Abstract
In small cell lung cancer (SCLC), acquired resistance to DNA-damaging therapy is challenging to study because rebiopsy is rarely performed. We used patient-derived xenograft models, established before therapy and after progression, to dissect acquired resistance to olaparib plus temozolomide (OT), a promising experimental therapy for relapsed SCLC. These pairs of serial models reveal alterations in both cell cycle kinetics and DNA replication and demonstrate both inter- and intratumoral heterogeneity in mechanisms of resistance. In one model pair, up-regulation of translesion DNA synthesis (TLS) enabled tolerance of OT-induced damage during DNA replication. TLS inhibitors restored sensitivity to OT both in vitro and in vivo, and similar synergistic effects were seen in additional SCLC cell lines. This represents the first described mechanism of acquired resistance to DNA damage in a patient with SCLC and highlights the potential of the serial model approach to investigate and overcome resistance to therapy in SCLC.
Investigation of serial PDX models reveals that translesion DNA synthesis mediates drug resistance and inhibition resensitizes.
INTRODUCTION
Small cell lung cancer (SCLC) is an aggressive neuroendocrine (NE) malignancy characterized by early metastasis and poor prognosis. SCLC is common, afflicting more than 30,000 people per year. Approximately 70% of these patients present with disseminated and incurable disease, termed “extensive stage” or ES-SCLC, which is treated with systemic therapy alone. In stark contrast with non-SCLC (NSCLC), where therapies that target oncogenic drivers or block immune checkpoints have revolutionized care, hallmark SCLC mutations such as loss of TP53 and RB1 are not targetable (1–3), and only a small fraction of patients benefit from immune checkpoint blockade (4, 5). As a result, DNA-damaging agents remain the backbone of systemic therapy for SCLC (6–8). Untreated ES-SCLC responds consistently and rapidly to etoposide plus cisplatin or carboplatin (EP) and may continue to respond to a second or third regimen following inevitable relapse. However, with each successive relapse, the fraction of patients that benefit from therapy drops markedly, and the responses that do occur are typically brief (9). SCLC tumors often evolve from a naïve chemo-responsive state to an acquired chemoresistant state.
For patients with metastatic NSCLC, next-generation sequencing of tumor samples has become the standard of care in the United States because of the extraordinary benefit of finding targetable oncogenic mutations (10). Targeted therapies such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors produce high response rates and durable clinical responses, but they are not curative (11, 12). At relapse, rebiopsy efforts have facilitated comparison of pretreatment and postrelapse samples to map the landscape of acquired resistance mechanisms and to develop therapies to overcome resistance (13–16). This paradigm has been so successful that it is now standard practice to diagnose acquired resistance in NSCLC with EGFR-sensitizing mutations through repeat testing of the progressing tumor (10). In contrast, there is no clinical role for rebiopsy in SCLC, and as a result, these paired pretreatment/postrelapse samples are exceedingly rare. Furthermore, the few postrelapse samples that have been obtained are either fixed or frozen. If the spectrum of resistance was well described, as for EGFR-mutant NSCLC, these nonviable postrelapse samples could be used to confirm the presence of specific resistance alterations, but without such a reference list, they can only be used to generate hypotheses.
Instead, dissection of resistance to DNA damage in SCLC has relied on models that lack clinical correlates. These models, either human or murine, are typically treated in the laboratory until they become drug resistant and compared with their untreated parents (17–20). However, laboratory drug regimens usually differ from clinical administration in dose and schedule, and without confirmation in samples or models from postrelapse patients, it is unclear whether these experimental systems recapitulate clinical resistance. Despite extensive research using this approach, no consensus or clinically actionable mechanisms of acquired resistance have been identified for any therapy in SCLC.
We established a platform to generate patient-derived xenograft (PDX) models from patients with SCLC at multiple clinical time points using circulating tumor cells (CTCs) as well as biopsy and effusion samples as source material (21). In SCLC following relapse, CTCs are relatively more accessible than biopsy samples and have been essential to generate a cohort of matched PDX pairs to discover chemotherapy escape mechanisms (22). PDX models of SCLC retain the histological features and genomic alterations of the original tumor, and they exhibit sensitivity to first-line EP that reflects the clinical history of the donor patients (21, 23, 24). In parallel, we conducted a single-arm phase 1/2 trial of combination therapy with the poly(adenosine diphosphate–ribose) polymerase (PARP) inhibitor olaparib plus the DNA alkylating agent temozolomide (TMZ; combination OT) in patients with relapsed SCLC. This regimen was well tolerated and resulted in a promising overall response rate of 41.7% (20 of 48 evaluable) (25), echoing similar results with a different PARP inhibitor, veliparib, in combination with TMZ (26). PDX models were established from OT trial participants, and similar to their clinical fidelity to EP, these models recapitulated the clinical response and resistance of their corresponding patients when treated in vivo with OT.
We derived both pre-OT treatment and post-OT relapse PDX models from two patients on the OT trial with durable responses, MGH1518 and MGH1528, and these serial models captured the transition from clinical sensitivity to acquired resistance (25). Here, we report a detailed analysis of these model pairs and describe the distinct ways in which they respond to OT-induced DNA damage. Whole-genome sequencing (WGS) confirmed the high mutation burden in these tumors and underscored the extreme difficulty of discovering mechanisms of resistance from unbiased genetic profiles. Taking an alternative first-principles approach, we deconstructed the cellular responses of these PDX models to OT into basic elements of DNA damage induction, repair, and tolerance. For patient MGH1518, we describe alterations in both cell cycle distribution and DNA replication that likely contribute to OT resistance. In particular, we find that up-regulation of translesion DNA synthesis (TLS) in MGH1518 following relapse is a key element of OT resistance during DNA replication. TLS is an error-prone form of DNA replication that can increase tolerance of DNA damage during S phase and may contribute to chemoresistance in diverse cancers, including NSCLC (27, 28). Using a novel TLS inhibitor, we were able to notably restore OT sensitivity in the postrelapse model. This combination treatment not only suppressed DNA replication and blocked cell proliferation in cultured cells, but it also caused regression of MGH1518-3 tumors in vivo. We conclude that, for a subset of patients with relapsed SCLC, TLS inhibitor combinations may be an effective therapeutic strategy.
RESULTS
Following response and relapse, OT-resistant SCLC has a massive increase in tumor mutational burden
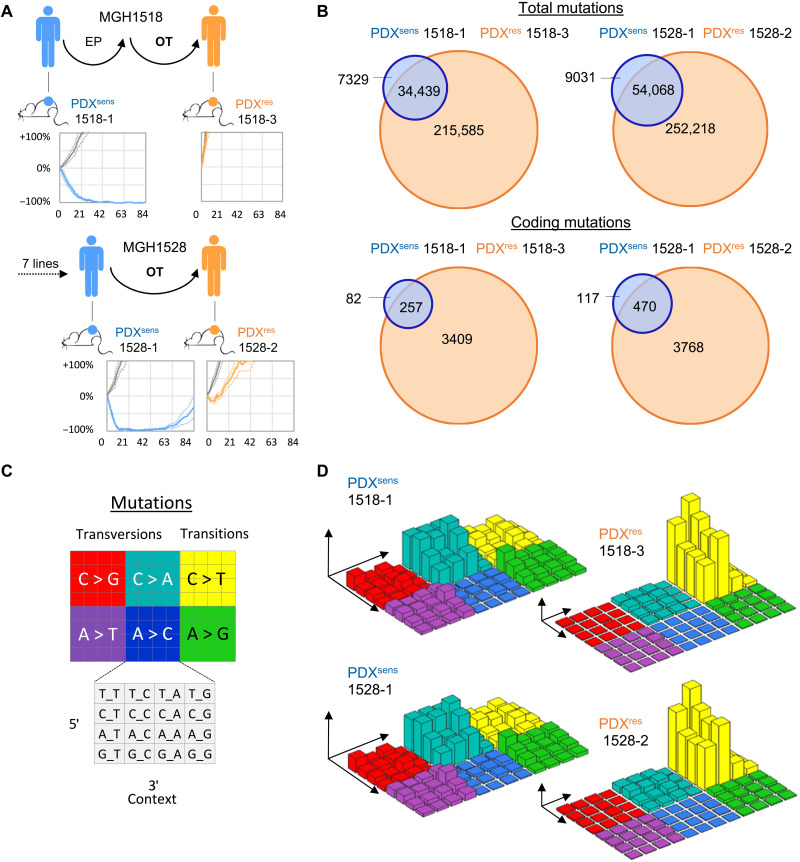
We derived serial PDX models from two patients with SCLC, MGH1518 and MGH1528, before treatment with OT and again at progression, after durable but partial responses [Fig. 1A; (25)]. The two pretreatment PDX models were highly sensitive to OT in vivo, recapitulating the clinical responses of their patients, and are hereafter called PDXsens 1518-1 and PDXsens 1528-1. In contrast, the models generated at clinical progression were both highly resistant to OT, hereafter called PDXres 1518-3 and PDXres 1528-2.
Fig. 1. OT-resistant SCLC models show a massive increase in tumor mutational burden after relapse.
(A) Schematized clinical histories and in vivo responses to OT of serial PDX models. Models from patients before treatment with OT (blue) and again at progression after a durable clinical response (orange), with vehicle-treated xenografts (gray). Average response in vivo to OT across three to six xenografts as percentage of ITV versus days after treatment (solid line) ± SEM (dashed lines). (B) Venn diagrams were constructed by comparing the shared and unique somatic variants of sequential biopsies. Coding somatic variants were identified as nonsynonymous mutations that fall within a coding region. (C and D) Three-dimensional bar plots (also called “Lego plots”) representing mutational signatures in a three-base context. (D) Left: Mutational signature identified in sensitive models showing C-to-A transversions that are associated with tobacco smoking [SBS4 in COSMIC (82)]. (D) Right: Mutational signature identified in resistant models containing tobacco smoking–associated mutations in addition to large numbers of C-to-T transitions associated with TMZ treatment [SBS11 in COSMIC (82)]. Axes and plots are not in scale, and smoking-associated mutations, which are still present in postrelapse PDXs, are overshadowed by the predominant TMZ signature.
Initially, to identify candidate mutations that could mediate acquired resistance to OT in SCLC, we pursued a comparative genomic approach that has been applied successfully to targeted therapy resistance in NSCLC. WGS was performed on the pairs of serial PDX models and their corresponding patient germline DNA samples. The total mutational burden and distribution across the genome were similar in the pretreatment models, with 41,768 total mutations in PDXsens 1518-1 and 63,099 mutations in PDXsens 1528-1, of which 339 in 1518-1 and 587 in 1528-1 occurred in coding regions (1.5 and 2.6% of all genes, respectively) (Fig. 1B). These mutations were highly enriched for C-to-A transversions characteristic of a tobacco smoking signature (Fig. 1, C and D, left) and were evenly distributed across all chromosomes (fig. S1, A and B). Notably, the mutational burden increased nearly 10-fold in both postrelapse models to 250,024 in PDXres 1518-3 (3666 coding, 15% of all genes) and 306,286 in PDXres 1528-2 (4238 coding, 16.6% of all genes) (Fig. 1B). This massive increase in mutational burden was almost entirely treatment related, as the predominant mutational signature switched to C-to-T transitions characteristic of TMZ-induced DNA damage, despite retention of the original smoking-related C-to-A mutations (Fig. 1, C and D, right). This large number of mutations was not unexpected, but it clearly precludes a candidate gene approach. Therefore, to identify potential mechanisms of resistance, we sought to understand the changes in the responses of the models to OT.
Olaparib and TMZ retain biochemical efficacy in OT-resistant SCLC
We first asked whether known mediators of resistance to either olaparib or TMZ could account for acquired OT resistance in the serial models. O6-methylguanine-DNA methyltransferase (MGMT) reverses TMZ-mediated alkylation of guanine residues to confer resistance; indeed, the silencing of MGMT by promoter methylation serves as a clinical biomarker for TMZ efficacy in glioblastoma (29). Multidrug resistance protein 1 (MDR1) can also confer TMZ resistance by drug efflux in glioblastoma (30, 31). Schlafen family member 11 (SLFN11) irreversibly inactivates stressed replication forks (32) and may be silenced to induce resistance to both chemotherapy and PARP inhibitors in SCLC (19, 33, 34). Neither MGMT nor MDR1 was expressed in either set of serial models, whereas SLFN11 was expressed in all four models and increased following relapse in PDXres 1518-3 (fig. S1, C and D). These results are consistent with our previously published RNA sequencing (RNA-seq) data (25) and suggest that none of these previously reported mechanisms (fig. S1, E and F), nor a change of NE status or SCLC subtype (fig. S1G), account for acquired resistance to OT in PDXres 1518-3 and PDXres 1528-2.
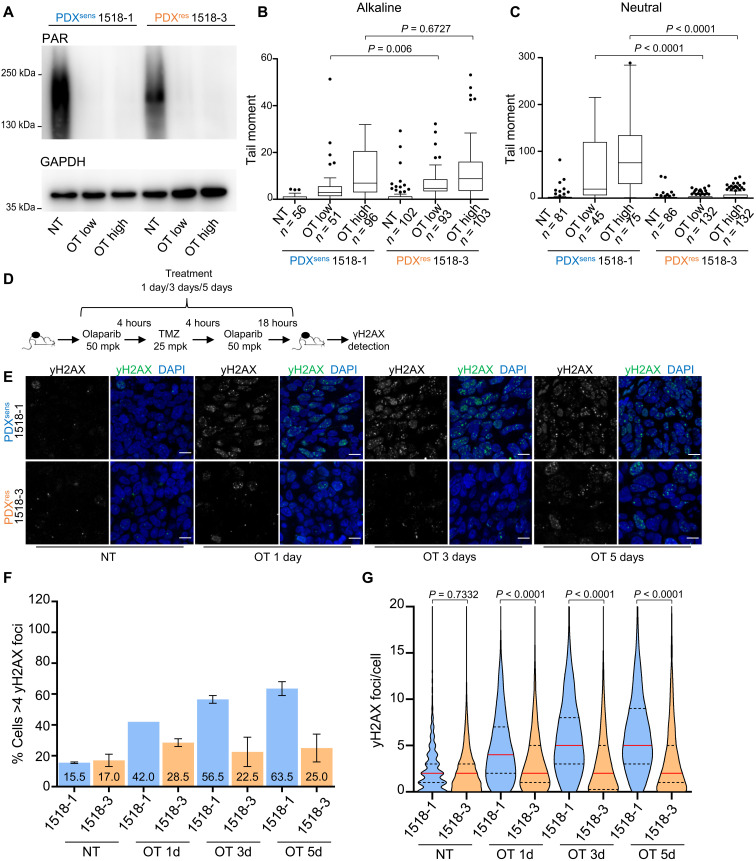
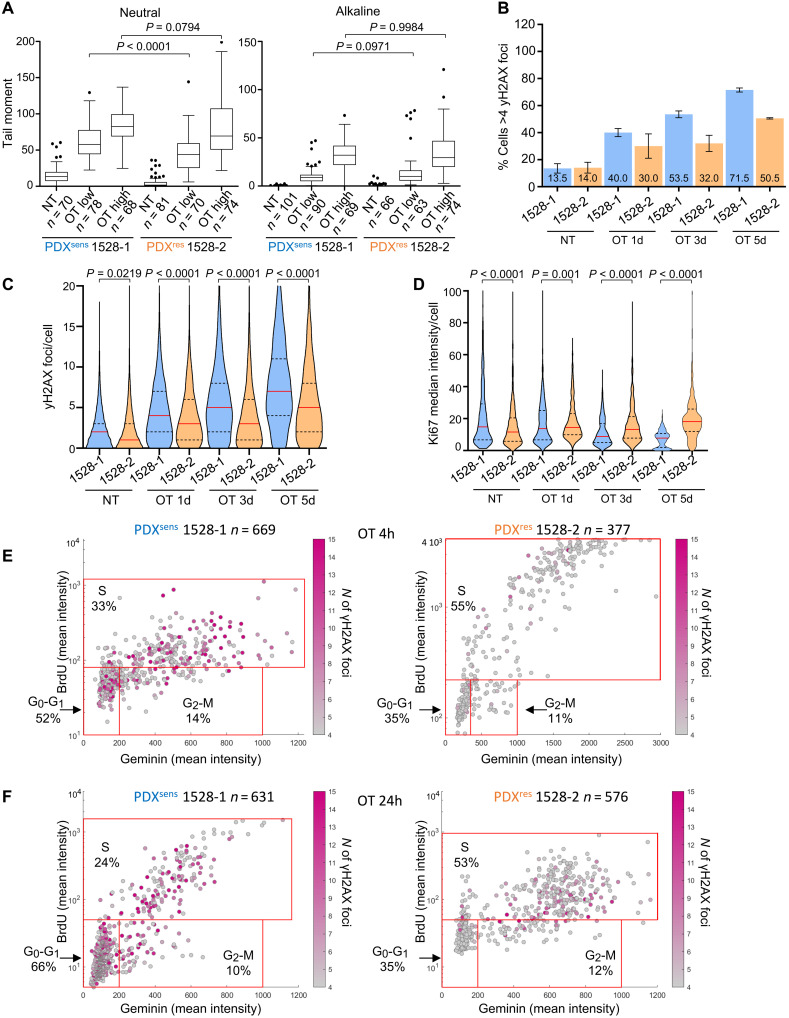
Drug-induced cell death requires target engagement, and resistance could be acquired through prevention of olaparib, TMZ, or the combination from acting on their intracellular molecular targets. There are many possible routes to pharmacodynamic failure, such as alteration of PARP1 complexes or up-regulation of drug efflux pumps, but all would reduce the initial accumulation of DNA damage. We generated short-term cultures from freshly dissected xenografts, treated in vitro with OT, and measured change in total protein PARylation and induction of DNA damage. Olaparib reduced PARylation to similar levels in short-term cultures derived from all four models (Fig. 2A and fig. S1H). To measure initial induction of DNA damage by OT, comet assays were performed under alkaline conditions in the same short-term cultures. The tail moments of OT-treated PDX cells increased in a dose-dependent and reproducible manner, and there was no measurable difference between PDXsens 1518-1 and PDXres 1518-3 (Fig. 2B). These results suggest that there is no significant difference in the capacity of olaparib to inhibit PARylation or TMZ to generate DNA damage following acquired resistance to OT.
Fig. 2. PDXres 1518-3 cells acquire fewer DNA double-strand breaks after OT treatment.
(A) Western blot analysis of protein extracts from isolated tumor cells. Total PARylation (PAR) was detected, and GAPDH was used as a loading control. (B and C) DNA damage assessment after OT treatment in isolated tumor cells. Treatment used: OT low (0.25 μM olaparib + 35 μM TMZ) and OT high (0.5 μM olaparib + 70 μM TMZ). NT, untreated samples. Quantification of the comet tail moment (% of DNA in the tail multiplied by the distance between means of the head and tail distributions) is shown. Box plots represent interquartile ranges, horizontal black lines denote the median, and points indicate outliers. Two-tailed Mann-Whitney statistical test and data from one representative experiment. (D) Schematic representation of time-course experiment. (E) γH2AX immunodetected on xenograft tissue sections and nuclei detected by DAPI. Scale bars, 10 μm. (F) Models and time points are indicated; numbers represent mean percentages. Data are from two independent experiments, and SEM bars are shown in black. (G) Truncated violin plot showing γH2AX foci number per cell, and medians are indicated by red lines and quartiles by black lines. Two-tailed Mann-Whitney statistical test and data from two independent experiments. (F and G) Apoptotic cells were identified by TUNEL (staining not shown) and excluded from the quantification. Numbers of analyzed cells are as follows: 1518-1, NT n = 2668, OT 1d n = 3687, OT 3d n = 3909, and OT 5d n = 3187; 1518-3, NT n = 2950, OT 1d n = 3844, OT 3d n = 4352, and OT 5d n = 3521.
Following relapse, OT-induced DNA lesions result in fewer DNA double-stranded breaks
Whereas comet assays performed under alkaline conditions detect many forms of DNA damage, including double- and single-stranded DNA breaks (DSBs and SSBs), assays performed under neutral conditions are more specific for DSBs (35). For PDXsens 1518-1, there was no difference between the neutral and alkaline comet tail moments, but interestingly, in OT-treated cells isolated from PDXres 1518-3, neutral comet assays failed to detect broken DNA fragments (Fig. 2C) despite their presence in alkaline assays (Fig. 2B). This distinction appears to be OT specific, as there was no difference between neutral and alkaline assay results following γ-irradiation (fig. S1I). OT induces SSBs in both MGH1518 models, but comet assays suggest that PDXres 1518-3 no longer converts OT-induced SSBs into DSBs.
The histone variant H2AX is phosphorylated by phosphatidylinositol 3-kinase–related kinases in response to DSBs and DNA replication stress. Upon the formation of DSBs or collapsed replication forks, discrete nuclear foci of phosphorylated H2AX (γH2AX) are readily detected. Thus, the levels of γH2AX foci provide an indirect means to follow the formation and repair of DSBs (36). We treated both PDXsens 1518-1 and PDXres 1518-3 in vivo with daily OT for increasing durations (1, 3, or 5 days), resected tumors following the final dose, detected γH2AX foci by immunofluorescence in fixed tumor sections (Fig. 2, D and E), and quantified both the percentage of cells with more than four foci and the number of foci per cell (Fig. 2, F and G). By either metric, there were markedly fewer γH2AX foci in PDXres 1518-3 than in PDXsens 1518-1 at all time points, suggesting a reduced γH2AX foci formation in these tumors. Together, these results suggest that OT alkylates DNA and inhibits protein PARylation to induce SSBs in both models, but in PDXres 1518-3, an acquired resistance mechanism prevents the accumulation of DSBs and γH2AX foci.
Relapsed PDX tumors are enriched for cells in either G0 or G1 that accumulate less DNA damage
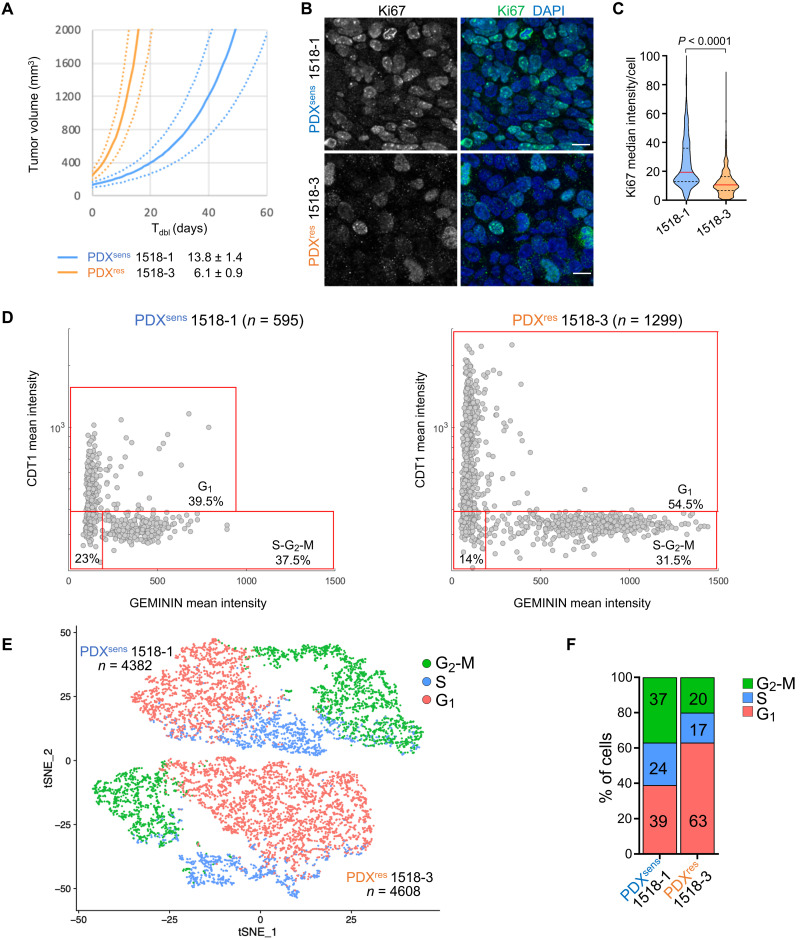
Sensitivity to cytotoxic agents varies with cell cycle progression, and DNA-damaging agents such as olaparib and TMZ are particularly cytotoxic during S phase due to interference with DNA replication forks. We investigated whether changes in cell cycle progression could account for the suppression of DSBs in PDXres 1518-3 cells by measuring the expression of Ki-67, a marker of active cell division (37). Across cancers, rapid tumor growth is usually associated with a high proportion of actively dividing cells as measured by Ki-67 expression, and PDXres 1518-3 tumors grew much faster than PDXsens 1518-1 (in vivo doubling time of 6.1 versus 13.8 days) (Fig. 3A). Therefore, it was unexpected to find that Ki-67 was markedly and significantly reduced in PDXres 1518-3 tissue sections, implying a lower proportion of dividing cells (Fig. 3, B and C). This was investigated further by probing for coexpression of the prereplication complex factor CDT1 and the DNA replication inhibitor GEMININ in fixed tissue sections. CDT1 is expressed during G1 and degraded in S phase, whereas GEMININ is expressed during S phase and degraded upon mitotic exit (38), permitting distinction of G1 from S-G2-M cells. This analysis revealed that 54.5% of cells in PDXres 1518-3 are in G1 phase while only 39.5% in PDXsens 1518-1, supporting the Ki-67 results (Fig. 3D). In parallel, single-cell RNA-seq (scRNA-seq) was performed on xenografts of each model following rapid dissociation. Cell cycle phase scores were calculated from the scRNA-seq data (39) and revealed similar fractions of G1 cells, 63% in PDXres 1518-3 versus 39% in PDXsens 1518-1 (Fig. 3, E and F, and fig. S2). While CDT1/GEMININ double-negative cells could be in very early G1 (Fig. 3D), they could also represent a nonreplicating subpopulation of G0 cells. Alternatively, the change in cell cycle distribution might indicate that progression through S and G2 phases has accelerated in PDXres 1518-3 more than progression through G1. This could reconcile the apparent inconsistency between short tumor doubling time and high proportion of G0-G1 cells, as relative acceleration of S-G2-M would increase G1 proportion by default. In either case, methodologically distinct approaches indicate that the cell cycle distribution in PDXres 1518-3 is strongly shifted toward cells in G0 or G1 despite an accelerated tumor growth rate in vivo.
Fig. 3. PDXres 1518-3 tumors are enriched for cells with cell cycle properties that differ from PDXsens tumor cells.
(A) Average growth curves for indicated models (solid), with SE estimates (dashed). Growth rates extrapolated from exponential regressions of tumor-volume measurements from 9 to 10 untreated xenografts. Model doubling times indicated in legend. (B) Ki67 (cell marker for active proliferative state) was immunodetected on xenograft tissue sections of the indicated models, and nuclei were detected by DAPI. Scale bars, 10 μm. (C) Truncated violin plot showing Ki67 median fluorescence intensity per cell; median values are indicated by red horizontal lines (19.4 for 1518-1 and 10.6 for 1518-3) and quartiles by black dashed lines. Data from two independent experiments are shown, and statistical analysis was performed with two-tailed Mann-Whitney test. Apoptotic cells were identified by TUNEL assay (staining not shown) and excluded from the quantification. Numbers of analyzed cells are as follows: 1518-1 n = 1402; 1518-3 n = 988. (D) CDT1 (cell cycle G1 phase marker) and GEMININ (cell cycle S-G2-M phase marker) were immunodetected on tumor tissue sections of the indicated models, and nuclei were detected by DAPI. Plots show the mean fluorescence intensity of both CDT1 and GEMININ for each cell represented by a gray dot. Percentage of cells in the specific cell cycle phase is shown. (E) tSNE plot of scRNA-seq data showing the predicted cell cycle phase of each analyzed cell from the two MGH1518 models. Each point represents a tumor cell. Prediction was performed using the Seurat CellCycleScoring package, and the percentages of cells in each cell cycle phase are shown in (F). (D and E) n indicates the number of analyzed cells for each PDX model.
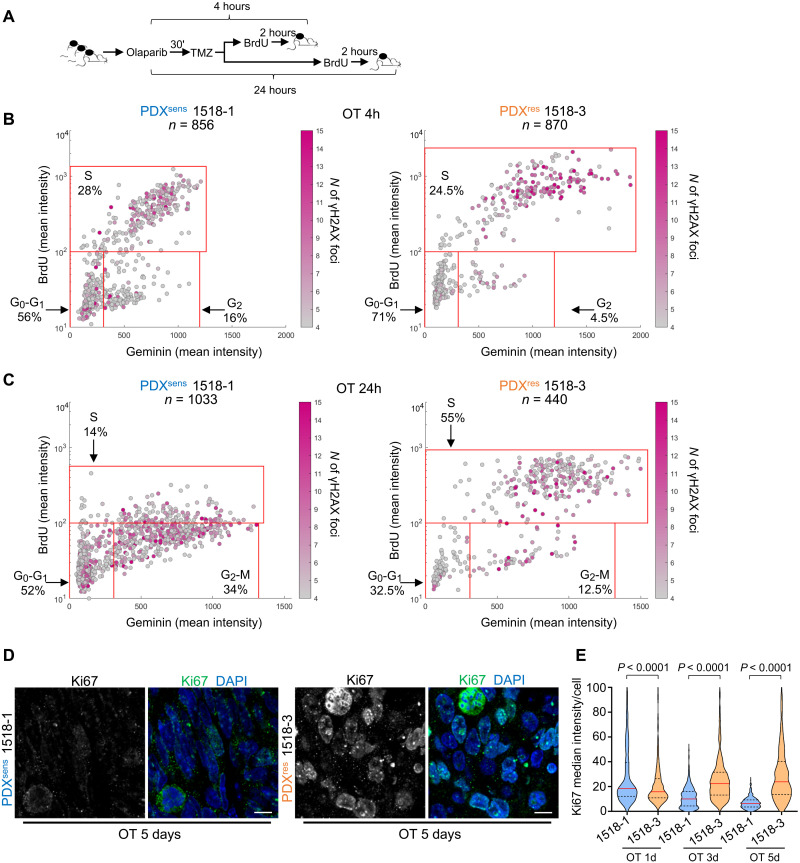
To test the hypothesis that cell cycle distribution contributes to acquired resistance to OT, we measured the susceptibility of cells in different phases to DNA damage in both sensitive and resistant models. Single doses of olaparib and TMZ were administered in vivo to both PDXsens 1518-1 and PDXres 1518-3, and tumors were resected either 4 or 24 hours after treatment to reveal the immediate and delayed cellular effects of OT. After treatment and 2 hours before resection, bromodeoxyuridine (BrdU) was injected intraperitoneally to label S-phase cells (Fig. 4A). Simultaneous immunofluorescence detection of BrdU, GEMININ, and γH2AX revealed whether OT caused DSBs (γH2AX+) in cells in G0-G1, S phase, or G2-M (BrdU− GEMININ−, BrdU+ GEMININ+, or BrdU− GEMININ+, respectively) (fig. S3). In PDXsens 1518-1, γH2AX foci were present in cells at all stages of division 4 hours after OT (Fig. 4B, left). In contrast, in PDXres 1518-3, nearly all the DNA damage was concentrated in the S-phase population, with little γH2AX accumulation in G1-G0 or G2-M cells (Fig. 4B, right). At 24 hours after treatment, the effects of OT on cell cycle distribution could be observed. In PDXsens 1518-1, there was a marked decrease in BrdU incorporation among GEMININ+ cells, suggesting that damaged cells either accumulated in G2 or failed to complete DNA replication. In contrast, the cell cycle profile of PDXres 1518-3 was largely unaffected by OT except for an increase in S-phase cells (Fig. 4C). This differential effect of OT on cell cycle distribution was magnified in longer time courses with daily administration of OT. In PDXsens 1518-1, the Ki-67 median intensity per cell decreased progressively with daily OT, and after 5 days of treatment, almost all tumor cells were nonproliferative. In contrast, in PDXres 1518-3, the Ki-67 intensity was largely unaffected after 1 day and increased after 3 and 5 days of treatment (Fig. 4, D and E). Together, these results suggest that PDXres 1518-3 cells in either G0 or G1 are resistant to OT and can promote tumor growth during and after treatment.
Fig. 4. PDXres 1518-3 cells accumulate less DNA damage and continue to replicate DNA during OT treatment.
(A) Schematic representation of time-course experiment. (B and C) BrdU (S phase marker), GEMININ (S-G2-M phase marker), and γH2AX (DNA damage marker) were immunodetected on tumor tissue sections of the indicated models and nuclei detected by DAPI (representative images are shown in fig. S3). Plots show the mean fluorescence intensity, and each cell is represented by a dot. γH2AX foci number in each cell is indicated by the magenta color intensity. Cells with ≤4 γH2AX foci were considered undamaged. Percentage of cells in the specific cell cycle phase is shown, and n indicates the number of analyzed cells. BrdU-GEMININ–positive cells are in S phase, BrdU-negative and GEMININ-positive cells are in G2-M phase, and BrdU-GEMININ–negative cells are in G1 phase. (D) Ki67 (cell marker for proliferative state) was immunodetected on xenograft tissue sections, and nuclei were detected by DAPI. Scale bars, 10 μm. (E) Truncated violin plot showing Ki67 median fluorescence intensity per cell, and median values are indicated by red horizontal lines and quartiles by black dashed lines. Data from two independent experiments are shown, and statistical analysis was performed with two-tailed Mann-Whitney test. Apoptotic cells were identified by TUNEL assay (staining not shown) and excluded from the quantification. Numbers of analyzed cells are as follows: 1518-1, OT-1d n = 628, OT-3d n = 488, and OT-5d n = 385; 1518-3, OT-1d n = 563, OT-3d n = 321, and OT-5d n = 536.
Mechanisms of acquired resistance to OT are different between patients
We used similar assays to quantify OT-induced DNA damage and cell cycle profiles in the MGH1528 serial models. Unlike the MGH1518 models, comet assays showed no difference in DNA damage generation including OT- and γ-irradiation–induced DSBs between PDXsens 1528-1 and PDXres 1528-2 (Fig. 5A and fig. S4A), and like PDXres 1518-3, PDXres 1528-2 manifested resistance to daily OT in vivo (25) with fewer γH2AX foci at all time points (Fig. 5, B and C, and fig. S4B) and no reduction in Ki-67 proliferation index after OT treatment (Fig. 5D and fig. S4C). The effects of acquired resistance on cell cycle distribution in untreated tumors and following OT treatment were also divergent in MGH1528 models. Unlike PDXres 1518-3, there was no shift toward G1-G0 in PDXres 1528-2 cells relative to PDXsens 1528-1 (figs. S4, D to F, and S5), and OT treatment induced far fewer γH2AX foci regardless of cell cycle phase (Fig. 5, E and F). Together, these results suggest that PDXres 1528-2 has acquired OT chemoresistance via mechanisms that are different from that of PDXres 1518-3.
Fig. 5. PDXres 1528-2 has a different acquired chemoresistance mechanism to OT treatment.
(A) DNA damage was assessed using comet assay as described in Fig. 2 (B and C). (B) Graph showing the percentage of cells with more than four γH2AX foci per cell in the indicated models and time points. Numbers represent mean percentages. Data are from two independent experiments. SEM bars are shown in black. (C) Truncated violin plot showing γH2AX foci number per cell, and median values are indicated by red horizontal lines and quartiles by black dashed lines. γH2AX was quantified in the following number of cells: 1528-1, NT n = 3530, OT 1d n = 3678, OT 3d n = 3592, and OT 5d n = 2490; 1528-2, NT n = 3050, OT 1d n = 3545, OT 3d n = 3814, and OT 5d n = 3186. (D) Truncated violin plot showing Ki67 median fluorescence intensity per cell, and median values are indicated by red horizontal lines and quartiles by black dashed lines. Ki67 was quantified in the following number of cells: 1528-1, NT n = 462, OT 1d n = 692, OT 3d n = 848, and OT 5d n = 547; 1528-2, NT n = 532, OT 1d n = 573, OT 3d n = 571, and OT 5d n = 769. (B to D) Apoptotic cells were identified by TUNEL assay and excluded from the quantification. Data from two independent experiments are shown. (B) Error bars in black. (C and D) Statistical analysis was performed with two-tailed Mann-Whitney test. (E and F) DNA damage and cell cycle analysis of PDXsens 1528-1 and PDXres 1528-2 as presented in Fig. 4 (B and C).
TLS inhibition synergizes with OT and induces cell death in resistant SCLC cells
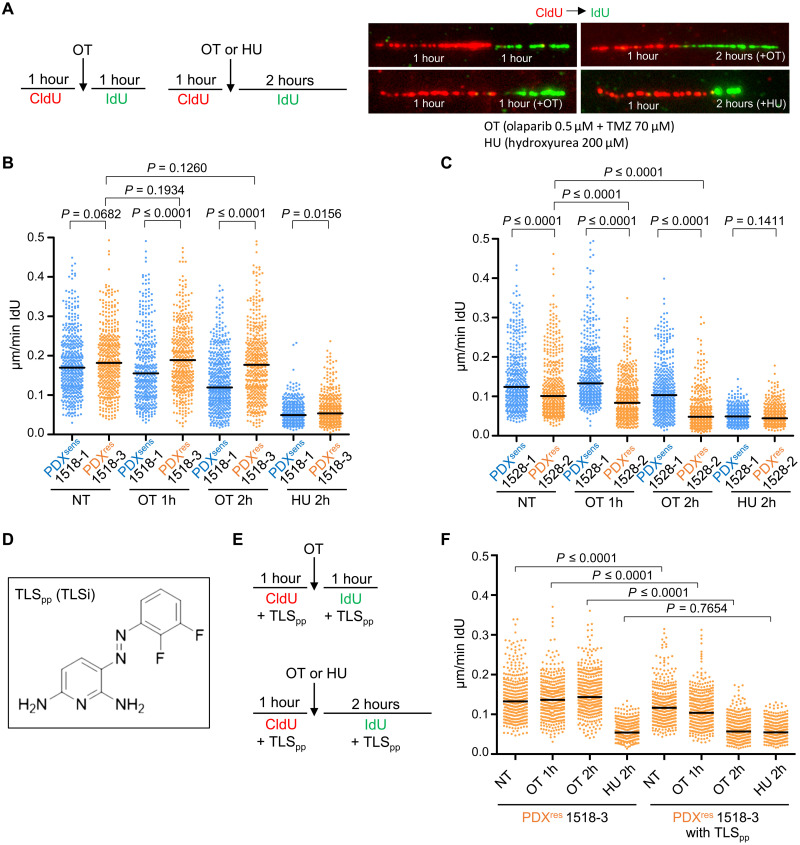
In addition to inhibiting DNA damage repair, PARP inhibition by olaparib can prevent the release of PARP1 protein from TMZ-damaged DNA (40). These trapped PARP1 complexes may be particularly disruptive to DNA replication, causing replication fork stalling and ultimate collapse (40, 41). In both pre-OT PDX models, the fraction of cells undergoing DNA replication declined after a single dose of OT. The change was modest in PDXsens 1528-1 and marked in PDXsens 1518-1. In contrast, this fraction was stable or increased in the postrelapse models (Figs. 4, B and C, and 5, E and F), suggesting a difference in DNA replication kinetics and fork stability. We investigated DNA replication kinetics in the absence or presence of OT by performing DNA fiber assays on PDX cells grown in short-term cultures (Fig. 6A). Cells were preincubated with the thymidine analog 5-chloro-2′-deoxyuridine (CldU) to label active replication forks and, at the starting time point, were switched to a second thymidine analog, 5-iodo-2′-deoxyuridine (IdU). For single-transition CldU-IdU fibers, the length of the IdU tract was divided by the IdU incubation time to yield replication fork speed. In untreated MGH1518 models, replication fork speeds were similar, but they diverged in the presence of OT, with progressive deceleration observed in PDXsens 1518-1 cells and virtually no change in PDXres 1518-3 cells (Fig. 6B). A different relationship was observed in MGH1528 models, as baseline replication fork speeds were slower in PDXres 1528-2 than in PDXsens 1528-1, but OT induced fork deceleration in both models (Fig. 6C). This again highlights the differences in mechanism of acquired resistance between these two patients. To determine the specificity of these replication fork properties, DNA fiber assays were performed in the presence of low-dose hydroxyurea (HU), which reversibly inhibits DNA replication through deoxynucleotide triphosphate deprivation but does not directly induce DNA damage. HU slowed replication forks equivalently in all four models, including PDXres 1518-3. This shows that PDXres 1518-3 replication forks are capable of deceleration, but they are not affected by OT.
Fig. 6. DNA replication forks do not stall/collapse in the presence of OT in PDXres 1518-3 cells, and inhibition of TLS can revert this phenomenon.
(A and E) Schematic showing the experimental setup. Incorporated thymidine analogs CldU (5-chloro-2′-deoxyuridine) and IdU (5-iodo-2′-deoxyuridine) were labeled with two different fluorophores, red and green, respectively, in the absence of drug or during OT or HU treatment. (B, C, and F) Quantification of DNA replication fork speed under unchallenged condition or in the presence of OT or HU. IdU tracts were quantified, and newly fired origins or terminations (tracts with only one label) and replicon merging (second label tract surrounded by two first label tracts) were excluded from the quantification. Each dot represents one fiber, and fibers were quantified from two independent experiments. Median values are indicated by black horizontal lines, and NT indicates not-treated samples. Statistical analysis was performed with two-tailed Mann-Whitney test. Numbers of quantified fibers are as follows: 1518-1, NT n = 460, OT 1h n = 394, OT 2h n = 486, and HU 2h n = 409; 1518-3, NT n = 443, OT 1h n = 416, OT 2h n = 448, and HU 2h n = 410; 1528-1, NT n = 431, OT 1h n = 434, OT 2h n = 428, and HU 2h n = 407; 1528-2, NT n = 434, OT 1h n = 442, OT 2h n = 454, and HU 2h n = 409. (F) Numbers of quantified fibers are: 1518-3, NT n = 379, OT 1h n = 405, OT 2h n = 407, and HU 2h n = 407; 1518-3 with TLSpp, NT n = 391, OT 1h n = 389, OT 2h n = 441, and HU 2h n = 408. (D) Chemical structure of the novel TLS inhibitor (TLSpp).
Theoretically, replication forks in PDXres 1518-3 could be insensitive to OT because no damage is induced, but alkaline comet assays of OT-treated cells show that this is not the case (Fig. 2B). Alternatively, replication forks may temporarily bypass OT-induced damage through tolerance mechanisms such as TLS, in which high-fidelity replicative DNA polymerases are replaced with lower-fidelity TLS polymerases that can bypass DNA lesions (42, 43). Supporting this possibility, among 30 PDX models of SCLC, PDXres 1518-3 demonstrated the highest expression of PDIP38 (POLDIP2) (44, 45) and POLD2 (46), which may mediate the replacement of replicative polymerases with TLS polymerases. PDXres 1518-3 also displays low expression of p21 (CDKN1A), which can suppress TLS through direct interaction with proliferating cell nuclear antigen (47, 48) [fig. S6; (25)]. To test whether preserved replication fork processivity despite OT is dependent on TLS, we repeated the DNA fiber assays in the presence of a novel phenazopyridine-based TLS inhibitor, hereafter called TLSpp, that blocks the interaction of REV1 with TLS polymerases (Fig. 6D) (49, 50). Treatment with TLSpp alone modestly slowed PDXres 1518-3 replication forks, but in combination with OT, it caused marked fork deceleration to speeds previously observed only with HU (Fig. 6, E and F).
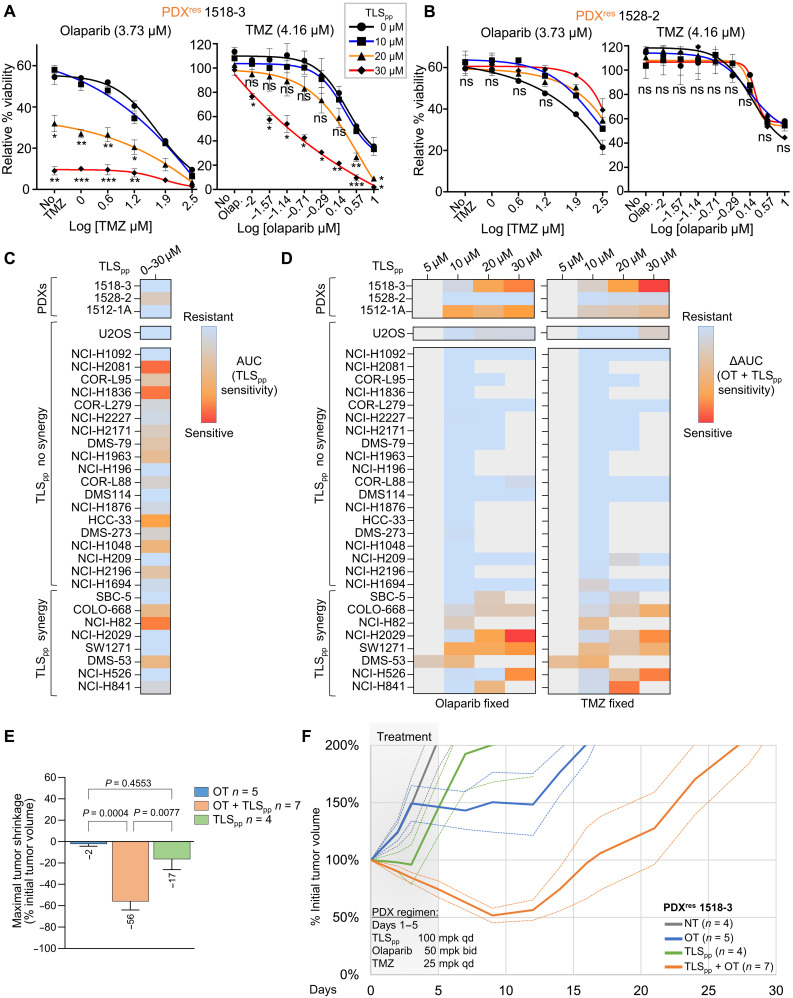
We then tested the capacity of TLS inhibition to resensitize SCLC models to OT. TLS inhibition sensitized PDXres 1518-3 cells to OT in vitro (Fig. 7A and fig. S7A) but did not sensitize PDXres 1528-2 cells (Fig. 7B and fig. S7B), consistent with our hypothesis that a different mechanism accounts for acquired resistance in PDXres 1528-2. Moreover, JH-RE-06, a structurally distinct and independently described TLS inhibitor that targets REV1 by binding the REV7 interaction surface and therefore inhibits a different protein-protein interaction (PPI), also sensitized PDXres 1518-3 to OT (fig. S7, C and D) (51, 52). We further tested the OT + TLSpp combination in 27 cell lines established from patients with SCLC and compared with an osteosarcoma cell line that is known to be insensitive to TLS activity inhibition, U2OS (53). Whereas the OT sensitivity of U2OS cells did not change by inhibiting TLS, SBC-5, COLO-668, NCI-H82, NCI-H2029, SW1271, DMS53, NCI-H526, and NCI-H841 cell lines were significantly more sensitive to the combination of OT + TLSpp than to OT alone (Fig. 7, C and D, and fig. S8). In NCI-H2029 and DMS53, this synergy was evident at multiple drug concentrations. TLSpp did not seem to synergize in any of the other SCLC lines. However, the in vitro sensitivity to the TLSpp alone observed in some lines (Fig. 7C) prevented the evaluation of higher TLSpp concentrations. These effects of TLSpp required REV1; the ability of TLSpp to increase sensitivity of SW1271 SCLC cell line to OT was abolished by REV1 knockdown. SCLC lines that are classified as NE subtype were significantly more sensitive to the treatment with TLSpp alone as compared to non-NE lines. However, NE subtype did not correlate with sensitivity to the OT + TLSpp combination (fig. S9A). Moreover, we found that cell line sensitivity to the OT + TLSpp combination negatively correlated with sensitivity to OT (fig. S9B).
Fig. 7. TLS inhibitor synergizes with OT and induces cell death in PDXres 1518-3 SCLC cells.
(A and B) Cell survival assays with either PDXres 1518-3 (A) or PDXres 1528-2 (B) treated with OT at indicated concentrations, in the presence of TLSpp (blue, orange, and red curves) or without TLSpp (black curve). Left: Cell viability when treated with a fixed concentration of olaparib and increasing concentration of TMZ. Right: Cell viability when treated with a fixed concentration of TMZ and increasing concentration of olaparib. Data represent the mean percentages of survival from at least two independent experiments. Statistical analysis was calculated with one-way ANOVA Tukey multiple comparisons, and all P values are described in the “Statistical analysis” section. (C and D) Heatmaps showing the sensitivity of SCLC cells to TLSpp single-agent treatment (C) and in combination with OT (D). Explanatory figure legends for AUC (area under the curve) and ΔAUC calculations are in fig. S8 (B and C). (E) Bar graph showing tumor shrinkage in PDX models treated with different regimens. Mean and SEM bars are shown, and regimens are indicated in the legend. Statistical analysis was calculated with one-way ANOVA Tukey multiple comparisons, and P values are shown. n indicates the number of xenografts analyzed for each regimen. (F) Solid curves represent the mean % ITV versus time (days) after a single cycle (days 1 to 5) of TLSpp (100 mpk qd), OT (Ola 50 mpk bid + TMZ 25 mpk qd), or TLSpp + OT. Dashed curves represent tumor volume curve ± SEM. Numbers of replicate xenografts per regimen are represented parenthetically. Grayed area represents the treatment cycle.
It is possible that in vivo additional SCLC tumor cells and patients would be highly sensitive to the OT + TLSpp combination. Therefore, we tested TLSpp with PDXres 1518-3 in vivo with or without concurrent OT. The results of this experiment confirmed that TLSpp increases the sensitivity of PDXres 1518-3 tumors to OT. TLSpp had only a minor effect when used as monotherapy (Fig. 7, E and F, and fig. S9C) but caused significant tumor shrinkage and extended time to relapse once the drug treatment ended. We also found that TLSpp increased the sensitivity to OT both in vitro and in vivo in a second PDX model derived from a resistant patient, 1512-1A (fig. S9, D to F). Together, these results strongly suggest that resistance to OT in SCLC can be acquired via up-regulation and/or hyperactivation of TLS and that, in some cases, sensitivity to OT can be partially restored by addition of a TLS inhibitor. We found that PDXres1518-3 tumors that grew back after OT + TLSpp treatment remained sensitive to the drug combination (fig. S9G). This suggests that regrowth of the PDXres 1518-3 tumor after the first cycle was likely due to incomplete cell killing rather than acquisition of chemoresistance to TLSpp. This suggests that it might be possible to further improve the in vivo efficiency of this novel TLSpp agent.
DISCUSSION
Over the past 40 years, SCLC has been treated almost exclusively with DNA-damaging agents. Although these regimens are effective initially, most patients with SCLC succumb to treatment failure due to acquired resistance. Although broad molecular trends in chemoresistant SCLC have been observed in preclinical models, such as loss of NE differentiation or up-regulation of WNT signaling, no mechanism of resistance has been identified directly in a sample obtained from a patient following relapse (54). We previously reported a high response rate to OT for patients with relapsed SCLC, and for two patients on the trial who responded to OT, we derived pretreatment and postrelapse PDX models that faithfully recapitulated clinical response and resistance (25). Here, we investigate the molecular drivers of acquired resistance to OT in patients with SCLC using these serial PDX models. For one patient, MGH1518, we found that following relapse, TLS increases tolerance of OT-induced damage during DNA replication, and we tested a novel TLS inhibitor to overcome this tolerance. In the same model, we found that cells in either G0 or G1 have also become resistant to OT, suggesting at least one additional mechanism of resistance (fig. S9H). In a second patient, MGH1528, the acquired resistance mechanisms differ from MGH1518, indicating the presence of both inter- and intratumoral heterogeneity.
In HR-deficient breast and ovarian cancers, drivers of acquired resistance to PARP inhibitors such as BRCA1/2 reversions or other mutations that restore HR can be recognized because the basis for initial drug sensitivity is known (55). In contrast, the basis for SCLC sensitivity to OT or other DNA-damaging agents has not been elucidated in sufficient detail to focus on a single pathway or set of genes. Without a focused list of candidates, a genome-wide comparison of pretreatment and postrelapse PDX models yields many possible candidates. Prior therapy with OT compounds this challenge by increasing the mutational burden 10-fold in the postrelapse models. Furthermore, the drivers of OT resistance might not be genomic, as a study of chemo-naïve SCLC PDX models treated to resistance with EP revealed alterations that were primarily epigenetic (19). After ruling out expression changes in previously reported candidate genes, such as loss of SLFN11 or gain of MGMT or drug efflux pumps (19, 29, 30, 33, 34), we adopted a methodical, first-principles approach to investigating acquired resistance to OT, comparing DNA damage induction, repair, and tolerance in the pretreatment and postrelapse models. This approach would not have been possible with fixed or frozen clinical samples because it requires the ability to assay cellular responses to OT.
In both models of OT-resistant SCLC, the biochemical efficacy and initial induction of DNA damage were preserved, but their responses to DNA damage diverged thereafter. For models from patient MGH1528, OT treatment resulted in the same initial number of DSBs before and after relapse, but for patient MGH1518 following relapse, the OT-induced lesions were no longer converted to DSBs, suggesting that mechanisms of acquired resistance differed between patients. Intertumoral heterogeneity has been found in resistance mechanisms to inhibitors of mitogenic pathways, such as EGFR inhibitors, and diverse resistance mechanisms may even be found within synchronous subclones of the same cancer (15, 56–59). However, we were surprised to find evidence of multiple adaptations to OT within the same cancer cell (fig. S9H). S-phase cells in PDXres 1518-3 were resistant to OT, and replication fork kinetics were no longer affected by treatment, suggesting increased tolerance of DNA damage. The underlying mechanism of tolerance, TLS, is specific to S-phase cells and may account for the failure of SSBs to convert to DSBs (60, 61). In addition, TLS may accelerate S phase, increasing the proportion of tumor cells in G0 or G1 despite the faster xenograft growth rate (Fig. 3). Since OT no longer slows S phase under TLS, treatment shifts the balance even further toward G0-G1 in PDXres 1518-3 tumors, and these cells are resistant to OT through a yet-unidentified mechanism (Fig. 4). It is possible that the uncontrolled proliferation of the resistant cells may activate a checkpoint in G1 that is independent of p53, pRB, and p21. The identity of this checkpoint is currently unknown, but one can imagine that cell cycle progression has become uncoupled from the production of a rate-limiting factor. In theory, the limiting factor could be any component that is needed for rapid growth (protein, lipid, or metabolite). A second possibility is that PDXres 1518-3 tumor cells might not be arrested in G1, but they may spend proportionally more time in G1 than in other stages of the cell cycle because they progress faster through S-G2-M. This model highlights an important difference between acquired resistance to inhibitors of mitogenic pathways and regimens such as OT that induce DNA damage and inhibit repair. Both the type of damage that results from OT and the repair pathway used by the cell may change with each phase of cell division, and as PDXres 1518-3 illustrates, fully acquired resistance may involve adaptations in multiple phases (fig. S9H).
Collision of replication forks with DNA lesions may compound OT-induced damage. TLS facilitates bypass of these lesions to prevent cell death. Tolerance of DNA damage through TLS has been implicated in resistance to TMZ as well as cisplatin, doxorubicin, and cyclophosphamide (28, 62–64). Changes in gene expression in PDXres 1518-3, such as suppression of p21 and increased POLDIP2, suggested that TLS may be favored in this model, but DNA fiber assays provided direct evidence of lesion bypass, as replication fork speeds were preserved in the presence of OT. This hypothesis was further supported by a novel small-molecule inhibitor of TLS, TLSpp, which disrupts the interaction of REV1 with other TLS DNA polymerases (49, 50). In PDXres 1518-3, TLSpp combined with OT slows DNA replication forks and induces cell death, whereas in PDXres 1528-2, where there was no evidence of OT tolerance via TLS, TLSpp had little effect.
Although up-regulation of TLS was first identified in PDXres 1518-3, experiments with a panel of SCLC cell line suggest that TLS plays a significant role in approximately 30% of lines and may be active to a lesser extent in many more. Together, these observations suggest that activation of TLS is not an unusual event in SCLC. It has been suggested that TLS activity may cause resistance to DNA replication gap-inducing therapies, such as PARP1 or ATR inhibition (65). Therefore, the combination of translesion DNA synthesis inhibitor (TLSi) with gap-inducing chemotherapies might markedly improve the efficacy of these types of treatments. Discovery of the underlying genetic or epigenetic drivers of TLS in this model will aid with identification of biomarkers to further deploy TLS inhibitors in SCLC. Notably, PDXres 1518-3 is resistant to both OT and first-line EP, and TLS can also confer resistance to cisplatin in models of ovarian cancer, NSCLC, and lymphoma (27, 28, 66). In some patients with relapsed SCLC, TLS may account for the acquired cross-resistance that causes ultimate treatment failure.
In contrast to PDXres 1518-3, we saw no sensitivity to TLSpp in PDXres 1528-2. In this model, γH2AX foci numbers were consistently reduced, and DNA replication forks were slower than in sensitive cells, even in the absence of DNA damage. It is possible that fork reversal occurs more frequently in PDXres 1528-2, and this facilitates repair of damage. Alternatively, replication forks may be more stable in PDXres 1528-2, increasing their tolerance of OT-induced DNA damage as it has been suggested in other systems (67), or resistant cells might have an enhanced ability to repair OT-induced damage independently of replication fork properties. In either case, further studies will be required to identify the precise molecular events that mediate acquired chemoresistance in this tumor. We note that, to date, we have been unable to identify a transcriptional signature associated with the effects of TLSpp in the SCLC models and cell lines. In the analysis of scRNA-seq data, cell clustering seemed to be driven first by patient of origin and second by cell cycle position. Gene set enrichment analysis confirmed the results previously obtained with bulk RNA-seq of these serial models (25). The most notable transcriptional change in PDXres 1518-3 is the up-regulation of MYC. It has been suggested that MYC can repress the transcription of p21 even in the absence of p53 (68, 69), and this may increase TLS in PDXres 1518-3 cells. Although MYC expression correlates with resistance to OT in SCLC PDX models (25), in these experiments, we saw no clear correlation between MYC expression and sensitivity to TLSpp, and it remains unclear how frequently MYC activation leads to a dependence on TLS activity.
To the best of our knowledge, up-regulation of TLS represents the first specific molecular mechanism to be identified in tumor cells directly derived from a patient with SCLC that confers acquired resistance to DNA damage. Identification of TLS required comparison of live tumor models derived from a therapy-responsive patient before treatment and again at progression. This approach to serial tumor sampling has revolutionized the management of NSCLC, as mechanisms of acquired resistance can be diagnosed in the clinic and then counteracted or bypassed. Although rebiopsy is not part of the current clinical paradigm for SCLC, the same approach could be applied provided that resistance mechanisms were known and readily identifiable with clinical biomarkers and that therapies to address resistance were developed. Interrogation of additional pairs of serial models will be crucial to reach this goal and capture the spectrum of resistance mechanisms that render SCLC refractory to treatment.
MATERIALS AND METHODS
Experimental design
The objective of this study was to identify and characterize the molecular mechanisms of acquired chemoresistance to the experimental therapy OT in PDX models of SCLC. Controlled laboratory experiments were conducted in tumor-harboring NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice, tumor cells isolated from freshly dissected tumors, or established cultured cell lines. For the in vivo experiments, sample sizes were determined on the basis of the observed variation in tumor progression from previous studies. For the in vitro experiments, sample sizes were calculated on the basis of the observed result variability obtained by performing the same experimental assays. We minimized physiological variation using mice of the same sex and age and using cell lines that were cultured under the same conditions and used at a similar passage number for each individual experiment. Final end points and collection of tumor material were based on the tumor size and/or animal health conditions. For all experiments, animals were allocated randomly across different treatment groups, and tumor volume measurements were performed in a blinded fashion, in which the investigator was unaware of the experimental condition. No data were excluded from this study, and each experiment was repeated at least two times.
PDX model WGS and analysis
We sequenced PDX-derived tumor and matched normal (peripheral blood) from two cases representing six samples of SCLC. PDX tumor tissue was dissociated, and tumor DNA and peripheral blood control DNA were extracted using the Qiagen DNeasy Blood & Tissue Kit (ref #69506). DNA was sequenced by Novogene to achieve between 48 and 57.7 average coverage (mean, 51.99) in tumors and between 27.4 and 30.2 average coverage (mean, 28.8) in normal samples. Reads were aligned to the human genome (build hg19) using bwa-mem (70) then processed through Picard tools to mark duplicates and GATK Base Recalibrator (GATK4 Broad Institute) to recalibrate base quality scores. To avoid mapping mouse reads, we further filtered the resulting bam files for only high-quality alignments (-nm 3, -as 50, -mg 30). Indel realignment was performed with GATK standard pipeline against Mills_and1000G_gold_standard.indels.no_chr.hg19.sites.vcf (GATK4 Broad Institute). Somatic mutations were called using MuTect (v1.1.7) (71), and insertion/deletions were called using Strelka (v2.6.6) (72). Called variants were further filtered through a panel of normals from the Broad Institute. To further eliminate mutations potentially due to mouse contamination, variants were filtered through mapexr (73). Mutations were annotated using snpEff (74). Mutational signatures were calculated by non-negative matrix fractionization in MATLAB (v2017b). Further analysis and figure preparations were performed using R (v4.0.2). Copy number variant calls and segmentation were generated from WGS data by using FACETS as previously described (75, 76).
PDX scRNA-seq methods
Sample dissociation
Freshly isolated tumors were extracted and dissociated using the human tumor dissociation kit (Miltenyi Biotec, catalog no. 130-095-929) with custom modifications. Tissue was placed into a 1.5-ml Eppendorf tube containing 420 μl of Dulbecco’s modified Eagle’s medium (DMEM) without fetal calf serum (FCS), 42 μl of enzyme H, 21 μl of enzyme R, and 5 μl of enzyme A (provided with the kit). Tissue was minced into small pieces using surgical scissors, and the tube was topped off with an additional 512 μl of DMEM to a total volume of 1 ml. The tissue was then incubated for 15 min in a thermomixer (Eppendorf, F1.5) at 37°C, 350 rpm. After the dissociation, cells were filtered through a 50-μm filter (Sysmex, catalog no. 04-004-2327) followed by an additional mincing step on the filter using a 1-ml syringe plunger and washes with 5 ml of media with 10% heat-inactivated FCS. Red blood cell lysis was performed using ACK buffer (Gibco, catalog no. A1049201), followed by a second filtration using a 30-μm filter (Sysmex, catalog no. 04-004-2326). Next, cells were washed with cold 1× phosphate-buffered saline (PBS) containing 1.5% FCS, spun down at 1500 rpm, 4°C for 5 min, resuspended, and counted for yield and viability with trypan blue using a manual hemocytometer (Bright-line, catalog no. 1492).
Cell fluorescence-activated cell sorting
Before staining cells with the live/dead Zombie Violet Dye (BioLegend, 423114), cells were washed three times in cold 1× PBS (without FCS). Next, Zombie violet dye was added, and cells were incubated for 15 min at room temperature according to the manufacturer’s guidelines. Cell surface labeling was then performed for 30 min at 4°C using standard protocol with the following antibody panel to identify and exclude mouse immune and stromal populations: human TrueStain FcX (BioLegend, catalog no. 422302); mouse TrueStain FcX (BioLegend, catalog no. 101320); APC anti-mouse CD45.1 (BioLegend, catalog no. 110714); APC anti-mouse CD45.2 (BioLegend, catalog no. 109814); fluorescein isothiocyanate (FITC) anti-mouse H-2Db (BioLegend, catalog no. 114606); FITC anti-mouse H-2Kb (BioLegend, catalog no. 116506); FITC anti-mouse H-2Kd (BioLegend, catalog no. 116606); FITC anti-mouse H-2Dd (BioLegend, catalog no. 114706); BV605 anti-human HLA-A,B,C (BioLegend, catalog no. 311432); and phycoerythrin (PE) anti-human EpCAM (BioLegend, catalog no. 324206). Sorting of single live SCLC cells (Zombielow, mCD45.1/2−, mH2-Db/Kb/Kd/Dd−) was performed using a Sony MA900 cell sorter. Cells were sorted into a 15-ml tube containing DMEM with 10% FCS. After sorting, tubes with sorted cells were vortexed briefly, spun down at 1500 rpm, 4°C for 5 min, resuspended, and counted for yield.
Single-cell RNA sequencing
Single-cell RNA libraries were generated using the 10x Genomics Chromium Single Cell V(D)J Reagent Kit using 5′ v1 chemistry (10x Genomics, catalog no. 1000006) according to the manufacturer’s protocol. Quality control (QC) assessment was performed after generating the complementary DNA and gene expression libraries by evaluating sample concentration using the Qubit dsDNA high sensitivity kit (Invitrogen, catalog no. Q32854) and fragment size using the high sensitivity BioA DNA kit (Agilent, catalog no. 5067-4626). All libraries that passed QC were sequenced on a NextSeq 500 sequencer (Illumina) using pair-end 38-base reads.
scRNA-seq data generation and filtration
FASTQ files were aligned to both the Homo sapiens genome assembly GRCh37 (hg19 v3.0.0) and the Mus musculus genome assembly GRCm38 (mm10 v3.0.0) from the Genome Reference Consortium using cellranger 3.1.0. Following individual alignments, the human and mouse BAM files were processed by XenofilteR (77) to produce a filtered human BAM file free of contamination from mouse reads. The filtered BAMs were converted back to FASTQ files using bamtofastq 1.2.0 from 10x Genomics and realigned to hg19 using cellranger. Filtered feature barcode matrices from cellranger were imported into R 4.1.0 using Seurat 4.0.2. Additional filtration was applied to exclude cells with fewer than 1500 or greater than 5500 genes, fewer than 4000 or greater than 35,000 RNA counts, and greater than 20% mitochondrial reads.
Dimensionality reduction and clustering
Data were normalized using the NormalizeData function, and expression values were scaled using the ScaleData function in Seurat. Principal components were selected for building the neighborhood graph, Uniform Manifold Approximation and Projection, and t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction according to the elbow plot method. Clustering was performed with the Louvain algorithm with an initial resolution of 1.0 for all samples combined or 0.7 for samples from one patient.
Differential expression and gene set enrichment analyses
Differential gene expression analysis was conducted in R using DESeq2. Gene set enrichment analysis was performed using the fgsea package using the DESeq2 test statistic as input gene ranks. Pathways were selected from mSigDB using the H:Hallmark gene sets.
Cell cycle scoring analyses
Cell cycle phase proportion estimates were calculated using the CellCycleScoring function in Seurat, which assigns each cell an S and G2-M score and subsequently assigns the cell to S (high S score), G2-M (high G2-M score), or G1 (low S and G2-M scores). Cell cycle estimates were performed using all cells derived from the same patient.
Drugs, inhibitors, and treatments
Olaparib (AZD2281, M1664, Abmole) was dissolved in dimethyl sulfoxide (DMSO) for in vitro experiments. In vivo dosing was performed with oral gavage, and the formulation for a single dose was 1.5 mg of olaparib in 200 μl [175 μl of 10% HPβCD in PBS (pH 7.2) + 25 μl of olaparib stock (60 mg/ml) in DMSO]. TMZ (M2129, Abmole) was dissolved in DMSO for in vitro experiments. In vivo dosing was performed with oral gavage, and the formulation for a single dose was 0.75 mg of TMZ in 200 μl of 1% O-carboxymethylcellulose in water. BrdU (B5002, Sigma-Aldrich) was dissolved in PBS at a concentration of 25 mg/ml; it was dosed by intraperitoneal injection of a 400-μl dose. Both JH-RE-06 (S8850, Selleckchem) and TLSpp (a novel phenazopyridine-based TLS inhibitor produced and gifted by M.K.H.) were dissolved in DMSO for in vitro experiments. JH-RE-06 and TLSpp are completely different structurally and that they inhibit REV1/TLS activity in slightly different ways. TLSpp specifically disrupts the PPI between REV1-C-terminal (REV1-CT) and the RIR domains of other TLS polymerases. RIR motifs are present in “inserter” and “extender” polymerases so binding to REV1-CT at this interface can disrupt multiple TLS PPIs responsible for multiple different aspects of TLS. JH-RE-06 inhibits REV1/TLS activity through an unexpected and unpredictable way. It binds to REV1-CT at the REV7-binding interface and induces dimerization with another REV1-CT domain, which prevents it from binding to REV7. Thus, both compounds bind to REV1-CT and inhibit PPIs between this domain and other TLS polymerases, but they inhibit different PPIs. γ-Irradiator used for comet assay experiments is a MARK-1 (J. L. Shepherd and Associates, San Fernando, CA) equipped with cesium-137 as source of radiations.
PDX model generation, treatment, and results evaluation
All tissue and blood samples from patients were collected as per Institutional Review Board–approved protocols with written informed consent from the patients and in accordance with the Declaration of Helsinki. All mouse studies were conducted through Institutional Animal Care and Use Committee–approved animal protocols in accordance with Massachusetts General Hospital institutional guidelines. The number next to “MGH” refers to the specific patient, and the numbers after the dash refer to the time point during patient treatment when the sample was collected. PDX models used for this study were initially generated in our laboratory as described previously (21). Scalpel-dissected xenograft fragments were either immediately implanted into the right flank of an NSG (Jackson Laboratories) mouse for passaging. After tumor emergence, palpable tumors were measured with electronic calipers weekly until tumors exceeded 1500 mm3, at which point, animals were euthanized, and tumors were resected. Tumor volume was estimated by using the formula: tumor volume = [(tumor length) × (tumor width2)] × 0.52. For estimation of the growth rate of a single xenograft, serial volume measurements were performed on tumors between 50 and 1500 mm3, and exponential regression was performed on these tumor-volume series. Model growth rates were extrapolated from the average of individual xenograft growth coefficients for 9 to 10 untreated xenografts. Resected tumors could be cryopreserved in cryomedium [RPMI + 10% fetal bovine serum (FBS) + 4 mM l-glutamine + penicillin-streptomycin (100 U/ml) + heparin (500 U/ml) + 10% DMSO] for later passaging, fixed in 3.7% formaldehyde for immunofluorescence studies, or fresh-frozen in liquid nitrogen for Western blot analysis. MGH1528-2 post-5dOT model was derived from an MGH1528-2 PDX (resistant to OT) that had been treated with OT for 5 days. Since this model was used for scRNA-seq, OT sensitivity studies in MGH1528-2 post-5dOT PDX model were performed to reconfirm the resistance of this model (fig. S7A). Both MGH1528-2 post-5dOT and MGH1518-3 PDX model treatment studies were initiated at xenograft volumes = 300 to 600 mm3 for at least four mice per model per treatment arm, and tumors were measured two times weekly. Drugs were used as follows: olaparib (50 mg/kg, oral gavage) for days 1 to 5, TMZ (25 mg/kg, oral gavage) for days 1 to 5, and TLSpp (100 mg/kg, intraperitoneal injection) for days 1 to 5. Treatment cycle of 5 days was performed only once for the experiments shown in Fig. 7 (E and F) and fig. S5A. One treatment cycle of 5 days followed by 3 weeks with no treatment and a final cycle of 5 days were used for the experiment in fig. S9G. For treatment metrics: response = change in tumor volume between initial tumor volume (ITV) and days 14 to 28 minimum; for end points: tumor volume > 2× ITV or 80 days after start of treatment.
Cell culture and cell lines
U2OS and all SCLC cell lines were obtained between 2017 and 2022 from the Massachusetts General Hospital Center for Molecular Therapeutics, which performs routine authentication by single-nucleotide polymorphism and short tandem repeat analyses and mycoplasma testing. SCLC cells were maintained in RPMI-HITES media + 2% FBS, while DMEM + 5% FBS was used for U2OS, and DMEM F12 + 10% FBS was used for DMS114 and SBC5 cells. All cell lines were passaged for less than 3 months before performing the experiment. All cells were grown in 1% penicillin-streptomycin and 1% l-glutamine at 37°C and 5% CO2.
Tumor tissue preparation and immunofluorescence
Freshly dissected xenograft samples were fixed for 2 hours at room temperature in fixing solution [3.7% formaldehyde and 100 mM NaPO4 (pH 7.4)] and rinsed in 70% ethanol. Dehydration and paraffin embedding were performed by standard method with an automated machine. Tissue sections of 4 μm in thickness were cut with a microtome. Sections were deparaffinized by incubating the sections 2 × 10 min in fresh 100% xylene. Rehydration was performed as follows: 2 × 5 min in 100% ethanol, 5 min in 96% ethanol, 5 min in 80% ethanol, 10 min in 70% ethanol, and 2 × 10 min in water. Antigen retrieval was performed in either citric buffer (pH 6.0) (C9999-100ML, Sigma-Aldrich) or tris-EDTA buffer (pH 9.0) (10064-758, VWR Diagnostic BioSystems) at 98°C for 20 min. Slides were washed in water, and sections were permeabilized for 45 min in 0.2% Triton X in PBS. Sections were blocked for 60 min with 10% goat serum, 0.05% Triton X, and 0.05% Tween 20 in PBS. Primary antibody incubation was carried out overnight in blocking solution at 4°C in a humid chamber followed by 3× washes for 10 min with 0.1% Tween 20 in PBS. Sections were then incubated with secondary antibodies for 90 min at room temperature in a humid chamber. After 3× washes with 0.1% Tween 20 in PBS, sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min and mounted using SlowFade Diamond Antifade Mountant with DAPI (S36964, Invitrogen). Slides were imaged with a Zeiss LSM 710 confocal microscope. For terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining and detection of apoptotic tumor cells, we used the ApopTag Red In Situ Apoptosis Detection Kit (S7165, Millipore) following the manufacturer’s directions. TUNEL was performed after a permeabilization step and followed by section blocking and immunofluorescence protocol as described above. We used the following antibodies: γH2AX (pH 6.0; 1:150; 9718, Cell Signaling Technology; or pH 6.0; 1:200; ab26350, Abcam), Ki67 (pH 6.0; 1:150; ab15580, Abcam), CDT1 (pH 9.0; 1:150; 8064, Cell Signaling Technology), GEMININ (pH 9.0; 1:100; Geminin-L-CE, Leica Microsystems; or pH 6.0; 1:350; 10802-1-AP, Proteintech), BrdU (pH 6.0; 1:100; ab6326, Abcam), and p21 (pH 6.0; 1:50; 2947, Cell Signaling Technology).
Automated image analysis and quantification
Images were analyzed, and fluorescence was quantified with either MATLAB or Cell Profiler (78). In brief, cell nuclei were segmented using a custom-made image processing pipeline that can distinguish them from the background. The pipeline identifies the nuclei based on DAPI staining, their size, and circularity. Nuclei that are close to image borders or that are too close and cannot be individually segmented were automatically removed by the software. Ki67, GEMININ, CDT1, and BrdU fluorescence total intensities were quantified by the software only within the segmented nuclei. For γH2AX staining and quantification, the software identifies the formed foci within the nucleus based on their size and intensity. Apoptotic nuclei are detected by the software based on TUNEL assay and excluded from the quantification.
Western blot
Freshly frozen xenograft tissue samples were lysed in radioimmunoprecipitation assay buffer using a TissueLyzer II (Qiagen) homogenizer at 4°C following the manufacturer’s instructions. SDS–polyacrylamide gel electrophoresis was performed by standard methods, and polyvinylidene difluoride membranes were blocked with bovine serum albumin (BSA) 2.5% and PBS Tween 0.5% for 60 min at room temperature and probed overnight at 4°C with the following antibodies: PAR (1:4000; 4336-BPC-100, Trevigen), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:40,000; MAB374, Millipore), MDR1 (1:200; sc-13131, Santa Cruz Biotechnology), MGMT (1:1000; 2739, Cell Signaling Technology), SLFN11 (1:500; sc-374339, Santa Cruz Biotechnology), p57 (1:500; ab75974, Abcam), p16 (1:2000; ab81278, Abcam) and p21 (1:1000; 2947, Cell Signaling Technology), CYCLIN A2 (1:1000; 4656, Cell Signaling Technology), CYCLIN B1 (1:1000; 12231, Cell Signaling Technology), H3pSer10 (1:1000; 06-570, Millipore), and MYC (1:500, sc-40, Santa Cruz Biotechnology). Membranes were imaged with a Syngene G:BOX, band densitometry was performed using FIJI, and ratio to loading control (GAPDH) was calculated.
Single-cell suspension preparation from PDX tumors
Xenografts were resected and collected in cold RPMI media, manually minced with scalpel, and dissociated using the Human Tumor Dissociation Kit (130-095-929, Miltenyi Biotec) with a gentleMACS Octo Dissociator (Miltenyi Biotec) following the manufacturer’s instructions. Live cells were enriched by centrifugation of 20 ml of cell suspension layered on top of 15 ml of Ficoll-Paque PLUS (17-1440-02, GE Life Sciences) in a 50-ml falcon tube [centrifugation conditions were 2271 rpm (1200 rcf) at 19°C with acceleration/deceleration = 1, for 30 min]. Tumor cells enriched at the interface were collected and depleted of murine cells with anti-mouse immunoglobulin G microbeads (130-104-694, Miltenyi Biotec) following the manufacturer’s instructions.
Cell viability assay
Isolated tumor cells were seeded in 96-well format in HITES media + 2% FBS + 10 μM ROC kinase inhibitor (Y-27632, Selleckchem). Titration of olaparib and TMZ was performed with a D300e digital drug dispenser (Tecan Life Sciences). Viability was assessed after 5 days using CellTiter-Glo 2.0 (G9243, Promega).
DNA fiber assay
DNA fiber analysis was performed as previously described (79) with slight modifications. Briefly, isolated tumor cells were first pulse-labeled with 50 μM CldU (Sigma-Aldrich, catalog no. C6891), washed twice with equilibrated PBS, and then labeled with 100 μM IdU (Sigma-Aldrich, catalog no. I7125) under the conditions specified in the figure legends. Collected cells were resuspended in cold PBS (1 × 106 cells/ml), and 2.5 μl was stretched on a glass slide (tilting the slide at a 15° angle) after mixing and incubating with 7.5 μl of spreading buffer [0.5% SDS, 200 mM tris-HCl (pH 7.4), and 50 mM EDTA] for 2 min. DNA fibers were fixed in methanol:acetic acid (3:1) for 2 min at room temperature and air-dried. Afterward, slides were denatured in 2.5 N HCl for 30 min at room temperature and blocked in 3% BSA/0.05% Tween 20 for 60 min at room temperature. CldU and IdU detection was performed using rat anti-BrdU (1:100; ab6326, Abcam) and mouse anti-BrdU (1:20; BD-347580, BD Biosciences) for 60 min at 37°C followed by Alexa 488 anti-mouse (1:100; 715-545-151, Jackson ImmunoResearch) and Alexa 594 anti-rat (1:100; A-21209, Thermo Fisher Scientific) for 30 min at room temperature. Slides were rinsed three times with 0.05% Tween in PBS, air-dried, and mounted with Prolong Gold at room temperature overnight. Fibers were imaged with an Echo Revolve microscope (40× objective), and fiber length was quantified using FIJI.
Comet assay
Cells were treated for 3 hours under the conditions specified in the figure legends, and comet assay was performed at the end of the treatment or immediately after irradiation. Single-cell gel electrophoresis was carried out by using Trevigen’s comet assay kit (catalog no. 4250-050-K) and following a previously described protocol (80) with modifications. Briefly, tumor cell suspension (5 × 105 cells/ml) and melted LM agarose (at 37°C) were mixed in 1:10 (v/v) ratio. A total of 50 μl of this solution was poured onto the comet slide. Slides were then kept at 4°C in the dark for 10 min and afterward at 37°C for 5 min for better agarose adhesion. The slides were placed in an ice-cold, freshly prepared lysing solution for 60 min at 4°C.
For alkaline comet assay, lysing solution was composed of 1.2 M NaCl, 100 mM EDTA, 0.1% sarcosyl, and 0.26 M NaOH (pH >13). After lysis, slides were immersed in freshly prepared alkaline unwinding-electrophoresis solution [300 mM NaOH and 1 mM EDTA (pH >13)] for 60 min at room temperature in the dark. Subsequently, the slides were placed in an electrophoresis chamber filled with cold alkaline unwinding-electrophoresis solution and run for 30 min at 15 V (1 V for each centimeter of distance between the two electrodes), at 4°C protected from the light. After the run, slides were washed twice in dH2O for 5 min and incubated in 70% ethanol for 5 min at room temperature. Slides were then dried at 37°C until agarose completely disappeared.
For neutral comet assay, lysing solution was composed of 2% sarcosyl, 0.5 M EDTA, and proteinase K (0.4 mg/ml; pH 8.0; 506-PKP, Viagen Biotech). After lysis, slides were immersed in freshly prepared neutral electrophoresis buffer [100 mM tris base and 300 mM sodium acetate (pH 9.0)] for 30 min at 4°C in the dark. Subsequently, the slides were placed in an electrophoresis chamber filled with cold neutral buffer and run for 60 min at 15 V (1 V for each centimeter of distance between the two electrodes), at 4°C protected from the light. After the run, slides were incubated in DNA precipitation solution (1 M ammonium acetate in 95% ethanol) for 30 min at room temperature and then incubated in 70% ethanol for 30 min at room temperature. Slides were dried at 37°C until agarose completely disappeared.
Comets were stained with SYBR Gold (S7563, Invitrogen) for 30 min at room temperature. Then, slides were rinsed in water and completely dried at 37°C. Last, slides were mounted with SlowFade Diamond Antifade Mountant (S36963, Invitrogen). Slides were imaged with an Echo Revolve microscope (10× objective), and pictures were analyzed with Open Comet ImageJ plug-in (81).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9, and specific statistical tests used are indicated in the figure legends. Statistical analysis used for WGS and scRNA-seq is described in the specific paragraph in Materials and Methods. All experiments were performed at least two times, and data shown in the figures either represent one of the replicates or pulled measurements from different replicates. For comparisons between sample pairs, datasets were analyzed by the nonparametric Mann-Whitney U test, and exact P values were reported. For multiple comparisons, datasets were analyzed by one-way analysis of variance (ANOVA) Tukey’s multiple comparison test with a single pooled variance. P values are displayed as not significant (ns) for P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
Acknowledgments
We are grateful to the patients and families who participated in these research studies. We thank D. Haber and S. Maheswaran for CTC isolation from patient samples. We thank the current and former members of the Dyson, Zou, Lan, and Elia laboratories for critical discussions and sharing reagents. We thank J. Minna for thoughtful comments on the manuscript. We thank A. Zheng for help and contribution with the cell line survey.
Funding: This work has been supported by the research fellowship grant from the Deutsche Forschungsgemeinschaft (M.S.), National Cancer Institute grant U01CA220323-A1 (N.J.D.), grant U24CA213274 (N.J.D.; P.I. C. Rudin, MSKCC), Cancer Prevention and Research Institute of Texas (CPRIT) grant RR20007 (B.J.D.), NIH career development award 1K08CA237832 (B.J.D.), a career development awards from the Lung Cancer Research Foundation (B.J.D.), R01CA247232 (M.K.H.), and Norwegian Cancer Society grant 182694 and CCR startup funds (J.F.W.).
Author contributions: Experimental design: M.S. and B.J.D. Data acquisition: M.S., J.Z., E.W., D.T.M., S.P., and T.J.L. Data analysis: M.S., T.J.L., J.F.W., and A.S. Critical materials: M.K.H. Funding acquisition: N.J.D. and B.J.D. Supervision: B.J.D., N.J.D., L.Z., and A.F.F. Critical discussion: B.J.D., N.J.D., L.Z., A.F.F., M.S.L., and M.S.-F. Writing: M.S., N.J.D., and B.J.D.
Competing interests: M.S.-F. receives funding from Bristol-Myers Squibb. B.J.D. receives consulting fees from AstraZeneca. A.F.F. is currently an employee of Novartis and owns stock in Novartis. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
References
REFERENCES AND NOTES
- 1.George J., Lim J. S., Jang S. J., Cun Y., Ozretic L., Kong G., Leenders F., Lu X., Fernandez-Cuesta L., Bosco G., Muller C., Dahmen I., Jahchan N. S., Park K. S., Yang D., Karnezis A. N., Vaka D., Torres A., Wang M. S., Korbel J. O., Menon R., Chun S. M., Kim D., Wilkerson M., Hayes N., Engelmann D., Putzer B., Bos M., Michels S., Vlasic I., Seidel D., Pinther B., Schaub P., Becker C., Altmuller J., Yokota J., Kohno T., Iwakawa R., Tsuta K., Noguchi M., Muley T., Hoffmann H., Schnabel P. A., Petersen I., Chen Y., Soltermann A., Tischler V., Choi C. M., Kim Y. H., Massion P. P., Zou Y., Jovanovic D., Kontic M., Wright G. M., Russell P. A., Solomon B., Koch I., Lindner M., Muscarella L. A., la Torre A., Field J. K., Jakopovic M., Knezevic J., Castanos-Velez E., Roz L., Pastorino U., Brustugun O. T., Lund-Iversen M., Thunnissen E., Kohler J., Schuler M., Botling J., Sandelin M., Sanchez-Cespedes M., Salvesen H. B., Achter V., Lang U., Bogus M., Schneider P. M., Zander T., Ansen S., Hallek M., Wolf J., Vingron M., Yatabe Y., Travis W. D., Nurnberg P., Reinhardt C., Perner S., Heukamp L., Buttner R., Haas S. A., Brambilla E., Peifer M., Sage J., Thomas R. K., Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin C. M., Durinck S., Stawiski E. W., Poirier J. T., Modrusan Z., Shames D. S., Bergbower E. A., Guan Y., Shin J., Guillory J., Rivers C. S., Foo C. K., Bhatt D., Stinson J., Gnad F., Haverty P. M., Gentleman R., Chaudhuri S., Janakiraman V., Jaiswal B. S., Parikh C., Yuan W., Zhang Z., Koeppen H., Wu T. D., Stern H. M., Yauch R. L., Huffman K. E., Paskulin D. D., Illei P. B., Varella-Garcia M., Gazdar A. F., de Sauvage F. J., Bourgon R., Minna J. D., Brock M. V., Seshagiri S., Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peifer M., Fernandez-Cuesta L., Sos M. L., George J., Seidel D., Kasper L. H., Plenker D., Leenders F., Sun R., Zander T., Menon R., Koker M., Dahmen I., Muller C., Di Cerbo V., Schildhaus H. U., Altmuller J., Baessmann I., Becker C., de Wilde B., Vandesompele J., Bohm D., Ansen S., Gabler F., Wilkening I., Heynck S., Heuckmann J. M., Lu X., Carter S. L., Cibulskis K., Banerji S., Getz G., Park K. S., Rauh D., Grutter C., Fischer M., Pasqualucci L., Wright G., Wainer Z., Russell P., Petersen I., Chen Y., Stoelben E., Ludwig C., Schnabel P., Hoffmann H., Muley T., Brockmann M., Engel-Riedel W., Muscarella L. A., Fazio V. M., Groen H., Timens W., Sietsma H., Thunnissen E., Smit E., Heideman D. A., Snijders P. J., Cappuzzo F., Ligorio C., Damiani S., Field J., Solberg S., Brustugun O. T., Lund-Iversen M., Sanger J., Clement J. H., Soltermann A., Moch H., Weder W., Solomon B., Soria J. C., Validire P., Besse B., Brambilla E., Brambilla C., Lantuejoul S., Lorimier P., Schneider P. M., Hallek M., Pao W., Meyerson M., Sage J., Shendure J., Schneider R., Buttner R., Wolf J., Nurnberg P., Perner S., Heukamp L. C., Brindle P. K., Haas S., Thomas R. K., Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44, 1104–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn L., Mansfield A. S., Szczęsna A., Havel L., Krzakowski M., Hochmair M. J., Huemer F., Losonczy G., Johnson M. L., Nishio M., Reck M., Mok T., Lam S., Shames D. S., Liu J., Ding B., Lopez-Chavez A., Kabbinavar F., Lin W., Sandler A., Liu S. V., First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N. Engl. J. Med. 379, 2220–2229 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Paz-Ares L., Dvorkin M., Chen Y., Reinmuth N., Hotta K., Trukhin D., Statsenko G., Hochmair M. J., Özgüroğlu M., Ji J. H., Voitko O., Poltoratskiy A., Ponce S., Verderame F., Havel L., Bondarenko I., Kazarnowicz A., Losonczy G., Conev N. V., Armstrong J., Byrne N., Shire N., Jiang H., Goldman J. W.; CASPIAN Investigators , Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet 394, 1929–1939 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Santarpia M., Daffina M. G., Karachaliou N., Gonzalez-Cao M., Lazzari C., Altavilla G., Rosell R., Targeted drugs in small-cell lung cancer. Transl. Lung Cancer Res. 5, 51–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byers L. A., Rudin C. M., Small cell lung cancer: Where do we go from here? Cancer 121, 664–672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shtivelman E., Hensing T., Simon G. R., Dennis P. A., Otterson G. A., Bueno R., Salgia R., Molecular pathways and therapeutic targets in lung cancer. Oncotarget 5, 1392–1433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farago A. F., Keane F. K., Current standards for clinical management of small cell lung cancer. Transl. Lung Cancer Res. 7, 69–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ettinger D. S., Wood D. E., Aisner D. L., Akerley W., Bauman J. R., Bharat A., Bruno D. S., Chang J. Y., Chirieac L. R., D’Amico T. A., Dilling T. J., Dowell J., Gettinger S., Gubens M. A., Hegde A., Hennon M., Lackner R. P., Lanuti M., Leal T. A., Lin J., Loo B. W. Jr., Lovly C. M., Martins R. G., Massarelli E., Morgensztern D., Ng T., Otterson G. A., Patel S. P., Riely G. J., Schild S. E., Shapiro T. A., Singh A. P., Stevenson J., Tam A., Yanagawa J., Yang S. C., Gregory K. M., Hughes M., NCCN guidelines insights: Non-small cell lung cancer, version 2.2021. J. Natl. Compr. Canc. Netw. 19, 254–266 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Mok T. S., Wu Y. L., Thongprasert S., Yang C. H., Chu D. T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J. J., Chewaskulyong B., Jiang H., Duffield E. L., Watkins C. L., Armour A. A., Fukuoka M., Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Mok T. S., Wu Y. L., Ahn M. J., Garassino M. C., Kim H. R., Ramalingam S. S., Shepherd F. A., He Y., Akamatsu H., Theelen W. S., Lee C. K., Sebastian M., Templeton A., Mann H., Marotti M., Ghiorghiu S., Papadimitrakopoulou V. A.; AURA3 Investigators , Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H., Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLOS Med. 2, e73 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi S., Boggon T. J., Dayaram T., Janne P. A., Kocher O., Meyerson M., Johnson B. E., Eck M. J., Tenen D. G., Halmos B., EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 352, 786–792 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Sequist L. V., Waltman B. A., Dias-Santagata D., Digumarthy S., Turke A. B., Fidias P., Bergethon K., Shaw A. T., Gettinger S., Cosper A. K., Akhavanfard S., Heist R. S., Temel J., Christensen J. G., Wain J. C., Lynch T. J., Vernovsky K., Mark E. J., Lanuti M., Iafrate A. J., Mino-Kenudson M., Engelman J. A., Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piotrowska Z., Isozaki H., Lennerz J. K., Gainor J. F., Lennes I. T., Zhu V. W., Marcoux N., Banwait M. K., Digumarthy S. R., Su W., Yoda S., Riley A. K., Nangia V., Lin J. J., Nagy R. J., Lanman R. B., Dias-Santagata D., Mino-Kenudson M., Iafrate A. J., Heist R. S., Shaw A. T., Evans E. K., Clifford C., Ou S. I., Wolf B., Hata A. N., Sequist L. V., Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET Fusion. Cancer Discov. 8, 1529–1539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirski S. E., Gerlach J. H., Cole S. P., Multidrug resistance in a human small cell lung cancer cell line selected in adriamycin. Cancer Res. 47, 2594–2598 (1987). [PubMed] [Google Scholar]
- 18.Twentyman P. R., Wright K. A., Mistry P., Kelland L. R., Murrer B. A., Sensitivity to novel platinum compounds of panels of human lung cancer cell lines with acquired and inherent resistance to cisplatin. Cancer Res. 52, 5674–5680 (1992). [PubMed] [Google Scholar]
- 19.Gardner E. E., Lok B. H., Schneeberger V. E., Desmeules P., Miles L. A., Arnold P. K., Ni A., Khodos I., de Stanchina E., Nguyen T., Sage J., Campbell J. E., Ribich S., Rekhtman N., Dowlati A., Massion P. P., Rudin C. M., Poirier J. T., Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell 31, 286–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böttger F., Semenova E. A., Song J.-Y., Ferone G., van der Vliet J., Cozijnsen M., Bhaskaran R., Bombardelli L., Piersma S. R., Pham T. V., Jimenez C. R., Berns A., Tumor heterogeneity underlies differential cisplatin sensitivity in mouse models of small-cell lung cancer. Cell Rep. 27, 3345–3358.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drapkin B. J., George J., Christensen C. L., Mino-Kenudson M., Dries R., Sundaresan T., Phat S., Myers D. T., Zhong J., Igo P., Hazar-Rethinam M. H., Licausi J. A., Gomez-Caraballo M., Kem M., Jani K. N., Azimi R., Abedpour N., Menon R., Lakis S., Heist R. S., Buttner R., Haas S., Sequist L. V., Shaw A. T., Wong K. K., Hata A. N., Toner M., Maheswaran S., Haber D. A., Peifer M., Dyson N., Thomas R. K., Farago A. F., Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov. 8, 600–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgkinson C. L., Morrow C. J., Li Y., Metcalf R. L., Rothwell D. G., Trapani F., Polanski R., Burt D. J., Simpson K. L., Morris K., Pepper S. D., Nonaka D., Greystoke A., Kelly P., Bola B., Krebs M. G., Antonello J., Ayub M., Faulkner S., Priest L., Carter L., Tate C., Miller C. J., Blackhall F., Brady G., Dive C., Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Williams J. A., Using PDX for preclinical cancer drug discovery: The evolving field. J. Clin. Med. 7, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidhyasagar V., Haq S. U., Lok B. H., Patient-derived xenograft models of small cell lung cancer for therapeutic development. Clin. Oncol. (R. Coll. Radiol.) 32, 619–625 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Farago A. F., Yeap B. Y., Stanzione M., Hung Y. P., Heist R. S., Marcoux J. P., Zhong J., Rangachari D., Barbie D. A., Phat S., Myers D. T., Morris R., Kem M., Dubash T. D., Kennedy E. A., Digumarthy S. R., Sequist L. V., Hata A. N., Maheswaran S., Haber D. A., Lawrence M. S., Shaw A. T., Mino-Kenudson M., Dyson N. J., Drapkin B. J., Combination olaparib and temozolomide in relapsed small-cell lung cancer. Cancer Discov. 9, 1372–1387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietanza M. C., Waqar S. N., Krug L. M., Dowlati A., Hann C. L., Chiappori A., Owonikoko T. K., Woo K. M., Cardnell R. J., Fujimoto J., Long L., Diao L., Wang J., Bensman Y., Hurtado B., de Groot P., Sulman E. P., Wistuba I. I., Chen A., Fleisher M., Heymach J. V., Kris M. G., Rudin C. M., Byers L. A., Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doles J., Oliver T. G., Cameron E. R., Hsu G., Jacks T., Walker G. C., Hemann M. T., Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 107, 20786–20791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie K., Doles J., Hemann M. T., Walker G. C., Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20792–20797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerson S. L., MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 4, 296–307 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Munoz J. L., Walker N. D., Scotto K. W., Rameshwar P., Temozolomide competes for P-glycoprotein and contributes to chemoresistance in glioblastoma cells. Cancer Lett. 367, 69–75 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Sharom F. J., ABC multidrug transporters: Structure, function and role in chemoresistance. Pharmacogenomics 9, 105–127 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Murai J., Tang S. W., Leo E., Baechler S. A., Redon C. E., Zhang H., Al Abo M., Rajapakse V. N., Nakamura E., Jenkins L. M. M., Aladjem M. I., Pommier Y., SLFN11 blocks stressed replication forks independently of ATR. Mol. Cell 69, 371–384.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lok B. H., Gardner E. E., Schneeberger V. E., Ni A., Desmeules P., Rekhtman N., de Stanchina E., Teicher B. A., Riaz N., Powell S. N., Poirier J. T., Rudin C. M., PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 23, 523–535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoppoli G., Regairaz M., Leo E., Reinhold W. C., Varma S., Ballestrero A., Doroshow J. H., Pommier Y., Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. U.S.A. 109, 15030–15035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olive P. L., Wlodek D., Banath J. P., DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 51, 4671–4676 (1991). [PubMed] [Google Scholar]
- 36.Marechal A., Zou L., DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X., Kaufman P. D., Ki-67: More than a proliferation marker. Chromosoma 127, 175–186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tada S., Cdt1 and geminin: Role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 12, 1629–1641 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai J., Zhang Y., Morris J., Ji J., Takeda S., Doroshow J. H., Pommier Y., Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J. Pharmacol. Exp. Ther. 349, 408–416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimoto K., Mizoguchi M., Hata N., Murata H., Hatae R., Amano T., Nakamizo A., Sasaki T., Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front. Oncol. 2, 186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisman A., Woodgate R., Translesion DNA polymerases in eukaryotes: What makes them tick? Crit. Rev. Biochem. Mol. Biol. 52, 274–303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee N., Walker G. C., Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maga G., Crespan E., Markkanen E., Imhof R., Furrer A., Villani G., Hubscher U., van Loon B., DNA polymerase δ-interacting protein 2 is a processivity factor for DNA polymerase λ during 8-oxo-7,8-dihydroguanine bypass. Proc. Natl. Acad. Sci. U.S.A. 110, 18850–18855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guilliam T. A., Bailey L. J., Brissett N. C., Doherty A. J., PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities. Nucleic Acids Res. 44, 3317–3329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y. S., Gregory M. T., Yang W., Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. U.S.A. 111, 2954–2959 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maya-Mendoza A., Moudry P., Merchut-Maya J. M., Lee M., Strauss R., Bartek J., High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Avkin S., Sevilya Z., Toube L., Geacintov N., Chaney S. G., Oren M., Livneh Z., p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell 22, 407–413 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Korzhnev D. M., Hadden M. K., Targeting the translesion synthesis pathway for the development of anti-cancer chemotherapeutics. J. Med. Chem. 59, 9321–9336 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Dash R. C., Ozen Z., Rizzo A. A., Lim S., Korzhnev D. M., Hadden M. K., Structural approach to identify a lead scaffold that targets the translesion synthesis polymerase Rev1. J. Chem. Inf. Model. 58, 2266–2277 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee N., Whitman M. A., Harris C. A., Min S. M., Jonas O., Lien E. C., Luengo A., Vander Heiden M. G., Hong J., Zhou P., Hemann M. T., Walker G. C., REV1 inhibitor JH-RE-06 enhances tumor cell response to chemotherapy by triggering senescence hallmarks. Proc. Natl. Acad. Sci. U.S.A. 117, 28918–28921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wojtaszek J. L., Chatterjee N., Najeeb J., Ramos A., Lee M., Bian K., Xue J. Y., Fenton B. A., Park H., Li D., Hemann M. T., Hong J., Walker G. C., Zhou P., A small molecule targeting mutagenic translesion synthesis improves chemotherapy. Cell 178, 152–159.e111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nayak S., Calvo J. A., Cong K., Peng M., Berthiaume E., Jackson J., Zaino A. M., Vindigni A., Hadden M. K., Cantor S. B., Inhibition of the translesion synthesis polymerase REV1 exploits replication gaps as a cancer vulnerability. Sci. Adv. 6, eaaz7808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herzog B. H., Devarakonda S., Govindan R., Overcoming chemotherapy resistance in SCLC. J. Thorac. Oncol. 16, 2002–2015 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Noordermeer S. M., van Attikum H., PARP inhibitor resistance: A tug-of-war in BRCA-mutated cells. Trends Cell Biol. 29, 820–834 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Suda K., Murakami I., Sakai K., Tomizawa K., Mizuuchi H., Sato K., Nishio K., Mitsudomi T., Heterogeneity in resistance mechanisms causes shorter duration of epidermal growth factor receptor kinase inhibitor treatment in lung cancer. Lung Cancer 91, 36–40 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Piotrowska Z., Niederst M. J., Karlovich C. A., Wakelee H. A., Neal J. W., Mino-Kenudson M., Fulton L., Hata A. N., Lockerman E. L., Kalsy A., Digumarthy S., Muzikansky A., Raponi M., Garcia A. R., Mulvey H. E., Parks M. K., DiCecca R. H., Dias-Santagata D., Iafrate A. J., Shaw A. T., Allen A. R., Engelman J. A., Sequist L. V., Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 5, 713–722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suda K., Murakami I., Katayama T., Tomizawa K., Osada H., Sekido Y., Maehara Y., Yatabe Y., Mitsudomi T., Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin. Cancer Res. 16, 5489–5498 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Suda K., Murakami I., Sakai K., Mizuuchi H., Shimizu S., Sato K., Tomizawa K., Tomida S., Yatabe Y., Nishio K., Mitsudomi T., Small cell lung cancer transformation and T790M mutation: Complimentary roles in acquired resistance to kinase inhibitors in lung cancer. Sci. Rep. 5, 14447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu D., Calvo J. A., Samson L. D., Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roos W., Baumgartner M., Kaina B., Apoptosis triggered by DNA damage O6-methylguanine in human lymphocytes requires DNA replication and is mediated by p53 and Fas/CD95/Apo-1. Oncogene 23, 359–367 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Wu F., Lin X., Okuda T., Howell S. B., DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 64, 8029–8035 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Roos W. P., Tsaalbi-Shtylik A., Tsaryk R., Guvercin F., de Wind N., Kaina B., The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs. Mol. Pharmacol. 76, 927–934 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Sakurai Y., Ichinoe M., Yoshida K., Nakazato Y., Saito S., Satoh M., Nakada N., Sanoyama I., Umezawa A., Numata Y., Shi-Xu J., Ichihara M., Takahashi M., Murakumo Y., Inactivation of REV7 enhances chemosensitivity and overcomes acquired chemoresistance in testicular germ cell tumors. Cancer Lett. 489, 100–110 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Nayak S., Calvo J. A., Cantor S. B., Targeting translesion synthesis (TLS) to expose replication gaps, a unique cancer vulnerability. Expert Opin. Ther. Targets 25, 27–36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okuda T., Lin X., Trang J., Howell S. B., Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin. Mol. Pharmacol. 67, 1852–1860 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Chen H., Chen H., Zhang J., Wang Y., Simoneau A., Yang H., Levine A. S., Zou L., Chen Z., Lan L., cGAS suppresses genomic instability as a decelerator of replication forks. Sci. Adv. 6, eabb8941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coller H. A., Grandori C., Tamayo P., Colbert T., Lander E. S., Eisenman R. N., Golub T. R., Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. U.S.A. 97, 3260–3265 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gartel A. L., Ye X., Goufman E., Shianov P., Hay N., Najmabadi F., Tyner A. L., Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U.S.A. 98, 4510–4515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cibulskis K., Lawrence M. S., Carter S. L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E. S., Getz G., Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim S., Scheffler K., Halpern A. L., Bekritsky M. A., Noh E., Källberg M., Chen X., Kim Y., Beyter D., Krusche P., Saunders C. T., Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Mannakee B. K., Balaji U., Witkiewicz A. K., Gutenkunst R. N., Knudsen E. S., Sensitive and specific post-call filtering of genetic variants in xenograft and primary tumors. Bioinformatics 34, 1713–1718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., Ruden D. M., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen R., Seshan V. E., FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin J. J., Langenbucher A., Gupta P., Yoda S., Fetter I. J., Rooney M., Do A., Kem M., Chang K. P., Oh A. Y., Chin E., Juric D., Corcoran R. B., Dagogo-Jack I., Gainor J. F., Stone J. R., Lennerz J. K., Lawrence M. S., Hata A. N., Mino-Kenudson M., Shaw A. T., Small cell transformation of ROS1 fusion-positive lung cancer resistant to ROS1 inhibition. npj Precis. Oncol. 4, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kluin R. J. C., Kemper K., Kuilman T., de Ruiter J. R., Iyer V., Forment J. V., Cornelissen-Steijger P., de Rink I., Ter Brugge P., Song J. Y., Klarenbeek S., McDermott U., Jonkers J., Velds A., Adams D. J., Peeper D. S., Krijgsman O., XenofilteR: Computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinformatics 19, 366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B. A., Karhohs K. W., Doan M., Ding L., Rafelski S. M., Thirstrup D., Wiegraebe W., Singh S., Becker T., Caicedo J. C., Carpenter A. E., CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Genois M. M., Gagne J. P., Yasuhara T., Jackson J., Saxena S., Langelier M. F., Ahel I., Bedford M. T., Pascal J. M., Vindigni A., Poirier G. G., Zou L., CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol. Cell 81, 784–800.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olive P. L., Banath J. P., The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Gyori B. M., Venkatachalam G., Thiagarajan P. S., Hsu D., Clement M. V., OpenComet: An automated tool for comet assay image analysis. Redox Biol. 2, 457–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tate J. G., Bamford S., Jubb H. C., Sondka Z., Beare D. M., Bindal N., Boutselakis H., Cole C. G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S. C., Kok C. Y., Noble K., Ponting L., Ramshaw C. C., Rye C. E., Speedy H. E., Stefancsik R., Thompson S. L., Wang S., Ward S., Campbell P. J., Forbes S. A., COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lord C. J., Ashworth A., BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
References