Abstract
Mycobacterium sp. strain CH1 was isolated from polycyclic aromatic hydrocarbon (PAH)-contaminated freshwater sediments and identified by analysis of 16S rDNA sequences. Strain CH1 was capable of mineralizing three- and four-ring PAHs including phenanthrene, pyrene, and fluoranthene. In addition, strain CH1 could utilize phenanthrene or pyrene as a sole carbon and energy source. A lag phase of at least 3 days was observed during pyrene mineralization. This lag phase decreased to less than 1 day when strain CH1 was grown in the presence of phenanthrene or fluoranthene. Strain CH1 also was capable of using a wide range of alkanes as sole carbon and energy sources. No DNA hybridization was detected with the nahAc gene probe, indicating that enzymes involved in PAH metabolism are not related to the well-characterized naphthalene dioxygenase gene. DNA hybridization was not detected when the alkB gene from Pseudomonas oleovorans was used under high-stringency conditions. However, there was slight but detectable hybridization under low-stringency conditions. This suggests a distant relationship between genes involved in alkane oxidation.
Polycyclic aromatic hydrocarbons (PAHs) are fused-ring aromatic compounds whose presence in contaminated soils and sediments poses a significant risk to the environment, and they have cytotoxic, mutagenic, and in some cases carcinogenic effects on human tissue (13, 18, 25). PAHs are hydrophobic compounds, whose persistence within ecosystems is due chiefly to their low aqueous solubility. Microbial biotransformation is a major environmental process affecting the fate of PAHs in both terrestrial and aquatic ecosystems. The microbial degradation of PAHs having two or three rings is well documented (5–7, 11). Only within the last decade have a number of bacteria that metabolize larger PAH molecules been isolated. These include Alcaligenes denitrificans (33), Rhodococcus sp. strain UW1 (32), several Pseudomonas species (20, 31), and various Mycobacterium species (1, 3, 12, 23).
Different degradative genes are involved in the metabolism of aromatic and aliphatic hydrocarbons (26). The genes coding for the enzymes involved in the degradation of alkanes (alk) and naphthalene (nah) have been extensively characterized (16, 17, 24). Assessment of hydrocarbon utilization capacity for a large number of environmental isolates indicated that strains were capable of mineralizing either aromatic or aliphatic hydrocarbon compounds. Although Foght et al. postulated that bacteria having multidegradative capacity might exist, no strains were found with the capability to degrade both classes of compounds (10). A more recent report has established that the abilities to degrade aliphatic and aromatic hydrocarbons are not necessarily mutually exclusive (27). Two Pseudomonas strains that can degrade both naphthalene and alkanes have been isolated. These pseudomonads possess both the alk and nah catabolic pathways for hydrocarbon biodegradation (34).
Recent reports (27, 34) that both naphthalene and alkanes can be degraded by a single bacterial species have raised the question whether bacteria capable of degrading larger (three- or four-ring) PAHs could also metabolize alkanes and, if so, whether they would possess well-characterized hydrocarbon degradation genes, such as nahAc and alkB. We report here the isolation and characterization of Mycobacterium sp. strain CH1. This strain is capable of mineralizing phenanthrene, pyrene, and fluoranthene. In addition to PAHs, strain CH1 could use branched alkanes and n-alkanes (dodecane, hexadecane, and pristane) that are liquids at 25°C and also solid-phase alkanes (octadecane, docosane, and octacosane) as the sole carbon and energy source. These degradative capabilities were found to be unrelated to the nahAc gene and only distantly related to the alkB gene.
MATERIALS AND METHODS
Chemicals.
[4,5,9,10-14C]pyrene (specific activity, 32.2 mCi/mmol; purity, >95% by high-pressure liquid chromatography [HPLC]), [9-14C]phenanthrene (specific activity, 8.3 mCi/mmol; purity, >98% by HPLC), [3-14C]fluoranthene (specific activity, 55 mCi/mmol; purity, >95% by HPLC), [ring-U-14C]anthracene (specific activity, 15.5 mCi/mmol; purity, >97% by HPLC), [9-14C]fluorene (specific activity, 11 mCi/mmol; purity, >98% by HPLC), [1-14C]naphthalene (specific activity, 10.1 mCi/mmol; purity, >98% by gas chromatography [GC] and HPLC), [1-14C]octadecane (specific activity, 3.6 mCi/mmol; purity, >98% by HPLC and GC), and [1-14C]dodecane (specific activity, 3.7 mCi/mmol; purity, >98% by GC) were purchased from Sigma Chemical Co. The radiochemical purity of each 14C-labeled PAH or linear alkane was assayed by either GC or HPLC, using an in-line radiometric flow cell detector. Unlabeled pyrene, phenanthrene, fluoranthene, fluorene, anthracene, naphthalene, octadecane, dodecane, docosane, hexadecane, octacosane (purity, >95% by HPLC or GC), and chloramphenicol also were obtained from Sigma Chemical Co. The branched alkane pristane (2,6,10,14-tetramethylpentadecane) was obtained from Fluka. Amplitaq DNA polymerase was purchased from Perkin-Elmer. [32P]dATP was purchased from New England Nuclear. Bacteriological media and reagents were purchased from Life Technologies. All solvents and chemicals used were analytical grade reagents.
Culture conditions.
The minimal mineral medium used for enrichment of PAH-degrading bacteria contained (per liter) 1.0 g of (NH4)2SO4, 5.0 g of KH2PO4, 0.1 g of MgSO4 · 7H2O, 5 mg of Fe(NH4)2(SO4)2, and 1.0 ml of sterile trace metal solution (3). The pH of the culture solution was adjusted to 7.0 with sodium hydroxide. Riverine sediments known to be heavily saturated with large PAHs were obtained from the Great Lakes Environmental Research Laboratory (Ann Arbor, Mich.). Pyrene-degrading bacteria were isolated from an enrichment culture obtained by adding riverine sediment samples to minimal medium containing 400 mg of pyrene per ml and were detected on pyrene-coated mineral medium agar plates (14). Initial identification of the bacterial strain as a Mycobacterium species was made by Gram staining and fatty acid analysis conducted by Microbial ID Inc. (Newark, Del.).
Hydrocarbon biodegradation.
Hydrocarbon mineralization by Mycobacterium sp. strain CH1 was measured as 14CO2 in triplicate bioreactors (15). Cells were pregrown on 5.0 mM acetate to stationary phase in either the presence or absence of 300 mg of PAHs per liter. The cells were collected by low-speed centrifugation (7,700 × g) at 5°C, washed, and resuspended in mineral medium before use. The growth of strain CH1 was monitored spectrophotometrically at 600 nm.
DNA amplification, sequence analysis, and probe hybridization.
DNA was extracted by a method developed for gram-positive bacteria (35). The 16S rDNA was amplified by PCR (Rapid-Cycler, Idaho Falls, Idaho). The forward-primer (50F) sequence was 5′-ACCACATGCAAGTGCAACG-3′, and the reverse-primer (1392R) sequence was 5′-ACGGGCGGTGTGTAC-3′. PCR was conducted by using a touchdown protocol from 65 to 55°C as previously described (21). The amplification product was purified by agarose gel electrophoresis and cloned into the pGEM-T vector (Promega Scientific, Santa Barbara, Calif.), and both strands were sequenced (Iowa State University Sequencing Facility). The sequences were compared to those in GenBank by using the Blast alignment tool (2).
Pseudomonas putida NCIB 9816 was used as the sequence source for the naphthalene dioxygenase gene probe. Total DNA was used, and amplification of the nahAc gene (1,245 bp) was carried out with 5′-AAGCACCTGATTCATGGCGATGA-3′ as the forward primer and 5′-GAACTCAGCCCAGTTGGAGCTGCTG-3′ as the reverse primer. P. oleovorans ATCC 29347 was used as the sequence source for the alkane hydroxylase gene probe. Amplification was carried out with 5′-GCAAATGAAACTGGTTGGG-3′ as the forward primer and 5′-GGGTAACCCGTCGGAAGAGC-3′ as the reverse primer to a 906-bp fragment. The gene probes were labeled with [32P]dATP in a PCR. P. putida NCIB 9816 DNA was used as a positive control for nahAc, and P. oleovorans DNA was used for alkB. Escherichia coli DNA was used as a negative control in the dot blot hybridization assays. A 10-μg portion of total DNA was denatured and applied to nylon membranes under low vacuum (Micron Separations, Inc.). The DNA was hybridized at 42°C in the presence of 50% formamide by standard procedures (19). The filters were washed under high-stringency (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.5% sodium dodecyl sulfate [SDS] at 68°C), intermediate-stringency (0.1× SSC plus 0.5% SDS at 37°C), or low-stringency (2.0× SSC plus 0.5% SDS at 23°C) conditions.
Nucleotide sequence accession number.
The 16S rDNA sequence has been deposited in GenBank (accession no. AF054278).
RESULTS AND DISCUSSION
Isolation and characterization of Mycobacterium sp. strain CH1.
The PAH-degrading strain was obtained from contaminated riverine sediment by standard culture enrichment techniques with pyrene as the sole source of carbon and energy. When colonies were grown on pyrene-coated agar plates, zones of clearing appeared, indicating pyrene degradation. Upon isolation, the bacterium was found to be gram positive with yellow pigmentation. Fatty acid analysis was conducted, and the isolate was found to contain large amounts of tuberculosteric acid (10Me-18:0), a defining characteristic of Mycobacterium species. DNA sequence analysis of PCR-amplified 16S rDNA was also performed and confirmed the genus identification of the pyrene-degrading microorganism as Mycobacterium. DNA sequence analysis also differentiates strain CH1 from the other PAH-utilizing mycobacteria, Mycobacterium sp. strains RJG11-135, PYR-1, and PAH 135.
Degradation of PAHs and polycyclic aliphatic hydrocarbons.
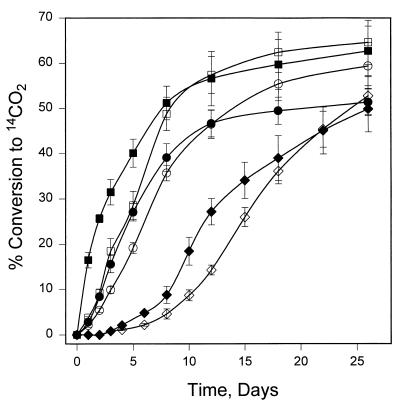
Biodegradation of PAHs was monitored by measuring the conversion of 14C-radiolabeled substrate to 14CO2. Previous studies have demonstrated that Mycobacterium sp. strain PYR-1 is subject to enzyme induction upon exposure to pyrene (12). The mineralization of pyrene and fluoranthene by Mycobacterium sp. strain CH1 when pregrown on acetate is shown in Fig. 1. Cells pregrown in the presence of pyrene or fluoranthene had a lag phase of less than 1 day prior to initiation of mineralization of either PAH. When cells are pregrown in the absence of PAHs, there is a significant lag phase prior to the onset of mineralization for both fluoranthene and pyrene degradation. The metabolism of other PAHs was examined by growing strain CH1 on acetate in the presence of phenanthrene, anthracene, fluorene, or naphthalene and measuring the mineralization of these PAHs. Phenanthrene was mineralized with a lag phase of less than 1 day, whereas anthracene, fluorene, and naphthalene were not mineralized (data not shown). To further investigate the expression of degradative enzymes, the effect of the protein synthesis inhibitor chloramphenicol on PAH mineralization was examined (data not shown). Chloramphenicol eliminated mineralization by cells grown on acetate only; however, CH1 cells grown in the presence of phenanthrene were still capable of converting this PAH to carbon dioxide in the presence of the inhibitor.
FIG. 1.
Effect of pregrowth conditions on the mineralization of pyrene and fluoranthene by Mycobacterium sp. strain CH1. Cells were resuspended in minimal salts medium at a concentration equivalent to 0.10 absorbance unit at 600 nm. Bioreactors were incubated in the presence of 25 mg of [14C]pyrene or [14C]fluoranthene per liter. The production of 14CO2 was measured as described in Materials and Methods. Pregrowth on acetate in the presence of pyrene is represented by ■ and •, and pregrowth on acetate in the presence of fluoranthene is represented by □ and ○. Cells pregrown on acetate in the absence of PAHs are represented by ◊ and ⧫. Mineralization of pyrene is represented by ■, □, and ⧫. Mineralization of fluoranthene is represented by •, ○, and ◊.
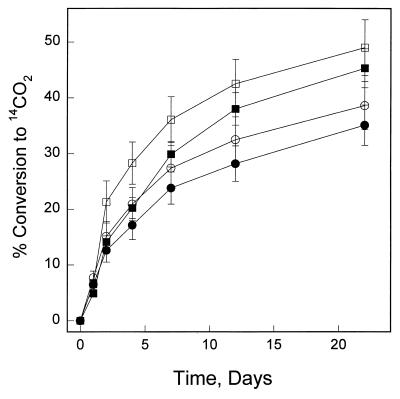
The ability of strain CH1 to metabolize liquid- and solid-phase long-chain aliphatic compounds was also investigated. The rate of mineralization of radiolabeled alkane is not directly related to the aqueous solubility of these sparingly soluble hydrocarbons (Fig. 2). Octadecane is mineralized at approximately the same rate as dodecane, although the aqueous solubility in distilled water of dodecane (0.0037 mg/liter) is greater than that of octadecane (0.0021 mg/liter) (28). The ability of both aromatic and aliphatic hydrocarbons to serve as the sole carbon and energy source for aerobic growth of Mycobacterium sp. strain CH1 was examined. Either pyrene or phenanthrene could be used as the sole carbon and energy source for growth. Although fluoranthene could be mineralized, it was unable to support cell growth. The liquid linear alkanes (dodecane and hexadecane) and the liquid branched-chain alkane (pristane), as well as the solid-phase long-chain alkanes (octadecane, docosane, and octacosane), were all able to serve as the sole carbon and energy source for growth. Cyclohexane was unable to support the growth of Mycobacterium sp. strain CH1 cells. The maximum growth attained, as measured by absorbance at 600 nm, for each substrate was as follows: phenanthrene, 0.82 ± 0.06; pyrene, 0.41 ± 0.08; dodecane, 0.80 ± 0.04; hexadecane, 0.80 ± 0.06; octadecane, 0.84 ± 0.11; docosane, 0.60 ± 0.04; octacosane, 0.48 ± 0.07; and acetate, 0.88 ± 0.03.
FIG. 2.
Mineralization of [14C]octadecane and [14C]dodecane by Mycobacterium sp. strain CH1. Cells were pregrown on acetate in the presence (□, ○) or absence (■, •) of octadecane. Cells were added to triplicate bioreactors at an initial concentration of 0.10 absorbance unit at 600 nm. Mineralization was measured by the evolution of 14CO2, as described in the text. □, ■, mineralization of octadecane; •, ○, mineralization of dodecane.
The nahAc gene, which codes for the large subunit of naphthalene dioxygenase from P. putida NCIB 9816, was used as a hybridization probe to assess the homology between naphthalene dioxygenase and PAH-degradative enzymes used by Mycobacterium sp. strain CH1 for PAH mineralization. Even under low-stringency conditions, there was no detectable hybridization to indicate that homology exists between the nahAc gene and genes encoding PAH-degradative enzymes in strain CH1. The alkB gene was used to assess homology between the aliphatic degradative pathway in strain CH1 and alkane hydroxylase from P. oleovorans. Weak hybridization under low-stringency conditions between CH1 DNA and the alkB gene was detected. This suggests slight homology between the genes involved in alkane oxidation (data not shown).
In the last decade, several bacteria species capable of degrading PAHs containing more than three rings have been isolated (1, 4, 12, 20, 23, 31–33). Of these microorganisms, few have been shown to utilize four-ring PAHs for growth in the absence of cofactors or surfactants (1, 4, 31–33). This capacity differentiates Mycobacterium sp. strain CH1 from both Mycobacterium sp. strain PYR-1 (12) and Mycobacterium sp. strain RJG II-135 (23), which require additional cofactors for cell growth on pyrene. Mycobacterium sp. strain BB1 can grow on fluoranthene, phenanthrene, and pyrene (4), differentiating it from Mycobacterium sp. strain CH1.
It has been demonstrated that genes coding for both aromatic and aliphatic hydrocarbon degradation exist in bacteria isolated from Alaskan shoreline sediments affected by the Exxon Valdez oil spill of 1989 (27). Additionally, two hydrocarbon-degrading psychrotolerant bacteria strains possess both alkane (alk) and naphthalene (nah) catabolic pathways (34). Recently, a PAH-degrading Mycobacterium species isolated from a PAH-contaminated site in New Zealand was shown to grow on dodecane and hexadecane (1). Mycobacterium sp. strain CH1 is capable of using liquid- and solid-phase, high-molecular-weight n-alkanes, as well as a branched-chain alkane (pristane), as the sole source of carbon and energy. These results suggest that the occurrence of both aromatic and aliphatic hydrocarbon degradative capacity within an individual strain may be more common than was previously thought. The lack of hybridization of DNA isolated from strain CH1 with the nahAc gene under low-stringency conditions indicates that the enzyme system involved in PAH degradation is unrelated to the well-characterized naphthalene dioxygenase pathway. The weak hybridization of the alkB gene, even under low-stringency conditions, suggests that there is only limited homology between the genes involved in alkane oxidation in P. oleovorans and Mycobacterium sp. strain CH1. The lack of hybridization of the nahAc and alkB gene probes by bacteria capable of degrading naphthalene and alkanes has been reported previously (22, 34). Our results reinforce the need for isolation of additional clones encoding enzymes involved in the initial oxidation step of three- and four-ring PAHs and alkanes. The isolation of these genes will provide an improved method for assessing the potential of microorganisms to degrade these sparingly soluble compounds in pristine and hydrocarbon-contaminated environments.
REFERENCES
- 1.Aislabie J, Hunter D W F, Lloyd-Jones G. Proceedings of the 4th International In Situ and On-Site Bioremediation Symposium. Richardland, Wash: Batelle Press; 1997. Polyaromatic compound-degrading bacteria from a contaminated site in New Zealand; pp. 219–224. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:401–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Beauchop T, Elsden S R. The growth of microorganisms in relation to their energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 4.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium species. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons. Adv Appl Microbiol. 1984;30:31–71. doi: 10.1016/s0065-2164(08)70052-2. [DOI] [PubMed] [Google Scholar]
- 6.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 7.Cerniglia C E, Heitcamp M A. Microbial degradation of polycyclic aromatic hydrocarbons (PAH) in the aquatic environment. In: Varanasi U, editor. Metabolism of polycyclic aromatic hydrocarbons in the aquatic environment. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 41–68. [Google Scholar]
- 8.Churchill P, Churchill S. Surfactant-enhanced biodegradation of solid alkanes. J Environ Sci Health. 1997;32:293–306. [Google Scholar]
- 9.Churchill P F, Dudley R J, Churchill S A. Surfactant-enhanced bioremediation. Waste Manage. 1995;15:371–377. [Google Scholar]
- 10.Foght J M, Fedorak P M, Westlake W S. Mineralization of [14C]-hexadecane and [14C]-phenanthrene in crude oil: specificity among bacterial isolates. Can J Microbiol. 1990;36:169–175. doi: 10.1139/m90-030. [DOI] [PubMed] [Google Scholar]
- 11.Gibson D I, Subrahamanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Mercel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 12.Heitcamp M A, Franklin W, Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol. 1988;54:2549–2555. doi: 10.1128/aem.54.10.2549-2555.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvin T R, Martin J E. In-vitro and in-vivo embryo toxicity of fluroanthrene, a major prenatal component of diesel soot. Teratology. 1987;35:65A. [Google Scholar]
- 14.Kiyohara H, Nagao K, Yama K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982;43:454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knaebel D B, Vestal J R. A comparison of double vial to serum bottle radiorespirometry to measure microbial mineralization in soils. J Microbiol Methods. 1988;1:309–317. [Google Scholar]
- 16.Kok M, Oldenhuin R, van der Linden M P G, Raatjes P, Kingman J, van Lelyeld P H, Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene. J Biol Chem. 1989;254:5435–5441. [PubMed] [Google Scholar]
- 17.Kurkela S, Lehvasiaiho H, Palva E T, Terri T H. Cloning, nucleotide sequence, and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB 9816. Gene. 1988;73:356–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 18.LaVoce E I, Hecht S S, Bedenko V, Hoffman D. Identification of the mutagenic metabolites of fluorene, 2-methylfluorene, and 3-methylfluoranthene. Carcinogenesis. 1982;3:841–846. doi: 10.1093/carcin/3.8.841. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 20.Mueller J G, Chapman P J, Blattman B O, Pritchard P H. Isolation and characterization of a fluoranthene-utilizing strain of Pseudomonas paucimobilis. Appl Environ Microbiol. 1990;56:1079–1086. doi: 10.1128/aem.56.4.1079-1086.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, De Waal E C, Aitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellizari V H, Berborodnikov S, Quensen J F, Tiedje J M. Evaluation of strains isolated by growth on naphthalene and biphenyl for hybridization of genes to dioxygenase probes and polychlorinated biphenyl-degrading ability. Appl Environ Microbiol. 1996;62:2053–2058. doi: 10.1128/aem.62.6.2053-2058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider J J, Grosser R, Jayasimhulw K, Xue W, Warshawsky D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RGJII-135, isolated from a former coal gasification site. Appl Environ Microbiol. 1996;62:13–19. doi: 10.1128/aem.62.1.13-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon M J, Osslund T P, Saunders R, Ensley B D, Sugges S, Harcourt A, Suen W C, Cruden D, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 25.Sims P, Grover P L. Involvement of dihydrodiols and diol epoxides in the metabolic activation of polycyclic hydrocarbons other than benzo(a)pyrene. In: Gelborn H V, Ts’o P O P, editors. Polycyclic hydrocarbons and cancer. Vol. 3. New York, N.Y: Academic Press, Inc.; 1981. pp. 117–181. [Google Scholar]
- 26.Singer J T, Finnerty N R. Genetics of hydrocarbon utilizing microorganisms. In: Atlas R M, editor. Petroleum microbiology. New York, N.Y: McGraw Hill Book Co.; 1984. pp. 299–354. [Google Scholar]
- 27.Sotsky J B, Greer C W, Atlas R M. Frequency of genes in aromatic and aliphatic hydrocarbon biodegradation pathways within bacterial populations from Alaskan sediments. Can J Microbiol. 1994;40:981–985. doi: 10.1139/m94-157. [DOI] [PubMed] [Google Scholar]
- 28.Sutton C, Calder J A. Solubility of higher molecular weight n-paraffins in distilled water and seawater. Environ Sci Technol. 1974;8:654–657. [Google Scholar]
- 29.Takizawa N, Kaida S, Torique S, Montani T, Sawada T, Satoh S, Kiyohara H. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J Bacteriol. 1994;176:2444–2449. doi: 10.1128/jb.176.8.2444-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang W C, White J C, Alexander M. Utilization of sorbed compounds by microorganisms specifically isolated for that purpose. Appl Microbiol Biotechnol. 1998;49:117–121. doi: 10.1007/s002530051147. [DOI] [PubMed] [Google Scholar]
- 31.Thibault S L, Anderson M, Frankenberger W T., Jr Influence of surfactants on pyrene desorption and degradation in soils. Appl Environ Microbiol. 1996;62:283–287. doi: 10.1128/aem.62.1.283-287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter U, Beyer M, Klein J, Rehm H J. Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol. 1991;34:671–676. [Google Scholar]
- 33.Weisenfels W D, Beyer M, Klein J. Degradation of phenanthrene, fluorene, and fluoranthene by pure bacterial cultures. Appl Microbiol Biotechnol. 1990;32:479–484. doi: 10.1007/BF00903787. [DOI] [PubMed] [Google Scholar]
- 34.Whyte L G, Bourbonniere L, Greer C W. Biodegradation of petroleum hydrocarbons by psychrotropic Pseudomonas strains possessing both the alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol. 1997;63:3719–3723. doi: 10.1128/aem.63.9.3719-3723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Burns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]