Abstract
Inflammasomes sense intrinsic and extrinsic danger signals to trigger inflammatory responses and pyroptotic cell death. Homotypic pyrin domain (PYD) interactions of inflammasome forming nucleotide-binding oligomerization domain (NOD)–like receptors with the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD) mediate oligomerization into filamentous assemblies. We describe the cryo–electron microscopy (cryo-EM) structure of the human NLRP3PYD filament and identify a pattern of highly polar interface residues that form the homomeric interactions leading to characteristic filament ends designated as A- and B-ends. Coupling a titration polymerization assay to cryo-EM, we demonstrate that ASC adaptor protein elongation on NLRP3PYD nucleation seeds is unidirectional, associating exclusively to the B-end of the filament. Notably, NLRP3 and ASC PYD filaments exhibit the same symmetry in rotation and axial rise per subunit, allowing a continuous transition between NLRP3 and ASC. Integrating the directionality of filament growth, we present a molecular model of the ASC speck consisting of active NLRP3, ASC, and Caspase-1 proteins.
The transition of NLRP3 PYD nucleation seeds to ASC filament elongation reveals the directionality of ASC speck growth.
INTRODUCTION
Innate immune cells are able to recognize invading pathogens or cellular damage by germline-encoded pattern recognition receptors (1). Among these, the absent in melanoma 2 (AIM2)–like receptors (ALRs) and nucleotide-binding oligomerization domain–like receptors (NLRs) form supramolecular complexes termed inflammasomes, which regulate the activation of highly proinflammatory cytokines and pyroptotic cell death (2). These large cytosolic assemblies consist of sensor, adaptor, and effector proteins and form within minutes after recognizing their specific triggers (3, 4).
Canonical inflammasome formation starts with the activation-induced oligomerization of ALR or NLR sensor proteins that, in turn, recruit pro–Caspase-1, either by direct homotypic protein domain interactions or via the adaptor protein ASC (apoptosis-associated speck-like protein containing a CARD). Proximity-induced autoproteolytic cleavage yields active Caspase-1 that mediates activation and release of highly proinflammatory cytokines of the interleukin-1 (IL-1) family and triggers a proinflammatory form of cell death called pyroptosis (5, 6). NLRP3 (NACHT, LRR and PYD domain-containing protein 3) is the best-studied inflammasome protein to date as it is involved in the recognition of a plethora of different activating ligands encountered during bacterial, viral, or fungal infections, as well as in sterile inflammation (4). While, during pathogen encounter, NLRP3 activation may be a beneficial event of the innate immune response, it confers detrimental effects during sterile inflammation, manifesting in autoinflammatory diseases such as atherosclerosis, gout, and Alzheimer’s disease (7–10). Moreover, several mutations in the NLRP3 gene that induce the undue activation of the NLRP3 inflammasome were linked to a group of rare, inherited, autoinflammatory diseases, which are summarized as cryopyrin-associated periodic syndromes (CAPS) (11).
NLRP3 inflammasome formation is thought to be a two-step process. In the first priming event, NLRP3 expression is up-regulated via nuclear factor κB–dependent signaling (12). At this stage, NLRP3 is present in an autoinhibited but signaling-competent conformational state. The engagement of an activating signal then initiates the second step of NLRP3 inflammasome formation (4). Activation-driven conformational changes allow for the oligomerization of NLRP3 with the participation of its central NACHT domain and the N-terminal pyrin domain (PYD). Oligomerized NLRP3 recruits the bipartite adaptor protein ASC through homotypic PYD-PYD domain interactions, serving as a nucleation platform for the formation of filamentous ASC assemblies (13, 14).These prion-like structures then cluster into large ASC specks that trigger Caspase-1 activation (15–17). Caspase recruitment domain (CARD) interactions mediate this assembly that bundles multiple ASC filaments into a large conglomerate called ASC speck and also perform heteromeric interactions between the ASC and the Caspase-1 CARDs (18). This self-propagating process based on a cascade of ordered interactions allows for a robust signal amplification mechanism (18, 19).
To date, two ASC-dependent inflammasome sensors, namely, AIM2 and NLRP6, have been shown to form PYD filaments that can nucleate ASC polymerization in vitro and in cells (20, 21). For NLRP3, previous in vitro studies suggested the need for the NACHT domain for successful ASC polymerization (13), while the PYD alone was sufficient to induce ASC speck formation in human embryonic kidney (HEK) 293T cells (22, 23). A filament structure of the PYD of the adaptor protein ASC has been determined previously (13, 14), and a model for ASC filament formation with the CARD domain serving as a signal amplification hub for inflammasome assembly has been proposed (18). However, the critical step necessary for inflammasome signal transduction, the PYD filament interaction between the inflammasome sensor and the adaptor ASC, remains poorly understood.
In this study, we determined the cryo–electron microscopy (cryo-EM) structure of the human NLRP3PYD filament at 3.6-Å resolution, identifying the same symmetry in rotation and axial rise per subunit compared to the ASCPYD filament (13). Each NLRP3PYD subunit is arranged in a hexagon-like assembly, creating three asymmetric interfaces that interact with six adjacent PYDs. The complementary interfaces form unique surfaces at the filament ends, designated as A- and B-ends. We developed an in vitro filament reconstitution assay that allows for the homotypic transition from an NLRP3PYD nucleation seed to ASC filament elongation. Using cryo-EM, we find that filament transition is unidirectional and exclusively occurs at the B-end of the NLRP3PYD filament. This defines the filament growth direction of ASC adaptor elongation, the assembly of ASCCARD domains into filament bundles for ASC specking, and the interactions with pro–Caspase-1 for its activation by autoproteolysis. Our observations reveal the dynamics of homotypic filament formation and have implications for the possible interference by antibodies or small molecules at the filament growing site.
RESULTS
Polymerization of recombinant NLRP3PYD and cryo-EM structure determination
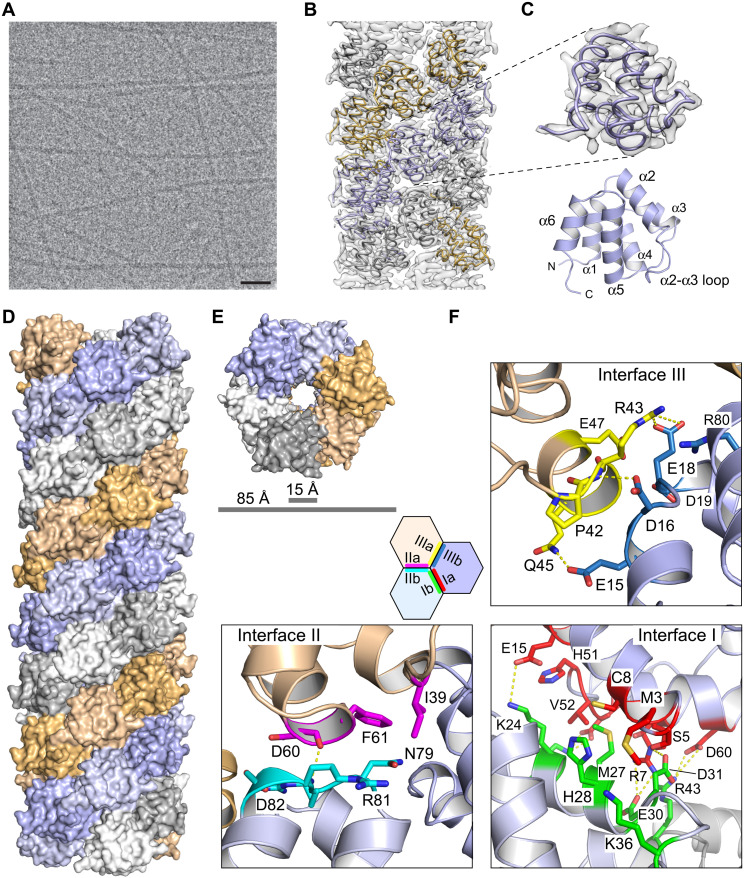
For structural analyses, human NLRP3PYD was purified with a tobacco etch virus (TEV) cleavable N-terminal glutathione S-transferase (GST) tag from Escherichia coli cells. The PYD filament formation was induced by removing the affinity tag, followed by incubation of the gel-filtered NLRP3PYD protein at 37°C, which resulted in long and straight individual filaments suitable for cryo-EM structure determination (Fig. 1A and fig. S1, A and B). Micrographs were recorded with a Titan Krios transmission electron microscope (TEM), equipped with a Falcon2 camera. Three-dimensional (3D) refinement, using the helical parameters of the ASCPYD filament [Protein Data Bank (PDB): 3J63] (13) as a starting point, yielded an electron density map with a resolution of 3.6 Å (fig. S1, C to E). Rigid body fitting of the NLRP3PYD crystal structure (3QF2) (24) into the cryo-EM map was followed by real-space refinement to achieve the final NLRP3PYD filament structure (Fig. 1B). The final model encompasses residues 3 to 94, exhibiting a root mean square deviation value of 0.54 Å with the crystal structure of NLRP3PYD (Fig. 1C). Within the six-helical bundle of the PYD (helices α1 to α6), only slight deviations between the filament and the globular structure are seen in the region at the end of helix α3 and the beginning of helix α4, whereas the overall arrangement is well preserved (fig. S2).
Fig. 1. Cryo-EM structure of the NLRP3PYD filament.
(A) Representative cryo-EM micrograph of NLRP3PYD sample after incubation at 37°C overnight. Scale bar, 50 nm. (B) Superposition of the NLRP3PYD filament model with the reconstructed electron density. (C) Close-up image of a single NLRP3PYD filament chain in the electron density map and ribbon representation of the NLRP3PYD structure. (D) Surface representation of the side view of the NLRP3PYD filament with a three-start helical symmetry. (E) Top view of the NLRP3PYD filament showing the three-start helical symmetry. (F) Detailed interactions in the type I, type II, and type III interfaces of the NLRP3PYD filament. A schematic diagram of the hexagonal assembly of the NLRP3PYD domain within the filament structure indicates the three asymmetric interfaces.
Architecture and interfaces of the NLRP3PYD filament
The NLRP3PYD filament is a cylindrical, hollow structure with an outer and inner diameter of ~85 and ~15 Å, respectively (Fig. 1, D and E). It displays a three-start helical symmetry of 54.9° right-handed rotation and an axial rise of 14.3 Å per subunit. The filament assembles as a triplicate by the Cn(3) helical symmetry. As for other death domain folds (25), the NLRP3PYD filament is composed of three major asymmetric interfaces, designated as type I, II, and III interfaces, with the opposing interacting surfaces named as a and b. In the NLRP3PYD filament, interface I is the largest in size, burying about 1020 Å2 of surface area (a- and b-sides together) on one PYD molecule. This interface, which is also the most dominant, is composed of polar residues that appear mostly conserved within the PYD family (26). The type II and III interfaces are more variable and bury about 430- and 500-Å2 surface areas in the NLRP3PYD filament. Because of the three-start helical symmetry, the type I interface mediates intrastrand interactions, whereas the type II and III interfaces mediate interstrand interactions (Fig. 1D). All three interfaces are constituted through electrostatic, polar, and hydrophobic interactions.
Interface I is mediated by interactions between residues of the α1 and α4 helices of one chain and residues of the α2 and α3 helices of the neighboring chain (Fig. 1F). Within a distance of 4 Å, there is a remarkable high number of eight salt bridges, involving each four charged residues on both interface sites, as well as five hydrogen bonds constituting this interface. Here, the guanidinium group of Arg7 is most prominent, forming tight electrostatic interactions with the side chains of Glu30 and Asp31. Interface II is mediated by interactions of α4 residues of one chain with the α5-α6 loop of a neighboring strand chain (Fig. 1F). However, only one hydrogen bond at a 3.1-Å distance is formed between the backbone carbonyl group of Asp60 and the backbone amine of Arg81. Last, interactions between helix α3 of one chain and the loop of helices α1 and α2 of the neighboring strand chain form the type III interface (Fig. 1F). Two salt bridges between Arg43 and Glu18 and three hydrogen bonds constitute the binding, besides many hydrophobic interactions. Unexpectedly, with 430 Å2, the buried surface area of interface II appears smaller than the concealed surface area of interface III with 500 Å2 in the NLRP3PYD filament structure. This is in marked difference to the ASCPYD filament, where interfaces II and III each bury 540 and 360 Å2 of surface area, respectively (13). Moreover, interactions between three subunits within the filament at the interface tips appear characteristic for the NLRP3PYD assembly, as multiple residues undergo interactions with more than one interface.
Tripartite interactions at the interface tips
The hexagonal assembly in the filament structure where every PYD molecule is surrounded by six other PYD molecules contains two unique corner sites. There is one remarkable charged interface at the joints of three molecules that appears unique to the NLRP3PYD filament. This interface involves three arginines in a staggered arrangement from three different interface sites, with Arg43IIIa from one molecule occupying the central position, headed by Arg7Ia from a second molecule at one site and Arg80IIb from a third molecule at the other site (Fig. 2A). The assembly of these three positively charged residues is facilitated by the compensation with six negatively charged residues in close vicinity to the arginines. A mixed salt bridge network from three different molecules is mediated by Asp60IIa, directly facing Arg43IIIa and backed up by Glu18IIIb (Fig. 2B). Glu30Ib and Asp31Ib from one molecule are close to Arg7Ia from a second molecule, and Arg80IIb is lastly interacting with Asp16IIIb and Asp82IIb within one molecule. The tripartite interaction in the filament assembly between Asp60, Arg43, and Glu18, however, from three different PYD molecules is unique to NLRP3 within the known structures of filament-forming PYDs of the inflammasome, as no other PYD sequence contains this charge network (Fig. 2A).
Fig. 2. Sequence conservation and interface sides in the NLRP3PYD filament.
(A) Structure-based sequence alignment of the four PYDs whose filament structures have been determined. Interface-forming residues are highlighted and shown below the secondary structure elements of NLRP3PYD. Residues are colored according to the respective interface sites as determined for the PYD filaments of NLRP3 (this study; 3 to 99), NLRP6 (6NCV; 14 to 104), AIM2 (6MB2; 1 to 94), and ASC (3J63; 1 to 91). (B) Tripartite interactions at the tips of the interfaces. Positively and negatively charged residues of all six interface sites assemble to a highly polar interaction. (C) Close-up image of the second unique interaction at the interface tips. (D) Schematic diagram of the hexagonal subunit assembly in the three-start helix and designation of the interface sites. The a- and b-sides of the three asymmetric interfaces assemble at the filament’s ends to characteristic surfaces, designated as A- and B-ends of the filament, respectively. (E) Surface representation of the NLRP3PYD filament model with the interfaces at the top and bottom layers colored according to the scheme in (A). The a- and b-sides merge at the filament ends to the A- and B-ends of the filament.
The second unique corner in the hexagon assembly of the NLRP3PYD filament structure is less pronounced but similarly involves the tripartite formation of a hydrogen bond network. Here, residues from five interface sites come together with Glu15IIIb in an exposed position close to Lys24Ib from a second molecule and Gln45IIIa from a third molecule (Fig. 2C). Asn79IIb from the second molecule and hydrophobic interactions with Phe61IIa and Pro42IIIa from the third molecule complement these homotypic interactions at the tip of the three protein interfaces. The three types of asymmetric interfaces with the opposing interaction sites a and b in the honeybee comb–like assembly of PYD subunits assemble into one continuous surface at the two ends of the filament (Fig. 2D). We therefore define these sites as the A-end and B-end of the NLRP3PYD filament, respectively (Fig. 2E).
Mutational analysis of interface forming residues
To study the impact of individual residues on the filament formation of NLRP3PYD, we performed site-directed mutagenesis analyses with recombinant proteins and in cells. Dynamic light scattering (DLS) experiments and negative-stain EM images were recorded to visualize filament formation. On the basis of the filament structure, seven residues from five different interface sites were selected, and the charge reversal mutations R7E, E15R, R43E, R80E, and R81E; the double-mutation K23E/K24E; and the mutation M27E were introduced. In addition, the four NLRP3PYD CAPS mutants D21H, D31V, H51R, and A77V were analyzed, which are known for their phenotype in Muckle-Wells disease (27–29). First, the kinetic profile of NLRP3PYD filament formation was monitored in a time course experiment using DLS. At a protein concentration of 48 μM and a temperature of 25°C, the saturation of the oligomerization process was achieved in about 40 min, as revealed by the normalized growth signal (Fig. 3A). The polymerization status of the NLRP3PYD interface and CAPS mutants was determined at a time point of 90 min when the polymerization had reached a stable plateau (Fig. 3B). All of the tested interface mutants lost their ability to oligomerize into ordered polymers, but instead formed unspecific aggregates (Fig. 3C), confirming the importance of each mutated residue for the homotypic filament formation. In contrast, of the tested CAPS mutants, only the D31V variant lost the ability to polymerize, while the A77V mutant formed short but ordered filaments (Fig. 3, B and C). The disease mutations D21H and H51R showed a strong polymerization behavior, which made analysis by gel filtration and subsequent DLS impossible. However, this phenotype was confirmed by negative-stain EM analysis.
Fig. 3. Mutational analysis of NLRP3PYD interface residues.
(A) NLRP3PYD polymerization in vitro monitored by DLS. Normalized intensity signals are recorded as a function of time and are shown here for one representative experiment. (B) Polymerization status of NLRP3PYD and mutants carrying additional interface and CAPS mutations monitored by DLS after an incubation period of 1.5 hours at 25°C. Normalized intensity signals are reported for each variant (n = 3). Boxes show the median and span from the 25th to the 75th percentiles. Whiskers mark minimum and maximum values, respectively. (C) Negative-stain electron micrographs of recombinant NLRP3PYD and interface-disrupting mutants (top two lines) or CAPS mutants (bottom line). CAPS mutations D21H and H51R even increase PYD filament formation. Scale bars, 200 nm. (D) Quantification of HeLa cells stably expressing ASC-mTurquoise transfected with full-length NLRP3-mCitrine, NLRP3PYD-mCitrine, or indicated NLRP3PYD mutants, 12 and 32 hours after transfection. Dots are representative of two (n = 2 ± SD; 12 hours) and five (n = 5 ± SEM; 32 hours) independent experiments for the two different time points, respectively. (E) Images of HeLa cells stably expressing ASC-mTurquoise transfected with NLRP3-mCitrine fusion constructs as used in (D). Scale bars, 10 μm. wt, wild type; kCnt, kilo-counts (kCounts).
To test the implications of the interface mutants in a physiological context, we performed ASC speck assays in HeLa cells. Here, full-length, wild-type NLRP3-mCitrine, or wild-type or mutant NLRP3PYD with mCitrine C-terminally fused to the protein was transfected into HeLa cells stably expressing ASC-mTurquoise. Priming of ASC polymerization by a filament nucleation factor induces the ASC speck formation that can be subsequently quantified (23). As a readout, the percentage of cells that develop ASC specks was determined in dependence on the amount of transfected DNA and analyzed for two different time points (Fig. 3D). Images of the speck formation of full-length NLRP3 and NLRP3PYD wild-type and mutant protein are shown in Fig. 3E and fig. S3A. The NLRP3PYD protein stably triggered ASC speck formation, albeit to a lesser extent than full-length NLRP3. The mutants E15R, K23E/K24E, and M27E most successfully abolished the ability of the NLRP3PYD to nucleate ASC polymerization. The mutants D31V, R43E, R80E, and R81E also impaired ASC speck formation, however, to a lesser extent, suggesting only partial defectiveness. The ASC specking ability of the CAPS mutant A77V was similar to the wild-type protein, whereas the CAPS mutants D21H and H51R showed an even increased efficacy of nucleating ASC, both as PYD-only and as full-length protein (Fig. 3D and fig. S3B). As a control, mutant proteins D21A and H51A behaved similar to the wild-type protein, while the charge reversal mutant H51E showed reduced ASC speck formation (fig. S3B). The CAPS mutation D31V instead completely abrogated the ability to induce an ASC speck.
The transition of the NLRP3 to ASC PYD filament is unidirectional
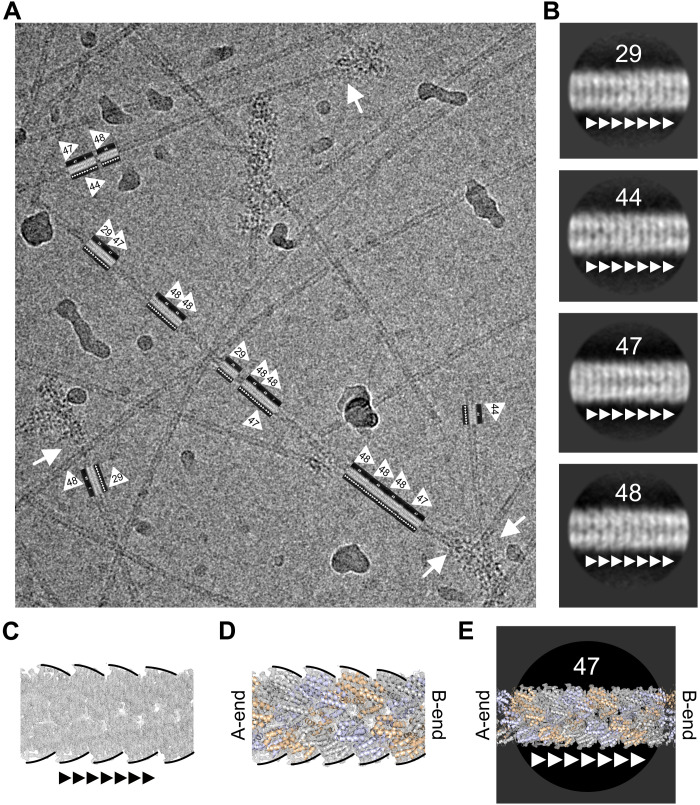
With the structure of the NLRP3PYD filament determined, we aimed at analyzing the transition to ASC adaptor protein elongation on a molecular level. ASC polymerization is triggered by NLRP3 nucleation seeds (13, 18), leading ultimately to the formation of large ASC specks. These specks are thought to arise from a continuous transition of NLRP3 and ASC PYD filaments and subsequent cross-linking of ASCPYD stems via CARD-CARD domain interactions (18, 30). Seeding of ASC filaments by NLRP3 in vitro was achieved by developing a polymerization protocol, where 50 μl of 50 μM monomeric NLRP3PYD at pH 3.8 was rapidly neutralized by the addition of 1 μl of 3 M tris buffer (pH 8) to initialize filament growth (Fig. 4A). After 3 min of incubation time at 25°C, monomeric, soluble ASC-mCherry at 16 μM concentration (pH 3.8) was added at a 1:50 molar ratio of ASC to NLRP3 and was incubated for 5 min. This step was repeated twice to slowly raise the concentration of ASC and allow the elongation of the NLRP3PYD-seeded ASC-mCherry filaments. To distinguish the two filament types, we used a 50-kDa ASC-mCherry fusion protein that is fourfold larger than the 12-kDa NLRP3PYD protein and, thus, appeared markedly thicker on the negative-stain micrographs compared to the thinner NLRP3PYD filaments (Fig. 4B). Transitions of the NLRP3PYD nucleation seeds to ASC filament elongation were observed in many instances (Fig. 4C). Following the gentle titration procedure that leads to a very dilute ASC concentration initially, only a few cases of spontaneous ASC polymerization events were seen, whereas, in approximately half of the cases, elongation of ASC on NLRP3PYD filaments occurred. ASC filament growth was observed only on one end of the NLRP3PYD filament, but never on both ends, suggesting directionality in the transition from the NLRP3 to ASC filament. Repeated titration of ASC-mCherry to the solution led to longer ASC filament growth, indicating a preference for the elongation of existing filaments compared to spontaneous de novo polymerization of ASC alone (Fig. 4D). Notably, the thicker ASC-mCherry filaments are always centered on the NLRP3PYD seeds, indicating that the transition is mediated by the N-terminal PYD of the ASC protein. The congruent helical symmetry between filaments assembled by AIM2PYD and the downstream adaptor ASCPYD has been already elegantly shown by the Egelman and Sohn laboratories (31, 32).
Fig. 4. ASC filament polymerization on NLRP3PYD nucleation seeds.
(A) Cartoon depiction of an NLRP3PYD-induced ASC filament polymerization assay. Filament formation of monomeric NLRP3PYD is induced by neutralization to pH 8.0. Stepwise titration of highly diluted ASC-mCherry leads to the elongation on NLRP3 filament seeds. (B) Negative-stain electron micrographs of NLRP3PYD (left) and ASC-mCherry filaments (right). Scale bars, 200 nm. (C) Elongation of ASC-mCherry filaments on NLRP3PYD filament seeds after the first two titration steps. Several thin NLRP3PYD filaments are seen out of which thicker ASC-mCherry filaments grow out. Note that, at the most, one transition is seen from an NLRP3 to an ASC filament, suggesting a directionality of filament growth. Scale bar, 200 nm. (D) Elongation of the ASC-mCherry filament on NLRP3PYD seeds after three titration steps. Increasing the ASC concentration induces ASC filaments to grow longer on the NLRP3PYD nucleation seeds. Scale bar, 200 nm.
ASC elongates at the B-end of the NLRP3 PYD filament
To identify the site of elongation on the NLRP3PYD filament and the directionality of ASC filament growth, we performed cryo-EM imaging, providing the resolution required for the determination of filament directionality. The titration protocol for the domain transition was adjusted to generate longer NLRP3PYD filaments that allowed for the analysis of straight filament stretches. In the cryo-EM micrographs, ASC-mCherry filaments appear more diffuse than in the negative-stain mode; nonetheless, they could be clearly identified (Fig. 5A). Only NLRP3PYD filaments merging into ASC-mCherry filament transitions were selected and used for subsequent image analysis. Average class sums calculated from 19,351 particles that were selected from 423 micrographs unambiguously revealed a sawtooth-shaped density, reflecting the directionality of the filaments. A selection of average class sums showing this characteristic sawtooth pattern is displayed in Fig. 5B. The electron density map of the filament confirms this silhouette, indicated by the black lines lining the envelope (Fig. 5C). Superposition of our filament structure onto those classes reveals the orientation of the filament in the average sums, as the sawtooth-shaped density arises from the bending of the C-terminal helix α6 of the PYD (Fig. 5D). Note that this characteristic pattern must not necessarily be visible in the average class sums, as the pitch of the PYD in the three-start helix leads to various projections within the short sections used for object averaging.
Fig. 5. ASC exclusively elongates at the B-end of the NLRP3PYD filament.
(A) Representative cryo-EM micrograph of NLRP3PYD filament–to–ASC-mCherry filament transitions. Long NLRP3PYD filaments were grown that allow for analysis of straight segments. The direction of the transition site to ASC-mCherry is indicated by small arrowheads in the boxes. Transition sites of NLRP3 to ASC filaments are indicated by white arrows. (B) Average class sums of individual segment particles for four representative classes. The direction toward the NLRP3–to–ASC PYD transition is indicated by small arrowheads. A sawtooth pattern at the edges and a perforation pattern in the filament center are characteristic for some classes. (C) Electron density map of the NLRP3PYD filament with the characteristic sawtooth pattern identifies the A- and B-ends of the filament. (D) Modeling of the filament ribbon structure into the electron density map. (E) Overlay of the filament structure of the average class sums identifies the directionality of the NLRP3–to–ASC PYD filament transition. Shown for the average class 47 is the characteristic sawtooth pattern, and the perforation identifies the B-end of the NLRP3 filament for the elongation with ASC.
For classes 29, 44, 47, and 48, a superposition was allowed to unambiguously correlate the NLRP3PYD filament direction with the given orientation of the class sums. This is shown as an example for class 47, where the left and the right end could be assigned to the A- and B-ends of the filament, respectively (Fig. 5E). To pin down the directionality of the NLRP3PYD filament seen in the micrographs, we first added a directionality flag into the class averages (white triangles). We then extracted the rotation and translation information for each of the underlying particles (figs. S4 and S5). We used this information to superimpose the flagged class averages onto the original micrographs. This procedure revealed that the white triangles on the picked particles reassigned to the original micrographs are always pointed toward the transition to the ASC filament (Fig. 5A). Of eight average class sums combined in a total of 560 boxed filament sections, none pointed in the opposing direction (fig. S4D). From these data analyses, we conclude that the transition from NLRP3PYD filament seeds to ASC filament elongation exclusively occurs at the B-end of the existing NLRP3PYD filament and that the direction of ASC filament growth is from the A- to the B-end.
Interactions in the NLRP3–ASC PYD transition interface
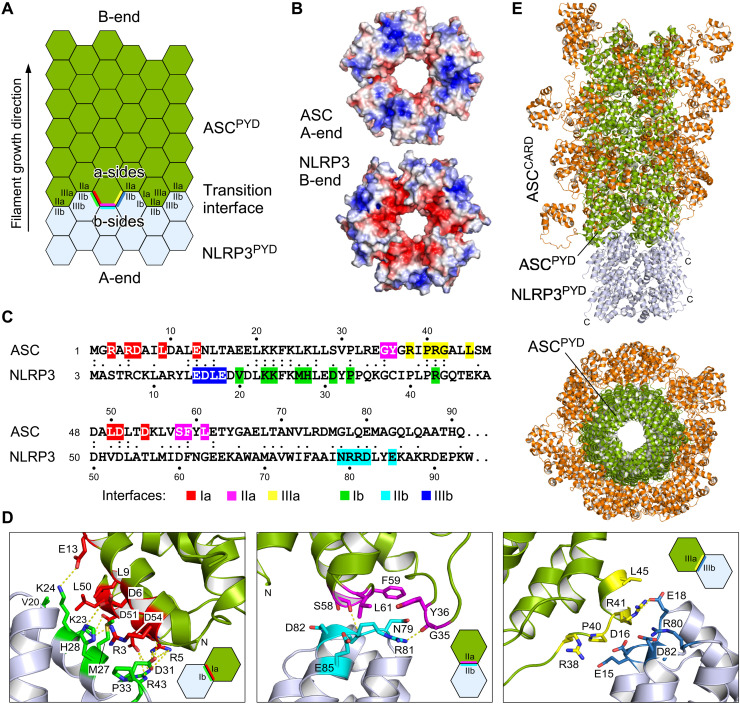
Knowing the ASC elongation site on NLRP3 nucleation seeds, we set out to investigate the transition interface between these two homotypic PYD filaments. Twelve subunits of NLRP3PYD and the ASCPYD filament (13) were superimposed and aligned according to the growth direction (Fig. 6A). As the filaments exhibit the same rotational symmetry and axial rise, a continuous transition is facilitated. Using the GROMACS molecular dynamics package (33), we optimized the binding interface between the B-end of the NLRP3PYD filament and the A-end of the ASCPYD filament elongation by energy minimization of a fully hydrated protein complex. The interfaces fit very well with complementary charge and hydrogen bond interactions with no atomic clashes (Fig. 6, B to D). In total, 33 residues constitute the hetero-PYD filament interface, of which 12 are identical and another 9 are similar in both ASC and NLRP3 domains (Fig. 6C). Overall, the sequence homology between the ASCPYD and the NLRP3PYD is unexpectedly low, with a sequence identity of 22% and a similarity of 43%. Residues in interface I are particularly well conserved, whereas interface II residues on the b-side diverge markedly between both PYDs.
Fig. 6. Transition of NLRP3PYD nucleation seeds to ASCPYD filament elongation.
(A) Cartoon of the filament transition from the nucleation seed NLRP3PYD to the elongation adaptor ASCPYD. The transition interface of the heteromeric PYD interactions is indicated. (B) Electrostatic surface potential of the complementary filaments from the ASCPYD A-end and the NLRP3PYD B-end, displayed from −4 Boltzmann constant (kBT) (red) to +4 kBT (blue). (C) Sequence alignment of the ASC and NLRP3 PYDs with the interface residues highlighted. Interacting residues from the a-side interfaces in ASC and the b-side interfaces in NLRP3 are colored as before. (D) Close-up images of the interactions in the three interface types I, II, and III for the energy-minimized transition interface between the NLRP3PYD filament (light blue) and the ASCPYD filament (green). Salt bridges (types I and III) and hydrogen bonds (type II) are indicated by dashed lines. (E) Model of an ASC filament (green) elongating on the NLRP3PYD filament nucleation seed (light blue). Whereas the PYD domain of ASC forms the filamentous stem structure, the CARD domain could adopt variable compositions at the surface of the filament.
The interactions within the heteromeric PYD assembly are visualized for all three interface types in Fig. 6D. The entire buried surface area in this transition segment comprises 930 Å2 for interface I followed by 560 and 430 Å2 for interfaces II and III, respectively. The increase in interface II compared to the NLRP3PYD filament alone (430 Å2) results from the a-side residue Leu61 in ASC, which is a small glycine in NLRP3, and an interaction of Arg81 in NLRP3 with residues Gly35 and Tyr36 of the α2-α3 loop in ASC forming a hydrogen bond with the Gly35 carboxyl group (Fig. 6D). This interaction has no correspondent in NLRP3 due to a different conformation and sequence composition of this loop. A similar interaction is also not seen in the ASCPYD filament, as the corresponding residue Gln79 does not reach out as far as Arg81 in NLRP3. The increase in interface area III compared to the ASCPYD filament alone (320 Å2), instead, results from b-side residues Glu18 and Arg80 in NLRP3, which correspond to small residues Thr16 and Leu78 in ASC. From the interface analysis, we did not identify a unique restriction mechanism that would prohibit elongation at the filament A-end of NLRP3 with the B-end of ASC. Instead, we speculate that the kinetics of filament growth drives the direction from the A-end to the B-end and, thus, determines the elongation directionality. To create a model of the NLRP3PYD nucleation seed to ASC adaptor transition, the NLRP3PYD filament was elongated with 32 ASCPYD subunits, which each were overlaid with a randomly selected nuclear magnetic resonance (NMR) model of the full-length ASC protein (34). This model provides a first glimpse on how the adaptor ASC could bridge from the NLRP3 inflammation sensor to the Caspase-1 pyroptosis effector (Fig. 6E).
DISCUSSION
The formation of an ASC speck is the hallmark of inflammasome activation in immune cells and the signal that initiates pyroptotic cell death. A cascade of sensor, adaptor, and effector protein interactions transmits and amplifies the signal of danger- or pathogen-induced triggers that ultimately leads to cytokine release as a systemic inflammatory response (2, 3). The human NLRP3 protein is one critical sensor of the innate immune system that induces ASC speck formation upon activation. Here, we show that the PYD of NLRP3 can assemble into helical filaments similarly as it has been described for the AIM2, NLRP6, and ASC PYD filaments (13, 20, 21, 32). The cryo-EM structure reveals the molecular assembly of the PYD in the filament using asymmetric type I, II, and III interfaces (Fig. 1). This assembly is similar to other filament-forming PYDs, despite the low sequence conservation of only 22% identity. In particular, electrostatic interactions in the NLRP3PYD as well as polar and hydrophobic contacts facilitate the assembly of individual PYDs into the right-handed helical filament. We show at the protein level that the NLRP3PYD filament can act as a nucleation seed for ASC filament elongation. This transition is exclusively mediated on one end of the NLRP3PYD filament, which we defined as the “B-end” according to the interface sites Ib, IIb, and IIIb that assemble to one surface and form this filament end (Fig. 2). The directionality of the homotypic PYD filament transition determines the growth direction of the ASC speck and sets the molecular basis for understanding these micrometer-large signaling assemblies.
The contribution of individual residues of the NLRP3PYD to form filaments was confirmed by site-directed mutagenesis with recombinant proteins and in cells. The charge reversal mutations introduced at the interface sites each abrogated the ordered formation of a filament (Fig. 3). However, some NLRP3PYD mutants such as E15R or M27E still showed a strong tendency for aggregation, indicating their ability for homomeric interactions, albeit in an uncoordinated manner. The two CAPS mutants D21H and H51R appear of particular interest, both showing a tendency for an increased filament polymerization activity compared to wild-type NLRP3PYD. This is seen not only in negative-stain EM but also by their ability to nucleate ASC in cells. It suggests that a well-coordinated PYD/PYD transition between the sensor and the adaptor molecule is critical for optimal inflammasome function. Our studies provide a biochemical basis of NLRP3 hyperactivity in patients carrying gain-of-function mutations in the PYD domain of NLRP3. Asp21 is at the beginning of helix α2 and not involved in any intermolecular salt bridge or hydrogen bond formation. The side chain is facing Asp60 (interface IIa) and Gln45 (IIIa) of an adjacent molecule at 5.8 and 6.1 Å distance, respectively. The mutation to the larger histidine may enhance the interaction to these two residues, forming additional hydrogen bonds in these two interfaces (fig. S3C). His51 instead contributes only a small van der Waals interaction to interface Ia with a portion of 28 of 500 Å2 of buried surface area. Its exchange to a larger arginine could form a new interaction network by reaching out with a hydrogen bond to Gln45 of the interface IIIa from a neighboring subunit and a salt bridge to Glu15 of the same molecule (fig. S3D). AIM2, which has an arginine at this position, contains instead a charge-repulsive lysine at the position of Gln45 and a smaller aspartate at the position of Glu15, prohibiting such interaction network. Our observation that the ability of the NLRP3PYD to form filaments was actually enhanced by the CAPS mutations D21H and H51R underlines how finely balanced the interplay between autoinhibition and effector function is in inflammasome regulation. Thus, the NLRP3PYD does not seem to be evolutionarily designed for the firmest filament formation but may represent a compromise to the requirements of multiple molecular states, including the homotypic transition to another molecule as ASCPYD.
Regulation of inflammasome formation also occurs at the level of posttranslational modifications (35). Phosphorylation of NLRP3 at Ser5 was shown to prevent NLRP3PYD oligomerization in HEK293T cells (23). The NLRP3PYD filament determined here shows that the hydroxyl group of Ser5 participates in the intrastrand interface type I through hydrogen bonding with Asp31. Introducing a bulky phosphorylation at Ser5 containing a negative charge repulsive to Asp31 is likely to disrupt the tight salt bridge and hydrogen bonding interplay of this interface (fig. S6A). Conversely, acetylation of NLRP3 at residues Lys23 and Lys24 was shown to trigger aging-associated chronic inflammation through promoting NLRP3 inflammasome formation (36). Residues Lys23 and Lys24 participate in type I interface interactions in both the NLRP3PYD filament and the modeled NLRP3PYD to ASC transition. Acetylation of NLRP3 residues Lys23 and Lys24 disrupts the salt bridges formed with ASC residues Asp51 and Glu13, respectively; however, it establishes a new network of hydrogen bonds that might facilitate increased ASC-to-NLRPPYD filament binding (fig. S6B).
The formation of filamentous structures in the cytosol is a very rare and typically tightly controlled process in mammalian cells. The, by far, best-characterized examples are the cytoskeleton structures of actin and microtubules. These two structures are reversible and highly regulated by numerous factors that control nucleation, elongation, capping, severing, and cross-linking (37, 38). Moreover, both are nucleotide-binding proteins, and their polymerization activity depends on nucleotide and magnesium concentrations (39, 40). The direction of filament growth is essential for understanding the dynamics of the filament structure and its dependency on (ligand-)concentration and steady-state equilibria. In contrast to the dynamic cytoskeleton, filament formation of the NLRP3PYD and ASC is irreversible. Both filaments form readily with recombinant protein in a concentration- and pH-dependent manner, but once filaments are formed, they do not disassemble upon dilution. A close interplay of on and off rates at the A- and B-ends, similar to the barbed and pointed end of the actin filament, leading to a treadmilling mechanism, does not seem to exist in the PYD assembly of NLRP3 and ASC, as far as we know today. Instead, the formation of the NLRP3PYD and ASC filaments appear as a “point of no return” that will ultimately lead to cell death and pyroptosis.
It is remarkable that ASC only assembles at one site of the NLRP3PYD filament, which defines the growth direction of the ASC speck (Figs. 4 and 5). From the EM data presented here, we cannot differentiate whether there is a structural constraint, e.g., in form of a steric clash, which prohibits the assembly of the ASCPYD to the A-end of the NLRP3PYD filament, or whether the on rate of the a-side PYD interfaces of monomeric ASC for binding to the B-end of the NLRP3PYD filament is so much in favor compared to binding in the opposite direction that virtually only one form of the homotypic domain transition occurs (Fig. 6). It is tempting to speculate that only the interaction kinetics determines the homotypic domain transition directionality and that the filament growth direction, e.g., of the NLRP3PYD alone, is unidirectional and is determined by the binding kinetics. PYD-only proteins (POPs) and CARD-only proteins (COPs) have been described as negative regulators of the inflammasome by acting on the ASCPYD or Caspase-1CARD domains, respectively (41, 42). The COP “inhibitor of CARD” was shown to block Caspase-1CARD polymerization in vitro by capping the filament through a defective interface (43). It remains to be elucidated whether the POPs associate only to one end of the ASCPYD filament, inducing a cap to the growing ASC speck.
It can also be envisioned that topological constraints allow only one growth and PYD transition direction. Inactive NLRP3 forms a pentamer or hexamer of dimers with the PYDs shielded in a hollow sphere (44, 45). Release of the PYDs upon activation, directing the C termini of the PYDs toward the core assembly, would expose the b-sides of the PYDs for filament extension. Active NLRC4 was shown to form a disc-like structure in an assembly of up to 11 subunits (46–49). The large leucine-rich repeat (LRR) domain mainly contributes to the planar assembly of the disc, while the death domain coalesces in the central hub. In addition, for NLRP3, a disc-like assembly with participation of the serine/threonine kinase NEK7 was proposed in its active state (50). Moreover, the NLRP3 basic region 131 to 147 following the NLRP3PYD (3 to 94) was shown to associate with lipid membranes (51). While it is not clear whether active NLRP3 launches the inflammasome formation from a membrane surface, it is well conceivable that filament growth is only possible in one direction if such sphere- or disc-like structure associates on a membrane surface.
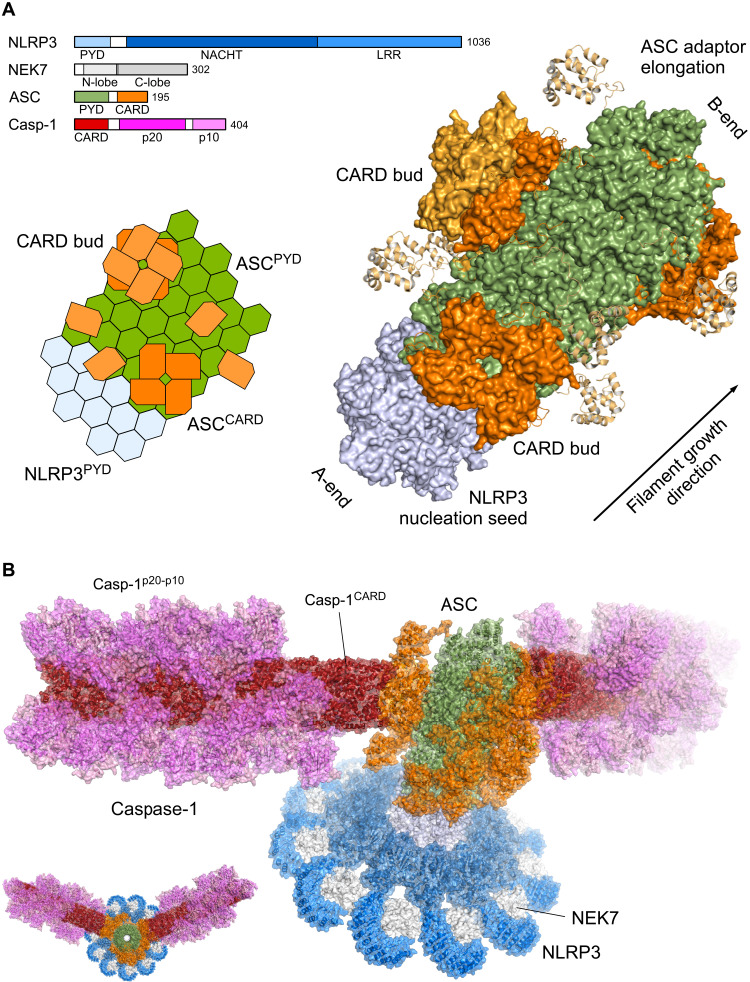
Knowing the direction of the homotypic NLRP3–to–ASC PYD transition and piecing together recent findings from other laboratories, we generated a molecular model of an ASC speck (Fig. 7). A comprehensive analysis of ASC filament formation, serving as a signal amplification mechanism for inflammasomes, was performed previously (18). It was shown that assembly of the ASC speck involves the oligomerization of ASCPYD into filaments, while cross-linking these filaments is mediated by the CARD (18, 30). Accordingly, ASC mutants with a nonfunctional CARD only assemble filaments but do not form specks. While NLRP3 is the sensor and its PYD is the nucleation seed for inflammasome formation, ASC is the adaptor that transmits as a filament elongation factor the information to the effector protein Caspase-1 (52, 53). This cascade ultimately leads to ASC specking, Caspase-1 activation, IL-1β maturation, and Gasdermin D cleavage. Notably, unidirectional growth was also observed for the transition of ASCCARD or NLRC4CARD filaments to Caspase-1CARD (54). Using nanogold-labeling experiments with biotinylated protein, filament elongation was seen only in one direction. The growth direction was predicted by a comparison of preferred interface sides between either ASC, NLRP1, NLRC4, or CARD8 CARDs and the Caspase-1 CARD to occur at the b-sides of the interfaces, although the left-handed one-start helical CARD filament is of different topology than the right-handed three-start PYD filament (54, 55).
Fig. 7. Molecular model of an ASC speck.
(A) Cartoon and atomistic model of the NLRP3PYD-ASC filament elongation. The molecular domain architecture of human NLRP3, NEK7, ASC, and Caspase-1 (Casp-1) is displayed in bar diagrams. The NLRP3PYD nucleation seed (light blue) and the elongation to the ASCPYD filament (green) are shown with the ASCCARD (orange) assemble on the stem. The linker between the PYD and CARD domains in ASC allows small ASCCARD nucleation seed (CARD buds) of one to three filament rings. (B) Molecular model of an ASC speck. The NLRP3-NEK7 complex forms the disc-like assembly (blue and gray) of the active NLR, with the PYD filament in the center core (light blue). The ASCPYD stem (green) with the CARD buds (orange) constitute the adaptor to Caspase-1 effector filament, with the Caspase-1CARD filament (red) and the zymogen p20-p10 dimer (violet and pink) attached. The structures used to generate this model assembly were the following: NLRP3-NEK7 complex [6NPY (50)], NLRP3PYD filament (this structure), ASCPYD filament [3J63 (13)] with an overlay with the ASC full-length NMR structure on the PYD subunit [2KN6 (34); 20 models], ASCCARD buds (assembly of one or two ASCCARD rings on the ASCPYD stem) [6N1H (54)], ASCCARD–Caspase-1CARD transition [7KEU (56)], Caspase-1CARD filament [5FNA (43)], and Caspase-1–zymogen p20-p10 dimer [3E4C (65)].
We generated a model of 11 NLRP3-NEK7 complexes assembling into the nucleation seed for ASC filament elongation. The ASCPYD forms the stem of the elongating ASC filament, while the CARDs assemble as a bud onto the surface of the stem. In the model, we placed 32 ASC molecules with the N-terminal PYD arranged in the filament structure, as determined by Lu et al. (13). The C-terminal CARD again is assembled in a new filament structure (54) or in any of the 20 conformers determined by NMR spectroscopy (34). The length of the 23-residue linker between the PYD and CARD of ASC easily allows for the formation of two rings of the four-leafed CARD filament on the ASCPYD stem (Fig. 7A). This “bud of CARDs” tethered to the PYD stem acts as a nucleation seed for Caspase-1 recruitment and subsequent Caspase-1CARD filament formation (52). Notably, the ASCCARD filament diameter is ~7 nm (54) and, thus, 1.5-nm smaller than the ASCPYD filament. The ASCCARD filament structure assembles through a left-handed one-start helical symmetry with about 3.6 subunits per turn (54). The transition of the ASCCARD to the Caspase-1CARD following the proposed directionality from b-sites to a-sites in this one-start helix (54–56) marks the last step in this domino-type NLRP3PYD:ASCPYD-ASCCARD:CaspCARD queue. There are at least 3.5-fold more Caspase-1 molecules found in the ASC speck than ASC molecules (13), giving rise to the assembly of 120 Caspase-1 proteins in our model. Onto the three ASCCARD buds with 4 to 10 subunits each, 54 to 66 Caspase-1 molecules were modeled, either as a full-length protein or in its cleaved form with CARD, p20, and p10 subunits (Fig. 7B) (57). The transition zone from the active NLRP3PYD filament seed to the ASCPYD filament in a functional inflammasome can be as short as one ASCPYD filament ring, as shown for the ASC (E80R) mutant that retained the ability to induce cell death (18). These data nicely confirm our experimentally determined directionality of NLRP3PYD-to-ASC filament growth, as the ASC E80R mutation is located on the b-side of the PYD interface surfaces, allowing for the transition of the NLRP3PYD filament B-end to the ASCPYD A-end, whereas subsequent ASC filament elongation is prohibited by this mutation (18). Accordingly, the ASCCARD bud for a functional inflammasome can be as short as only six ASC subunits. The model of an ASC speck shown here displays the minimal assembly of NLRP3, ASC, and Caspase-1 proteins in a rough 1:3:10 stoichiometry. While this model holds for an approximately 80-nm length, the ASCCARD-mediated cross-linking (18, 30) of multiple such assemblies may generate the micrometer-like structure seen in fluorescence microscopy for the ASC speck (58–60).
The NLRP3PYD filament structure and the determination of the transition directionality to ASC filament elongation reveal the molecular basis for NLRP3-mediated ASC specking. Our observations have implications for the possible interference with antibodies or small molecules at the filament growing site. Capping the inflammasome at the filament growth direction could be an effective mechanism to block these higher-order signalosomes and lead to new therapeutic intervention in inflammation.
MATERIALS AND METHODS
NLRP3PYD cloning, expression, and purification
Human NLRP3 (UniProt accession number Q96P20; residues 3 to 110; C108S) was cloned into a pGEX-4T1 expression vector, providing an N-terminal GST tag followed by a TEV protease cleavage site. The C108S mutation was introduced to prevent disulfide bond formation with residue C8, which was observed in one of two chains in the crystal structure (24) of NLRP3PYD. The construct was transformed and expressed in E. coli cells, strain BL21(DE3), by growing the culture at 37°C to an OD600 (optical density at 600 nm) of 0.8 and by inducing with 0.3 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) overnight (o/n) at 20°C. Cells were collected by centrifugation and lysed by sonication in a lysis buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP). The cell lysate was centrifuged at 20,000g for 30 min, and the supernatant was administered to a pre-equilibrated GSTrap column using an ÄKTA prime FPLC system (GE Healthcare). The column was washed with 10 column volumes of lysis buffer, and the protein was eluted in the same buffer supplemented with 15 mM l-glutathione. The affinity-purified protein was TEV-cleaved o/n at 4°C and subsequently subjected to gel filtration on an equilibrated Superdex 75 gel filtration column 16/600 (GE Healthcare). Monomer fractions were pooled and concentrated to 1 mg/ml, corresponding to a concentration of 79 μM, and filament formation was induced with fresh protein by incubation at 37°C o/n.
Protein variants carrying mutations in the PYD-PYD assembly interfaces (R7E, E15R, K23E/K24E, M27E, R43E, D60R, R80E, and R81E) and CAPS mutants (D21H, D31V, H51R, and A77V) were generated in the NLRP3PYD (residues 3 to 110; C108S) construct using the megaprimer mutagenesis method. The NLRP3PYD interface and CAPS mutant proteins were expressed and purified as GST fusion proteins following the same procedure as described above. For kinetic measurements, the generation of monomeric, soluble NLRP3PYD included an additional dialysis step after TEV protease cleavage into a buffer containing 50 mM glycine (pH 3.8) and 150 mM NaCl. The dialyzed protein was further purified on a pre-equilibrated Superdex 75 gel filtration column (GE Healthcare), and fractions containing monomeric soluble NLRP3PYD were concentrated to 1 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C.
Preparation of recombinant ASC
Full-length human ASC, followed by a TEV protease cleavage site and mCherry, was cloned with Nde I/Xho I sites at the 5′ and 3′ ends, respectively, into a pET-23a expression vector providing a C-terminal hexa-histidine tag. This construct was transformed and expressed in E. coli cells, strain BL21(DE3), by growing the culture at 37°C to an OD600 of 0.8 and induced with 0.1 mM IPTG for 4 hours at 37°C. Cells were collected by centrifugation and lysed by sonication in lysis buffer A containing 20 mM tris (pH 8.0), 500 mM NaCl, and 5 mM imidazole. Cell lysates were centrifuged at 20,000g for 30 min, and the pellet was dissolved in buffer A supplemented with 4 M Gdn-HCl for 1 hour at 4°C. Subsequently, the suspension was centrifuged at 20,000g for 30 min, and the supernatant was administered onto a pre-equilibrated HisTrap column using an ÄKTA Prime FPLC system (GE Healthcare). The column was washed with 10 column volumes of solubilization buffer, supplemented with 20 mM imidazole, and the protein was eluted in the same buffer supplemented with 300 mM imidazole. The pooled elution fractions’ pH was decreased to 3.8 and dialyzed against 50 mM glycine buffer (pH 3.8) and 150 mM NaCl. The protein was further purified on a pre-equilibrated Superdex 75 gel filtration column (GE Healthcare). Fractions containing monomeric, soluble ASC-mCherry protein were pooled, concentrated to 1 mg/ml, aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C.
Negative-stain EM
For negative-stain EM, 4 μl of protein sample was applied onto a glow-discharged copper grid coated with a continuous carbon layer (PLANO). The protein sample was incubated for 1 min before blotting away the excess sample with a filter paper. The sample-coated side of the grid was then washed by dipping it into three individual 20-μl drops of protein buffer with a blotting step in between. Last, the sample was negatively stained with 2% uranylformiate for 30 s, the excess sample was blotted away, and the grid was air-dried. Negative-stained EM girds were imaged using a JEOL JEM-2200FS TEM operating at 200 kV.
Cryo-EM sample preparation, data collection of NLRP3PYD filaments, and NLRP3PYD–to–ASC-mCherry filament transitions
To enhance filament binding, R1.2/1.3 Cu 300 grids (Quantifoil) were glow-discharged and coated with graphene oxide (0.2 mg/ml in ddH2O), followed by coating with poly-l-lysine (1 mg/ml in ddH2O) before sample application. Four microliters of polymerized NLRP3PYD (1 mg/ml) or NLRP3PYD:ASC-mCherry transitions (molar ratio of 10:1) supplied with 0.01% of the surfactant octyl maltoside was then applied onto the freshly pretreated grids. Samples were blotted for 3 to 4 s at 80% humidity and 20°C and subsequently plunge-frozen in liquid ethane using an EM GP blotter (Leica Microsystems). Grids containing NLRP3PYD filaments were imaged using a Krios Titan TEM (Thermo Fisher Scientific), operated at 300 kV and equipped with a modified Falcon2 direct electron detector, enabling frame acquisition. Grids containing the NLRP3PYD:ASC-mCherry transition filaments were imaged with a Krios Titan TEM equipped with a Falcon3 direct electron detector. For NLRP3PYD, 3000 frame movies with a total dose of 60 e−/A and 70 frames each were collected at a defocus range of −1 to −2.5 μm. For NLRP3PYD:ASC-mCherry, 1668 frame movies with a total dose of 60 e−/A and 40 frames each were collected in counting mode at a defocus range of −1.8 to −3 μm.
Cryo-EM data processing and model building of the NLRP3PYD filament
Cryo-EM data processing was done in RELION3 (61). Frame movies were aligned using RELION’s own implementation of the MotionCor2 algorithm. Motion-corrected but non–dose-weighted micrographs were used to determine contrast transfer function (CTF) parameters using CTFFIND 4.1. For further processing, 724 high-quality micrographs were chosen, and the start and end coordinates of filaments were manually identified using the EMAN2 interface (62). In total, 100,821 particles were extracted in RELION from the motion-corrected and dose-weighted micrographs with a box size of 220 pixels, assuming a helical rise of 14 Å. Particles were subjected to reference-free 2D classification, which yielded 13 2D classes showing high-resolution features accounting for a total of 60,333 particles. This subset was selected for further processing. An initial model was calculated on the basis of the ASCPYD filament structure (PDB: 3J63) (13). For this, the atomic model was first converted into a simulated electron density, low-pass–filtered to 8 Å, and then symmetrized assuming a helical twist of 54° and an axial rise of 14 Å. 3D refinement was then performed using the same values as the initial search parameters for the helical symmetry. 3D alignment parameters were used to re-extract centered particle images from the micrographs. These were subjected to an intermediate round of 2D classification, which yielded 10 classes with high-resolution features accounting for 26,172 particles.
Next, a subsequent round of 3D refinement was performed using these particles. For this, the map generated from the 60,333 particles was used to calculate a solvent mask. Helical parameters converged to a helical twist of 54.44° and an axial rise of 14.16 Å and yielded a resolution of 3.7 Å. Next, CTF refinement and Bayesian polishing were performed. A final round of 3D refinement produced a map at a resolution of 3.6 Å, with a helical twist of 54.88° and an axial rise of 14.32 Å. Chain A of the dimeric NLRP3PYD crystal structure (PDB: 3QF2) (24) served as initial model for model building of the NLRP3PYD filament in Coot (63). Real-space refinement was done in Phenix (64). The final model encompasses residues 3 to 94 with clear densities of most side chains seen in the electron density map (fig. S1E). A multiple sequence alignment of all 14 human NLRP PYDs displays the degree of sequence conservation in correlation to the secondary structure (fig. S7).
Determination of NLRP3PYD filament ASC-mCherry elongation directionality by cryo-EM
To determine the directionality of NLRP3PYD filament–seeded elongation by ASC-mCherry, we recorded a cryo-EM dataset of NLRP3PYD:ASC-mCherry transitions. The 1668 recorded movies were aligned using RELION’s own implementation of the MotionCor2 algorithm. Motion-corrected, non–dose-weighted micrographs were subjected to CTF estimation using CTFFIND 4.1. In total, 423 micrographs that allowed a clear distinction of individual transition events were chosen for further processing. Start and end coordinates were always picked in the same direction relative to the filament transition site. The start coordinate was located at the transition distant, and the end coordinate was located at the transition proximal end of the generated particle box. In total, 19,351 particles were extracted in RELION with a box size of 220 pixels and used for reference-free 2D classification. Four 2D classes (29, 44, 47, and 48) showed high-resolution features and a characteristic sawtooth pattern. Only these “good classes” were used in the following procedure. The directionality of the class averages with respect to the PYD filament structure was determined by manual superposition of the electron density onto the class averages. The directionality of the class average was then flagged by overlaying arrows onto the class average.
To identify the directionality of the NLRP3PYD-ASCPYD transition, we extracted the x and y coordinates, psi, and psiprior angles from each of the particles that constituted the four good classes. The directionality-flagged class averages were then aligned onto the extracted xy positions in the CTF- and motion-corrected micrographs and rotated according to the extracted psi angle (fig. S4). The process was repeated for 100 micrographs, and, in each case, the B-end of the class average pointed toward the ASC-mCherry filament transition. In addition, we noted that because the particle picking was always performed toward the ASC-mCherry transition, the psi and psiprior angles of the particles were always very similar. Figure S4 shows histograms of the Δpsi angles (psiprior − psi) of all particles in the good classes. Because of our described picking procedure of picking toward the ASC-mCherry transition, the histograms are centered around 0°. This means that RELION did not change the overall direction of any of the particles during the formation of the class averages. Any filaments with a reversed directionality would have led to peaks at 180° in the histogram. Very few instances of +360° or −360° occurred for filaments that were almost horizontal in the micrograph and thereby aligned along the x axis [e.g., 179.2° − (−178.7°) = 357.9°], which is, however, in agreement with the identified transition directionality.
NLRP3PYD filament polymerization monitored by DLS
Before the kinetic analysis of NLRP3PYD polymerization, a sample containing monomeric, soluble NLRP3PYD [50 mM glycine (pH 3.8), 150 mM NaCl, and 0.5 mM TCEP] was rapidly thawed, centrifuged at 13,000 rpm for 5 min, and filtered through a 0.1-μm syringe filter (Whatman). The protein concentration was adjusted to 0.6 mg/ml using the sample buffer, and filament formation was induced by adjusting the sample to pH 8.0 through addition of 3 M tris (pH 8.0) to a final concentration of 60 mM. Filament polymerization was monitored by batch DLS with a DynaPro NanoStar instrument (Wyatt). Data were acquired at 25°C in a time course experiment of 100 min in 60-s intervals by averaging three runs of 20 s until a plateau of filament polymerization was reached.
For polymerization analysis of single-point mutants, recombinant NLRP3PYD and mutant proteins were purified in a buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, and 0.5 mM TCEP as described above, and size exclusion chromatography fractions containing monomeric recombinant protein were pooled and concentrated to 0.6 mg/ml. Filamentation was induced by incubation at 25°C for 1.5 hours, and the polymerization status of the samples was subsequently monitored by batch DLS. Data were acquired by averaging three runs of 20 s. Three technical replicates of each experiment were performed.
Polymerization of NLRP3PYD and point mutants analyzed by negative-stain EM
In addition to DLS, the polymerization behavior of wild-type NLRP3PYD and point mutants was analyzed by negative-stain EM. Here, GST-NLRP3PYD or GST-NLRP3PYD point mutants at a concentration of 1 mg/ml were subjected to TEV protease cleavage (molar ratio TEV protease:GST-NLRP3PYD of 1:25) for 1.5 hours at 25°C and were subsequently analyzed by negative-stain EM as described above.
ASC specking experiments in HeLa cells
Five thousand HeLa cells stably overexpressing ASC-mTurquoise were seeded and transfected in duplicates with increasing amounts (0, 3.1, 6.3, 12.5, 25, 50, 100, or 200 ng) of plasmids encoding wild-type and point mutant NLRP3 (1-95)–mCitrine fusion proteins. Twelve hours or 36 hours after transfection, cells were fixed, and nuclei were stained using a phosphate-buffered saline (PBS) solution containing both paraformaldehyde (4%) and DRAQ5 (1:2000). Ten images per well were taken using filter sets to detect cyan fluorescent protein (i.e., filter set 47 from Zeiss: excitation BP 436/20, beam splitter FT 455, and emission BP 480/40) and DRAQ5 (i.e., filter set 50 from Zeiss: excitation BP 640/30, beam splitter FT 660, and emission BP 690/50) using the 20× objective of a Zeiss observer Z1 microscope. Images were analyzed using the CellProfiler 2.2.0 software to count nuclei and ASC specks. For each well, the ratio of ASC specks/nuclei was calculated. Ratios extracted from 20 independent images (10 images per well, conditions in duplicates) were averaged to calculate the final ratio of ASC speck/nuclei for every condition.
ASC specking experiments in HEK293T cells
HEK293T cells stably expressing an ASC-BFP fusion were seeded in 24-well plates at a density of 75,000 cells per well and incubated at 37°C for 24 hours. Per well, 100 ng of a doxycycline-inducible TetO6-NLRP3-hPGK-TetON3G-T2A-mCherry construct, encoding full-length NLRP3 (wild-type or indicated mutants), was transfected using 0.5 μl of Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Eighteen hours after transfection, NLRP3 expression was induced by adding doxycycline to a final concentration of 10 ng/ml, followed by an incubation of 6 hours at 37°C and addition of nigericin (10 μM) for another 1 hour. Cells were harvested by trypsinization, followed by washing with Dulbecco’s PBS (DPBS) and resuspension in 100 μl of flow buffer (DPBS supplemented with 2 mM EDTA and 0.5% bovine serum albumin). Flow cytometry analysis of the samples was carried out using an LSRFortessa II cell analyzer at medium flow rate. Transfected cells were gated for mCherry, which was coexpressed from the NLRP3-encoding plasmid. The gate for mCherry-positive (mCherry+) cells was set to low levels of expression (mCherry low) according to an mCherry-negative control. In addition, the mCherry gate was adjusted between experiments to yield similar levels of baseline ASC specks in the wild-type control samples while maintaining similar levels of mCherry+ cells. The level of ASC specks formed was determined as a percentage of the mCherry+ population.
NLRP3PYD seeding ASC filament elongation assays
To induce filament formation for homotypic PYD transition experiments, 50 μl of monomeric NLRP3PYD protein [50 mM glycine (pH 3.8), 150 mM NaCl, and 0.5 mM TCEP] at a concentration of 50 μM was adjusted to pH 8.0 by adding 1 μl of 3 M tris (pH 8.0) to a final concentration of 60 mM. The solution was incubated for 3 min at 25°C, allowing for the formation of short NLRP3PYD filaments. Monomeric soluble ASC-mCherry [50 mM glycine (pH 3.8), 150 mM NaCl, and 0.5 mM TCEP) was added to the NLRP3PYD sample to reach a molar ratio of 1:100 (ASC-mCherry to NLRP3PYD), and the sample was incubated for 5 min. This step was repeated two more times at increasing ASC volumes to reach consecutive molar ratios of ASC-mCherry to NLRP3PYD of 1:50 and 1:25. After each addition and subsequent incubation with the ASC-mCherry protein, the protein solution was applied onto EM grids, negatively stained, and imaged using a JEOL JEM-2200FS microscope to visualize binary complex assemblies containing ASC-mCherry filaments topping on NLRP3PYD filament seeds.
For the cryo-EM procedure, the titration protocol to generate NLRP3PYD:ASC-mCherry filament transitions was adjusted to receive longer initial NLRP3PYD filaments that could be used for subsequent particle extraction. The boxed particles were used for determining the directionality of ASC-mCherry elongation on NLRP3PYD filaments. Here, 50 μl of monomeric NLRP3PYD protein [50 mM glycine (pH 3.8), 150 mM NaCl, and 0.5 mM TCEP] at a concentration of 50 μM was adjusted to pH 8.0 as described above, and the solution was incubated for 6 min at 25°C. Monomeric soluble ASC-mCherry [50 mM glycine (pH 3.8), 150 mM NaCl, and 0.5 mM TCEP] was added to the NLRPPYD sample to reach a molar ratio of 1:50 (ASC-mCherry to NLRP3PYD) followed by 5-min incubation. In two consecutive steps, the molar ratio of ASC-mCherry to NLRP3PYD was increased to 1:25 and 1:10, each followed by 5-min incubation. The sample of the last incubation step was used for cryo-EM sample preparation.
Molecular modeling of an ASC speck
A model of an 11-mer NLRP3-NEK7 complex was created by separate structural alignment of the NACHT and LRR-NEK7 subunits of (PDB: 6NPY) (50) to each subunit of the disc-like NLRC4 structure (3JBL) (49). This ring-shaped structure was placed at the A-end of a 11-subunit NLRP3PYD filament (our structure; 7PZD). At its B-end, this filament was elongated by 32 subunits of an ASCPYD filament (3J63) (13), and the transition between both filaments was energy-minimized by GROMACS (33). The NMR ensemble of full-length ASC (2KN6) (34) was split into 20 individual structures. Each ASCPYD in the filament was overlaid with a randomly selected NMR structure to create a “full-length” model (with respect to the ASC polypeptide) of the ASC filament. Because of the 24–amino acid linker between the ASCPYD and ASCCARD domains, it was possible to model eight-subunit large “buds” of an ASCCARD filament at three different positions of the ASCPYD filament. The arrangement of the subunits in these buds was modeled by using the ASCCARD filament structure (6N1H) (54) as a template. A superposition of the ASCCARD–to–Caspase-1CARD transition structure (7KEU) (56) onto the buds created the transition to the Caspase-1CARD filament (5FNA) (43). Last, structural models of the Caspase-1–zymogen (p20-p10 dimer; 3E4C) (65) were placed at each junction of two Caspase-1CARD molecules in the filament.
Acknowledgments
We thank the research center caesar, Bonn, and the Rudolf-Virchow-Zentrum at Universität Würzburg for EM measurement time as well as J. Moecking for supervision of HEK cell assays.
Funding: This work was supported by a grant from the Else Kröner-Fresenius-Stiftung to M.G. (2014_A203). M.G. and E.L. are funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy–EXC2151–390873048.
Author contributions: I.V.H. purified proteins and performed biochemical experiments. I.V.H. and H.B. prepared the cyro-EM samples. I.V.H. and E.B. processed the cryo-EM data. I.V.H. and G.H. built the model. I.V.H., G.H., and M.G. interpreted the data. I.V.H. and A.K. established the NLRPPYD seeding assay for ASC filament elongation. J.F.R.-A. performed ASC specking assay in HeLa cells that E.L. supervised. I.V.H. performed ASC specking assay of CAPS mutants in HEK293T cells. I.V.H. and M.G. wrote the paper. All authors contributed to editing the manuscript and supporting the conclusions.
Competing interests: M.G. and E.L. are cofounders and consultants of IFM Therapeutics. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. No materials used in this study are subject to a material transfer agreement. The cryo-EM structures have been deposited in the Electron Microscopy Data Bank (EMDB) with accession number EMD-13727 for the NLRP3PYD filament. The atomic coordinates and structure factors have been deposited in the PDB with accession number 7PZD.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
References
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Martinon F., Burns K., Tschopp J., The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Swanson K. V., Deng M., Ting J. P.-Y., The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franchi L., Muñoz-Planillo R., Núñez G., Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 13, 325–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latz E., Xiao T. S., Stutz A., Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friker L. L., Scheiblich H., Hochheiser I. V., Brinkschulte R., Riedel D., Latz E., Geyer M., Heneka M. T., β-Amyloid clustering around asc fibrils boosts its toxicity in microglia. Cell Rep. 30, 3743–3754.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ising C., Venegas C., Zhang S., Scheiblich H., Schmidt S. V., Vieira-Saecker A., Schwartz S., Albasset S., McManus R. M., Tejera D., Griep A., Santarelli F., Brosseron F., Opitz S., Stunden J., Merten M., Kayed R., Golenbock D. T., Blum D., Latz E., Buée L., Heneka M. T., NLRP3 inflammasome activation drives tau pathology. Nature 575, 669–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strowig T., Henao-Mejia J., Elinav E., Flavell R., Inflammasomes in health and disease. Nature 481, 278–286 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Venegas C., Kumar S., Franklin B. S., Dierkes T., Brinkschulte R., Tejera D., Vieira-Saecker A., Schwartz S., Santarelli F., Kummer M. P., Griep A., Gelpi E., Beilharz M., Riedel D., Golenbock D. T., Geyer M., Walter J., Latz E., Heneka M. T., Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552, 355–361 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D., Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301–305 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., Latz E., Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. Baltim. 183, 787–791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu A., Magupalli V. G., Ruan J., Yin Q., Atianand M. K., Vos M. R., Schröder G. F., Fitzgerald K. A., Wu H., Egelman E. H., Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sborgi L., Ravotti F., Dandey V. P., Dick M. S., Mazur A., Reckel S., Chami M., Scherer S., Huber M., Böckmann A., Egelman E. H., Stahlberg H., Broz P., Meier B. H., Hiller S., Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 112, 13237–13242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baroja-Mazo A., Martín-Sánchez F., Gomez A. I., Martínez C. M., Amores-Iniesta J., Compan V., Barberà-Cremades M., Yagüe J., Ruiz-Ortiz E., Antón J., Buján S., Couillin I., Brough D., Arostegui J. I., Pelegrín P., The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 15, 738–748 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Cai X., Chen J., Xu H., Liu S., Jiang Q.-X., Halfmann R., Chen Z. J., Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin B. S., Bossaller L., De Nardo D., Ratter J. M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S. R., Al-Amoudi A., Mangan M. S., Zimmer S., Monks B. G., Fricke M., Schmidt R. E., Espevik T., Jones B., Jarnicki A. G., Hansbro P. M., Busto P., Marshak-Rothstein A., Hornemann S., Aguzzi A., Kastenmüller W., Latz E., The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat. Immunol. 15, 727–737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick M. S., Sborgi L., Rühl S., Hiller S., Broz P., ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7, 11929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruland J., Inflammasome: Putting the pieces together. Cell 156, 1127–1129 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lu A., Li Y., Yin Q., Ruan J., Yu X., Egelman E., Wu H., Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 1, 15013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C., Lu A., Xie W. J., Ruan J., Negro R., Egelman E. H., Fu T.-M., Wu H., Molecular mechanism for NLRP6 inflammasome assembly and activation. Proc. Natl. Acad. Sci. U.S.A. 116, 2052–2057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marleaux M., Anand K., Latz E., Geyer M., Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly. FEBS Lett. 594, 2383–2395 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Stutz A., Kolbe C.-C., Stahl R., Horvath G. L., Franklin B. S., van Ray O., Brinkschulte R., Geyer M., Meissner F., Latz E., NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214, 1725–1736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae J. Y., Park H. H., Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J. Biol. Chem. 286, 39528–39536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrao R., Wu H., Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 22, 241–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park H. H., PYRIN domains and their interactions in the apoptosis and inflammation signaling pathway. Apoptosis Int. J. Program. Cell Death. 17, 1247–1257 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Hu J., Zhu Y., Zhang J.-Z., Zhang R.-G., Li H.-M., A novel mutation in the pyrin domain of the NOD-like receptor family pyrin domain containing protein 3 in Muckle-Wells syndrome. Chin. Med. J. 130, 586–593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martorana D., Bonatti F., Mozzoni P., Vaglio A., Percesepe A., Monogenic autoinflammatory diseases with mendelian inheritance: Genes, mutations, and genotype/phenotype correlations. Front. Immunol. 8, 344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turunen J. A., Wedenoja J., Repo P., Järvinen R.-S., Jäntti J. E., Mörtenhumer S., Riikonen A. S., Lehesjoki A.-E., Majander A., Kivelä T. T., Keratoendotheliitis fugax hereditaria: A novel cryopyrin-associated periodic syndrome caused by a mutation in the nucleotide-binding domain, leucine-rich repeat family, pyrin domain-containing 3 (NLRP3) gene. Am J. Ophthalmol. 188, 41–50 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Schmidt F. I., Lu A., Chen J. W., Ruan J., Tang C., Wu H., Ploegh H. L., A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 213, 771–790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matyszewski M., Zheng W., Lueck J., Mazanek Z., Mohideen N., Lau A. Y., Egelman E. H., Sohn J., Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome. Nat. Commun. 12, 2735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrone S. R., Matyszewski M., Yu X., Delannoy M., Egelman E. H., Sohn J., Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat. Commun. 6, 7827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C., GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 34.de Alba E., Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC). J. Biol. Chem. 284, 32932–32941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., Liu Z., Xiao T. S., Post-translational regulation of inflammasomes. Cell. Mol. Immunol. 14, 65–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He M., Chiang H.-H., Luo H., Zheng Z., Qiao Q., Wang L., Tan M., Ohkubo R., Mu W.-C., Zhao S., Wu H., Chen D., An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 31, 580–591.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodson H. V., Jonasson E. M., Microtubules and microtubule-associated proteins. Cold Spring Harb. Perspect. Biol. 10, a022608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollard T. D., Borisy G. G., Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 (2003). [DOI] [PubMed] [Google Scholar]
- 39.O’Brien E. T., Salmon E. D., Walker R. A., Erickson H. P., Effects of magnesium on the dynamic instability of individual microtubules. Biochemistry 29, 6648–6656 (1990). [DOI] [PubMed] [Google Scholar]
- 40.Swenson A. M., Trivedi D. V., Rauscher A. A., Wang Y., Takagi Y., Palmer B. M., Málnási-Csizmadia A., Debold E. P., Yengo C. M., Magnesium modulates actin binding and ADP release in myosin motors. J. Biol. Chem. 289, 23977–23991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matusiak M., Van Opdenbosch N., Lamkanfi M., CARD- and pyrin-only proteins regulating inflammasome activation and immunity. Immunol. Rev. 265, 217–230 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Indramohan M., Stehlik C., Dorfleutner A., COPs and POPs patrol inflammasome activation. J. Mol. Biol. 430, 153–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu A., Li Y., Schmidt F. I., Yin Q., Chen S., Fu T.-M., Tong A. B., Ploegh H. L., Mao Y., Wu H., Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol. 23, 416–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochheiser I. V., Pilsl M., Hagelueken G., Moecking J., Marleaux M., Brinkschulte R., Latz E., Engel C., Geyer M., Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604, 184–189 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Andreeva L., David L., Rawson S., Shen C., Pasricha T., Pelegrin P., Wu H., NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184, 6299–6312.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diebolder C. A., Halff E. F., Koster A. J., Huizinga E. G., Koning R. I., Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: Implications for NLR activation. Struct. Lond. Engl. 23, 2349–2357 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.-W., Sui S.-F., Chai J., Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Tenthorey J. L., Haloupek N., López-Blanco J. R., Grob P., Adamson E., Hartenian E., Lind N. A., Bourgeois N. M., Chacón P., Nogales E., Vance R. E., The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L., Chen S., Ruan J., Wu J., Tong A. B., Yin Q., Li Y., David L., Lu A., Wang W. L., Marks C., Ouyang Q., Zhang X., Mao Y., Wu H., Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharif H., Wang L., Wang W. L., Magupalli V. G., Andreeva L., Qiao Q., Hauenstein A. V., Wu Z., Núñez G., Mao Y., Wu H., Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., Chen Z. J., PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu A., Wu H., Structural mechanisms of inflammasome assembly. FEBS J. 282, 435–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tapia-Abellán A., Angosto-Bazarra D., Alarcón-Vila C., Baños M. C., Hafner-Bratkovič I., Oliva B., Pelegrín P., Sensing low intracellular potassium by NLRP3 results in a stable open structure that promotes inflammasome activation. Sci. Adv. 7, eabf4468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Fu T.-M., Lu A., Witt K., Ruan J., Shen C., Wu H., Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl. Acad. Sci. U.S.A. 115, 10845–10852 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong Q., Robinson K., Xu C., Huynh P. T., Chong K. H. C., Tan E. Y. J., Zhang J., Boo Z. Z., Teo D. E. T., Lay K., Zhang Y., Lim J. S. Y., Goh W. I., Wright G., Zhong F. L., Reversade B., Wu B., Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8. Nat. Commun. 12, 188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robert Hollingsworth L., David L., Li Y., Griswold A. R., Ruan J., Sharif H., Fontana P., Orth-He E. L., Fu T.-M., Bachovchin D. A., Wu H., Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun. 12, 189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boucher D., Monteleone M., Coll R. C., Chen K. W., Ross C. M., Teo J. L., Gomez G. A., Holley C. L., Bierschenk D., Stacey K. J., Yap A. S., Bezbradica J. S., Schroder K., Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 215, 827–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandes-Alnemri T., Wu J., Yu J.-W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E. S., The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14, 1590–1604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.I. M. Glück, G. P. Mathias, S. Strauss, T. S. Ebert, C. Stafford, G. Agam, S. Manley, V. Hornung, R. Jungmann, C. Sieben, D. C. Lamb, Nanoscale organization of the endogenous ASC speck. bioRxiv 2021.09.17.460822 [Preprint]. 18 September 2021. 10.1101/2021.09.17.460822. [DOI] [PMC free article] [PubMed]
- 60.Kagan J. C., Magupalli V. G., Wu H., SMOCs: Supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 14, 821–826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elliott J. M., Rouge L., Wiesmann C., Scheer J. M., Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation*. J. Biol. Chem. 284, 6546–6553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin T., Chuenchor W., Jiang J., Cheng J., Li Y., Fang K., Huang M., Smith P., Xiao T. S., Design of an expression system to enhance MBP-mediated crystallization. Sci. Rep. 7, 40991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eibl C., Grigoriu S., Hessenberger M., Wenger J., Puehringer S., Pinheiro A. S., Wagner R. N., Proell M., Reed J. C., Page R., Diederichs K., Peti W., Structural and functional analysis of the NLRP4 pyrin domain. Biochemistry 51, 7330–7341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinheiro A. S., Proell M., Eibl C., Page R., Schwarzenbacher R., Peti W., Three-dimensional structure of the NLRP7 pyrin domain: Insight into pyrin-pyrin-mediated effector domain signaling in innate immunity. J. Biol. Chem. 285, 27402–27410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eibl C., Hessenberger M., Wenger J., Brandstetter H., Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions. Acta Crystallogr. D Biol. Crystallogr. 70, 2007–2018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hiller S., Kohl A., Fiorito F., Herrmann T., Wider G., Tschopp J., Grütter M. G., Wüthrich K., NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure 11, 1199–1205 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Su M.-Y., Kuo C.-I., Chang C.-F., Chang C.-I., Three-dimensional structure of human NLRP10/PYNOD pyrin domain reveals a homotypic interaction site distinct from its mouse homologue. PLOS ONE 8, e67843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
References
Data file S1