Abstract
Biosilicification—the formation of biological structures composed of silica—has a wide distribution among eukaryotes; it plays a major role in global biogeochemical cycles, and has driven the decline of dissolved silicon in the oceans through geological time. While it has long been thought that eukaryotes are the only organisms appreciably affecting the biogeochemical cycling of Si, the recent discoveries of silica transporter genes and marked silicon accumulation in bacteria suggest that prokaryotes may play an underappreciated role in the Si cycle, particularly in ancient times. Here, we report a previously unidentified magnetotactic bacterium that forms intracellular, amorphous silica globules. This bacterium, phylogenetically affiliated with the phylum Nitrospirota, belongs to a deep-branching group of magnetotactic bacteria that also forms intracellular magnetite magnetosomes and sulfur inclusions. This contribution reveals intracellularly controlled silicification within prokaryotes and suggests a previously unrecognized influence on the biogeochemical Si cycle that was operational during early Earth history.
Detection of intracellular SiO2 in MTB suggests a role of prokaryotes in the biogeochemical Si cycle in early Earth history.
INTRODUCTION
Silicon is the second most abundant element in Earth’s crust after oxygen. It is an essential element for life, both on land and in the ocean (1, 2). Several groups of eukaryotes such as diatoms, radiolarians, choanoflagellates, siliceous sponges, and higher plants take up large amounts of silicon from the environment as soluble, monomeric silicic acid (orthosilicic acid, Si[OH]4) to build their skeletal structures composed of silica (SiO2) (3, 4). This formation of biological structures composed of silica is called biosilicification. It has been proposed that the radiations of biosilicifying eukaryotes during the Phanerozoic drove secular decreases in dissolved silicon (DSi) in the global ocean, ultimately resulting in the low DSi concentrations seen today (5, 6). Recent studies, however, find that biosilicification also occurs in the cells of certain bacteria, such as Bacillus cereus and its close relatives (7), and some cyanobacteria affiliated to the Synechococcus genus (8). Subsequent field studies indicate that biosilicification of these bacteria could make a nonnegligible contribution to the global silicon cycle (9–11). Moreover, the detection of genes involved in bacterial silicon-related metabolisms suggests that prokaryotic biosilicification might have been present in the oceans as early as the Archean (3, 12, 13).
Despite these suggestive findings, an understanding of the distribution and mechanism of biosilicification within prokaryotes remains far less advanced than those in eukaryotes. For instance, the cyanobacterium Synechococcus may deposit silicon in the form of Mg-containing hydrated silicates, which is therefore distinct from amorphous silica (biogenic opal-A) biosilicified by diatoms (14, 15). Within B. cereus, DSi uptake occurs at the early stationary phase of bacterial growth and is closely coupled with spore formation. Silica is deposited on the spore coat as a layer of nanometer-sized particles to enhance resistance to acids (7). This indicates that at least some species of bacteria can uptake silicic acid from undersaturated solutions, as it is the case for biosilicifying eukaryotes (9). Although the possibility of biosilicification in prokaryotes has continued to attract research interest over the past 160 years (16), strong evidence for such a process has remained elusive (3, 13).
Magnetotactic bacteria (MTB) may be among the earliest microorganisms on Earth that perform intracellular biomineralization of iron and sense the geomagnetic field (17). They are phylogenetically diverse and ubiquitous in aquatic environments including freshwater and marine ecosystems, where they are dominantly distributed at or just below the oxic-anoxic transition zone (OATZ) (18). Despite both their phylogenetic and morphological diversity, MTB share the ability to produce intracellular nanocrystals of magnetite (Fe3O4) or greigite (Fe3S4) under strict genetic control in a subcellular compartment called the magnetosome (19). In the past decade, it has been increasingly shown that MTB also have the capability to form diverse types of intracellular inclusions such as sulfur (S0) globules, Ca/Mg-rich polyphosphate granules, and calcium carbonate or phosphate particles (20–24). This suggests that this group of ecologically important prokaryotes may drive biogeochemical elemental cycling of C, N, P, S, and Fe within aquatic OATZ by coupling geomagnetic-guided up-down shuttling with redox-related metabolic dynamics (20–22).
In this study, we found one particular group of magnetotactic cocci in sediments collected from Lake Weiyanghu (WYH) in China (Fig. 1, A to C, and movie S1). A phylogenetic analysis based on 16S ribosomal RNA (rRNA) gene sequences indicates that this bacterium, tentatively named strain WYHC-5, is affiliated with the phylum Nitrospirota (previously known as Nitrospirae) and is therefore a member of the deep-branching MTB group. Advanced electron microscopy analyses show that strain WYHC-5 can accumulate a large amount of silicon within its cells in the form of amorphous silica globules besides forming magnetite-type magnetosomes and sulfur inclusions. Metagenomic analysis of the WYHC-5 genome reveals the presence of a key gene, possibly involving in silica deposition. This study therefore provides previously unrecognized structural and molecular evidence for intracellularly controlled biosilicification within prokaryotes. It suggests a possible influence on the biogeochemical silicon cycle during early Earth history.
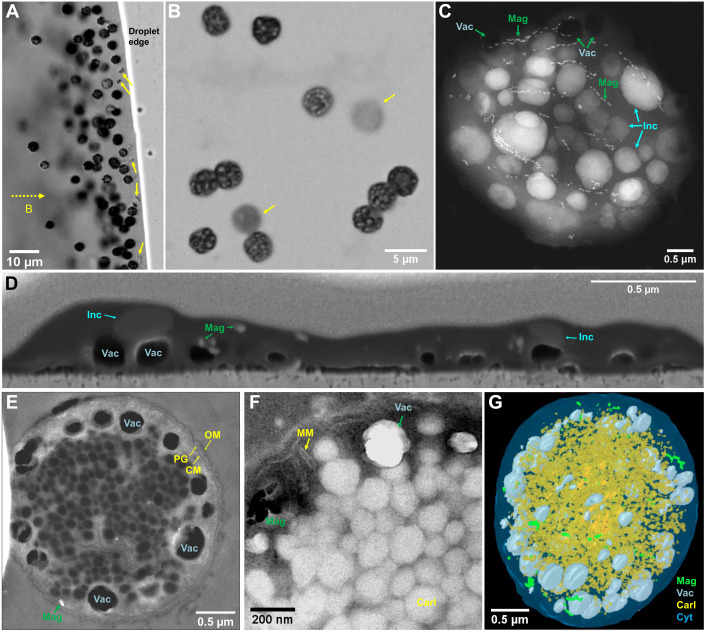
Fig. 1. Morphology and ultrastructure of the magnetotactic strain WYHC-5.
(A) Optical micrograph showing living coccoidal MTB (WYHC-5). They swim out from the sediment located to the left of the image (sediment not shown) along the external applied magnetic field lines (dashed line with arrow) and gather on the water droplet edge. A few other types of small MTB are indicated by yellow arrows. (B) Optical micrograph of WYHC-5 cells magnetically collected from sediments in laboratory microcosms. The bacteria were stained by crystal violet before optical microscopy observations. Two out-of-focus cells are indicated by yellow arrows. (C) STEM image of one typical WYHC-5 cell acquired in the STEM-HAADF showing multiple chains of magnetosome magnetite (Mag), numerous electron-dense inclusions (Inc), and electron-transparent vacuoles (Vac). (D) SEM image of air-dried WYHC-5 cell selected from a series of FIB sections showing that the vacuoles, magnetosome magnetite, and electron-dense inclusions are relatively independent from each other within the cell. The empty vacuoles remain approximately spherical in shape, suggesting that they have a rigid enclosed structure. (E) STEM-HAADF image of an ultrathin section of a freeze-substituted WYHC-5 cell showing (i) the typical Gram-negative cell wall (CM, cytoplasmic membrane; OM, outer membrane; PG, peptidoglycan) and (ii) vacuoles adjacent but separated from the CM. (F) Bright-field TEM image of an ultrathin section of a freeze-substituted WYHC-5 cell showing empty magnetosome membrane (MM; the inside magnetite crystal might have been lost during sample preparation), and the aggregate of polyhedral carboxysome-like bodies (Carl). (G) 3D FIB-SEM reconstruction and visualization of one freeze-substituted WYHC-5 cells [green, magnetosome magnetite; gray, vacuole; yellow, carboxysome-like body; light blue, cytoplasm (Cyt)]. The FIB-SEM sections and 3D reconstructions for air-dried and freeze-substituted WYHC-5 cells are provided as movies S2 and S3, respectively.
RESULTS
Morphological, structural, and physiological features of WYHC-5
Transmission electron microscopy (TEM) observations show that WYHC-5 cells have an average diameter of 3.58 μm (SD = 0.42 μm, n = 28) and form bullet-shaped magnetite-type magnetosomes with a mean length and width of 70.4 (SD = 27.0 nm) and 27.5 nm (SD = 6.3 nm), respectively (n = 503). These magnetosomes are organized into loosely packed chains (fig. S1 and table S1).
In addition to the magnetosomes, two membrane-encapsulated subcellular structures were identified using a combination of two-dimensional (2D) and 3D imaging by TEM and focused ion beam scanning electron microscopy (FIB-SEM) tomography (Fig. 1, fig. S1, and movies S2 and S3). One type consists of electron-transparent spherical vacuoles with an average diameter of 216.6 nm (SD = 64.7 nm, n = 30). They appear as rigid structures adjacent to the cytoplasmic membrane (Fig. 1, C to E, and table S1). The other type of structure consists of polyhedral inclusions clustering in piles within the cytoplasm (Fig. 1, E and F). These polyhedral inclusions have an average diameter of 107.6 nm (SD = 14.8 nm, n = 103) (table S1) and are morphologically identical to carboxysomes (i.e., carboxysome-like body), protein-based microcompartments found in all cyanobacteria and in some chemolithoautotrophic bacteria (25).
Carboxysomes are composed of thousands of subunits including hexameric and pentameric proteins that form a shell encapsulating ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and a carbonic anhydrase enzyme. They function to fix CO2 as part of the Calvin-Benson-Bassham (CCB) cycle (25, 26). Basic Local Alignment Search Tool (BLAST) searches showed that the WYHC-5 genome contains the genes encoding RuBisCO and carbonic anhydrase (data files S1 and S2). In contrast, the genes encoding the carboxysome shell proteins were not detected from the WYHC-5 genome, possibly due to the incompleteness of sequencing depth (e.g., genome completeness of ~95.3%). Alternatively, strain WYHC-5 might have distinctive molecular mechanism that regulates the formation of carboxysome-like bodies. In addition, the WYHC-5 genome contains key enzymes involved in the fixation of CO2 via the reductive acetyl coenzyme A [Wood-Ljungdahl (WL)] pathway (data file S2). The presence of key genes of both the CCB cycle and the WL pathway suggests that strain WYHC-5 has the potential to fix CO2 via a chemoautotrophic growth.
Identification of intracellular silica globules within WYHC-5
WYHC-5 cells also contain numerous intracellular electron-dense inclusions, which are morphologically different from the three above-mentioned subcellular structures (Fig. 1, C and D). These inclusions are lost upon chemical treatments during high-pressure freezing and freeze substitution (Fig. 1, E and F). Elemental chemistry analyses achieved by energy-dispersive x-ray spectrometry (EDXS), coupled with imaging by high-angle annular dark-field scanning TEM (HAADF-STEM), revealed that the intracellular electron-dense inclusions could be divided in two types (Fig. 2 and fig. S2).
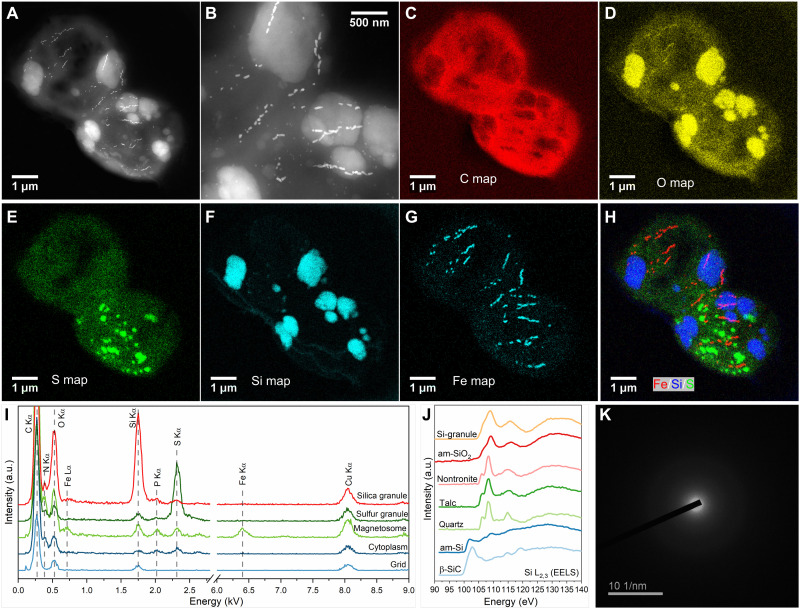
Fig. 2. Characterization of the magnetotactic strain WYHC-5 producing intracellular amorphous silica globules.
(A) STEM-HAADF image of two WYHC-5 cells. (B) High-magnification STEM-HAADF image of intracellular particles and granules from the cells shown in (A). (C to H) STEM-EDXS elemental maps of WYHC-5 cells shown in (A). (C) Carbon (C Kα). (D) Oxygen (O Kα). (E) Sulfur (S Kα). (F) Silicon (Si Kα). (G) Iron (Fe Kα). (H) Red-green-blue map of Fe (red), Si (blue), and S (green). (I) STEM-EDX spectra obtained on different regions of interest: silica granule, sulfur granule, magnetosome, cytoplasm, and TEM grid. a.u., arbitrary unit. (J) TEM-based electron energy loss spectrum (EELS) measured at Si L2,3 edges on an intracellular silicon-rich granule (Si-granule). This spectrum is compared with spectra of reference β-silicon carbide (β-SiC), pure amorphous Si (am-Si), quartz, talc, nontronite, and amorphous silica (am-SiO2). (K) SAED pattern of an intracellular silicon-rich granule.
Type I inclusions had a mean size of 0.53 μm (SD = 0.34 μm, n = 113) and contained Si and O as major elements, while type II inclusions had a mean size of 0.38 μm (SD = 0.23 μm, n = 56) and contained S as a major element (table S1). TEM-based electron energy-loss spectroscopy (EELS) analyses at the Si L2,3 edge showed that type I inclusions yield a spectrum identical to that of amorphous silica (27) but markedly different from other Si-rich reference compounds (Fig. 2J) (27). Selected-area electron diffraction (SAED) analysis further demonstrated a broad ring characteristic of an amorphous phase (Fig. 2K). This indicates that type I inclusions are composed of amorphous silica. Synchrotron-based scanning transmission x-ray microscopy (STXM) coupled with near-edge x-ray absorption fine structure (NEXAFS) spectroscopy (21, 28) indicate that type II inclusions are elemental sulfur (S0) globules possibly with linear polymeric structures (fig. S3). A semiquantitative calculation of the STEM-EDXS data showed that the averaged concentrations (weight %) of iron, silicon, and sulfur within the WYHC-5 cells are 0.4% (SD = 0.3%), 5.1% (SD = 5.0%), and 3.2% (SD = 6.2%), respectively (n = 15) (fig. S2).
Phylogenetic distribution of WYHC-5 and other Si-related prokaryotes
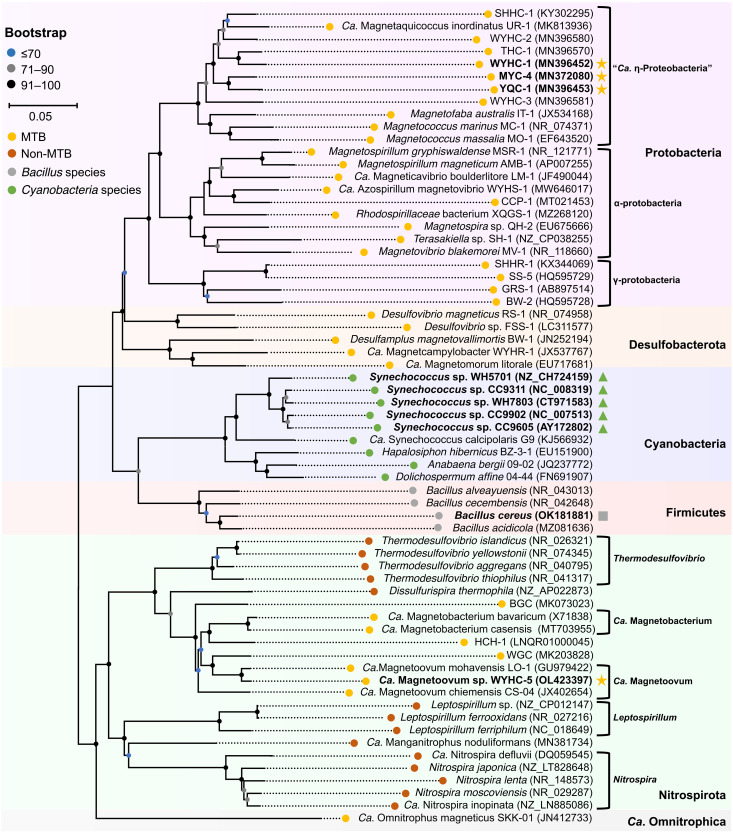
Our molecular analyses based on 30 full-length 16S rRNA gene sequences from the magnetic collection of WYHC-5 cells indicate that two sequences share high identity at >97% with the Leptothrix sp. oral clone AW043, a sheathed filamentous Fe-oxidizing bacterium that is common in freshwater environments (29). They could be derived from a contamination during magnetic separation of MTB (30). In contrast, most of the sequences (i.e., 28 of the 30) were almost identical between each other (>99%; table S2). They share a ~98.3% sequence identity with the Candidatus Magnetoovum mohavensis LO-1 strain, which has been detected in a number of freshwater and slightly brackish aquatic environments in the Mohave Desert in southern Nevada (31). We imaged cells with an oligonucleotide probe (WYHC5-215) specific of the 28 sequences by coupling fluorescence in situ hybridization and SEM (FISH-SEM) (32). Only WYHC-5–like spherical bacteria were hybridized by both the universal bacterial probe EUB338 and the oligonucleotide probe WYHC5-215 (fig. S3, A and B). Correlative SEM observations confirmed that these spherical bacteria were WYHC-5 based on their characteristic bullet-shaped magnetosome chains and granular-shaped cells (fig. S3, C to E). Overall, strain WYHC-5 may represent a novel species of the Ca. Magnetoovum genus (i.e., Ca. Magnetoovum sp. WYHC-5) based on a 98.7% criterion for species (33). Both species in the Ca. Magnetoovum genus, together with Candidatus Magnetobacterium bavaricum and Ca. Magnetobacterium casensis in the Ca. Magnetobacterium genus, belong to the phylum Nitrospirota (31), a deeply-branching MTB group (17).
We also assessed the presence of intracellular silicon-rich inclusions within some typical MTB strains, which have been well characterized by STEM-EDXS mapping using HAADF mode. Of a total of 17 MTB strains scattered throughout the phylogenetic tree of MTB (table S3), three magnetotactic cocci affiliated with “Candidatus Etaproteobacteria” in the phylum Proteobacteria (e.g., strains WYHC-1, YQC-1, and MYC-4) were found to precipitate intracellular submicrometer-sized globules composed of Si and O (figs. S5 to S7). This demonstrates that several MTB other than the WYHC-5 strain are also able to form intracellular Si-O–rich globules. Although intracellular silicification is not a systematic feature of MTB, it can be found in several species scattered in separate branches (Fig. 3).
Fig. 3. Phylogenetic position of strain WYHC-5 and other silicifying bacteria.
The maximum likelihood phylogenetic tree was constructed on the basis of 16S rRNA gene sequences of 39 MTB throughout the phylogenetic tree of MTB (yellow nodes), 14 non-MTB species affiliated with the phylum NItrospirota (blue nodes), four Bacillus species affiliated with the Firmicutes phylum (gray nodes), and six cyanobacteria species (green nodes). Silicifying bacteria are labeled in bold. Intracellular biomineralization of silica globules within the MTB system, formation of silica granules on the spore coat of B. cereus, and silicification within cyanobacteria are indicated with yellow stars, a gray square, and green triangles, respectively. Bootstrap values at nodes are given as percentages of 1000 replicates. GenBank accession numbers are given in parentheses.
We further measured the concentration of DSi in the solutions where the above-mentioned MTB were grown. The aqueous chemistry analyses showed that DSi ranges from ~10 to 45 parts per million (ppm) (table S4), which is notably lower than the saturation value for amorphous silica in distilled water (~115 ppm at 25°C) (34). It can therefore be inferred that an active uptake of DSi occurs across the cell membrane in intracellularly silicifying MTB, resulting in an increased DSi concentration within the intracellular silicification compartments so that silica deposition can take place.
Identification of Si-related gene within strain WYHC-5
To further understand the molecular mechanism of silica precipitation within strain WYHC-5, we first reviewed major genes or proteins involved in silicon transportation and silica deposition within several silicifying eukaryotes (e.g., diatoms, siliceous sponges, and higher plants) (data files S3 and S4) and then used them as query sequences to identify potential Si-related genes in the WYHC-5 genome. The bioinformatics analysis showed that the WYHC-5 genome contains a gene encoding a protein highly similar to silicatein (Fig. 4A), a well-characterized silica deposition protein that serves as both an enzymatic catalyst and an organic template for biosilicification in sponges (35, 36). This gene of WYHC-5, tentatively named silicatein-1, locates on a 2414–base pair (bp) contiguous containing five (predicted) genes. Silicatein-1 is 248–amino acid length, sharing an identity of 38.3% and a coverage of 86.3% with the silicatein of siliceous sponge species Mycale phyllophila.
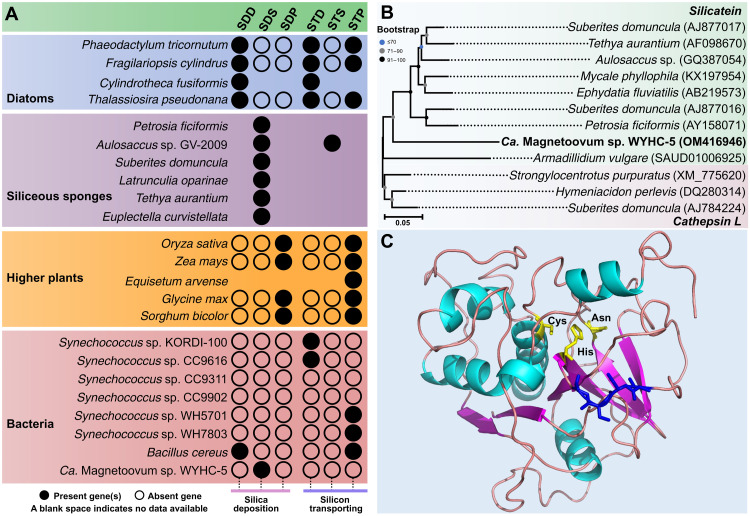
Fig. 4. Prediction of Si-related gene and protein in strain WYHC-5.
(A) Distribution of silica deposition (SD) and silicon transporting (ST) genes in typical silicifying eukaryotes and prokaryotes (including strain WYHC-5). The SDD, SDS, and SDP represent the silica deposition proteins identified in diatoms [including silaffin, silacidin, silicalemma-associated proteins (SAP), silicanin, and cingulin], siliceous sponges (including silicatein, glassin, and silintaphin), and higher plants (siliplant1). STD, STS, and STP represent the silicon-transporting proteins identified in diatoms [including SIT (silicon transporter) and SIT-like], siliceous sponges [Na+/HCO3− [Si(OH)4] co-transporter (NBCSA)] and higher plants (including lsi1, lsi2, lsi2-like, lsi3, lsi6, OsCASP1, NIP2, and NIP3). More detailed data are showed in data files S3 to S5. (B) Cluster analysis of the cDNA sequences of silicatein (light green background) in siliceous sponges, A. vulgare, strain WYHC-5, and cathepsin L (light red background) in siliceous sponges. Bootstrap values at nodes are given as percentages of 1000 replicates. GenBank accession numbers are given in parentheses. (C) Modeling of the 3D structure of the WYHC-5 silicatein-1. The GMQE and QMEAN scores are 0.71 and 0.69 ± 0.05, respectively, indicating that the model structure is accurate and shows good agreement with the template. The catalytic center of silicatein-1, comprising the amino acids Cys, His, and Asn (yellow), and the localization of the serine cluster (blue) are marked.
Since silicatein is homologous with the cathepsin L protein, a lysosomal endopeptidase expressed in most eukaryotic cells (36–38), we performed a cluster analysis of the complementary DNA (cDNA) sequences of silicatein in siliceous sponge species, Armadillidium vulgare (39), and strain WYHC-5 (this study), as well as cathepsin L in siliceous sponge species via the maximum likelihood method (40). The result showed that the silicatein-1 gene of WYHC-5 clusters with the silicatein branches with a high bootstrap value of the node (>90) (Fig. 4B). This supports the hypothesis that the WYHC-5 silicatein-1 may represent a gene encoding a silica deposition enzyme rather than cathepsin L protein. To further affirm the function of the WYHC-5 silicatein-1, we predicted the amino acids forming the catalytic center by multiple sequence alignment (fig. S8). Protein structure modeling revealed that the WYHC-5 silicatein-1 has a catalytic triad of Cys-His-Asn in the center and a serine cluster on the molecular surface (Fig. 4C). Such a catalytic domain structure is highly identical with that of the silicatein in siliceous sponges (38). We therefore propose that silicatein-1 may function as a template for silica deposition within strain WYHC-5.
We did not detect genes encoding silicon transporting proteins in the WYHC-5 genome (Fig. 4A). One possibility is that it was missing from the genome due to its incompleteness (e.g., genome completeness of ~95.3%). Alternatively, strain WYHC-5 might have a silicon-transporting mechanism different from that found in silicifying eukaryotes.
DISCUSSION
The silicification mode varies among different prokaryotic groups, although the exact process and molecular mechanism has yet to be defined. They can take up silicic acid from an undersaturated external solution into the cell, as is the case for biosilicifying eukaryotes. This is different from bacterial (and archeal) extracellular silicification occurring in supersaturated silicic acid in geothermal environments where the bacteria/archea facilitate the aggregation of colloidal polysilicic acids by providing organic surfaces (41, 42). Experimental results show that silicification is tightly coupled with spore formation in the genus Bacillus (43). The incorporated silicic acid is deposited in the form of a silica layer encapsulating the spores of Bacillus species and might contribute to spore survival under acidic conditions (7). It is still unclear how Synechococcus manages silicon deposition intracellularly or extracellularly (8), although silicon accumulation appears to be a common feature of many Synechococcus species (8, 9). By contrast, MTB strain WYHC-5 accumulates silicon intracellularly in the form of amorphous silica globules because of an estimated DSi concentration about ~3500-fold higher than that in surrounding water used for experiments (table S4), and it therefore represents both a novel and a highly effective mode of prokaryotic biosilicification.
Intracellular silica precipitation by MTB strain WYHC-5 likely influences or regulates its ecology and biology. First, these silica globules must alter cell buoyancy by increasing the average cell density. In nature, MTB occur predominantly at or just below the OATZ in sediments or chemically stratified water columns (18, 44). Therefore, in contrast with gas vesicles functioning as flotation devices in planktonic cyanobacteria and other prokaryotes (45), intracellular silica globules might serve as a ballast for bacteria to control their benthic position in sediments or the water column (23, 46). Silica has a density of ~2.3 g/cm3 at 25°C and a standard atmospheric pressure, providing greater weight for a given volume. A test of this hypothesis in the future may be achieved by analyzing any potential relationship between the vertical distribution of the cells and their amount of intracellular silica globules. Second, both experimental studies and genomic predictions indicate that many extant MTB have the potential to grow either heterotrophically or autotrophically (21, 47, 48). Whether strain WYHC-5 could grow autotrophically remains an open question, but the presence of many carboxysome-like bodies may provide a potential compartment for CO2 fixation or other metabolic purposes (25). Fixation of CO2 derived from HCO3− consumes protons and therefore can result in a potential pH increase (49). Silica precipitation could act as a pH buffer system for the efficient carbon concentration required for subsequent CO2 fixation. Such a pH buffering function for biogenic silica has also been well documented in diatoms (50).
This study expands the record of silicifying prokaryotes from the genus Bacillus belonging to the Firmicutes phylum and the cyanobacterium Synechococcus to several MTB species affiliated with the phyla Nitrospirota and Proteobacteria (Fig. 3). However, as mentioned above, the biosilicification pattern (e.g., the form and location of silicified products) is obviously different among bacterial groups. Such diversity of biosilicification in prokaryotes may be related to the variations in the composition and homology of Si-related genes among different bacterial groups and even species/strains. For instance, the deposition of silica globules in strain WYHC-5 may be mediated by silicatein, a silica-synthesizing, catalytic triad hydrolase that was found in siliceous sponges. By contrast, in B. cereus, it is the expression of the spore coat protein CotB1 that correlates well with the time course of biosilicification, and its presence correlates with the location of the deposited silica (43). This protein shows notable similarity in its C-terminal region (50% identity over 16 amino acids and E value of 8.5) to silacidin A, a silica-forming peptide that is isolated from cell walls of the diatom Thalassiosira pseudonana (51). Moreover, B. cereus genome contains a silicon transporter gene with some similarity with the higher plant Lsi2 sequences (3, 52). By contrast, no gene homolog to the ones known to be involved in silica deposition has yet been detected in the cyanobacterium Synechococcus (Fig. 4A and data file S5). Different strains of the cyanobacterium Synechococcus contain their own silicon transporter genes with some similarity to diatom SIT (silicon transporter) or higher plant Lsi2 sequences (Fig. 4A and data file S5). The patchy distribution of silicifying bacteria over a broad phylogenetic range and the variability of Si-related genes among different bacterial groups including at the species/strains level suggest that (i) prokaryotic biosilicification appeared in an early ancestor and was lost by most lineages, (ii) the silicon-related genes responsible for silicic acid uptake and subsequent biosilicification have multiple evolutionary origins, or (iii) they might have experienced horizontal gene transfer (HGT) possibly from different silicifying eukaryotes (3, 53). In the latter case, Si-related genes in prokaryotes could have originated more recently than the middle Archean. A wider search for intracellular biosilicification within more bacteria and a systematic molecular study on the Si-related metabolism genes (i.e., key genes involved in silicon transport and uptake and silica deposition) within them will be critical for gaining a better understanding of the origin and evolution of biosilicification within prokaryotes and even eukaryotes. Moreover, cultures of diverse MTB strains with the capability of biosilicification will be necessary for exploring the molecular mechanisms of biosilicification in prokaryotes.
This study suggests that prokaryotes may play a role in driving the biogeochemical Si cycle in modern environments via intracellularly controlled silicification (8, 9, 11). Moreover, if prokaryotic biosilicification has an ancient origin rather than a recent HGT, then it may have contributed to biogenic silica production during early Earth history. Cherts are very common siliceous sedimentary rocks throughout Earth history (54). Phanerozoic cherts are mostly composed of silicified skeletons (e.g., diatoms, radiolarians, and sponge spicules) and thus are biological in origin. However, the origin of Precambrian cherts has long been an enigma due to a lack of evidence that silica-secreting organisms were present in the Precambrian in sufficient abundance to have had an appreciable influence on the Si cycle (54). While evidence from the fossil record (e.g., magnetofossils) reveals that MTB have existed as far back as ca. 1.9 billion years (Ga) (55), molecular clock dating suggests that MTB might have originated as early as ca. 3.38 to 3.2 Ga (17). Furthermore, all MTB alive today are microaerobic and anaerobic and versatile in their C, N, S, P, and Fe metabolisms (18, 20, 21). They are also environmentally ubiquitous, living in various freshwater, brackish, marine, and hypersaline habitats and even in hot springs (18). Overall, this supports the possibility that MTB lived in hypoxic environments earlier in Earth history in the Archean ocean (17, 56). Last, by forming intracellular silica globules, WYHC-5 cells accumulate as much as ~10.9% of the cell weight as silicon, which is comparable to the amounts of silica accumulated extracellularly by some silicifying eukaryotes such as the diatoms Thalassiosira weissflogii (9.6%) and Amphora coffeaeformis (18.0%) (57). As a result, before eukaryotes, these MTB forming intracellular silica may have contributed to the biogeochemical cycle of silicon since the middle Archea. Future analyses of the Precambrian cherts, involving a comprehensive identification of lipid biomarkers, magnetosome fossils, and silica bodies in microfossils may enable to test the possibility of a widespread occurrence and biogeochemical contribution of these microbes.
MATERIALS AND METHODS
The following sections describe methods and materials for field sampling, setup of laboratory microcosms, and collection of MTB; TEM analyses; high-pressure freezing and freeze substitution for electron microscopy analyses; FIB-SEM tomography experiments; synchrotron-based STXM analyses; 16S rRNA gene sequencing and phylogenetic analysis; coupled FISH-SEM experiments; genome sequencing, assembly, and annotation; identification of Si-related gene and protein; and aqueous chemistry analysis.
Field sampling, setup of laboratory microcosms, and collection of MTB
Surface sediments (~10 cm in thickness) were scraped and taken with a steel bucket held by a long string, at a ~1- to 2-m water depth in Lake Weiyanghu (34°24′16.3″N, 108°59′16.0″E). This freshwater lake is located in the north of Xi’an City in northwest China. The sampling site was characterized by a salinity of 0.58 per mil, a pH of 8.38, and a temperature of ~18°C at the time of sampling (September 2015) as measured by a HQ40d portable multiparameter meter (Hach Company, USA). The muddy water mixture was put into a 500-ml wide-mouth reagent bottles with the sludge water ratio as 2:1. All the bottles were immediately sealed and shipped back to the laboratory in Beijing within 72 hours. Once at the laboratory, the bottles were loosely capped and stored in darkness at ambient temperature (~20°C) to set up laboratory microcosms comprising MTB (30). Living MTB within the microcosms were checked routinely with the hanging-drop method (58) under an Olympus BX51 microscope. For each test, about ~50 μl of slurry was collected from the sediments at a 1 to 2 cm depth below the water-sediment interface. Magnetotaxis was detected by rotating a bar magnet 180° on the microscope stage. The motility and magnetotactic behavior of the MTB cells were analyzed and recorded under the Olympus BX51 microscope equipped with a phase-contrast fluorescence and a DP70 digital camera system (Olympus Corp., Tokyo, Japan).
We have studied the bacterial phylogeny and magnetosome biomineralization of uncultured MTB within the laboratory microcosms of freshwater sediments collected from Lake Weiyanghu. Our previous studies evidenced the morphological diversity of MTB inhabiting in this lake sediments, including spirilla (e.g., WYHS-1) (20), cocci (WYHC-1, WYHC-2, and WYHC-3) (30), and rod-shaped bacteria (e.g., WYHR-1) (59), affiliated with the class Alphaproteobacteria, the “Ca. Etaproteobacteria” class, and the phylum Desulfobacterota (formerly the class Deltaproteobacteria), respectively.
In July 2017, we found that one group of magnetotactic cocci, named strain WYHC-5, was blooming in two microcosms (of the 13 bottles). They were conspicuously larger than a few other magnetotactic cocci, vibrio, and rod-shaped bacteria (Fig. 1A). Strain WYHC-5 showed a north-seeking magnetotactic behavior when they were checked with the hanging-drop method (58) with an optical microscope. Almost all the WYHC-5 cells preferentially swam out from the sediments mounted on the surface of a glass slide to gather around the droplet edge along the external magnetic field lines toward the S-pole of a bar magnet. With time, some cells also escaped from the droplet edge to the opposing direction. At different distances, they decelerate, stop, and swim again to the droplet edge (movie S1). Such escape motility, also known as “ping-pong” motion, has been observed within multicellular magnetotactic prokaryotes (known as MMPs) and some of the big rod-shaped MTB (e.g., Magnetobacterium bavaricum) (60, 61). The cell density of WYHC-5 in the microcosms was approximately 1 × 105 cells/ml as estimated by counting the cells clustered around the droplet edge under optical microscope.
To obtain WYHC-5 cells in sufficient amounts for molecular and microscopy analyses, living MTB cells were extracted magnetically from about ~200 ml of slurry from the two microcosms with a homemade magnetic separation apparatus, as previously described (30). The harvested MTB cells were concentrated into a 1.5-ml Eppendorf tube, washed three times with Milli-Q water, and resuspended in ~200 μl of Milli-Q water. The cell density in the suspension was ~1.0 × 107 cells/ml as estimated by counting all the WYHC-5 cells within a ~0.2 μl of the cell suspension with the Olympus BX51 microscope. Optical microscopy observations of cells stained with crystal violet showed that most of the harvested MTB cells were WYHC-5 cells (Fig. 1B), and other bacteria accounted for less than 1% (n = 300) of the total population. The cell suspension was stored at −20°C before subsequent experiments.
TEM analyses
For TEM analyses, about 5 μl of the cell suspension was deposited onto carbon-supported TEM copper grids, washed three times with Milli-Q water, and dried in air. The specimens were manipulated under a nitrogen atmosphere and then kept frozen to prevent possible oxidation before experiments.
TEM experiments were performed with a JEM-2100F microscope (JEOL Ltd., Tokyo, Japan) at 200 kV at the Institut de Minéralogie, de Physique des Matériaux et Cosmochimie (Paris, France). This instrument is equipped with a field emission gun, an ultrahigh resolution pole piece, a Gatan GIF 200 energy filter, and a STEM device allowing Z-contrast imaging in HAADF mode. EELS were obtained using a dispersion of 0.3 eV per channel to record spectra in the 100 to 190 eV range with an energy resolution of ~1.3 eV as measured by the full width at half maximum of the zero-loss peak. The dwell time was optimized to acquire sufficient signal intensity and to limit beam damage. Spectra were corrected from plural scattering using the Egerton procedure available with the EL/P program (Gatan). EDXS mapping of elements was performed in HAADF-STEM mode with a JEOL EDXS detector with an ultrathin window allowing the detection of light elements. STEM-EDXS data analyses were performed with the JEOL Analysis Station software, and the semiquantitative chemical microanalyses of individual WYHC-5 cells relied on the use of k factors (62, 63). Compared with EDXS spot microanalyses using a conventional TEM, STEM-EDXS mapping using HAADF mode has advantages in identifying intracellular biomineralization of nanoparticles or submicron particles because it simultaneously provides Z-contrast image and elemental maps that show morphological and chemical features down to the nanometric scale (62).
High-pressure freezing and freeze substitution for electron microscopy analyses
To study the cellular ultrastructure by electron microscopy, we prepared the cell samples using high-pressure freezing and freeze substitution. Compared with traditional chemical fixation, cryofixation followed by freeze substitution better preserves samples and prevents loss of highly soluble organic and inorganic components for subsequently electron microscopy observations (64). The freeze substitution experiments were performed at the Institute of Biophysics, Chinese Academy of Sciences (Beijing, China) following the procedure below. Before high-pressure freezing, about 50 μl of the cell suspension was aspirated into a small copper tube (inner diameter of ~0.35 mm) and then frozen by a high-pressure freezer [Leica electron microscope (EM) HPM100]. Freeze substitution was performed in a substitution unit (Leica EM AFS2) in a substitution medium consisting of acetone with 2% osmium tetroxide, 1% uranyl acetate, and 0.1% lead citrate at −90°C for 72 hours and then gradually warming up to −60°C for 8 hours, −20°C for 8 hours, and 0°C for 2 hours. After washing with dry acetone at 0°C for 30 min, the samples were warming up to room temperature, infiltration with acetone and epoxy resin for several hours, then embedded into resin in an embedding mold, and put in oven at 60°C for 24 hours to cure completely.
For electron microscopy observations, the top of the epoxy resin block above the cells was carefully trimmed with a Leica EM trimmer until the black cells in the resin block could be obtained. For TEM observations, the trimmed resin was sectioned with microtome (Leica EM UC6) at room temperature. After collection, sections, measuring approximately 70 nm in thickness, were examined by a JEM-2100 microscope (JEOL Ltd., Tokyo, Japan) at 200 kV at the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGG-CAS, Beijing, China) and a JEM-F200 microscope (JEOL Ltd., Tokyo, Japan) at 200 kV at the Institute of Physics, Chinese Academy of Sciences (Beijing, China).
FIB-SEM tomography experiments
To provide further comprehensive 3D information on the cell ultrastructures at the nanometer scale (65), FIB-SEM tomography experiments were performed on both samples of air-dried WYHC-5 cells mounted on the surface of TEM grid and freeze-substituted WYHC-5 cells that remained at the surface of the epoxy resin block after microtome sectioning. Before FIB-SEM experiments, the resin block was glued on a 45° aluminum stub with electrically conductive colloidal silver (Ted Pella, lot no. 169773610), followed by coating with about 10 nm of carbon using a sputter coater (Leica, ACE200), while the TEM grid sample was directly mounted on a TEM sample holder specialized for FIB-SEM analysis without a carbon coating.
The FIB-SEM experiments were carried out with a ZEISS Crossbeam 550 FIB-SEM with a Gemini II column incl double condenser and two in-lens detectors (ZEISS Microscopy Customer Center, Beijing). Before starting the FIB-SEM tomography experiment, about 300 nm of platinum was deposited over regions of interest (i.e., the surfaces of target cells for FIB-SEM tomography) as a protective layer for subsequent FIB milling. This was achieved using an ion beam–(Ga+)–assisted Pt deposition system. Then a cube containing the target cells was prepared by using the FIB milling with focused gallium ions at 30 kV and 3 to 30 nA. Three trenches (~30 μm in length, 12 μm in width, and ~ 22 μm in depth) were milled around the cube to enable access of the electron beam to the imaging plane and to reduce shadowing effects in the lower part of the images (66). A tomographic dataset on one premilled cube was subsequently acquired using the “slice and view” technique. In each slicing step, about 10 nm of the cell or resin-embedded cell was removed with focused gallium ions at 30 kV and 300 pA, and the cross section was imaged by SEM. SEM images were recorded with an aperture of 60 μm in the high current mode at an accelerating voltage of 2 kV using both the secondary electron detector and in-lens EsB (energy selective backscattered) detector. The pixel dimensions of a recorded image were 4096 × 4096 pixel, and the time to acquire one frame was 90 s. Alignment and filtering of the FIB-SEM stacks were performed using Dragonfly (2021 version) imaging processing software package. Segmentation of the magnetite magnetosomes and the cytoplasm membrane were done semiautomatically based on their relatively high contrast compared with other cytoplasm regions. The vacuole- and carboxysome-like bodies within the cells were segmented manually with the brush tool.
STXM analyses
We investigated the chemical composition of the inclusions within WYHC-5 with the use of synchrotron-based STXM. STXM allows imaging and acquisition of NEXAFS spectra at high spectral (~0.1 eV) and spatial (~25 nm) resolution (28), which is able to provide detailed mapping of biological components (28). For STXM analyses, about 5 μl of the cell suspension was deposited onto the surface of silicon nitride windows (1.5 mm by 1.5 mm, 50-nm-thick SiN membrane window centered within a 5 mm by 5 mm, 200-μm-thick silicon frame; Norcada Inc., Canada). STXM experiments were carried out on the 10ID-1 soft x-ray spectromicroscopy beamline at the Canadian Light Source (28), as described previously (21). The microscope chamber was pumped down to 100 mtorr after sample insertion and then backfilled with one-sixth atm of helium gas. Monochromated x-rays were focused on a ~30-nm spot on the sample by a Fresnel zone plate. Images were generated by x-y raster scanning while recording the intensity of transmitted x-rays. A sequence of images or stack, which covers the energy region of interest (e.g., 158 to 180 eV for S 2p, 280 to 320 eV for C 1s, and 700 to 735 eV for Fe 2p), was recorded for spectroscopy analysis. Data analysis was performed using the aXis2000 software package (http://unicorn.mcmaster.ca/aXis2000.html).
16S rRNA gene sequencing and phylogenetic analysis
About 5 μl of the cell suspension was used to amplify the 16S rRNA gene of WYHC-5 cells by polymerase chain reaction with the universal bacterial primers 27F (5-′AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (24), as described in detail by Li et al. (32). The amplification products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Germany), ligated with the pMD19-T vector (TaKaRa, Japan), and then cloned in Escherichia coli (strain DH5α)–competent cells (Huada, Beijing, China) according to the manufacturer’s instructions. A total of 30 clones were picked randomly and were sequenced using the vector primers M13-47 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and RV-M (5′-GAGCGGATAACAATTTCACACAGG-3′) at the Beijing Huada Genomics Research Center (Beijing, China).
After discarding sequences of insufficient length (<1300 bp), remaining sequences were aligned with close relatives using the Gblocks 0.91b online algorithm with default parameters to eliminate poorly aligned positions (67). The maximum likelihood method phylogenetic tree was constructed using the MEGA Software package (version 7.0) (68). Bootstrap values were calculated with 1000 replicates.
Coupled fluorescence and SEM experiments
To confirm that the derived 16S rRNA gene sequences belong to WYHC-5 cells, about 75 μl of the cell suspension was centrifuged and resuspended in 75 μl of a 0.22 μm–filtered phosphate-buffered saline (PBS) buffer (0.1 M PBS, pH 7.4). Twenty-five microliters of 4% paraformaldehyde was added for fixation at 4°C. After 12 hours of fixation, the cells were centrifuged, suspended in a mixed solution of ~200 μl of 0.1 M PBS buffer (pH 7.4) and 200 μl of ethanol (100%), and, lastly, stored at −20°C for coupled fluorescence and SEM experiments.
For FISH experiments, a species-specific oligonucleotide probe, WYHC5-215 (5′-GCTCAGCTCCAGGCAGATTA-3′), was designed to target all the 28 16S rRNA gene sequences. The probe specificity was evaluated by using the online probe evaluation tool ProbeMatch; no other 16S rDNA sequences in the Ribosomal Database Project database were able to match it (69). The universal bacteria probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) was used as a positive control probe of bacteria for FISH experiments (32). Probe EUB338 was synthesized and fluorescently labeled with fluorescein phosphoramidite (FAM) at the 5′ end, while the species-specific probe WYHC5-215 was synthesized and labeled fluorescently with hydrophilic sulfoindocyanine dye Cy3 at the 5′ end. Magnetospirillum magneticum AMB-1 cells were added in appropriate amounts as inner control cells for FISH analyses. The bacteria hybridized with the EUB338 probe or WYHC5-215 probe, or both, were subsequently observed by epifluorescence microscopy and SEM according to the protocol described previously by Li et al. (32). Epifluorescence microscopy experiments were performed with an Olympus BX51 microscope. After epifluorescence microscope observations, the same sample was carbon-coated with a Leica ACE200 Low Vacuum Sputter Coater (Leica Microsystems, Wetzlar, Germany) and then observed with a Zeiss Ultra 55 field-emission gun SEM (Carl Zeiss, Germany) operating at 5 kV at the IGG-CAS (Beijing).
Genome sequencing, assembly, and annotation of strain WYHC-5
About 1.6 μl of the cell suspension was diluted 10 times with 14.4 μl of 0.1 M PBS (pH 7.4) buffer. Four subsamples (~4 μl for each) were used to parallelly amplify the whole genome DNA with the REPLI-g Single Cell Kit (QIAGEN, Germany) according to the manufacturer protocol. To purify the amplified DNA, the QIAEX II Gel Extraction Kit (QIAGEN, Germany) was used to discard protein residues and small fragments (e.g., shorter than 4000 bp). The purified genomic DNA was sequenced using the Illumina NovaSeq 6000 system with the paired-end strategy of 150-bp reads with an average of 270-bp insert size (Annoroad, Beijing, China). Raw Illumina reads were trimmed to remove the adapter sequences and low-quality bases using Trimmomatic-0.38 software (70). Then, the clean reads were assembled using IDBA software (71) with kmer = 20, 40, 60, 80, 100, and 120, respectively. The assembled scaffolds were binned using the MetaBAT2 (version 2.12.1), MaxBin2 (version 2.2.4), and CONCOCT software packages in the MetaWRAP pipeline (72).
The three binning results were refined with Bin_refinement and Reassemble_bins of MetaWRAP (72) with the completeness of ≥90% and contamination of ≤5%, resulting in three reconstructed genomes. One refined 3.18-Mb genome draft, containing 307 scaffolds with the GC (guanine-cytosine) content of 38%, was found to contain a partial sequence of 16S rDNA (1496 bp), which shares a sequence identity of 99.6% with the 16S rDNA sequence of strain WYHC-5. This suggests that it should be derived from the genome of strain WYHC-5. We then evaluated the quality of this genome with the lineage-specific marker genes and default parameters in CheckM v.1.0.12 (73). The results showed that the completeness and contamination of the obtained WYHC-5 genome were approximately 95.3 and 0.91%, respectively. Reconstructed genomes were annotated with the online database GeneMarkS (http://topaz.gatech.edu/GeneMark/genemarks.cgi) (74).
BLAST searches, bioinformatic, and cluster analysis
Using known proteins or enzymes as query sequences (data files S1, S3, and S4), BLAST searches were performed to identify genes or proteins related to carboxysomes, the WL pathway, and silica deposition within strain WYHC-5 using the offline BLAST software with the identification of ≥30% and coverage of ≥60% (75). Cluster analysis of cDNA sequences between silicatein and cathepsin L was carried out using the IQ-TREE software (version 1.6.12) (76) and the GTR + G + I model (general time-reversible model with gamma-distributed plus invariant sites) via the maximum likelihood method (40). Bootstrap values were calculated with 1000 replicates.
Modeling of the 3D structure of the WYHC-5 silicatein
To predict the active sites of enzyme, the silicatein protein from strain WYHC-5 is aligned with the silicatein proteins from siliceous sponge species and A. vulgare using ClustalW algorithm (version 1.2.2) (77). Then, the 3D structure model of silicatein protein from strain WYHC-5 was constructed with SWISS-MODEL online server (https://swissmodel.expasy.org/) using related protein structure as modeling template (78). The quality of 3D protein model was assessed by the GMQE (Global Model Quality Estimation) score, and QMEAN score was calculated automatically by SWISS-MODEL online server. The GMQE score is expressed as a number between 0 and 1, reflecting the expected accuracy of a model built with that alignment and template and the coverage of the target. Higher numbers indicate higher reliability. QMEAN score of around 0 indicate good agreement between the model structure and template structure of similar size.
Aqueous chemistry analysis
To characterize the chemical environment where MTB, including WYHC-5 cells, live, we measured the DSi concentrations in the water column of some selected laboratory microcosms, where sediments were collected from freshwater, marine, and hypersaline environments. In each case, about 150 ml of surface water was collected and filtered by acetate cellulose membrane with a pore size of ≤0.45 μm and sent to the Aoshi Analysis Inspection and Testing (Shanghai) Co. Ltd. (Beijing branch) for water chemistry analysis. The concentration of water-soluble silica was measured with the molybdosilicate method according to APHA 23rd, 4500-SiO2 C (2017) (www.standardmethods.org/doi/abs/10.2105/SMWW.2882.095) using a PE 2600 ultraviolet-visible spectrophotometer (Shimadzu Corporation, Japan). The detection limit for water-soluble silica was 0.1 ppm.
Acknowledgments
We thank R. Mitchell for constructive comments. We also thank L. Liu and X. Sun at the ZEISS Microscopy Customer Center (Beijing) for providing access to SEM-FIB facilities and X. Sha for assisting in FIB-SEM tomography experiments. Synchrotron STXM experiments (CLS proposal 23-7537) were performed at the 10ID-1 SM beamline, Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council of Canada, the Canadian Institutes of Health Research, the province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. We thank J.-M. Guigner for technical support on for the TEM facility at IMPMC.
Funding: This work was supported financially by the National Natural Science Foundation of China (grants no. 41890843, 41920104009, and 41621004), by Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (no. 311021003), and the Senior User Project of RVKEXUE2019GZ06 (Center for Ocean Mega-Science, Chinese Academy of Sciences).
Author contributions: J.L. and Y.P. designed this research. J.L., P.L., L.S., L.F., and J.H. prepared the samples and performed the molecular biology experiments. J.L., N.M., K.B., F.M., L.G., and E.L. carried out the TEM and FIB-SEM experiments. J.L. and J.W. conducted the synchrotron STXM experiments. J.L. wrote the manuscript with input from P.L., K.B., X.Z., Y.Z., X.C., and Y.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. 16S rRNA gene sequence and genome of strain WYHC-5 have been assigned to GenBank accession numbers OL423397 and JAKKUN000000000, respectively. Genes for the enzymes RuBisCO (accession no. OM416947), carbonic anhydrase (accession no. OM416948), and silicatein (accession no. OM416946) of strain WYHC-5 are published in GenBank.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S4
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
Data S1 to S5
REFERENCES AND NOTES
- 1.De Tombeur F., Turner B. L., Laliberté E., Lambers H., Mahy G., Faucon M.-P., Zemunik G., Cornelis J.-T., Plants sustain the terrestrial silicon cycle during ecosystem retrogression. Science 369, 1245–1248 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Tréguer P. J., De La Rocha C. L., The world ocean silica cycle. Ann. Rev. Mar. Sci. 5, 477–501 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Marron A., Ratcliffe S., Wheeler G., Goldstein R., King N., Not F., de Vargas C., Richter D., The evolution of silicon transport in eukaryotes. Mol. Biol. Evol. 33, 3226–3248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. H. Knoll, B. Kotrc, Protistan skeletons: A geologic history of evolution and constraint, in Evolution of Lightweight Structures: Analyses and Technical Applications, C. Hamm, Ed. (Springer, 2015), pp. 1–16. [Google Scholar]
- 5.Maliva R. G., Knoll A. H., Simonson B. M., Secular change in the Precambrian silica cycle: Insights from chert petrology. Geol. Soc. Am. Bull. 117, 835–845 (2005). [Google Scholar]
- 6.Kidder D. L., Erwin D. H., Secular distribution of biogenic silica through the Phanerozoic: Comparison of silica-replaced fossils and bedded cherts at the series level. J. Geol. 109, 509–522 (2001). [Google Scholar]
- 7.Hirota R., Hata Y., Ikeda T., Ishida T., Kuroda A., The silicon layer supports acid resistance of Bacillus cereus spores. J. Bacteriol. 192, 111–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baines S. B., Twining B. S., Brzezinski M. A., Krause J. W., Vogt S., Assael D., McDaniel H., Significant silicon accumulation by marine picocyanobacteria. Nat. Geosci. 5, 886–891 (2012). [Google Scholar]
- 9.Ikeda T., Bacterial biosilicification: A new insight into the global silicon cycle. Biosci. Biotechnol. Biochem. 85, 1324–1331 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Leblanc K., Cornet V., Rimmelin-Maury P., Grosso O., Hélias-Nunige S., Brunet C., Claustre H., Ras J., Leblond N., Quéguiner B., Silicon cycle in the tropical South Pacific: Contribution to the global Si cycle and evidence for an active pico-sized siliceous plankton. Biogeosciences 15, 5595–5620 (2018). [Google Scholar]
- 11.Krause J. W., Brzezinski M. A., Baines S. B., Collier J. L., Twining B. S., Ohnemus D. C., Picoplankton contribution to biogenic silica stocks and production rates in the Sargasso Sea. Global Biogeochem. Cycles 31, 762–774 (2017). [Google Scholar]
- 12.Moore K. R., Pajusalu M., Gong J., Sojo V., Matreux T., Braun D., Bosak T., Biologically mediated silicification of marine cyanobacteria and implications for the Proterozoic fossil record. Geology 48, 862–866 (2020). [Google Scholar]
- 13.Conley D. J., Frings P. J., Fontorbe G., Clymans W., Stadmark J., Hendry K. R., Marron A. O., De La Rocha C. L., Biosilicification drives a decline of dissolved Si in the oceans through geologic time. Front. Mar. Sci. 4, 397 (2017). [Google Scholar]
- 14.Ohnemus D. C., Krause J. W., Brzezinski M. A., Collier J. L., Baines S. B., Twining B. S., The chemical form of silicon in marine Synechococcus. Mar. Chem. 206, 44–51 (2018). [Google Scholar]
- 15.Brzezinski M. A., Krause J. W., Baines S. B., Collier J. L., Ohnemus D. C., Twining B. S., Patterns and regulation of silicon accumulation in Synechococcus spp. J. Phycol. 53, 746–761 (2017). [DOI] [PubMed] [Google Scholar]
- 16.M. Schultze, Die Structur der Diatomeenschale verglichen mit gewissen aus Fluorkiesel künstlich darstellbaren Kieselhäuten, in Verhandlungen des naturhistorischen Vereines der preussischen Rheinlande und Westphalens, W. Co, Ed. (Cohen, 1863), pp. 1–41. [Google Scholar]
- 17.Lin W., Patersona G. A., Zhu Q., Wang Y., Kopylovad E., Li Y., Knight R., Bazylinski D. A., Zhu R., Kirschvink J. L., Pan Y., Origin of microbial biomineralization and magnetotaxis during the Archean. Proc. Natl. Acad. Sci. U.S.A. 114, 2171–2176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefèvre C. T., Bazylinski D. A., Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bazylinski D. A., Frankel R. B., Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Li J., Liu P., Tamaxia A., Zhang H., Liu Y., Wang J., Menguy N., Zhao X., Roberts A. P., Pan Y. X., Diverse intracellular inclusion types within magnetotactic bacteria: Implications for biogeochemical cycling in aquatic environments. J. Geophys. Res. Biogeosci. 126, e2021JG006310 (2021). [Google Scholar]
- 21.Li J., Liu P., Wang J., Roberts A. P., Pan Y., Magnetotaxis as an adaptation to enable bacterial shuttling of microbial sulfur and sulfur cycling across aquatic oxic-anoxic interfaces. J. Geophys. Res. Biogeosci. 125, e2020JG006012 (2020). [Google Scholar]
- 22.Schulz-Vogt H. N., Pollehne F., Jürgens K., Arz H. W., Beier S., Bahlo R., Dellwig O., Henkel J. V., Herlemann D. P. R., Krüger S., Leipe T., Schott T., Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J. 13, 1198–1208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteil C. L., Benzerara K., Menguy N., Bidaud C. C., Michot-Achdjian E., Bolzoni R., Mathon F. P., Coutaud M., Alonso B., Garau C., Jézéquel D., Viollier E., Ginet N., Floriani M., Swaraj S., Sachse M., Busigny V., Duprat E., Guyot F., Lefèvre C. T., Intracellular amorphous Ca-carbonate and magnetite biomineralization by a magnetotactic bacterium affiliated to the Alphaproteobacteria. ISME J. 15, 1–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P., Liu Y., Ren X., Zhang Z., Zhao X., Roberts Andrew P., Pan Y., Li J., A novel magnetotactic Alphaproteobacterium producing intracellular magnetite and calcium-bearing minerals. Appl. Environ. Microbiol. 87, e01556-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeates T. O., Kerfeld C. A., Heinhorst S., Cannon G. C., Shively J. M., Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Y. G. Liao, Practical Electron Microscopy and Database (GlobalSino, 2013); www.globalsino.com/EM.
- 28.Wang J., Li J. H., Scanning transmission x-ray microscopy at the Canadian Light Source: Progress and selected applications in geosciences. At. Spectrosc. 43, 84–98 (2022). [Google Scholar]
- 29.Singh V. K., Singh A. L., Singh R., Kumar A., Iron oxidizing bacteria: Insights on diversity, mechanism of iron oxidation and role in management of metal pollution. Environ. Sustain. 1, 221–231 (2018). [Google Scholar]
- 30.Liu P., Liu Y., Zhao X., Roberts A. P., Zhang H., Zheng Y., Wang F., Wang L., Menguy N., Pan Y., Li J., Diverse phylogeny and morphology of magnetite biomineralized by magnetotactic cocci. Environ. Microbiol. 23, 1115–1129 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Lefèvre C. T., Frankel R. B., Abreu F., Lins U., Bazylinski D. A., Culture-independent characterization of a novel, uncultivated magnetotactic member of the Nitrospirae phylum. Environ. Microbiol. 13, 538–549 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Li J., Zhang H., Menguy N., Benzerara K., Wang F., Lin X., Chen Z., Pan Y., Single-cell resolution of uncultured magnetotactic bacteria via fluorescence-coupled electron microscopy. Appl. Environ. Microbiol. 83, e00409-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., Whitman W. B., Euzéby J., Amann R., Rosselló-Móra R., Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Gunnarsson I., Arnórsson S., Amorphous silica solubility and the thermodynamic properties of H4SiO4 in the range of 0° to 350°C at Psat. Geochim. Cosmochim. Acta 64, 2295–2307 (2000). [Google Scholar]
- 35.Müller W. E. G., Boreiko A., Wang X., Belikov S. I., Wiens M., Grebenjuk V. A., Schloβmacher U., Schröder H. C., Silicateins, the major biosilica forming enzymes present in demosponges: Protein analysis and phylogenetic relationship. Gene 395, 62–71 (2007). [DOI] [PubMed] [Google Scholar]
- 36.K. Shimizu, D. E. Morse, “Silicatein: A unique silica-synthesizing catalytic triad hydrolase from marine sponge skeletons and its multiple applications,” in Methods in Enzymology, B. S. Moore, Ed. (Academic Press, 2018), vol. 605, pp. 429–455. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu K., Cha J., Stucky G. D., Morse D. E., Silicatein α: Cathepsin L-like protein in sponge biosilica. Proc. Natl. Acad. Sci. U.S.A. 95, 6234–6238 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder H. C., Wang X., Tremel W., Ushijima H., Müller W. E. G., Biofabrication of biosilica-glass by living organisms. Nat. Prod. Res. 25, 455–474 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Chebbi M. A., Becking T., Moumen B., Giraud I., Gilbert C., Peccoud J., Cordaux R., The genome of Armadillidium vulgare (Crustacea, Isopoda) provides insights into sex chromosome evolution in the context of cytoplasmic sex determination. Mol. Biol. Evol. 36, 727–741 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Nei M., Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Konhauser K. O., Jones B., Phoenix V. R., Ferris G., Renaut R. W., The microbial role in hot spring silicification. Ambio 33, 552–558 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Orange F., Disnar J.-R., Westall F., Prieur D., Baillif P., Metal cation binding by the hyperthermophilic microorganism, Archaea Methanocaldococcus Jannaschii, and its effects on silicification. Palaeontology 54, 953–964 (2011). [Google Scholar]
- 43.Motomura K., Ikeda T., Matsuyama S., Abdelhamid M. A., Tanaka T., Ishida T., Hirota R., Kuroda A., The C-terminal zwitterionic sequence of CotB1 is essential for biosilicification of the Bacillus cereus spore coat. J. Bacteriol. 198, 276–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busigny V., Mathon F. P., Jézéquel D., Bidaud C. C., Viollier E., Bardoux G., Bourrand J.-J., Benzerara K., Duprat E., Menguy N., Monteil C. L., Lefèvre C. T., Mass collection of magnetotactic bacteria from the permanently stratified ferruginous Lake Pavin, France. Environ. Microbiol. 24, 721–736 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Pfeifer F., Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol. 10, 705–715 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Klaas C., Archer D. E., Association of sinking organic matter with various types of mineral ballast in the deep sea: Implications for the rain ratio. Global Biogeochem. Cycles 16, 63-1–63-14 (2002). [Google Scholar]
- 47.Lin W., Deng A., Wang Z., Li Y., Wen T., Wu L.-F., Wu M., Pan Y., Genomic insights into the uncultured genus “Candidatus Magnetobacterium” in the phylum Nitrospirae. ISME J. 8, 2463–2477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams T. J., Zhang C. L., Scott J. H., Bazylinski D. A., Evidence for autotrophy via the reverse tricarboxylic acid cycle in the marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 72, 1322–1329 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Wever A., Benzerara K., Coutaud M., Caumes G., Poinsot M., Skouri-Panet F., Laurent T., Duprat E., Gugger M., Evidence of high Ca uptake by cyanobacteria forming intracellular CaCO3 and impact on their growth. Geobiology 17, 676–690 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Milligan A. J., Morel F. M. M., A proton buffering role for silica in diatoms. Science 297, 1848–1850 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Wenzl S., Hett R., Richthammer P., Sumper M., Silacidins: Highly acidic phosphopeptides from diatom shells assist in silica precipitation in vitro. Angew. Chem. Int. Ed. 47, 1729–1732 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Arif S., Liaquat F., Yang S., Shah I. H., Zhao L., Xiong X., Garcia D., Zhang Y., Exogenous inoculation of endophytic bacterium Bacillus cereus suppresses clubroot (Plasmodiophora brassicae) occurrence in pak choi (Brassica campestris sp. chinensis L.). Planta 253, 25 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Hendry K. R., Marron A. O., Vincent F., Conley D. J., Gehlen M., Ibarbalz F. M., Queguiner B., Bowler C., Competition between silicifiers and non-silicifiers in the past and present ocean and its evolutionary impacts. Front. Mar. Sci. 5, 22 (2018). [Google Scholar]
- 54.Perry C. C., Silicification: The processes by which organisms capture and mineralize silica. Rev. Mineral. Geochem. 54, 291–327 (2003). [Google Scholar]
- 55.Chang S. B. R., Stolz J. F., Kirschvink J. L., Awramik S. M., Biogenic magnetite in stromatolites. II. Occurrence in ancient sedimentray environments. Precambrian Res. 43, 305–315 (1989). [Google Scholar]
- 56.Amor M., Mathon F. P., Monteil C. L., Busigny V., Lefèvre C. T., Iron-biomineralizing organelle in magnetotactic bacteria: Function, synthesis and preservation in ancient rock samples. Environ. Microbiol. 22, 3611–3632 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Xu H., Shi Z., Zhang X., Pang M., Pan K., Liu H., Diatom frustules with different silica contents affect copepod grazing due to differences in the nanoscale mechanical properties. Limnol. Oceanogr. 66, 3408–3420 (2021). [Google Scholar]
- 58.Schüler D., The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5, 209–214 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Li J., Zhang H., Liu P., Menguy N., Roberts A. P., Chen H., Wang Y., Pan Y., Phylogenetic and structural identification of a novel magnetotactic Deltaproteobacterium strain WYHR-1 from a freshwater lake. Appl. Environ. Microbiol. 85, e00731-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W. J., Wu L.-F., Flagella and swimming behavior of marine magnetotactic bacteria. Biomolecules 10, 460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greenberg M., Canter K., Mahler I., Tornheim A., Observation of magnetoreceptive behavior in a multicellular magnetotactic prokaryote in higher than geomagnetic fields. Biophys. J. 88, 1496–1499 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J., M. Oliver I., Cam N., Boudier T., Blondeau M., Leroy E., Cosmidis J., Skouri-Panet F., Guigner J.-M., Férard C., Poinsot M., Moreira D., Lopez-Garcia P., Cassier-Chauvat C., Chauvat F., Benzerara K., Biomineralization patterns of intracellular carbonatogenesis in cyanobacteria: Molecular hypotheses. Minerals 6, 10 (2016). [Google Scholar]
- 63.M. Abad, F. Nieto, “Quantitative EDX analysis in TEM. Practical development, limitations and standards,” in Science, Technology and Education of Microscopy: An Overview, A. Méndez-Vilas, Ed. (Formatex, 2003), pp. 687–694. [Google Scholar]
- 64.Blondeau M., Sachse M., Boulogne C., Gillet C., Guigner J.-M., Skouri-Panet F., Poinsot M., Ferard C., Miot J., Benzerara K., Amorphous calcium carbonate granules form within an intracellular compartment in calcifying cyanobacteria. Front. Microbiol. 9, 1768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villinger C., Gregorius H., Kranz C., Höhn K., Münzberg C., von Wichert G., Mizaikoff B., Wanner G., Walther P., FIB/SEM tomography with TEM-like resolution for 3D imaging of high-pressure frozen cells. Histochem.Cell Biol. 138, 549–556 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Kizilyaprak C., Bittermann A. G., Daraspe J., Humbel B. M., FIB-SEM tomography in biology. Methods Mol. Biol. 1117, 541–558 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O., Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim Y. J., Teletia N., Ruotti V., Maher C. A., Chinnaiyan A. M., Stewart R., Thomson J. A., Patel J. M., ProbeMatch: Rapid alignment of oligonucleotides to genome allowing both gaps and mismatches. Bioinformatics 25, 1424–1425 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Uritskiy G. V., DiRuggiero J., Taylor J., MetaWRAP–A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Besemer J., Lomsadze A., Borodovsky M., GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nguyen L.-T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li J., Menguy N., Gatel C., Boureau V., Snoeck E., Patriarche G., Leroy E., Pan Y., Crystal growth of bullet-shaped magnetite in magnetotactic bacteria of the Nitrospirae phylum. J. R. Soc. Interface 12, 20141288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karunakaran C., Prange A., Wang J., Lu Y., Hitchcock A. P., Hormes J., Speciation of sulphur globules in a single bacterial cell by sulphur 2p x-ray spectromicroscopy. Canadian Light Source Res. Rep. , 46–47 (2009). [Google Scholar]
- 81.Teodorescu C.-M., Gravel D., Choi J., Pugmire D., Dowben P. A., Fominykh N., Pavlychev A. A., Rühl E., Inner-shell excitation and fragmentation of sulfur aggregates. J. Electron. Spectrosc. Relat. Phenom. 101, 193–198 (1999). [Google Scholar]
- 82.Benzerara K., Yoon T. H., Tyliszczak T., Constantz B., Spormann A. M., Brown G. E., Scanning transmission x-ray microscopy study of microbial calcification. Geobiology 2, 249–259 (2004). [Google Scholar]
- 83.Chen S.-Y., Gloter A., Zobelli A., Wang L., Chen C.-H., Colliex C., Electron energy loss spectroscopy and ab initio investigation of iron oxide nanomaterials grown by a hydrothermal process. Phys. Rev. B. 79, 104103 (2009). [Google Scholar]
- 84.Li J., Menguy N., Leroy E., Roberts A. P., Liu P., Pan Y., Biomineralization and magnetism of uncultured magnetotactic coccus strain THC-1 with non-chained magnetosomal magnetite nanoparticles. J. Geophys. Res. Solid Earth 125, e2020JB020853 (2020). [Google Scholar]
- 85.Zhang H., Menguy N., Wang F., Benzerara K., Leroy E., Liu P., Liu W., Wang C., Pan Y., Chen Z., Li J., Magnetotactic coccus strain SHHC-1 affiliated to Alphaproteobacteria forms octahedral magnetite magnetosomes. Front. Microbiol. 8, 969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J., Pan Y., Chen G., Liu Q., Tian L., Lin W., Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: Transmission electron microscopy and magnetic observations. Geophys. J. Int. 177, 33–42 (2009). [Google Scholar]
- 87.Taoka A., Kondo J., Oestreicher Z., Fukumori Y., Characterization of uncultured giant rod-shaped magnetotactic Gammaproteobacteria from a freshwater pond in Kanazawa, Japan. Microbiology 160, 2226–2234 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Byrne M. E., Ball D. A., Guerquin-Kern J. L., Rouiller I., Wu T. D., Downing K. H., Vali H., Komeili A., Desulfovibrio magneticus RS-1 contains an iron- and phosphorus-rich organelle distinct from its bullet-shaped magnetosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 12263–12268 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qian X.-X., Liu J., Menguy N., Li J., Alberto F., Teng Z., Xiao T., Zhang W., Wu L.-F., Identification of novel species of marine magnetotactic bacteria affiliated with Nitrospirae phylum. Environ. Microbiol. Rep. 11, 330–337 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Liu P., Tamaxia A., Liu Y., Qiu H., Pan J., Jin Z., Zhao X., Roberts A. P., Pan Y., Li J., Identification and characterization of magnetotactic Gammaproteobacteria from a salt evaporation pool, Bohai Bay, China. Environ. Microbiol. 24, 938–950 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4
References
Movies S1 to S3
Data S1 to S5