Abstract
Community-acquired pneumonia requiring hospital admission is a prevalent and potentially serious infection, especially in high-risk patients (e.g., those requiring ICU admission or immunocompromised). International guidelines recommend early aetiological diagnosis to improve prognosis and reduce mortality. Syndromic panels that detect causative pathogens by molecular methods are here to stay. They are highly sensitive and specific for detecting the targets included in the test. A growing number of studies measuring their clinical impact have observed increased treatment appropriateness and decreased turnaround time to aetiological diagnosis, need for admission, length of hospital stay, days of isolation, adverse effects of medication and hospital costs. Its use is recommended a) per a pre-established protocol on making the diagnosis and managing the patient, b) together with an antimicrobial stewardship programme involving both the Microbiology Service and the clinicians responsible for the patient, and c) the final evaluation of the whole process. However, we recall that microbiological diagnosis with traditional methods remains mandatory due to the possibility that the aetiological agent is not included among the molecular targets and to determine the antimicrobial susceptibility of the pathogens detected.
Keywords: molecular diagnostic techniques, rapid diagnosis, high-throughput nucleotide sequencing / methods, respiratory tract infections, pneumonia / diagnosis, pneumonia / microbiology
INTRODUCTION
According to recent data from the World Health Organisation, lower respiratory tract infections, including pneumonia, are the third leading cause of death worldwide and are the most deadly infectious diseases.
Aetiological diagnosis is a challenge due to the difficulty in obtaining representative samples of the lower respiratory tract, except in intubated patients, and the low positivity of blood cultures.
Appropriate early antimicrobial treatment is essential to reduce mortality and improve patient outcomes.
Syndromic diagnostic panels that allow the detection of multiple microbial targets with short turnaround times have been available in recent years. In this brief review, we update the available evidence on their use.
CURRENT EPIDEMIOLOGY AND AETIOLOGY OF SEVERE COMMUNITY-ACQUIRED PNEUMONIA REQUIRING HOSPITAL ADMISSION
The severity of pneumonia ranges from mild to severe and is particularly dangerous in patients at the extremes of age, those with comorbidities (e.g., COPD) or immunocompromised.
Severe adult community-acquired pneumonia (CAP) is defined as pneumonia occurring in such patients who have not been hospitalised in the previous month.
CAP of moderate severity is usually treated in the inpatient ward (20-40% of cases) [1]. However, up to 1-10% of patients may require admission to an intensive care unit for management.
Several pathogens cause pneumonia, including viruses, bacteria and fungi. Traditionally, bacteria include Streptococcus pneumoniae, Haemophilus influenzae and atypical cases of pneumonia (e.g., Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella spp.). The most prevalent viruses are influenza A and B, respiratory syncytial virus (RSV), rhinoviruses, parainfluenza virus, and SARS-CoV-2. Less frequently, Nocardia spp. and mycobacteria are also found. The development of the disease largely depends on the host immune response, with pathogen characteristics having a less prominent role [2].
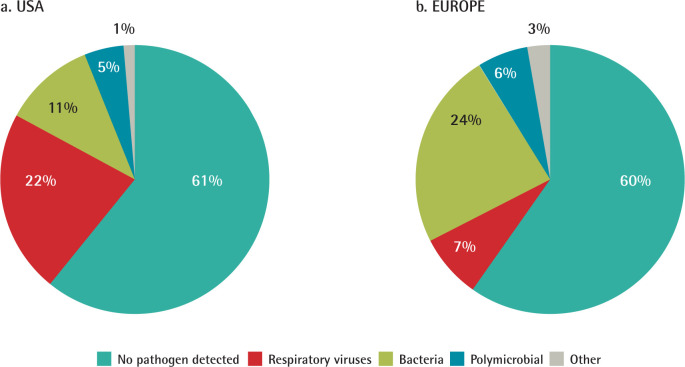
The prevalence of these pathogens varies between geographical regions. A recent study by Torres et al. shows these differences between Europe and the USA (Figure 1). In both cases, the aetiology was not discovered in up to 60% of patients. However, in Europe, bacteria predominate (24%), and, in the USA, respiratory viruses come in the first place (22%) [3]. Differences may be explained by the difficulty in obtaining valid respiratory samples to establish the diagnosis, antibiotics before sampling, and the sensitivity of diagnostic tests.
Figure 1.
Aetiology of CAP in the USA and Europe
Modified from Torres et al. [3]. Aetiology of CAP in adults in the USA from 2010-2012 (2,488 cases). Aetiology of CAP in adults in Europe from 2003-2014 (3,854 cases).
Schlaberg et al. studied viral diagnosis in children hospitalised with CAP without a previously identified aetiology by next-generation sequencing (RNA-seq) and pan viral group PCR for 19 viral families. These techniques were able to identify additional viruses in one-third of the patients. Human bocavirus, Coxsackieviruses, human parainfluenza virus 4, and human rhinoviruses C and A were more commonly detected in children with CAP compared with control subjects, but only human bocavirus was more common than in control subjects (19%; aOR 9.1, CI 1.6-103). This suggests that these pathogens may have played an etiologic role in CAP.
We have briefly reviewed the aetiology of CAP to show that the pathogens responsible for CAP are still the commonly recognised ones. Furthermore, these microorganisms are the ones that should be included in syndromic diagnostic panels using molecular methods to identify most of the targets of clinical interest.
TRADITIONAL MICROBIOLOGICAL DIAGNOSIS OF PNEUMONIA
International guidelines recommend reaching an aetio-logical diagnosis [4,5]. In addition, the appropriateness of early antibiotic or antiviral treatment leads to a decrease in mortality in this entity [6].
From the point of view of the Microbiology Service, the traditional techniques for the diagnosis of conventional infections by typical pathogens (Gram stain and culture of good quality samples from the lower respiratory tract, with identification of the potential pathogen and performance of antibio-gram tests) are not very sensitive and slow. Despite this, they continue to be used as the gold standard against which new and emerging diagnostic techniques are compared.
For viral infections, molecular techniques have taken over from older techniques (e.g. antigen detection, direct immunofluorescence, viral culture) and are considered the gold standard for diagnosing this group of microorganisms.
In addition to sputum sampling (for Gram stain and culture), blood cultures, urine for pneumococcal antigen and Legionella spp. detection, and nasopharyngeal exudate for SARS- CoV-2 detection, are obtained in most patients with moderate CAP admitted to the internal medicine ward [7].
WHAT ARE THE SYNDROMIC PANELS FOR THE DIAGNOSIS OF THIS ENTITY? WHAT IS THEIR DIAGNOSTIC YIELD?
Several syndromic panels are available for the etiological diagnosis of CAP (https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests ), which are summarised in Table 1. They differ in terms of the diagnostic technique used, the pathogens detected, the type of sample that can be used, the sample volume required, the time to results and the kind of result (e.g. qualitative or quantitative).
Table 1.
Syndromic panels for the diagnosis of community-acquired respiratory infections
| Diagnostic assay | Microorganisms detected | Type of sample | Turn-around-time |
|---|---|---|---|
| Verigene, Luminex | 6 viruses 3 bacteria |
Nasopharyngeal swab | <2h |
| NxTAG, Luminex | 18 viruses 3 bacteria |
NF, BAL, nasal aspirate, TA, sputum, FA | 5-6 h |
| DiagCore,Quiagen | 19 viruses 3 bacteria |
All types of samples | 1 h |
| Clart Pneumovir 2, Genomica | 18 viruses | NF, nasopharyngeal lavage, BAL | 2 h |
| Xpert Xpress SARS-CoV-2/Flu/RSV, Cepheid | 4 viruses | NF, nasal exudate, nasal lavage/aspiration | 36 min |
| ePlex Respiratory Pathogen 2, GenMark | 16 viruses 2 bacteria |
NF | 90 min |
| Unyvero, Curetis | 20 bacteria P. jirovecii 17 resistance markers |
Sputum, TA, BAL | <5 h |
| Anyplex II RV16, Seegene | 16 viruses | NF, nasopharyngeal aspirate, BAL | 4,5 h |
| RespiFinder 2SMART, PathoFinder | 20 viruses 4 bacteria |
Sputum, BAL, NF, nasopharyngeal aspirate | 2,5 h |
| bioFire FilmArray 2.0 Pneumonia plus, bioMerieux | 18 bacteria 9 viruses 7 Resistance markers |
Sputum, TA, BAL | <1 h |
| bioFire Respiratory Panel 2.1 Plus, bioMerieux | 4 bacteria 19 viruses |
NF | 45 minutes |
NF: nasopharyngeal exudate. TA: tracheal aspirate. FA: pharyngeal exudate. BAL: bronchoalveolar lavage
Challenges faced by these diagnostic panels include, among others, the prevalence of the aetiology for the choice of microorganisms included in the assay, the over-information that may occur for the prescribing doctor (e.g., risk of additional diagnostic studies and unnecessary treatments), interpretation of co-infection detection, the definition of the reference standard, rapid communication of results from the Microbiology Service (24/7), microbiological quality control of samples (e.g., prior Gram staining), use as point-of-care testing (POCT) outside the Microbiology Service and, finally, replacement or complement of other traditional diagnostic methods.
These assays have excellent diagnostic sensitivity and specificity [8-15]. The advantages of these syndromic panels lie in their ease of execution, the small sample volume required and the short time to result [16].
Disadvantages include that the panel composition is predefined, making it impossible to diagnose microorganisms not covered; diagnostic performance depends on the type of sample to be tested; some systems allow processing of individual samples, as they arrive in the laboratory, while others require batch testing; results are usually qualitative, except for Film Array®, which gives semi-quantitative results; it is not easy to differentiate between colonisation and infection by S. pneumoniae and H. influenzae; turn-around-time varies between commercial as-says; and, finally, the cost of these tests is high and has a direct impact on the budget of the Microbiology Service.
WHAT IS THE AVAILABLE EVIDENCE ON THE IMPACT OF USING THESE PANELS?
Since its appearance on the market, scientific evidence has been generated about its impact on the different process and outcome indicators [17,18]. The former include time to optimisation of antibiotic treatment and duration of antibiotic therapy. The latter include the need for hospital admission, length of stay, clinical cure, readmission or 30-day mortality, adverse drug reactions and hospital costs.
Among the publications available, we have selected the following that we consider to be of interest. Rappo et al. studied the impact of rapid diagnosis of respiratory viruses in adults using BIORFIRE Respiratory Panel® compared to standard diagnosis by viral antigen detection [19]. The study is a retrospective quasi-experimental work in the 2010-11 (standard diagnosis) and 2012 (PCR diagnosis) seasons. They included 339 patients diagnosed with a viral infection. The use of PCR allowed for shorter turnaround time (1.7 h vs 7.7 h), fewer admissions (50% vs 61%, p=0.046), shorter length of stay (38.8 h vs 49.8 h, p=0.040), shorter duration of antibiotic administration (23.7 h vs 48.1 h, p=0.032) and ordering fewer chest X-rays in this population (p=0.005). However, these differences did not hold when the diagnosis was of a respiratory virus other than influenza.
Rogers et al. found similar results in a subsequent study evaluating standard molecular diagnosis of influenza A/B in the 2011-12 season using BIO FIRE Respiratory Panel® in the following season (2012-13). This was a retrospective quasi-experimental study with 1136 children older than three months included, with a pneumonia prevalence of 32%. When using the syndromic panel, the authors found a shorter response time (6.4 h vs 18.7 h, p<0.001) and a higher number of patients diagnosed in the emergency department before admission (52% vs 13%, p<0.001). In terms of antibiotic use, there were no differences in the indication for antibiotic use. Still, when the result was received in less than four h, the duration of antibiotic treatment was shorter (p<0.003). Furthermore, in patients with a positive result, hospital stay and respiratory isolation duration were shorter (p=0.03 in both cases).
In a pragmatic, open-label, randomised controlled trial, Brendish et al. included 714 adult patients within 24 h or presenting to the emergency department with acute respiratory illness of fever over two winter seasons [20]. The routine use of molecular POCT for respiratory viruses did not reduce the proportion of patients treated with antibiotics significantly. However, many patients were started on antibiotics before the results of POCT could be made available. Despite this, more patients in the POCT group received single doses or brief antibiotics courses than patients in the control group (17% vs 9%, p=0.0047). POCT was also associated with a reduced length of stay (5.7 d vs 6.8 d, p=0.0443) and improved the use of anti-virals against influenza (91% vs 65%, p=0.0026) and was safe. We found equivalent results in other papers [21-23], some even with decreased hospital costs [22,24].
The impact of the BioFire FilmArray Pneumonia® panel on 259 BAL samples from adult inpatients was evaluated by Buchan et al. [14]. The use of this assay resulted in a 63.3% increase in specimens reported as positive. Over 99% of culture-negative discordant results were positive using an alternative molecular test or were below the culture threshold for reporting, suggesting that these were not false-positive detections. A review of patient medical records revealed the potential for antibiotic adjustment in 70.7% of patients, including discontinuation or de-escalation in 48.2% of patients, resulting in an average savings of 6.2 antibiotic days/ patient.
This increase in the number of aetiological diagnoses has also been highlighted in other works, with adjustment of antibiotic treatment in a high number of patients and increased de-escalation [15].
The above mentioned studies were carried out before the COVID-19 pandemic. In a more recent work, Barrasa et al. studied the prevalence of co-infections and secondary infections in COVID-19 patients using traditional cultures and the BIOFIRE® FILMARRAY® Pneumonia Panel plus (FA-RP). They included 92 consecutive adult patients admitted to the ICU at the Araba University Hospital in Vitoria-Gasteiz (Spain) with the diagnosis of severe pneumonia caused by SARS-CoV-2 between March 4th - June 2nd 2020 (first wave) [25]. In 63 patients, BAL or tracheal aspirates were collected for microbiologic culture, and in 33 (52%), the BioFire panel was also used (turn-around-time of about 67 min). None of the 33 FA-RP tests (14 performed on admission) identified other respiratory viruses. At admission or in the first 48 h of ICU stay, 32 microbial isolates were found in 24 patients (co-infections, 26%, 24/92). In these patients, concordant results between the FA-RP (≥ 104 DNA copies/ml) and cultures (BAL with a cut-off of 104 CFU/ ml) were obtained in 11 of 14 patients (overall agreement = 78%, kappa = 0.59 [95% CI 0.21–0.96]). Discordant results were obtained in three samples (Moraxella catarrhalis, Proteus spp and Streptococcus agalactiae). Conversely, 125 microbial isolates were found in 43 patients (secondary infections, 47%, 43/92) during ICU admission. Most samples were respiratory (52%), followed by urine (22%), blood (18%) and catheter tips (8%). The most commonly isolated microorganisms were P. aeruginosa, E. faecium and Enterobacterales, which represented half of the isolates in all secondary infections. Concordant results between the FA-RP and cultures were obtained in 12 out 19 patients (overall agreement = 63%, kappa = 0.31 [95% CI -0.05–0.67]), and discordant results were obtained in 6 samples, Enterococcus faecalis [2], Aspergillus fumigatus [2], Enterococcus faecium [1] and Candida albicans [1], targets not included in the panel. These results point to the need for microbiological diagnosis using real-time PCR and traditional cultures.
In summary, the use of syndromic multiplex polymerase chain reaction testing, coupled with antimicrobial stewardship, increases the timeliness of antiviral prescription in influenza patients and the rapid appropriateness of antibiotic treatment.
It is recommended to implement these systems together with the education of prescribing physicians.
The use of a pre-established diagnostic and management algorithm is recommended for the most significant clinical benefit. And monitoring end-user compliance with the algorithm to optimise patient management protocols.
These systems have been shown to support decision making (joint assessment of results and other diagnostic tests - e.g. procalcitonin).
These results justify the need for Clinical Microbiology Services to work 24-h a day, 7 days a week (24/7) [12,26-32].
WHAT ARE THE CURRENT RECOMMENDATIONS FOR THEIR USE
The use of syndromic pneumonia panels is recommended in symptomatic patients, those with new or worsening radiographic infiltrates, moderate to severe pneumonia, previous empirical antibiotic treatment, when there is concern about the presence of multi-resistant microorganisms or co-infections, for the detection of both bacteria and viruses (e.g., if viruses are not underdiagnosed), or in patients with certain diagnostic needs (e.g., immunocompromised patients) [2,5,18]. Currently, molecular diagnostics is the gold standard for the diagnosis of viral respiratory infections [5,33,34].
The 2019 American Thoracic Society guideline recommends [4]:
Do not obtain sputum for Gram stain and culture in patients with CAP who are not admitted (strong recommendation with a low level of evidence).
Perform etiological diagnosis in patients with CAP who are admitted, especially if they require mechanical ventilation, and with risk factors for MDR (e.g. MRSA, Pseudomonas aeruginosa) (strong recommendation with a very low level of evidence).
In an excellent review by Cilloniz, Torres et al., the authors propose to use them in patients with CAP with clinical suspicion of influenza or P. aeruginosa, or severe CAP requiring ICU admission [1]. And in all patients with clinical suspicion of hospital- and ventilator-associated pneumonia.
CONCLUSIONS
In patients with respiratory infections requiring hospital-isation, syndromic panels have been shown to increase aetiological diagnosis due to their higher sensitivity, shorten the turnaround time, decrease the duration of antibiotic treatment and increase the use of antivirals against influenza, shorten time to respiratory isolation of the patient and days of isolation, allow for de-escalation of antibiotics, and reduce the cost of antibiotic treatment and hospitalisation. It is recommended to use them in conjunction with an antimicrobial stewardship programme and a predetermined management algorithm.
FUNDING
This study was partially financed by PI20/01201 of the Carlos III Health Institute (ISCIII), Madrid Spain, partially financed by the by the European Regional Development Fund (FEDER) ‘A way of making Europe’
References
- 1.Cilloniz C, Liapikou A, Torres A. Advances in molecular diagnostic tests for pneumonia. Curr Opin Pulm Med. 2020;26(3):241-8. [DOI] [PubMed] [Google Scholar]
- 2.Azoulay E, Russell L, Van de Louw A, Metaxa V, Bauer P, Povoa P, et al. Diagnosis of severe respiratory infections in immunocompromised patients. Intensive Care Med. 2020;46(2):298-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres A, Cilloniz C, Niederman MS, Menendez R, Chalmers JD, Wunderink RG, et al. Pneumonia. Nat Rev Dis Primers. 2021;7(1):25. [DOI] [PubMed] [Google Scholar]
- 4.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45-e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson KE, Azar MM, Banerjee R, Chou A, Colgrove RC, Ginocchio CC, et al. Molecular Testing for Acute Respiratory Tract Infections: Clinical and Diagnostic Recommendations From the IDSA’s Diagnostics Committee. Clin Infect Dis. 2020;71(10):2744-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassetti M, Rello J, Blasi F, Goossens H, Sotgiu G, Tavoschi L, et al. Systematic review of the impact of appropriate versus inappropriate initial antibiotic therapy on outcomes of patients with severe bacterial infections. Int J Antimicrob Agents. 2020;56(6):106184. [DOI] [PubMed] [Google Scholar]
- 7.Julian-Jimenez A, Adan Valero I, Beteta Lopez A, Cano Martin LM, Fernandez Rodriguez O, Rubio Diaz R, et al. [Recommendations for the care of patients with community-acquired pneumonia in the Emergency Department]. Rev Esp Quimioter. 2018;31(2):186-202. [PMC free article] [PubMed] [Google Scholar]
- 8.Jarrett J, Uhteg K, Forman MS, Hanlon A, Vargas C, Carroll KC, et al. Clinical performance of the GenMark Dx ePlex respiratory pathogen panels for upper and lower respiratory tract infections. J Clin Virol. 2021;135:104737. [DOI] [PubMed] [Google Scholar]
- 9.Ozongwu C, Personne Y, Platt G, Jeanes C, Aydin S, Kozato N, et al. The Unyvero P55 ‘sample-in, answer-out’ pneumonia assay: A performance evaluation. Biomol Detect Quantif. 2017;13:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann C, Hirsch HH. Comparing Luminex NxTAG-Respiratory Pathogen Panel and RespiFinder-22 for multiplex detection of respiratory pathogens. J Med Virol. 2016;88(8):1319-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popowitch EB, O’Neill SS, Miller MB. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013;51(5):1528-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keske S, Ergonul O, Tutucu F, Karaaslan D, Palaoglu E, Can F. The rapid diagnosis of viral respiratory tract infections and its impact on antimicrobial stewardship programs. Eur J Clin Microbiol Infect Dis. 2018;37(4):779-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locher K, Roscoe D, Jassem A, Wong T, Hoang LMN, Charles M, et al. FilmArray respiratory panel assay: An effective method for detect-ing viral and atypical bacterial pathogens in bronchoscopy specimens. Diagn Microbiol Infect Dis. 2019;95(4):114880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchan BW, Windham S, Balada-Llasat JM, Leber A, Harrington A, Relich R, et al. Practical Comparison of the BioFire FilmArray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J Clin Microbiol. 2020;58(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monard C, Pehlivan J, Auger G, Alviset S, Tran Dinh A, Duquaire P, et al. Multicenter evaluation of a syndromic rapid multiplex PCR test for early adaptation of antimicrobial therapy in adult patients with pneumonia. Crit Care. 2020;24(1):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R. Syndromic Panel-Based Testing in Clinical Microbiology. Clin Microbiol Rev. 2018;31(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan KV. Rapid Molecular Panels: What Is in the Best Interest of the Patient? A Review of Patient Outcome Studies for Multiplex Panels Used in Bloodstream, Respiratory, and Neurological Infections. Clin Microbiol Newsl. 2017;39(16):125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanella MC, Meylan P, Kaiser L. Syndromic panels or ‘panel syndrome’? A perspective through the lens of respiratory tract infections. Clin Microbiol Infect. 2020;26(6):665-8. [DOI] [PubMed] [Google Scholar]
- 19.Rappo U, Schuetz AN, Jenkins SG, Calfee DP, Walsh TJ, Wells MT, et al. Impact of Early Detection of Respiratory Viruses by Multiplex PCR Assay on Clinical Outcomes in Adult Patients. J Clin Microbiol. 2016;54(8):2096-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brendish NJ, Malachira AK, Armstrong L, Houghton R, Aitken S, Nyimbili E, et al. Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir Med. 2017;5(5):401-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews D, Chetty Y, Cooper BS, Virk M, Glass SK, Letters A, et al. Multiplex PCR point of care testing versus routine, laboratory-based testing in the treatment of adults with respiratory tract infections: a quasi-randomised study assessing impact on length of stay and antimicrobial use. BMC Infect Dis. 2017;17(1):671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shengchen D, Gu X, Fan G, Sun R, Wang Y, Yu D, et al. Evaluation of a molecular point-of-care testing for viral and atypical pathogens on intravenous antibiotic duration in hospitalized adults with lower respiratory tract infection: a randomized clinical trial. Clin Microbiol Infect. 2019;25(11):1415-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark TW, Beard KR, Brendish NJ, Malachira AK, Mills S, Chan C, et al. Clinical impact of a routine, molecular, point-of-care, test-and-treat strategy for influenza in adults admitted to hospital (FluPOC): a multicentre, open-label, randomised controlled trial. Lancet Respir Med. 2021;9(4):419-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guna Serrano MR, Larrosa Escartin N, Marin Arriaza M, Rodriguez Diaz JC. Microbiological diagnosis of bacteraemia and fungaemia: Blood cultures and molecular methods. Enferm Infecc Microbiol Clin. 2019;37(5):335-40. [DOI] [PubMed] [Google Scholar]
- 25.Barrasa H, Martin A, Maynar J, Rello J, Fernandez-Torres M, Aguirre-Quinonero A, et al. High rate of infections during ICU admission of patients with severe SARS-CoV-2 pneumonia: A matter of time? J Infect. 2021;82(5):186-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branche AR, Walsh EE, Jadhav N, Karmally R, Baran A, Peterson DR, et al. Provider Decisions to Treat Respiratory Illnesses with Antibiotics: Insights from a Randomized Controlled Trial. PLoS One. 2016;11(4):e0152986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbas S, Bernard S, Lee KB, Pakyz A, Doern C, Doll M, et al. Rapid respiratory panel testing: Impact of active antimicrobial steward-ship. Am J Infect Control. 2019;47(2):224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe CF, Payne M, Puddicombe D, Mah A, Wong D, Kirkwood A, et al. Antimicrobial stewardship for hospitalized patients with viral respiratory tract infections. Am J Infect Control. 2017;45(8):872-5. [DOI] [PubMed] [Google Scholar]
- 29.Srinivas P, Rivard KR, Pallotta AM, Athans V, Martinez K, Loutzen-heiser S, et al. Implementation of a Stewardship Initiative on Respiratory Viral PCR-based Antibiotic Deescalation. Pharmacotherapy. 2019;39(6):709-17. [DOI] [PubMed] [Google Scholar]
- 30.Moradi T, Bennett N, Shemanski S, Kennedy K, Schlachter A, Boyd S. Use of Procalcitonin and a Respiratory Polymerase Chain Reaction Panel to Reduce Antibiotic Use via an Electronic Medical Record Alert. Clin Infect Dis. 2020;71(7):1684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rader TSt, Stevens MP, Bearman G. Syndromic Multiplex Polymer-ase Chain Reaction (mPCR) Testing and Antimicrobial Steward-ship: Current Practice and Future Directions. Curr Infect Dis Rep. 2021;23(4):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bou G, Calbo E, Crespo M, Cantón R, Álvarez de Luna FF, García Rodríguez J, et al. Justification for 24/7 clinical microbiology services. Enferm Infecc Microbiol Clin. 2022;40(1):1-4. [DOI] [PubMed] [Google Scholar]
- 33.Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;62(10):e51-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis. 2018;67(6):e1-e94. [DOI] [PMC free article] [PubMed] [Google Scholar]