Abstract
A lysis module encoded by the temperate bacteriophage φO1205 was identified. This lysis module contains a lysin gene, designated lyt51, and two putative holin-encoding genes, designated lyt49 and lyt50. lyt51 encodes a lytic enzyme specifically directed against streptococcal cell walls. Similar to other phage-encoded lysins, Lyt51 appears to have a modular design in which the N-terminal portion corresponds to its enzymatic activity while the C-terminal region is responsible for its substrate binding specificity. The two putative holin-encoding genes, lyt49 and lyt50, located immediately upstream of lyt51, were identified on the basis of their homology to other identified holin-encoding genes. Expression of lyt49 or lyt50 in Escherichia coli was shown to cause cell death and leakage of the intracellular enzyme isocitrate dehydrogenase into the growth medium without apparent lysis of the cells. Southern blotting experiments demonstrated that at least one of the three components of the identified lysis module is present in all members of a large collection of bacteriophages, indicating that components of this lysis module are widespread among bacteriophages infecting Streptococcus thermophilus.
Bacteriophage infection of lactic acid bacteria (LAB) used as starter cultures has long been a serious concern for the milk fermentation industry (16, 42). Bacteriophage attack of dairy fermentations can yield products of inferior quality or even result in complete product failure. Streptococcus thermophilus, a thermophilic component of starter cultures commonly employed for the manufacture of certain dairy products, such as yogurt, Swiss and Italian-type cheeses, and short-method cheddar cheese (12), is known to be susceptible to such bacteriophage attacks (3, 10, 39).
There is an increasing body of knowledge regarding phage-host interactions as well as molecular aspects of bacteriophages of LAB, in particular those infecting Lactococcus species (for reviews, see, among others, references 22, 28, 29, 37, and 45), Lactobacillus species (48), and, to a lesser extent, Leuconostoc species (6). In recent years, S. thermophilus has also become the subject of molecular genetic research, and a significant amount of sequence data for various S. thermophilus phages has become available (11, 13, 40, 56). Genomes of small, isometric-headed phages infecting S. thermophilus were reported to exhibit extensive similarities to those of other LAB-infecting phages of identical morphology, although the functions of the majority of the deduced gene products have yet to be elucidated (11, 13, 14, 40, 56).
One of the most dramatic events following bacteriophage infection is phage-induced lysis of the host cell. With many bacteriophages, this process has been shown to require the action of two phage-encoded proteins (64). The first is a so-called holin, a small transmembrane protein which oligomerizes to form lesions in the cytoplasmic membrane. These lesions function as pores to allow the nonspecific release of the second protein, an enzyme with peptidoglycan-hydrolyzing activity (64, 65). Lysins can only reach their substrate after passage through the cytoplasmic membrane, and there are no reports to date of a phage-encoded endolysin synthesized with a secretory signal sequence to mediate its release across the cytoplasmic membrane (64, 65). As in lambdoid phages, the genetic arrangement of the holin-lysin cassette in small, isometric-headed phages infecting LAB appears to be very specific; i.e., the small holin-encoding gene immediately precedes the lysin gene, and in most cases both genes overlap by one or more base pairs (64). In prolate-headed phages infecting LAB, however, the holin gene appears to be separated from the lysin gene by several kilobases of DNA (33, 47).
Holin proteins identified to date are relatively small, and they are predicted to contain the following structural features: two membrane-spanning α-helix domains separated by a β-turn, a short hydrophilic N terminus, and a highly charged C terminus. Young and Bläsi (65) divided holin proteins into three classes; the lambdoid class II holin, which is predicted to contain only two putative membrane-spanning domains separated by a β-turn while possessing a positively charged N-terminal region; the lambdoid class I holin, which, in addition to having the features of the class II holins, is characterized by the presence of a predicted third membrane-spanning domain; and the class III holin, i.e., the T4 t holin. The largest holin described to date is HolTW of the Staphylococcus aureus phage Twort (32), consisting of 185 amino acids due to an apparently extended C-terminal domain and belonging to class II (64). A number of holin genes have a dual-start motif allowing the synthesis of two products of slightly different lengths, one of which acts as the inhibitor of the other. This inhibitor, or antiholin, is believed to be of crucial importance for the exact timing of the lysis event (64, 65).
The genes encoding phage lysins (and their deduced protein products) of several LAB-infecting phages have been characterized at the molecular and biochemical levels (for a review, see reference 23); these include the lysins of lactococcal phages POO1 (24), φUS3 (43), c2 (62), vML3 (49), φLC3 (4), Tuc2009 (2, 51), and rlt (38), as well as that of the Lactobacillus delbrueckii phage LL-H (60). Many lysins have been shown to be composed of two distinct domains: the N-terminal domain, dictating the catalytic activity of the lysin, and the C-terminal region, determining the substrate binding and, therefore, the specificity of the enzyme (for a review, see reference 63). The N-terminal domain determines the category into which the lysin is classified; the glucosaminidases and lysozymes hydrolyze the glycosidic linkage between the amino sugars of the peptidoglycan, amidases hydrolyze the N-acetylmuramoyl-l-alanine amide linkage between the glycan strand and the cross-linking peptide, and endopeptidases hydrolyze the interpeptide bridge linkage.
In this work, we present the characterization of the lytic module of an S. thermophilus bacteriophage, which consists of three adjacent genes. The two 5′ proximally located genes specify two putative holins, whereas the 3′ distally located gene was shown to encode a protein with lytic activity. This lytic module’s products appear to be different from those of previously characterized lysin modules of LAB phages because of the presence of two holins and the apparently limited substrate specificity of the lysin.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and growth media.
Bacteriophages, bacterial strains, and plasmids used in this study are listed in Table 1. The 29 different S. thermophilus bacteriophages chosen for Southern blotting analysis (see Table 3) are described by Le Marrec et al. (31) and are not included in Table 1. S. thermophilus strains were grown at 43°C in either Elliker medium (Difco Laboratories, Detroit, Mich.) supplemented with 10 g of beef extract (Difco) liter−1 or M17 medium (59) supplemented with 10 g of lactose liter−1 (LM17). Escherichia coli was cultivated in Luria-Bertani (LB) broth as described by Sambrook et al. (46). Streptococcus mutans and Bacillus coagulans were grown at 37°C in tryptic soy broth (Difco) supplemented with 0.7% (wt/vol) yeast extract (Difco). Enterococcus faecalis and Lactobacillus paracasei were grown at 37°C in MRS broth (Difco). For selection of E. coli transformants, ampicillin and tetracycline were used at concentrations of 100 and 12.5 μg ml−1, respectively. Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were used, where appropriate, at concentrations of 0.5 mM and 40 μg ml−1, respectively. φO1205 was induced from its lysogenic host S. thermophilus CNRZ1205 with mitomycin C (Sigma Chemical Co., St. Louis, Mo.) at a final concentration of 0.2 μg ml−1. Phage particles were isolated as described by Sambrook et al. (46) with modifications as described by Stanley et al. (56).
TABLE 1.
Phages, bacterial strains, and plasmids used in this study
| Bacterium, phage, or plasmid | Relevant characteristics or genotype | Source or referencea |
|---|---|---|
| S. thermophilus | ||
| CNRZ1205 | Traditional strain used in yogurt production; lysogenic for φO1205 | 17 |
| CNRZ1205-3 | Prophage-cured derivative of CNRZ1205 | 55a, 56 |
| CNRZ1205-3A | Relysogenized CNRZ1205 | 55a, 56 |
| UCCY110 | Industrial strain used for yogurt production | UCC culture collection |
| NIZO440 | No information available | NIZO collection |
| NIZO702 | No information available | NIZO collection |
| NIZORR | No information available | NIZO collection |
| NIZO1443 | No information available | NIZO collection |
| NIZO1ST | No information available | NIZO collection |
| CNRZ302 | Traditional Gruyere cheese starter | INRA collection |
| CNRZ160 | Industrial strain used for yogurt production | INRA collection |
| CNRZ447 | Industrial strain | INRA collection |
| Other bacterial strains | ||
| E. coli XLI-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)] | Stratagene |
| Enterococcus faecalis | Wild-type strain | UCC culture collection |
| S. mutans | Wild-type strain | UCC culture collection |
| B. coagulans | Wild-type strain | UCC culture collection |
| L. paracasei | Wild-type strain | UCC culture collection |
| S. thermophilus phages | ||
| φO1205 | Temperate phage induced from CNRZ1205 | 17 |
| φ7201 | Lytic phage | NIZO collection |
| Plasmids | ||
| pQE60 | Ampr | Qiagen |
| pMS49 | pQE60 derivative containing lyt49 | This work |
| pMS50 | pQE60 derivative containing lyt50 | This work |
| pMS51 | pQE60 derivative containing lyt51 | This work |
NIZO, Nederlands Instituut voor Zuivelonderzoek, Ede, The Netherlands; INRA, Institut National de la Recherche Agronomique, Domaine de Vilvert, Jouy-en-Josas, France; UCC, University College Cork.
TABLE 3.
Presence of DNA homologues of φO1205 lyt49, lyt50, and lyt51 in other S. thermophilus phagesa
| Phage | Group | Hybridization profile withb:
|
||
|---|---|---|---|---|
| lyt49 | lyt50 | lyt51 | ||
| φ83 | cos | +++ | − | +++ |
| φ117 | cos | +++ | − | +++ |
| PO | cos | − | + | − |
| φ71 | cos | ++ | ++ | ++ |
| φ124 | cos | ++ | ++ | +++ |
| φ47 | cos | ++ | ++ | − |
| st2 | cos | +++ | ++ | +++ |
| BaS19 | cos | ++ | ++ | ++ |
| Q5 | cos | ++ | + | +++ |
| φ7201 | cos | +++ | + | +++ |
| φ7203 | cos | +++ | + | +++ |
| φ7205 | cos | +++ | − | +++ |
| φ7206 | cos | +++ | + | +++ |
| φ7209 | cos | +++ | + | +++ |
| φ8FN | cos | +++ | + | ++ |
| φ33 | cos | + | ++ | ++ |
| c20 | cos | ++ | ++ | +++ |
| BaS265 | cos | + | +++ | +++ |
| Q6 | cos | ++ | ++ | +++ |
| P4 | pac | ++ | ++ | +++ |
| φ31 | pac | + | ++ | +++ |
| φ1 | pac | +++ | ++ | +++ |
| φ4FN | pac | +++ | + | +++ |
| φ45 | pac | − | + | − |
| φO1205 | pac | +++ | +++ | +++ |
| 447-B4 | pac | ++ | ++ | − |
| Q1 | pac | +++ | − | − |
| Q3 | pac | +++ | + | +++ |
| Q7 | pac | +++ | − | − |
| Q10 | pac | +++ | − | − |
The phages are grouped on the basis of their packaging mechanisms (31).
+++, intense band; ++, moderately intense band; +, faint band; −, no hybridization evident.
DNA manipulations.
Restriction enzymes, T4 DNA ligase, Taq DNA polymerase, and the EXPAND long-template PCR system were obtained from Boehringer GmbH (Mannheim, Germany) and used according to the manufacturer’s instructions. Deoxyribonucleotides were obtained from Pharmacia LKB Biotechnology AB (Uppsala, Sweden). Plasmid DNA was isolated by the method of Birnboim and Doly (5) or by using a QIAPrep Spin Plasmid Miniprep kit (Qiagen, Hilden, Germany). Synthetic oligonucleotides (Oligo 1000M; Beckman Instruments) were used as primers for PCR and/or DNA sequencing. DNA fragments generated by PCR were purified by using a High Pure PCR product purification kit (Boehringer). Chromosomal DNA was isolated as described by Sambrook et al. (46) with modifications according to Stanley et al. (56).
PCR amplification, using chromosomal DNA of the lysogenic S. thermophilus strain CNRZ1205 as a template, was performed to generate DNA fragments encompassing lyt49 (455 bp), lyt50 (282 bp), or lyt51 (881 bp). Oligonucleotide primer pairs were based on the published sequence of φO1205 (56) and had the following sequences: 5′-GGGAATTCAGAGGTGCGTGCAATG-3′ and 5′-GGAAGCTTATTAATCATTTTCTTAT-3′ (to generate a DNA fragment encompassing lyt49), 5′-GGGAATTCAGAGGAAGAAGAATG-3′ and 5′-CCAAG CTTCTATTTTCCTTC-3′ (to amplify lyt50), and 5′-GGGAATTCGAAGGAAAATAGTATG-3′ and 5′-GGAAGCTTCGTGGTCTATTTG-3′ (to amplify lyt51). The single-underlined sequences (representing EcoRI restriction sites) and the double-underlined sequences (representing HindIII restriction sites) were introduced during the PCR amplification to facilitate cloning of the PCR fragments.
To allow their expression, the presumptive Shine-Dalgarno regions (52) of PCR-amplified lyt49, lyt50, and lyt51 were retained. After restriction, the amplified DNA fragments were isolated by using a Geneclean II kit (BIO101, Vista, Calif.) and were cloned into the expression vector pQE60, an IPTG-inducible, high-level E. coli expression vector (Qiagen). Transformation of E. coli was performed by the RbCl method (46).
DNA sequence analysis.
Recombinant plasmids were sequenced by using a model 373A automated DNA sequencer (Applied Biosystems Inc., Foster City, Calif.) to ensure that no mutations had occurred during DNA amplification or cloning procedures. Database searches were performed by using the FASTA (41), BLASTP, and TBLASTN (1) programs with the nonredundant-sequence databases located at http://genome.eerie.fr/bin/fasta-guess.cgi and http://www.ncbi.nlm.nih.gov/. Sequence alignments were performed by the Clustal method with the MEGALIGN release 3.06 program of the DNASTAR 1996 release software package. Computer analyses of the amino acid sequences of Lyt49 and Lyt50 were performed with the GOR Secondary Structure program of Garnier et al. (20), located at http://molbiol.soton.ac.uk/compute/GOR.html, and the Transmembrane Prediction program of Hofmann and Stoffel (27), located at http://ulrec3.unil.ch.software/TMPred_form.html.
Southern hybridization.
Following separation of digested DNA on an agarose gel, restriction fragments were transferred to a Hybond-N+ nucleic acid transfer membrane (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) by Southern blotting (55). An enhanced chemiluminescence kit (ECL; Amersham) was used for homology detection under low-stringency conditions as specified by the manufacturer.
Growth and viability assays.
Bacterial growth was monitored at 20-min intervals for 2 h by measuring the optical density at 600 nm (OD600); viable-cell numbers were determined by spread plating on LB agar plates containing ampicillin and tetracycline at concentrations of 100 and 12.5 μg ml−1, respectively.
Preparation of cell extracts and whole-cell substrates.
Cell extracts were prepared as follows. Bacterial cells (10 ml) were grown to late log phase (OD600 ≈ 1.5), harvested by centrifugation, resuspended in 800 μl of sodium phosphate buffer (pH 6.5), and broken by sonication (five pulses for 10 s each, with 1-min cooling intervals in an ice bath). These extracts were subsequently centrifuged (14,000 rpm for 15 min in an Eppendorf centrifuge [model 5415c]) to remove insoluble cell debris. Protein concentrations were determined by the method of Bradford (9), using bovine serum albumin as a standard. To prepare crude cell walls as an immobilized substrate in renatured polyacrylamide gels for in situ detection of lytic activities (see below), a bacterial culture (22) was grown to late log phase, harvested by centrifugation (7,500 rpm for 10 min in a Beckman centrifuge [model J2-21]), frozen for 2 h, and dried overnight under vacuum, after which the cells were dissolved in water, sterilized (15 min at 121°C and 15 lb/in2), and stored at −20°C until use.
Biological activity of lysin.
The in vivo effect of the lytic enzyme Lyt51 was determined by the addition of increasing amounts (18.57 to 371.2 μg of protein) of a crude extract of lyt51-overexpressing E. coli cells to growing cells of 10 different S. thermophilus strains and monitoring the OD600.
Isocitrate dehydrogenase assay.
Intracellular enzyme isocitrate dehydrogenase activity in the growth media of several E. coli strains was measured by using an isocitrate dehydrogenase kit (Sigma) in accordance with the manufacturer’s instructions; assays were repeated twice. The assay is based on isocitrate dehydrogenase-mediated oxidative decarboxylation of l-isocitrate in the presence of manganese ions, during which NADP is reduced to NADPH. The latter chemical conversion can be monitored spectrophotometrically at 340 nm. The activity of isocitrate dehydrogenase is proportional to the concentration of NADPH formed and is expressed in sigma units, with 1ς being the amount of sample that produces 1 nmol of NADPH in 1 h at 25°C.
SDS-PAGE.
Proteins were denatured at 100°C for 5 min with sample buffer before being subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described by Laemmli (30), on a 12.5% polyacrylamide gel (Mini Protean II; Bio-Rad Laboratories, Richmond, Calif.). After electrophoresis, gels were fixed with a solution containing 10% acetic acid and 25% isopropanol, stained with Coomassie blue R-250 (Sigma), and destained with 7% acetic acid. Rainbow prestained low-molecular-weight protein markers (Amersham) were used to deduce the molecular weight of the protein exhibiting lytic activity. In situ detection of lytic activity was performed, as described by Potvin et al. (44), with 12.5% polyacrylamide gels containing 0.2% (wt/vol) crude cell wall preparation (see above). Unless stated otherwise, after electrophoresis, gels were washed at 43°C for 24 to 48 h in a Tris-HCl wash buffer (10 mM Tris-HCl [pH 7.5], 0.1% [vol/vol] Triton X-100, 1 mM CaCl2, 1 mM MgCl2, and 0.5 mM dl-dithiothreitol) for protein renaturation, stained with 1% (wt/vol) methylene blue (Sigma) in 0.01% (wt/vol) KOH, and destained with deionized water.
RESULTS
Sequence similarities of ORF49, ORF50, and holins.
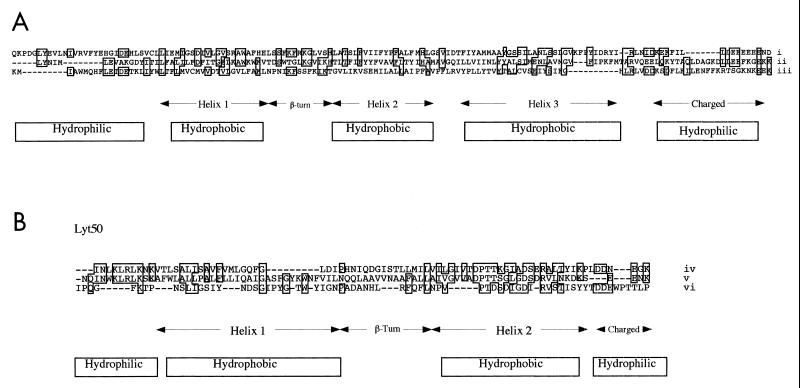
Open reading frame 49 (ORF49) and ORF50 of φO1205 were previously tentatively identified as genes specifying so-called holin proteins as inferred from their homology to holin genes of other bacteriophages infecting gram-positive bacteria (56). Holin proteins are small in size and usually contain two putative membrane-spanning domains separated by a β-turn linker region, as well as a hydrophilic N terminus and a highly charged C terminus (65). Although the products of ORF49 and ORF50, designated here as lyt49 and lyt50, respectively, show significant sequence similarity to other phage-encoded holins, they are not similar to each other. The deduced product of lyt49 consists of 141 amino acid residues and has the structural features of a lambdoid class I holin (65), i.e., a hydrophilic N terminus, three predicted membrane-spanning α-helix domains, a β-turn separating α-helices 1 and 2, and a highly charged C terminus (Fig. 1A). Database searches for proteins with similarity to the predicted amino acid sequence of lyt49 revealed that proteins encoded by ORFs of S. thermophilus phages φSfi21, φSfi19, and φSfi11 were highly similar (98.2% overall identity to ORF61 of φSfi21 [11] and ORF141 of φSfi19 [13] and 96.5% overall identity to ORF141 of φSfi11 [34]). In addition, significant similarities were observed between Lyt49 and the holin of the pneumococcal phage Cp-1 (36) (27.1% identity within a 70-amino-acid overlap) as well as the B. subtilis phage φ29 holin (21) (24.2% identity within a 95-amino-acid overlap) (Fig. 1A). The predicted protein product of lyt50 (Fig. 1B) consists of 80 amino acid residues and resembles members of the lambdoid class II holins (65). Lyt50 is highly similar to the suspected holin encoded by the S. thermophilus phage φSfi21 (11), exhibiting 78.2% identity over a length of 72 amino acids, in addition to being significantly similar to the holin of the lactococcal phage φLC3 (4) and the holin of the S. pneumoniae phage Ej-1 (15) (43.8% identity over a length of 29 amino acids and 40.9% identity over a length of 29 amino acids, respectively) (Fig. 1B).
FIG. 1.
(A) Multiple sequence alignment between Lyt49 (i) and the holins of pneumococcal phage Cp-1 (ii) and B. subtilis phage φ29 (iii). Identical amino acids are boxed. The approximate positions of three predicted transmembrane helices, the highly charged C-terminal domains, and the β-turn are indicated, as are areas of hydrophilicity and hydrophobicity. Dashes represent introduced gaps to maximize alignment. (B) Multiple sequence alignment between Lyt50 (iv) and the holins of lactococcal phage φLC3 (v) and pneumococcal phage Ej-1 (vi). Predicted secondary structures are indicated as described above.
Cloning and expression of lyt49 and lyt50 in E. coli.
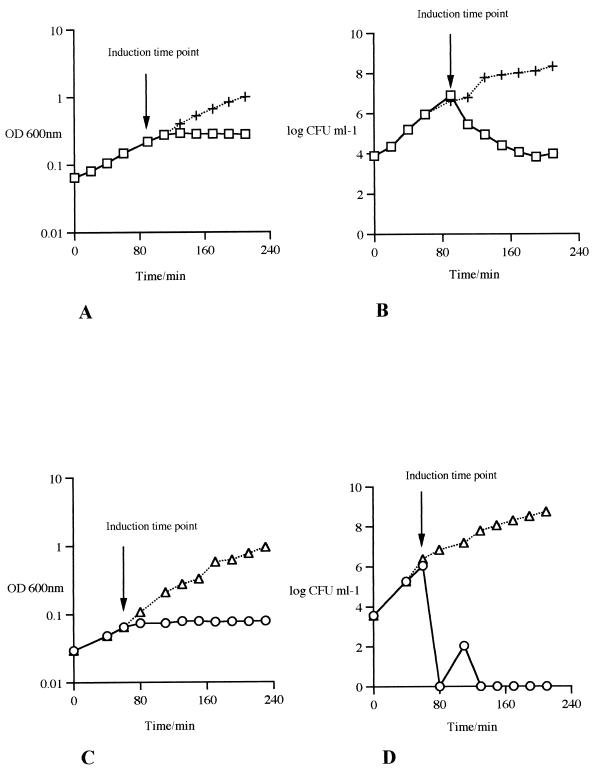
The similarities between several phage-encoded holin proteins and the proteins specified by lyt49 and lyt50 suggested that the latter may also function as holins. It has been shown for other phages that expression of a holin gene in either a heterologous or a homologous host system causes cell death through the formation of nonspecific lesions in the cytoplasmic membrane (4, 15, 26, 35, 38, 58). If lyt49 and lyt50 were indeed specifying holins, it was expected that their expression would therefore be lethal for the host cell. To test this expectation, DNA fragments encompassing either lyt49 or lyt50 were cloned into the E. coli inducible expression vector pQE60, resulting in plasmids pMS49 and pMS50, respectively (see Materials and Methods). Growth and viability of strains expressing Lyt49 and Lyt50 were measured following induction with IPTG. When the expression of lyt49 was induced in cells harboring pMS49, growth was arrested (Fig. 2A), and the viability of the E. coli cells was reduced by 3 orders of magnitude within 110 min after the addition of IPTG (Fig. 2B). Similarly, when the expression of lyt50 was induced, growth was also inhibited (Fig. 2C), and the cell viability was reduced by 5 to 6 orders of magnitude within 20 min after induction (Fig. 2D). In contrast, growth and viability of E. coli cells harboring the expression vector pQE60 were shown to be unaffected by the addition of IPTG (results not shown). These results indicate that lyt49 and lyt50 are able to induce a rapid and lethal effect on E. coli cells. To determine whether the gene products of lyt49 and lyt50 would be capable of forming cytoplasmic-membrane lesions large enough to allow the release of intracellular enzymes into the growth medium, the activity of the enzyme isocitrate dehydrogenase in the growth medium of E. coli cells carrying plasmid pMS49 (containing lyt49), pMS50 (containing lyt50), or pQE60 (control) was monitored. Table 2 clearly illustrates that induced expression of lyt49 or lyt50 in E. coli cells indeed resulted in a substantial leakage of this intracellular enzyme compared to the control. The finding that the medium containing uninduced cells carrying pMS50 appeared to exhibit a high background isocitrate dehydrogenase activity compared to that of the control strain was possibly due to a low-level expression of lyt50 in the absence of the inducer, resulting in leakage of intracellular enzymes without apparently affecting cell viability.
FIG. 2.
Effect of the expression of lyt49 or lyt50 on growth and viability of E. coli cells. Induction of a specific culture was performed by the addition of IPTG during the logarithmic phase of growth (the specific time points are indicated by arrows). (A and C) Growth of E. coli cells harboring the plasmids pMS49 (+ [uninduced], □ [induced]) or pMS50 (▵ [uninduced], ○ [induced]) was monitored by measuring the OD600. (B and D) The viability (CFU ml−1) of E. coli cells harboring plasmid pMS49 or pMS50 (annotation as for panels A and C) was determined by plating appropriate dilutions on LB agar plates at 20-min time intervals.
TABLE 2.
Isocitrate dehydrogenase activity assay of the growth media of various E. coli strains grown under equivalent conditions in the presence or absence of IPTG
| Sample | Induction with IPTG | Avg activity ± SD (ς units/ml) |
|---|---|---|
| pMS49 | No | 10.43 ± 0.32 |
| pMS49 | Yes | 38.19 ± 8.79 |
| pMS50 | No | 22.87 ± 0.32 |
| pMS50 | Yes | 67.21 ± 4.00 |
| pQE60 | No | 9.32 ± 3.14 |
| pQE60 | Yes | 9.10 ± 0.94 |
Attempts to visualize the protein products of lyt49 and lyt50 on SDS-PAGE gels were unsuccessful. This may be due to the small quantities of holin protein required to cause sufficient membrane damage to arrest all cytoplasmic activities, including further production of the holin.
lyt51 encodes a protein with cell wall-degrading activity.
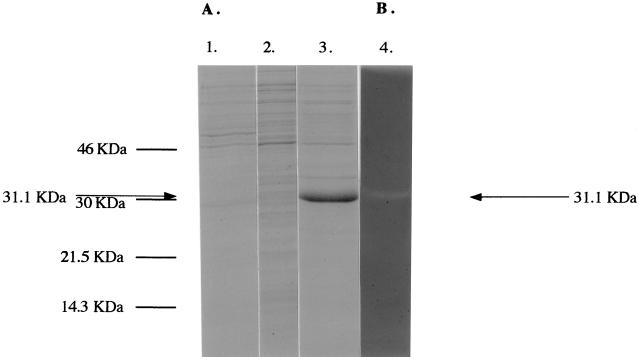
The deduced product of ORF51 (designated here as lyt51) of φO1205 exhibited significant similarity to several proteins in the sequence databases; in particular, high degrees of sequence similarity to the protein products of the corresponding ORFs of the S. thermophilus phages φSfi21 (11), φSfi19 (13), and φSfi11 (34) were evident (83, 81, and 83% overall identity, respectively). Similarities to characterized bacteriophage-encoded lytic enzymes such as the Pal lysin of the S. pneumoniae bacteriophage Dp-1 (50), previously characterized as an N-acetyl-muramoyl-l-alanine amidase (19), and the lysin of the lactococcal bacteriophage BK5-T (8, 26a) were also determined. The similarities of Lyt51, Pal, and the lysin of BK5-T appeared to be limited to the N-terminal portion of each of these proteins (Fig. 3A), the region which determines the catalytic activity. Additional sequence similarity, but confined to the C-terminal portion, was observed between Lyt51 and zoocin A (Fig. 3B), a bacteriocin-like inhibitory substance encoded by zooA of Streptococcus zooepidemicus (53). The latter protein has been reported to cause cellular lysis by cleavage of a hexaglycine substrate in the cell wall (54). These similarities suggest that lyt51 encodes a lytic activity, which is in agreement with the finding that genes encoding lytic enzymes are frequently located immediately downstream of the holin-encoding genes (23, 65). To determine whether the protein product of lyt51 does in fact confer lytic activity, lyt51 was cloned into the E. coli expression vector pQE60, resulting in the plasmid pMS51 (see Materials and Methods). When E. coli cells harboring this plasmid were induced, overexpression of a protein with an estimated molecular mass of 31,100 Da was observed upon SDS-PAGE. The calculated molecular mass of this protein, which is absent from an E. coli control strain, is in good agreement with the predicted protein size based on the amino acid sequence encoded by lyt51, 31,100 Da (Fig. 4). Unlike expression of lyt49 or lyt50, expression of lyt51 did not appear to have an adverse effect on the host cells, as indicated by normal growth and the absence of lysis of the induced recombinant E. coli strain.
FIG. 3.
(A) Multiple sequence alignment of the N-terminal domain (N-term) of Lyt51 and the corresponding regions of the Pal lysin of pneumococcal phage Dp-1 (50) and the predicted product of ORF259 from the lactococcal phage BK5-T (8). Identical amino acids are boxed; numbers refer to the amino acid positions of the published sequences. (B) Protein alignment of the C-terminal domains of Lyt51 and the bacteriocin-like inhibitory substance ZooA of S. zooepidemicus (53). Vertical dashes indicate identical residues, colons represent conserved amino acids, and numbers refer to the amino acid positions of the published sequences. Horizontal dashes represent introduced gaps to maximize alignment.
FIG. 4.
Expression of lyt51 in E. coli. Lane 1, SDS-PAGE of an extract (2.68 μg of protein) of E. coli cells containing plasmid pQE60 which were grown in the presence of an inducer (control); lane 2, SDS-PAGE of an extract (3 μg of protein) of cells harboring pMS51 which were grown in the absence of an inducer (lyt51 was not expressed); lane 3, SDS-PAGE of an extract (3.2 μg of protein) of pMS51-containing E. coli cells that were grown in the presence of inducer (lyt51 was expressed); lane 4, renaturing SDS-PAGE (with copolymerized crude cell walls of S. thermophilus CNRZ1205 as a substrate [see Materials and Methods]) of an E. coli cell extract (1.5 μg of protein) containing pMS51; the cleared area represents the position where the incorporated cell walls have been hydrolyzed through the action of a lytic activity. The sizes of the molecular mass markers are indicated on the left; the position of the lytic activity/Lyt51 protein is indicated by arrows.
To determine whether lyt51 of φO1205 indeed encodes a protein with lytic activity, an in situ detection assay (44), involving SDS-PAGE of a crude cell wall extract of S. thermophilus CNRZ1205-3, was performed. When cell extracts of E. coli cells containing plasmid pMS51 were used in this assay, a lytic activity corresponding to a protein with a molecular mass of 31 kDa was observed (Fig. 4, lane 4). This activity was absent from E. coli cells containing the control plasmid pQE60, which proves that the lytic activity is specified by lyt51. Preliminary biochemical characterization of Lyt51 has shown that the pH optimum for its lytic activity is between pH 7.0 and 7.5 and that addition of a bivalent cation (calcium or magnesium) or a reducing agent enhances the lytic activity of this protein. When crude cell wall preparations of E. coli, Micrococcus luteus, Enterococcus faecalis, L. paracasei, or B. coagulans were used for this in situ assay, no lytic activity was observed with extracts of E. coli cells containing plasmid pMS51, which suggested that Lyt51 has a very limited substrate specificity (results not shown). In contrast, when a cell wall preparation of S. mutans was used, a lytic activity corresponding to the Lyt51 protein was observed (results not shown).
Lyt51 causes in vivo lysis of growing S. thermophilus cells.
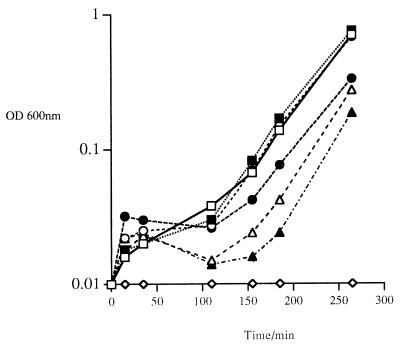
To determine the effect of addition of Lyt51 protein to a growing bacterial culture, various amounts of an extract of sonicated E. coli cells overexpressing Lyt51 were added to cultures of S. thermophilus CNRZ1205 at the time of inoculation. Following an initial short period of growth, inhibition was observed within 20 min. The level of inhibition was related to the amount of cell extract added. In addition, after 150 min, the growth of the cells resumed and a growth rate comparable to that of the control was achieved (Fig. 5). Similarly, an aliquot of sonicated extract containing 92.8 μg of protein was sufficient to inhibit the growth of growing cultures of 10 different S. thermophilus strains when added to 10 ml of liquid culture medium, showing that each of these strains is susceptible to the action of Lyt51 (results not shown).
FIG. 5.
Effect of addition of increasing amounts of crude E. coli cell extract containing Lyt51 on the logarithm of the OD600 of a 10-ml culture of growing cells of S. thermophilus CNRZ1205. □, control; ■, 18.6 μg of protein; ○, 92.8 μg of protein; •, 185.7 μg of protein; ▵, 278.55 μg of protein; ▴, 371.2 μg of protein; ◊, 464 μg of protein.
Presence of homologues of the lyt49, lyt50, and lyt51 genes in other bacteriophages of S. thermophilus.
To determine whether elements of the lysis module of φO1205 are also present in other S. thermophilus bacteriophages, DNA of 29 different phages (Table 3) was digested with EcoRV, separated on a 0.7% agarose gel, and blotted onto a nylon membrane. PCR-generated DNA fragments exactly encompassing lyt49, lyt50, or lyt51 of φO1205 were labelled, and each was used as a probe for Southern hybridization. The obtained results showed that 28 of the phages appear to contain a lyt49 homologue, that lyt50 hybridized with 24 of the phages, and that lyt51 showed homology to 23 of the phage genomes (Table 3). These results clearly demonstrate that elements of the identified lysis module of φO1205 are widespread in S. thermophilus phages. The presence of homologues of components of the lysis module of φO1205 is not restricted to other pac-containing phages alone but appears to be widespread among the cos-containing group of phages as well (Table 3) (see reference 31).
DISCUSSION
In this work, we describe the characterization of a lytic module isolated from a temperate bacteriophage of S. thermophilus. This system consists of two putative holin-encoding genes, lyt49 and lyt50, whose gene products were shown to have a bacteriocidal effect on E. coli cells, in addition to lyt51, a gene specifying a lytic enzyme which appears to be specifically active against the cell walls of S. thermophilus and S. mutans strains.
In addition to exhibiting similarity to uncharacterized ORFs of three different S. thermophilus bacteriophages (11, 13, 34), as well as to the holins of pneumococcal phage Cp-1 (36) and B. subtilis phage φ29 (58), the predicted secondary structure of the deduced protein product of lyt49 displays the conserved features typical of other class I holins (65). Similarly, the predicted secondary structure of the amino acid sequence of the lyt50 gene product (Fig. 1B) possesses the criteria set by Young and Bläsi (65) for the identification of holin-encoding genes and shows significant sequence similarity to the class II holins encoded by phages α80 (7) and φ11 (64) and the nearly identical holins encoded by the lactococcal phages Tuc2009 (2) and φLC3 (4).
In addition to the observed similarities, there are several experimental observations which strongly suggest that both lyt49 and lyt50 encode holins. First, the expression of lyt49 or lyt50 in E. coli is lethal but does not cause cell lysis (Fig. 2), a feature also observed with holins of other phages (4, 15, 26, 35, 38, 58). In addition, the expression of lyt49 and lyt50 in E. coli cells causes leakage of intracellular proteins, as determined by the detection of high levels of isocitrate dehydrogenase in the culture media, presumably due to the formation of nonspecific lesions in the cytoplasmic membrane. We conclude, therefore, that φO1205 possesses two adjacent, but separate, genes encoding different holins. Interestingly, an identical organization is also present in three other S. thermophilus bacteriophages, φSfi21 (11), φSfi19 (13), and φSfi11 (34). Although the exact role of each of the two holins in host lysis remains to be established, they may resemble certain gram-negative phages that are capable of producing two holin proteins, one of which acts as an inhibitor of the other (65). In several bacteriophages the holin and its inhibitor, the so-called antiholin, are encoded by one and the same gene, with the antiholin being a slightly shorter version of the holin. The lambdoid bacteriophage P2 (66), however, appears to employ two separate genes for the production of the holin and its antagonist (64). Perhaps the gene products of lyt49 and lyt50 of φO1205 constitute a similar pair of holin-antiholin proteins responsible for the accurate timing of the release of the lysin, an event which is of critical importance to the bacteriophage for the prevention of premature lysis of its propagating host. A hint as to the specific action of Lyt49 and Lyt50 may be derived from the observation that the expression of lyt50 appears to have a more lethal effect on the E. coli cells than the expression of lyt49 (Fig. 2). This observation is reminiscent of the situation in bacteriophage lambda, where the antiholin (S107) has a more lethal effect on the host cells than does the holin (S105) (57).
When the sequence of lyt51 of φO1205 was first described (56), no clear similarity to any other sequences available in the databases was observed. As indicated by more recent database searches and substantiated by experimental analysis, lyt51 encodes the lysin of φO1205. Lyt51 appears to have a modular design, exhibiting distinct amino- and carboxy-terminal regions (Fig. 3). The observed similarity between Lyt51 and the Pal lysin of Dp-1 (50) resides within the N-terminal region, which for the latter protein was shown to specify N-acetyl-muramoyl-l-alanine amidase activity (19). The C-terminal portion of Lyt51 resembles the equivalent region of zoocin A, a bacteriocin-like inhibitory substance produced by S. zooepidemicus (53). Similar comparative observations have been reported for ORF288 of the S. thermophilus phage φSfi19 (13). Since the C-terminal portion of phage lysins is usually involved in substrate recognition and binding (18, 22), it is tempting to speculate that these two proteins recognize the same substrate. The finding that Lyt51 is active against S. thermophilus and S. mutans cell walls but not against those of various other bacteria examined suggests that the cell walls of these two Streptococcus species contain a unique feature which may act as the substrate binding site of this lytic protein. An obvious candidate for this structural feature is the interpeptide bridge of the peptidoglycan layer, whose amino acid sequence in S. thermophilus, S. mutans, and S. zooepidemicus is lysine-alanine2–3 (25). This structural feature distinguishes the cell walls of these three bacteria from those of the other bacteria used in this study.
Southern blot analyses demonstrated extensive homology between the φO1205 genes involved in lysis and almost all of the S. thermophilus phage genomes examined. It appears, therefore, that essentially all S. thermophilus phages have a lysis module with a high degree of similarity to the lysis module characterized in this study. In fact, ORFs exhibiting a high degree of similarity to the components of the φO1205 lysis system have been identified by sequencing of other S. thermophilus phage genomes, such as those of φSfi21 (11), φSfi19 (13), φSfi11 (34), and φ7201 (55a). Although the putative lysis modules in φSfi21, φSfi19, φSfi11, and φ7201 also consist of three genes, which are similarly organized and homologous to lyt49, lyt50, and lyt51 of φO1205, it is apparent that some sequence divergence has occurred among the individual components of the lysis module. For example, lyt50 of φO1205 and its φ7201 homologue exhibit only 64.5% identity at the DNA level, which resulted in only a weak hybridization signal in a Southern blot (Table 3 and unpublished data). Lower but still significantly high percentages of sequence identity may therefore have resulted in the absence of a detectable hybridization signal (at least under the hybridization conditions used), and hybridization data for the lyt50 homologue of φ7201 have indeed corroborated this assumption (49a). It may therefore be that most, if not all, bacteriophages infecting S. thermophilus contain a lysis module consisting of three genes, each with various levels of homology, encoding two holin-like proteins and a protein with a lytic activity specifically active against streptococcal cell walls. This possible lack of diversity among these bacteriophages is in sharp contrast to phages infecting L. lactis (for reviews, see references 22 and 23) and may be a reflection of the low degree of genetic evolution among the former group of phages (13, 14, 34), which may be a result of their relatively recently acquired or limited ability to infect their streptococcal hosts.
ACKNOWLEDGMENTS
We are grateful to Clara Husson for advice, Sinéad Geary for DNA sequencing, Áine Healy for the synthesis of oligonucleotides, Linda Walsh for providing φ7201 DNA, the Nederlands Instituut voor Zuivelonderzoek and the Institut National de la Recherche Agronomique for generously providing bacterial strains, Liam Burgess for photographic work, and Alan J. Hillier for providing unpublished data.
D.V.S. is a recipient of a long-term European Molecular Biology Organization fellowship (ALTF-341-1995).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benbadis L, Faelen M, Slos P, Fazel A, Mercenier A. Characterisation and comparison of virulent bacteriophages of Streptococcus thermophilus isolated from yogurt. Biochimie. 1990;72:855–862. doi: 10.1016/0300-9084(90)90002-x. [DOI] [PubMed] [Google Scholar]
- 4.Birkeland N-K. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of the lactococcal bacteriophage φLC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boizet B, Mata M, Mignot O, Ritzenthaler P, Sozzi T. Taxonomic characterisation of Leuconostoc mesenteroides and Leuconostoc oenos bacteriophages. FEMS Microbiol Lett. 1992;90:211–216. [Google Scholar]
- 7.Bon J, Mani N, Jayaswal R K. Molecular analysis of lytic genes of bacteriophage 80α of S. aureus. Can J Microbiol. 1997;43:612–616. doi: 10.1139/m97-087. [DOI] [PubMed] [Google Scholar]
- 8.Boyce J D, Davidson B E, Hillier A J. Sequence analysis of the Lactococcus lactis temperate bacteriophage BK5-T and demonstration that the phage DNA has cohesive ends. Appl Environ Microbiol. 1995;61:4089–4098. doi: 10.1128/aem.61.11.4089-4098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–256. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Brüssow H, Frémont M, Bruttin A, Sidoti J, Constable A, Fryder V. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl Environ Microbiol. 1994;60:4537–4543. doi: 10.1128/aem.60.12.4537-4543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruttin A, Desiere F, Lucchini S, Foley S, Brüssow H. Characterization of the lysogeny DNA module from the temperate Streptococcus thermophilus bacteriophage φSfi21. Virology. 1997;233:136–148. doi: 10.1006/viro.1997.8603. [DOI] [PubMed] [Google Scholar]
- 12.Dellaglio F. Starters for fermented milks. 3. Thermophilic starters. Bull Int Dairy Fed. 1988;227:27–33. [Google Scholar]
- 13.Desiere F, Lucchini S, Brüssow H. Evolution of Streptococcus thermophilus bacteriophage genomes by modular exchanges followed by point mutations and small deletions and insertions. Virology. 1998;241:345–356. doi: 10.1006/viro.1997.8959. [DOI] [PubMed] [Google Scholar]
- 14.Desiere F, Lucchini S, Bruttin A, Zwahlen M-C, Brüssow H. A highly conserved DNA replication module from Streptococcus thermophilus phages is similar in sequence and topology to a module from Lactococcus lactis phage. Virology. 1997;234:372–382. doi: 10.1006/viro.1997.8643. [DOI] [PubMed] [Google Scholar]
- 15.Díaz E, Munthali M, Lünsdorf H, Höltje J V, Timmis K N. The two step lysis system of pneumococcal bacteriophage Ej-1 is functional in gram-negative bacteria: triggering of the major pneumococcal autolysin in E. coli. Mol Microbiol. 1996;19:667–681. doi: 10.1046/j.1365-2958.1996.399929.x. [DOI] [PubMed] [Google Scholar]
- 16.Everson T C. Control of phage in the dairy plant. Bull Int Dairy Fed. 1991;263:24–28. [Google Scholar]
- 17.Fayard B, Haefliger M, Accolas J P. Interactions of temperate bacteriophages of Streptococcus salivarius ssp. thermophilus with lysogenic indicators affect phage DNA restrictions patterns and host range. J Dairy Res. 1993;60:385–399. [Google Scholar]
- 18.García P, García J L, García E, Sánchez-Puelles J M, López R. Modular organisation of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene. 1990;86:81–88. doi: 10.1016/0378-1119(90)90116-9. [DOI] [PubMed] [Google Scholar]
- 19.García P, Méndez E, García E, Ronda C, López R. Biochemical characterization of a murein hydrolase induced by bacteriophage Dp-1 in Streptococcus pneumoniae: comparative study between bacteriophage-associated lysin and the host amidase. J Bacteriol. 1984;159:793–796. doi: 10.1128/jb.159.2.793-796.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implication of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 21.Garvey K J, Saedi M S, Ito J. Nucleotide sequence of Bacillus phage Phi29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 1986;14:10001–10008. doi: 10.1093/nar/14.24.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvey P, van Sinderen D, Twomey D P, Hill C, Fitzgerald G F. Molecular genetics of bacteriophage and natural phage defence systems in the genus Lactococcus. Int Dairy J. 1995;5:905–947. [Google Scholar]
- 23.Gasson M J. Lytic systems in lactic acid bacteria and their bacteriophages. Antonie Leeuwenhoek. 1996;10:147–159. doi: 10.1007/BF00395931. [DOI] [PubMed] [Google Scholar]
- 24.Geiss A. Cloning and DNA sequence analysis of a lysin gene of the lactococcal bacteriophage POO1. Kiel, Germany: Federal Dairy Research Centre; 1992. [Google Scholar]
- 25.Hardie J M, Whiley R A. The genus Streptococcus. In: Wood B J B, Holzapfel W H, editors. The lactic acid bacteria. 2. The genera of lactic acid bacteria. Glasgow, United Kingdom: Chapman and Hall; 1995. pp. 59–124. [Google Scholar]
- 26.Henrich B, Binishofer B, Bläsi U. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage φadh. J Bacteriol. 1995;177:723–732. doi: 10.1128/jb.177.3.723-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Hillier, A. J. Personal communication.
- 27.Hofman K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem. 1993;347:166–170. [Google Scholar]
- 28.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I B, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 29.Klaenhammer T R, Fitzgerald G F. Bacteriophages and bacteriophage resistance. In: Gasson M J, de Vos W, editors. Genetics and biotechnology of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic and Professional; 1994. pp. 106–168. [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Le Marrec C, van Sinderen D, Walsh L, Stanley E, Vlegels E, Moineau S, Heinze P, Fitzgerald G, Fayard B. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl Environ Microbiol. 1997;63:3246–3253. doi: 10.1128/aem.63.8.3246-3253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loessner M J, Gaeng S, Wendlinger G, Maier S J, Scherer S. The two component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- 33.Lubbers M W, Waterfield N R, Beresford T P J, Le Page R W F, Jarvis A W. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl Environ Microbiol. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucchini S, Desiere F, Brüssow H. The structural gene module in Streptococcus thermophilus bacteriophage φSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology. 1998;246:63–73. doi: 10.1006/viro.1998.9190. [DOI] [PubMed] [Google Scholar]
- 35.Martín A C, López R, García P. Functional analysis of the two-gene lysis system of the pneumococcal phage Cp-1 in homologous and heterologous host cells. J Bacteriol. 1998;180:210–217. doi: 10.1128/jb.180.2.210-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martín A C, López R, García P. Analysis of the complete nucleotide sequence and functional organization of the genome of Streptococcus pneumoniae bacteriophage Cp-1. J Virol. 1996;70:3678–3687. doi: 10.1128/jvi.70.6.3678-3687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mata M, Ritzenthaler P. Present state of lactic acid bacteria phage taxonomy. Biochimie. 1988;70:395–399. doi: 10.1016/0300-9084(88)90213-1. [DOI] [PubMed] [Google Scholar]
- 38.Nauta A. Molecular characterisation and exploitation of the temperate Lactococcus lactis bacteriophage rlt. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 39.Neve H, Krusch U, Teuber M. Classification of virulent bacteriophages of Streptococcus salivarius subsp. thermophilus isolated from yogurt and Swiss-type cheese. Appl Microbiol Biotechnol. 1989;30:624–629. [Google Scholar]
- 40.Neve H, Zenz K I, Desiere F, Koch A, Heller K J, Brüssow H. Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and φSfi21: implications for the modular theory of phage evolution. Virology. 1998;241:61–72. doi: 10.1006/viro.1997.8960. [DOI] [PubMed] [Google Scholar]
- 41.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peitersen N. Practical phage control. Bull Int Dairy Fed. 1991;263:1–43. [Google Scholar]
- 43.Platteeuw C, de Vos W M. Location, characterisation and expression of the lytic enzyme encoding gene, lytA, of Lactococcus lactis bacteriophage φUS3. Gene. 1992;118:115–120. doi: 10.1016/0378-1119(92)90257-p. [DOI] [PubMed] [Google Scholar]
- 44.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214:214–218. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 45.Sable S, Lortal S. The lysins of bacteriophages infecting lactic acid bacteria. Appl Microbiol Biotechnol. 1995;43:1–6. doi: 10.1007/BF00170613. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Schouler C, Erlich S D, Chopin M C. Sequence and organisation of the lactococcal prolate-headed bIL67 phage genome. Microbiology. 1994;140:3061–3069. doi: 10.1099/13500872-140-11-3061. [DOI] [PubMed] [Google Scholar]
- 48.Sechaud L, Cluzel P J, Rousseau M, Baumgartner A, Accolas J P. Bacteriophages of Lactobacillus. Biochimie. 1988;70:401–410. doi: 10.1016/0300-9084(88)90214-3. [DOI] [PubMed] [Google Scholar]
- 49.Shearman C A, Jury K L, Gasson M J. Controlled expression and structural organization of a Lactococcus lactis bacteriophage lysin encoded by two overlapping genes. Appl Environ Microbiol. 1994;60:3063–3073. doi: 10.1128/aem.60.9.3063-3073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Sheehan, M., G. Fitzgerald, and D. van Sinderen. Unpublished data.
- 50.Sheehan M M, García J L, López R, García P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol Microbiol. 1997;25:717–725. doi: 10.1046/j.1365-2958.1997.5101880.x. [DOI] [PubMed] [Google Scholar]
- 51.Sheehan M M, García J L, López R, García P. Analysis of the catalytic domain of the lysin of the lactococcal bacteriophage Tuc2009 by chimeric gene assembling. FEMS Microbiol Lett. 1996;140:23–28. doi: 10.1111/j.1574-6968.1996.tb08309.x. [DOI] [PubMed] [Google Scholar]
- 52.Shine J, Dalgarno L. The 3′-terminal sequence of E. coli 16S rRNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmonds R S, Simpson W J, Tagg J R. Cloning and sequencing of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene. 1997;189:255–261. doi: 10.1016/s0378-1119(96)00859-1. [DOI] [PubMed] [Google Scholar]
- 54.Simmonds R S, Pearson L, Kennedy R C, Tagg J R. Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl Environ Microbiol. 1996;62:4536–4541. doi: 10.1128/aem.62.12.4536-4541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 55a.Stanley, E., D. van Sinderen, and G. Fitzgerald. Unpublished data.
- 56.Stanley E, Fitzgerald G F, LeMarrec C, Fayard B, van Sinderen D. Sequence analysis and characterisation of φO1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology. 1997;143:3417–3429. doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 57.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 58.Steiner M, Lubitz W, Bläsi U. The missing link in the phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terzaghi B E, Sandine W E. Improved media for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasala A, Välkkilä M, Caldentey J, Alatossava T. Genetic and biochemical characterization of the Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H lysin. Appl Environ Microbiol. 1995;61:4004–4011. doi: 10.1128/aem.61.11.4004-4011.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Wilkinson B J, Jayaswal R K. Sequence analysis of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. Gene. 1991;102:105–109. doi: 10.1016/0378-1119(91)90547-o. [DOI] [PubMed] [Google Scholar]
- 62.Ward L J H, Beresford W P J, Lubbers M W, Jarvis B D W, Jarvis A W. Sequence analysis of the lysin gene region of the prolate lactococcal bacteriophage c2. Can J Microbiol. 1993;39:767–774. doi: 10.1139/m93-113. [DOI] [PubMed] [Google Scholar]
- 63.Weerakoon L K, Jayaswal R K. Sequence analysis of the region upstream of a peptidoglycan hydrolase-encoding gene from bacteriophage φ11 of Staphylococcus aureus. FEMS Microbiol Lett. 1995;133:9–15. doi: 10.1111/j.1574-6968.1995.tb07853.x. [DOI] [PubMed] [Google Scholar]
- 64.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 66.Ziermann R, Bartlett B, Calendar R, Christie G E. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J Bacteriol. 1994;176:4974–4984. doi: 10.1128/jb.176.16.4974-4984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]