/e 175 (CH3)3SiO+⩵CHCH2CO2CH3; m/e 133, (C5H13SiO2)+; m/e 131, (C5H11SiO2)+; m/e 89, (CH3)3SiO+; m/e 73, (CH3)3Si+.
Effect of the alk promoter–phaC1–VSVG-tag inducer DCPK on gene expression and polymer content.
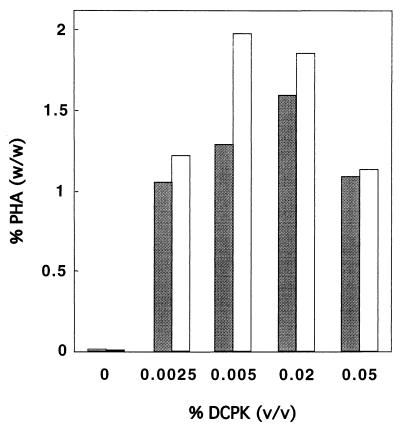
Since application of an inducible expression system for phaC gene expression can result in significantly increased PHA accumulation compared to the wild-type expression system in recombinant E. coli (32), we used the inducible alk promoter–phaC1–VSVG-tag system to determine the optimal PHA polymerase level for maximum MCL PHA production in JMU193 recombinants. E. coli JMU193(pET702, pCK01-alkS, pCY323), containing alk promoter–phaC1–VSVG-tag, alkS, and ′tesA, was grown in E2 minimal medium with gluconate as the carbon source and 0.01% (wt/vol) arabinose as the inducer. Cells were induced in the early exponential phase with DCPK in concentrations ranging from 0 to 0.05% (vol/vol). The PHA content and monomer composition were determined 25 and 44 h after the cells reached the stationary phase. If no DCPK was added to the cultures, only trace amounts of PHA (<0.1%) were accumulated. With increasing DCPK concentrations, more PHA was produced. The optimal DCPK concentration for maximum PHA production in JMU193 recombinants was between 0.005 and 0.02% (vol/vol), resulting in a maximum PHA concentration of 2.0% of cdw 44 h after the cells reached the stationary phase. Further increases of the DCPK concentration resulted in a decrease of the polymer content to half of this level (Fig. 2). These data are in accordance with previous results showing that PHA polymerase production and PHA accumulation are proportional only at low inducer concentrations. At medium and high inducer concentrations, PHA polymerase production shows a saturation profile, while in E. coli recombinants PHA accumulation decreases at higher polymerase levels (32). Longer incubation times in the stationary phase (44 h compared to 25 h) resulted in higher maximum PHA concentrations (2.0% compared to 1.3% of cdw). Variation of the DCPK concentration had only minor effects on the monomer composition of the PHA produced. Depending on the DCPK concentration, the synthesized polymer consisted of 27 to 33 mol% of 3-hydroxyhexanoate, 62 to 67 mol% of 3-hydroxyoctanoate, and 5 to 6 mol% of 3-hydroxydecanoate (Table 3). Interestingly the predominant monomer in the PHA of JMU193 recombinants was 3-hydroxyoctanoate, unlike the 3-hydroxydecanoate found in P. putida (20).
FIG. 2.
PHA accumulation by E. coli JMU193(pET702, pCK01-alkS, pCY323) as a function of DCPK concentration. Cells were cultivated in E2 minimal medium with 1% (wt/vol) gluconate, antibiotics, 0.01% (wt/vol) arabinose, and DCPK as indicated. Samples were obtained 25 h (shaded bars) and 44 h (open bars) after the cells reached the stationary phase, lyophilized, and analyzed by GC.
TABLE 3.
PHA content and monomer composition of E. coli JMU193(pET702, pCK01-alkS pCY323)
| Inducer and concna | Amt of PHA (%, wt/wt) | PHA monomer compositionb (mol%)
|
||
|---|---|---|---|---|
| C6 | C8 | C10 | ||
| DCPK | ||||
| 0 | <0.1 | —c | — | — |
| 0.0025 | 1.2 ± 0.02 | 29 | 65 | 6 |
| 0.005 | 2.0 ± 0.01 | 27 | 67 | 6 |
| 0.02 | 1.9 ± 0.04 | 30 | 65 | 5 |
| 0.05 | 1.1 ± 0.01 | 33 | 62 | 5 |
| Arabinose | ||||
| 0 | <0.1 | — | — | — |
| 0.001 | 0.26 ± 0.006 | 33 | 64 | 3 |
| 0.01 | 1.24 ± 0.04 | 23 | 69 | 8 |
| 0.1 | 0.19 ± 0.06 | 21 | 71 | 8 |
| 2 | <0.1 | — | — | — |
Concentrations are percentages (vol/vol) for DCPK and percentages (wt/vol) for arabinose.
Cells were cultivated in E2 minimal medium with 1% gluconate, antibiotics, DCPK, and arabinose at the concentrations indicated. When varying the arabinose concentration, 0.01% DCPK was added, and when varying the DCPK concentration, 0.01% arabinose was added. Cells were harvested 25 h (arabinose variation) or 46 h (DCKP variation) after reaching the stationary phase and lyophilized, and the PHA composition was measured by GC. C6, 3-hydroxyhexanoate; C8, 3-hydroxyoctanoate; C10, 3-hydroxydecanoate.
—, not analyzed due to the low overall PHA content (<0.1%, wt/wt).
Effect of bad promoter-′tesA inducer arabinose on ′thioesterase I activity and polymer content.
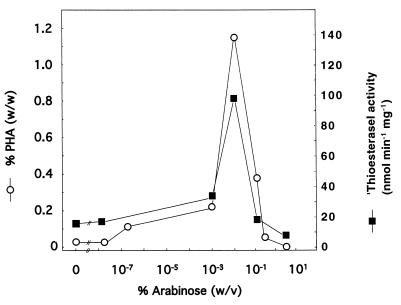
Excessively high levels of ′thioesterase I were expected to inhibit flux through the β-oxidation pathway via cleavage of the acyl-CoA thioesters (7). Therefore, we used a ′tesA expression system which allows fine-tuned regulation, to determine if there is a specific level of ′thioesterase I that is optimal for PHA synthesis. We chose a vector in which ′tesA was cloned downstream of the bad promoter, allowing low-level expression and modulation of gene expression over a wide range of inducer concentrations. The concentrations of arabinose that might permit the accumulation of PHA by cleavage of acyl-ACP esters were chosen on the basis of the data of Guzman et al. (14). We cultivated JMU193(pET702, pCK01-alkS, pCY323) on gluconate with the addition of appropriate antibiotics, 0.01% (wt/vol) DCPK, and arabinose concentrations ranging from 0 to 2% (wt/vol). At 25 h after the cells reached the stationary phase, they were harvested and the PHA content was determined. The PHA content increased with increasing arabinose concentration from trace amounts (<0.1% of cdw), when no arabinose was added, to a maximum of 1.2% of cdw at an arabinose concentration of 0.01% (wt/vol). Higher concentrations of arabinose resulted in a decreased polymer content (0.1% of cdw). ′Thioesterase I activity measurements in crude extracts from cells harvested 25 h after the cells reached the stationary phase showed low enzyme activities (20 mU/mg) when no arabinose was added, increasing to 104 mU/mg at 0.01% (wt/vol) arabinose and decreasing to 27 mU/mg at higher arabinose concentrations. As indicated in Fig. 3, the PHA accumulation profile in recombinant E. coli JMU193 correlated with the ′thioesterase I activity as a function of the arabinose concentration. From this and the fact that ′tesA-negative recombinants did not produce PHA, we conclude that there is a direct involvement of ′thioesterase I in MCL PHA biosynthesis in E. coli JMU193(pET702, pCK01-alkS, pCY323).
FIG. 3.
Correlation of PHA accumulation and ′thioesterase I activity. E. coli JMU193(pET702, pCK01-alkS, pCY323) was cultivated in E2 minimal medium with 1% (wt/vol) gluconate, antibiotics, 0.01% (wt/vol) DCPK, and arabinose as indicated. The cells were harvested 25 h after reaching the stationary phase and lyophilized, and the PHA content was determined by GC (open circles). ′Thioesterase I activity (solid squares) was determined in crude extracts by spectrophotometric measurements and corrected by subtracting the ′thioesterase activity of crude extracts of E. coli JMU193 lacking the ′thioesterase I-encoding gene, from crude extracts of E. coli JMU193 expressing the ′thioesterase I-encoding gene.
DISCUSSION
We have generated recombinant E. coli strains capable of producing MCL PHA from gluconate. To this end, we equipped an E. coli strain blocked in β-oxidation with the PHA polymerase encoded by the phaC1 or phaC2 gene from P. oleovorans and the cytosolic ′thioesterase I-encoding ′tesA gene from E. coli. The thioesterase hydrolyzes acyl-ACPs, producing enhanced intracellular levels of free fatty acids (6), which can then be channelled into the β-oxidation and used by the PHA polymerase as substrates for incorporation into PHA.
MCL PHA was detected only in recombinant E. coli JMU193 harboring both the phaC- and ′tesA-containing plasmids. The involvement of ′thioesterase I in PHA biosynthesis in JMU193 recombinants is further indicated by the direct correlation between ′thioesterase I activity and PHA accumulation. Additional indirect evidence for the importance of ′thioesterase I for PHA production is the fact that PHA accumulation started in the stationary phase. This is in agreement with findings of Cronan and coworkers, who reported that expression of ′tesA results in an increase in the total fatty acid synthesis, particularly in the stationary phase, whereas it is known that the overall rate of lipid synthesis is inhibited in cultures lacking ′thioesterase I (6, 8). Thus, MCL PHA biosynthesis in E. coli JMU193(pET702, pCK01-alkS, pCY323) may be assumed to include the following steps (Fig. 4). ′Thioesterase I-producing cells generate free fatty acids, which can accumulate to concentrations that are 15-fold higher than those seen in parallel cultures of strains lacking ′thioesterase I (6). The free fatty acids are produced by hydrolysis of the thioester bond linking acyl-ACPs generated during de novo fatty acid synthesis. Intracellular fatty acids are activated by action of the acyl-CoA synthase (FadD), resulting in acyl-CoAs, which can be channelled into the β-oxidation pathway. Because of the deficient β-oxidation cycle in E. coli JMU193, 3-hydroxyacyl-CoAs and 2-trans-enoyl-CoAs accumulate and are channelled into PHA by involvement of the PHA polymerase (32). Following our pathway hypothesis, we assume that recombinant E. coli JMU193 accumulated MCL PHA as a result of functioning of de novo fatty acid synthesis and certain steps of the β-oxidation cycle linked by the ′thioesterase I. Pseudomonas has also been shown to accumulate MCL PHA by simultaneous functioning of both fatty acid metabolic routes (19). Both PHA polymerase and ′thioesterase I were produced by fine-tuned gene expression systems. For induction of the alk promoter, which produces the PHA polymerase, an optimum inducer concentration of 0.01% DCPK was determined, which is in agreement with previous data and reflects the fact that only small amounts of PHA polymerase are sufficient for maximum PHA production (32). For the bad promoter, expressing the ′tesA gene, we found the inducer concentration which resulted in maximum PHA accumulation to be 0.01% arabinose. As expected, we found that no induction or induction with low inducer concentrations (≤10−7% arabinose) yielded low ′thioesterase I activity. The fact that ′thioesterase I activity could be detected in the repressed state (zero induction) is in accordance with the results of Guzman et al. (14). They found that although the bad promoter is tightly controlled, protein levels in the repressed states are not always zero. An increase of the inducer concentration led to an increased ′thioesterase I activity (140 mU at 10−2% arabinose), which correlated with maximum PHA production. Unexpectedly, with a further increase of the inducer concentration, the ′thioesterase I activity decreased; the reason for this is still unknown. Since we were not able to detect inclusion bodies of the ′thioesterase I protein (data not shown), the decrease in activity might well be due to feedback inhibition by free fatty acids. Further experiments must be carried out to optimize the ′thioesterase I activity in order to increase the PHA amount accumulated in the cell.
We have found that E. coli JMU193 equipped with the PhaC1 polymerase from P. oleovorans accumulates 3-hydroxyoctanoate as the predominant monomer of PHA when grown on gluconate. These observations are in agreement with previous findings showing that E. coli JMU193 harboring either PhaC1 or PhaC2 polymerase of P. oleovorans accumulated polymer with 3-hydroxyoctanoate as the predominant monomer when grown on fatty acids (32). In contrast, it was reported that when GPp104, an MCL PHA-negative mutant of P. putida KT2442 harboring either the PhaC1 or PhaC2 polymerase-encoding gene from P. oleovorans, is cultivated on glucose, the main constituent of the polyester is 3-hydroxydecanoate (20). Furthermore it was shown that many Pseudomonas species, including P. putida, P. aeruginosa, P. aureofaciens, and P. mendocina, accumulate a polymer with 3-hydroxydecanoate as the major constituent when cells are grown on gluconate or carbohydrate substrates (16, 20, 38). Similarly, when E. coli LS 1298, deficient in β-oxidation, is equipped with the PhaC1 polymerase-encoding gene from P. aeruginosa, and PHA is accumulated from fatty acids, 3-hydroxydecanoate is also the predominant monomer (25). The change of the predominant monomer in the MCL PHA when the PhaC polymerases of P. oleovorans are used in the P. putida mutant GPp104 and in E. coli JMU193 reflects the importance of the varying concentration profiles of 3-hydroxyacyl-CoAs for PHA monomer composition in these different strains.
In summary, this is the first report of production of MCL PHA in engineered E. coli grown on gluconate. In comparison to recombinant E. coli strains that are able to accumulate MCL PHA from fatty acids (25, 32), our strains have the economic advantage that they can use inexpensive carbohydrate or carbohydrate-derived substrates such as gluconate as the sole carbon source for MCL PHA production. Although the PHA content of engineered E. coli must be increased significantly before this method can be used for biotechnological application this study demonstrates that MCL PHA production from cheap carbon sources in E. coli, an important goal for the commercial application of these polymers, is basically feasible. Moreover, the strategy of MCL PHA production presented here might be relevant with respect to PHA synthesis in plants, for which several projects from industry- and university-related groups are under way.
ACKNOWLEDGMENTS
We thank John E. Cronan, Jr., for providing plasmids pBAD22, pHC122, and pCY322/3; Sven Panke for providing plasmid pCK01-alkS; and Wouter Duetz for making helpful suggestions.
This work was supported by grants from the Swiss Federal Office for Education and Science (BBW no. 96.0348) to S.K. and Q.R.
REFERENCES
- 1.Ackermann J U, Babel W. Approaches to increase the economy of the PHB production. Polym Degrad Stabil. 1998;59:183–186. [Google Scholar]
- 2.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagdasarian M, Lurz R, Ruckert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 4.Barnes E M, Wakil S W. Studies on the mechanism of fatty acid synthesis. J Biol Chem. 1968;243:2955–2962. [PubMed] [Google Scholar]
- 5.Byrom D. Polymer synthesis by microorganisms: technology and economics. Trends Biotechnol. 1987;5:246–250. [Google Scholar]
- 6.Cho H, Cronan J E., Jr Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J Biol Chem. 1995;270:4216–4219. doi: 10.1074/jbc.270.9.4216. [DOI] [PubMed] [Google Scholar]
- 7.Cronan J E., Jr In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J Bacteriol. 1997;179:1819–1823. doi: 10.1128/jb.179.5.1819-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronan J E., Jr Phospholipid alterations during growth of Escherichia coli. J Bacteriol. 1968;95:2054–2061. doi: 10.1128/jb.95.6.2054-2061.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Smet M J, Eggink G, Witholt B, Kingma J, Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol. 1983;154:870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi Y. Microbial polyesters. New York, N.Y: VCH; 1990. [Google Scholar]
- 11.Ellman G L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 12.Fahy P C, Persley G C. Plant bacterial diseases: a diagnostic guide. New York, N.Y: Academic Press, Inc.; 1983. [Google Scholar]
- 13.Fidler S, Dennis D. Polyhydroxyalkanoate production in recombinant Escherichia coli. FEMS Microbiol Rev. 1992;103:231–236. doi: 10.1016/0378-1097(92)90314-e. [DOI] [PubMed] [Google Scholar]
- 14.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn S K, Chang Y K, Lee S Y. Recovery and characterization of poly(3-hydroxybutyric acid) synthesized in Alcaligenes eutrophus and recombinant Escherichia coli. Appl Environ Microbiol. 1995;61:34–39. doi: 10.1128/aem.61.1.34-39.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haywood G W, Anderson A J, Ewing D F, Dawes E A. Accumulation of a polyhydroxyalkanoate containing primarily 3-hydroxydecanoate from simple carbohydrate substrates by Pseudomonas sp. strain NCIMB 40135. Appl Environ Microbiol. 1990;56:3354–3359. doi: 10.1128/aem.56.11.3354-3359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepner L. Cost analysis of fermentation processes. Chimia. 1996;50:442–443. [Google Scholar]
- 18.Hrabak O. Industrial production of poly-β-hydroxybutyrate. FEMS Microbiol Rev. 1992;103:251–256. [Google Scholar]
- 19.Huijberts G N M, de Rijk T C, de Waard P, Eggink G. 13C nuclear magnetic resonance studies of Pseudomonas putida fatty acid metabolic routes involved in poly(3-hydroxyalkanoate) synthesis. J Bacteriol. 1995;176:1661–1666. doi: 10.1128/jb.176.6.1661-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huijberts G N M, Eggink G, de Waard P, Huisman G W, Witholt B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl Environ Microbiol. 1992;58:536–544. doi: 10.1128/aem.58.2.536-544.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huisman G W, de Leeuw O, Eggink G, Witholt B. Synthesis of poly-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:1949–1954. doi: 10.1128/aem.55.8.1949-1954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 23.Kessler B, Witholt B. Synthesis, recovery and possible application of medium-chain-length polyhydroxyalkanoates: a short overview. Macromol Symp. 1998;130:245–260. [Google Scholar]
- 24.Lageveen R G, Huisman G W, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langenbach S, Rehm B H A, Steinbüchel A. Functional expression of the PHA synthase gene PhaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/s0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee E Y, Choi C Y. Gas chromatography mass spectrometric analysis and its application to a screening procedure for novel bacterial polyhydroxyalkanoic acids containing long chain saturated and unsaturated monomers. J Ferment Bioeng. 1995;80:408–414. [Google Scholar]
- 27.Lee S Y. Bacterial polyhydroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 28.Page W J. Production of poly-β-hydroxybutyrate by Azotobacter vinelandii strain UWD during growth on molasses and other complex carbon sources. Appl Microbiol Biotechnol. 1989;31:329–333. [Google Scholar]
- 29.Poirier Y, Dennis D E, Klomparens K, Somerville C. Polyhydroxybutyrate, a biodegradable thermoplastic, produced in transgenic plants. Science. 1992;256:520–523. doi: 10.1126/science.256.5056.520. [DOI] [PubMed] [Google Scholar]
- 30.Poirier Y, Nawrath C, Somerville C. Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Bio/Technology. 1995;13:142–150. doi: 10.1038/nbt0295-142. [DOI] [PubMed] [Google Scholar]
- 31.Rehm B H A, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The PHAG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme A transferase. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 32.Ren Q. Biosynthesis of medium chain length poly-3-hydroxyalkanoates: from Pseudomonas to Escherichia coli. Ph.D. thesis. Zürich, Switzerland: ETH Zürich; 1997. [Google Scholar]
- 33.Rhie H G, Dennis D. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvaleriate) synthesis in recombinant pha+Escherichia coli. Appl Environ Microbiol. 1995;61:2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schaad N W. Laboratory guide for identification of plant pathogenic bacteria. St. Paul, Minn: American Phytopathological Society; 1988. [Google Scholar]
- 36.Steinbüchel A. PHB and other polyhydroxyalkanoic acids. In: Rehm H-J, Reed G, editors. Biotechnology. Vol. 6. Weinheim, Germany: VCH; 1996. pp. 405–464. [Google Scholar]
- 37.Steinbüchel A. Polyhydroxyalkanoic acid. In: Byrom D, editor. Biomaterials. Novel materials from biological sources. Basingstoke, United Kingdom: Macmillan Publishers Ltd.; 1991. pp. 123–213. [Google Scholar]
- 38.Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Leij F R, Witholt B. Strategies for the sustainable production of new biodegradable polyesters in plants: a review. Can J Microbiol. 1995;41:222–238. [Google Scholar]
- 40.Witholt B. Method for isolating mutants overproducing nicotinamide adenine dinucleotide and its precursors. J Bacteriol. 1972;109:350–364. doi: 10.1128/jb.109.1.350-364.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Obias V, Gonyer K, Dennis D. Production of polyhydroxyalkanoates in sucrose-utilizing recombinant Escherichia coli and Klebsiella strains. Appl Environ Microbiol. 1994;60:1198–1205. doi: 10.1128/aem.60.4.1198-1205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]