Abstract
Lactobacillus curvatus CECT 904 and Lactobacillus sake CECT 4808 were selected on the basis of their proteolytic activities against synthetic substrates. Further, the effects of whole cells, cell extracts, and a combination of both enzymatic sources on muscle sarcoplasmic proteins were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and reverse-phase high-performance liquid chromatography analyses. Strains of both species displayed proteinase activities on five sarcoplasmic proteins. The inoculation of whole cells caused a degradation of peptides, whereas the addition of cell extracts resulted in the generation of both hydrophilic and hydrophobic peptides. This phenomenon was remarkably more pronounced when L. curvatus was involved. Whole cells also consumed a great amount of free amino acids, while the addition of intracellular enzymes contributed to their generation. L. sake accounted for a greater release of free amino acids. In general, cell viability and also proteolytic events were promoted when cell suspensions were provided with cell extracts as an extra source of enzymes.
Lactic acid bacteria are essential agents of the meat fermentation process that contribute to the hygienic and sensory qualities of meat products. This quality is achieved mainly by the metabolic activities of these bacteria on carbohydrates and proteins, resulting in sugar depletion, pH reduction, and the generation of flavor compounds (5, 12). Most attention has been focused on the development of starter cultures with adequate fermentation characteristics, the number of studies of the proteolytic activities of lactic acid bacteria is limited. The protein breakdown that takes place during the ripening of dry fermented sausages leads to an increase in the concentration of peptides and free amino acids (6, 9, 15, 20). This increase is the result of the proteolytic activities of both endogenous and microbial enzymes, although the main role of microorganisms seems to be confined to the secondary hydrolysis of oligopeptides and small peptides (32).
Lactobacillus sake and Lactobacillus curvatus are the most prevalent microorganisms in dry fermented sausages, and their use as starter cultures is also widespread (12, 14). However, a detailed study of their proteolytic systems is lacking, and only a dipeptidase (21), a tripeptidase (25), and an aminopeptidase (26) of L. sake have been purified and characterized. The activities of those purified peptidases have also been studied under the effects of curing agents and other technologic factors involved in the manufacture process (27–29). Nevertheless, a few studies have dealt with the real effects of proteolytic enzymes on muscle proteins and derived peptides. Only exogenous enzymes have been tested as potential enhancers of proteolytic rates (4, 8, 11). The aim of this study was to determine the proteolytic activities of different enzyme combinations from L. curvatus and L. sake strains on sarcoplasmic proteins to predict the suitability of these strains and their proteolytic enzymes as starter cultures or additives, respectively, for the processing of dry fermented sausages.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following Lactobacillus strains were used: L. curvatus CECT 904 and NCDO 2739 and L. sake CECT 4808 (24), NCDO 2714, and L110, a commercial starter culture (Texel, Groupe Rhone-Poulenc, Courbevoir, France). Cells of all strains were routinely grown in MRS broth (Merck, Darmstadt, Germany) at 30°C for 24 h and then maintained at either 4°C or −80°C in 15% (vol/vol) glycerol. For enzymatic assays growth medium was inoculated with a microorganism (1%, vol/vol) that had been previously subcultured twice and incubated for 16 h at 30°C.
Preparation of cell suspensions and extracts.
The endoproteolytic activity against casein-fluorescein isothiocyanate (FITC) was assayed in whole-cell suspensions. Cells were harvested by centrifugation (10,000 × g for 20 min at 4°C), washed twice in 0.085% (wt/vol) NaCl containing 20 mM CaCl2, and resuspended in a 2% initial volume of 50 mM Tris-HCl (pH 6.5). The optical densities of cell suspensions were determined at 660 nm, and the corresponding dry weights were deduced from a calibration curve.
Aminopeptidase activity was assayed in cell extracts (CE) obtained by a modification of the procedure described by Sanz and Toldrá (26). Cells were collected as stated above, washed twice in 20 mM phosphate buffer (pH 7.0), and resuspended in the same buffer (10% of initial volume) containing 1 mg of lysozyme (Sigma, St. Louis, Mo.) per ml. Cells were also supplemented with 0.6 M sucrose and 5 mM MgCl2 for L. curvatus strains or 0.45 M sucrose for L. sake strains. After incubation at 30°C for 1 h, the cell wall fraction was removed by centrifugation (15,000 × g for 20 min at 4°C). The pellet was washed in 20 mM phosphate buffer (pH 7.0), resuspended in the same buffer, and sonicated for 15 min. Cell debris was removed by centrifugation (20,000 × g for 20 min at 4°C), and the supernatant constituted the CE.
Assay of proteinase and aminopeptidase activities.
Proteinase activity was determined, with casein-FITC type II (Sigma) as the substrate, by a modification of the procedure described by Twining (31). The reaction mixture, consisting of 70 μl of 50 mM Tris-HCl (pH 6.5) (containing 0.4% FITC–casein and 20 mM CaCl2) and 100 μl of whole-cell suspension, was incubated at 37°C for 1 h. The resulting fluorescence was measured in a multiscan fluorimeter (Fluoroskan II; Labsystems, Helsinki, Finland) at 485 and 538 nm as excitation and emission wavelengths, respectively. One unit of activity was defined as the amount of enzyme hydrolyzing 1 μmol of substrate per h at 37°C. Proteinase activity was expressed as units per milligram of dry weight.
Aminopeptidase activity was measured against several aminoacyl-7-amido-4-methyl coumarin (AMC) derivatives (l-Ala-, l-Lys-, l-Ser-, l-Phe-, l-Val-, l-Arg-, l-Gly-, l-Leu-, l-Tyr-, l-Pro-, and l-Pyr-AMC [Sigma]) and l-Glu-1-4-p-nitroanilide (Fluka Biochemika, Buchs, Switzerland) according to the method of Sanz and Toldrá (26, 27). Reaction mixtures were incubated at 37°C for 15 min, except for the chromogenic substrate, which was incubated for 1 h. One unit of activity was defined as described above, and aminopeptidase activity was expressed as units per milligram of protein. Quadruplicate assays were performed, with four samples and controls measured for each experimental point.
Determination of protein concentration.
The protein concentration was determined by the bicinchoninic acid method with BCA protein assay reagent (Pierce, Rockford, Ill.). Bovine serum albumin was used as the standard.
Activity on muscle protein extracts. (i) Extraction of muscle proteins.
Sarcoplasmic proteins were extracted according to the method of Molina and Toldrá (19) but with 20 mM phosphate buffer, pH 6.5, for homogenization. The final extract was filter sterilized through a 0.22-μm-pore-size filter (Millipore, Bedford, Mass.). The sterility of the extract was confirmed by determining the absence of bacterial growth in Plate Count Agar (Merck) as described below. The protein content of the sarcoplasmic extract was 1.8 mg/ml.
(ii) Enzymatic mixtures.
Three independent assays were carried out with as the enzymatic sample either whole-cell suspensions, CE, or a combination (1:1) of both for each of the strains tested (L. curvatus CECT 904 and L. sake CECT 4808). The reaction mixture consisted of 6 ml of whole-cell suspension or CE aseptically added to 30 ml of protein extract. In the assay of the mixture of whole-cell suspensions and CE, each part of the mixture was obtained as described above but in a half volume (3 ml) and afterwards mixed and assayed for activity. The mixtures were incubated at 37°C in a shaken water bath and sampled initially (0 h) and after 96 h for further analyses.
(iii) Bacterial count and pH measurement.
Bacterial counts were determined on Plate Count Agar (Merck) and MRS agar (Merck) after incubation at 37 and 30°C, respectively, for 48 h. The pH values of the reaction mixtures were monitored by using a model 2001 pH meter (Crison Instrument S.A., Barcelona, Spain).
(iv) Gel electrophoresis.
The hydrolysis of muscle proteins was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (18) with 12% (wt/vol) polyacrylamide gels and staining with Coomassie brilliant blue R-250. The proteins used as standards were myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase b (97 kDa), serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21 kDa), lysozyme (14 kDa), and aprotinin (6 kDa) from Bio-Rad (Richmond, Calif.).
(v) Peptide analyses.
The evolution of the peptide profiles of protein extracts was analyzed by reverse-phase high-performance liquid chromatography (HPLC) with a model 1050 liquid chromatograph (Hewlett-Packard, Palo Alto, Calif.), equipped with a multiwavelength UV detector and an automatic injector. Two milliliters of each sample was deproteinized with 5 ml of acetonitrile. The supernatant was concentrated by evaporation to dryness and resuspended in 200 μl of solvent A (0.1% [vol/vol] trifluoracetic acid in MilliQ water). Samples (25 μl) were applied onto a Symmetry C18 column (4.6 mm [inside diameter] by 250 mm [length]; Waters Corporation, Mildford, Mass.). The eluate system consisted of solvent A, described above, and solvent B (acetonitrile-water [60:40] with 0.085% [vol/vol] trifluoroacetic acid). The elution was performed at a 0.9-ml/min flow rate (40°C) isocratically in 1% solvent B for 5 min and then with a linear gradient (from 1 to 100%) of solvent B for 25 min. Peptides were detected at 214 nm.
(vi) Amino acid and natural dipeptide analyses.
The changes in amino acids and natural dipeptide contents in muscle extracts were also monitored. Five hundred microliters of each sample plus 50 μl of an internal standard (0.325 mg of hydroxyproline per ml) was deproteinized with 1.375 ml of acetonitrile. Two hundred microliters of the supernatant was derivatized to its phenylthiocarbamyl derivatives according to the method of Bidlingmeyer et al. (3). The derivatized amino acids were analyzed by reverse-phase HPLC according to the method described by Aristoy and Toldrá (1).
RESULTS
Proteolytic activities against synthetic substrates.
The endoproteinase and aminopeptidase activities of the five Lactobacillus strains studied are shown in Table 1. Endoproteolytic activities, measured as the hydrolysis of casein-FITC, were considerably low in all cases. However, we observed broad substrate specificities for the intracellular exopeptidases of the studied strains, with high activities being detected against all the assayed substrates except pyroglutamic acid-AMC. Strains of both species showed the highest activities against alanine, valine, and leucine. L. sake CECT 4808 was selected on the basis of its generally higher endo- and exoproteolytic activities for further studies of muscle proteins. L. curvatus CECT 904 was also selected mainly for its greater ability to hydrolyze casein-FITC; such hydrolysis may be a limiting activity for further utilization of generated small peptides by other intracellular enzymes.
TABLE 1.
Proteinase and aminopeptidase activities of Lactobacillus strains against synthetic substrates
| Substrate | Activitya of:
|
||||
|---|---|---|---|---|---|
|
L. curvatus
|
L. sake
|
||||
| CECT 904 | NCDO 2739 | CECT 4808 | NCDO 2714 | L110 | |
| Casein-FITC | 0.008 ± 0.001 | 0.002 ± 0.001 | 0.011 ± 0.005 | 0.009 ± 0.005 | 0.006 ± 0.005 |
| Ala-AMC | 9.25 ± 1.35 | 62.49 ± 5.70 | 41.53 ± 6.20 | 19.01 ± 1.12 | 22.14 ± 5.10 |
| Lys-AMC | 0.68 ± 0.10 | 0.52 ± 0.01 | 0.25 ± 0.05 | 1.06 ± 0.16 | 0.42 ± 0.19 |
| Ser-AMC | 0.22 ± 0.06 | 0.17 ± 0.05 | 0.43 ± 0.09 | 0.10 ± 0.01 | 0.34 ± 0.03 |
| Phe-AMC | 0.42 ± 0.05 | 0.41 ± 0.015 | 0.42 ± 0.05 | 0.23 ± 0.01 | 0.42 ± 0.04 |
| Val-AMC | 1.15 ± 0.17 | 8.36 ± 0.02 | 12.24 ± 0.55 | 3.49 ± 0.42 | 11.45 ± 0.04 |
| Arg-AMC | 1.23 ± 0.06 | 1.22 ± 0.11 | 0.98 ± 0.09 | 2.13 ± 0.55 | 0.67 ± 0.03 |
| Gly-AMC | 0.12 ± 0.004 | 0.19 ± 0.05 | 0.25 ± 0.04 | 0.06 ± 0.01 | 0.19 ± 0.007 |
| Leu-AMC | 7.29 ± 0.49 | 18.30 ± 1.08 | 20.29 ± 2.66 | 5.80 ± 0.4 | 16.53 ± 0.42 |
| Tyr-AMC | 0.09 ± 0.42 | 0.15 ± 0.021 | 0.17 ± 0.01 | 0.09 ± 0.01 | 0.19 ± 0.01 |
| Pyro-AMC | NHb | NH | NH | NH | NH |
| Pro-AMC | 0.59 ± 0.04 | 0.77 ± 0.013 | 0.74 ± 0.27 | 0.33 ± 0.02 | 0.85 ± 0.07 |
| Glu-pNA | 0.22 ± 0.01 | 0.16 ± 0.05 | 1.46 ± 0.01 | NH | 0.05 ± 0.01 |
Proteinase activity was measured against casein-FITC and expressed as units per milligram of dry weight. Aminopeptidase activity was measured against amino acid-AMC or –l-Glu-1-4-p-nitroanilide substrates and expressed as units per milligram of protein. The values shown are the means of results of four experiments ± standard errors of the means.
NH, nonhydrolyzed.
Bacterial counts and pH evolution in muscle protein mixtures.
The viability of bacteria during the incubation with sarcoplasmic proteins revealed that bacterial counts significantly decreased at the end of the incubation period when whole cells of both L. curvatus CECT 904 and L. sake CECT 4808 were used as the enzymatic source. The combined use of whole cells and CE resulted in a higher final viability, especially for L. curvatus, remaining at levels of 105 CFU/ml. As expected, no growth was detected when only the CE was added to the muscle protein extracts. The pH values remained in the range of 6.5 to 7.0 during the complete incubation period.
SDS-PAGE analyses.
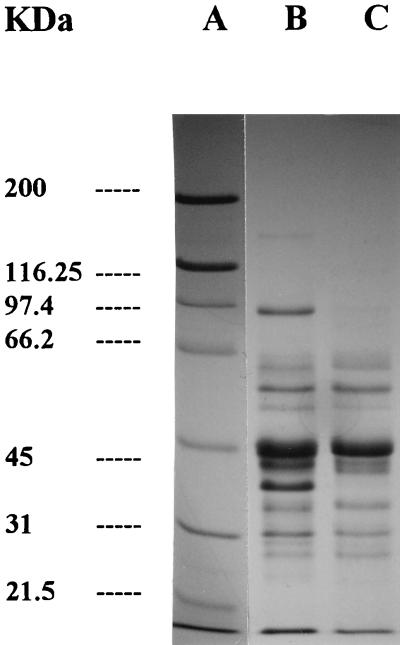
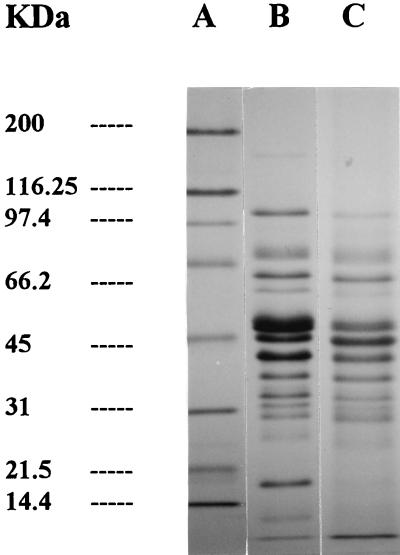
The protein patterns resulting from the action of L. curvatus CECT 904 and L. sake CECT 4808 on sarcoplasmic proteins are shown in Fig. 1 and 2, respectively. Control samples did not reflect major proteolytic changes as a result of the possible endogenous activity (data not shown). The activities of whole cells from both strains studied resulted in the disappearance and/or decrease in intensity of protein bands at approximately 160, 97, 43, 37, and 26 kDa (Fig. 1). No proteolytic changes were observed when the CE of L. curvatus CECT 904 was used (data not shown). In contrast, the mixture of whole cells and CE of L. sake CECT 4808 (Fig. 2) caused a severe degradation of bands of about 160 and 97 kDa, while other bands were partially hydrolyzed (45 kDa).
FIG. 1.
SDS-PAGE of sarcoplasmic protein hydrolysis by L. curvatus CECT 904. Lane A, standards; lane B, samples containing whole cells at 0 h; lane C, samples containing whole cells at 96 h of incubation.
FIG. 2.
SDS-PAGE of sarcoplasmic protein hydrolysis by L. sake CECT 4808. Lane A, standards; lane B, samples containing whole cells plus CE at 0 h; lane C, samples containing whole cells plus CE at 96 h of incubation.
Peptide analyses.
Peptide mappings resulting from proteinase action of L. curvatus CECT 904 or L. sake CECT 4808 on sarcoplasmic proteins are shown in Fig. 3 and 4, respectively.
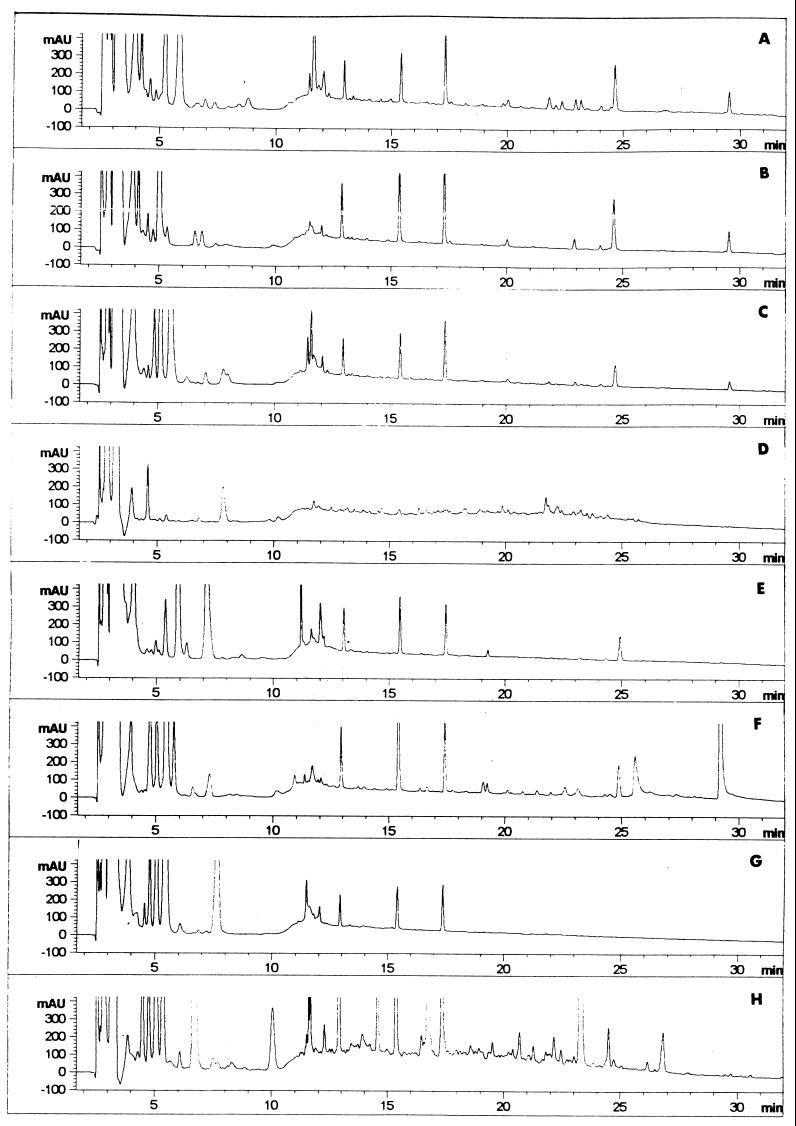
FIG. 3.
Reverse-phase HPLC patterns of soluble peptides contained in meat extracts treated with L. curvatus CECT 904 at 0 and 96 h of incubation. The profiles of control samples (A and B), samples containing whole cells (C and D), samples containing CE (E and F), and samples containing whole cells plus CE (G and H) are shown.
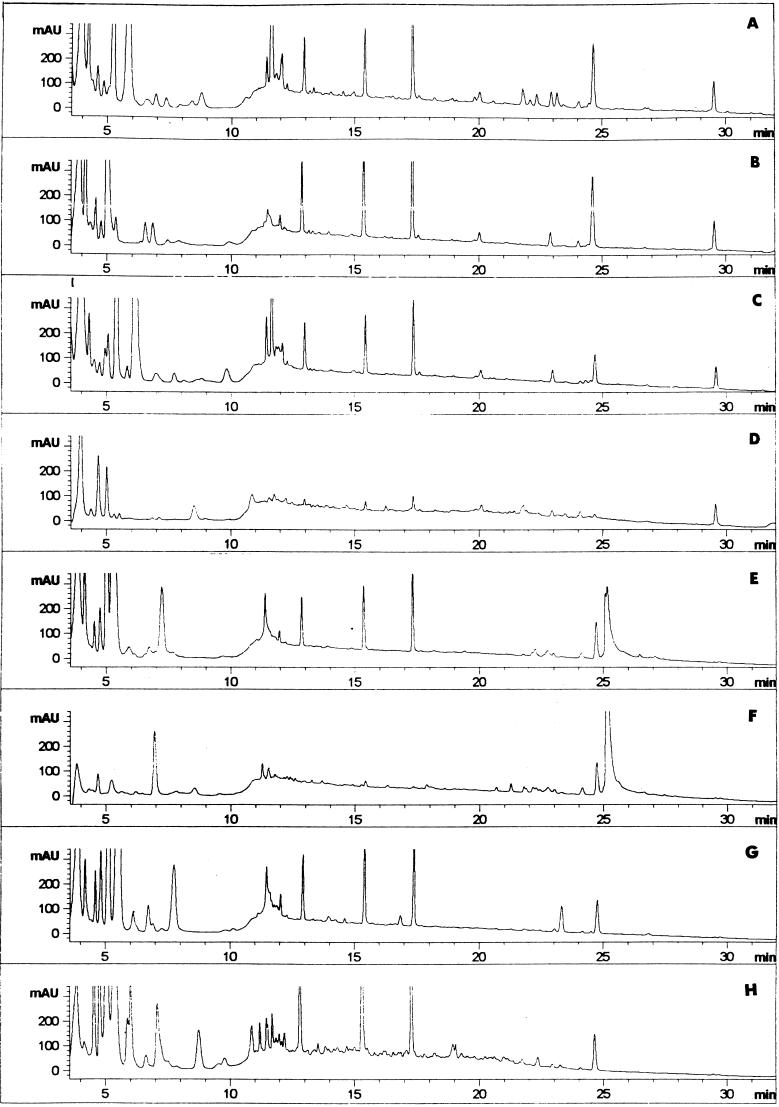
FIG. 4.
Reverse-phase HPLC patterns of soluble peptides contained in meat extracts treated with L. sake CECT 4808 at 0 and 96 h of incubation. The profiles of control samples (A and B), samples containing whole cells (C and D), samples containing CE (E and F), and samples containing whole cells plus CE (G and H) are shown.
The control profiles presented some minor changes, such as the disappearance of peaks eluting at 5.8, 11.4 to 12.0, and 21.8 to 23.2 min, after the incubation period (Fig. 3A and B and 4A and B). When whole cells of L. curvatus and L. sake strains were inoculated, most of the peaks, eluting at more than 5 min, disappeared (Fig. 3C and D and 4C and D, respectively). However, the activity of the CE from L. curvatus (Fig. 3E and F) resulted in an increase in the number of peaks (retention times of 4.7 to 5.1, 25.8, and 29.3 min) and in the intensities of others already present (retention times of 13, 15.5, 17.5, and 25 min). Different peptide mappings were observed with the CE from L. sake (Fig. 4E and F). In this case, the hydrolysis of most peptides in the regions eluting in the ranges 0 to 6 and 11.0 to 17.5 min was observed. When whole cells and CE from L. curvatus were added together, a high number of new peaks were detected in the range from 6 to 27 min of retention (Fig. 3G and H). The combination of whole cells and CE from L. sake was not as relevant but also contributed to new peptides in the region from 6.0 to 12.5 min (Fig. 4G and H).
Amino acid analyses.
The net increment in free amino acids and natural meat dipeptides after incubation with L. curvatus CECT 904 and L. sake CECT 4808 is shown in Table 2. Whole cells of both L. curvatus and L. sake strains consumed the free amino acids present in the meat extracts, especially carnosine and glutamine, resulting in net decreases in their concentrations with respect to those of the control, except for threonine. However, there were increases in the total amino acid contents of 53.14 mg/100 ml for L. sake and 29.58 mg/100 ml for L. curvatus when only the CE was added as the enzyme. The kinds and rates of amino acids released varied. So, the activity of L. curvatus resulted mainly in the generation of high amounts of glutamic acid, β-alanine, histidine, alanine, arginine, and lysine while the activity of L. sake raised the levels of glutamic acid, β-alanine, γ-aminobutyric acid, threonine, alanine, leucine, phenylalanine, and ornithine. The results differed considerably when the combination of whole cells and CE was added. With L. curvatus, an important consumption of certain compounds, such as proline, alanine, glycine, taurine, carnosine, and anserine, as a result of its higher survival in the extracts (Table 1), and only slight increases in β-alanine, γ-aminobutyric acid, threonine, phenylalanine, and tyrosine were detected. L. sake contributed to the increase of almost all amino acids, especially glutamic acid, alanine, and β-alanine. The total amount of generated free amino acid content was negative for L. curvatus as a result of its metabolizing activity, while it was positive for L. sake (104.59 mg/100 ml).
TABLE 2.
Generation of free amino acids and natural dipeptides in sarcoplasmic protein extracts
| Substrate | Amt of amino acid or dipeptide generated (mg/100 ml of extract)a
|
||||||
|---|---|---|---|---|---|---|---|
| Control |
L. curvatus
|
L. sake
|
|||||
| Whole cells | CE | Cells + CE | Whole cells | CE | Cells + CE | ||
| Aspartic acid | 6.41 | 0.02 | −0.05 | 9.30 | −0.29 | −0.27 | 0.52 |
| Glutamic acid | −22.89 | −6.79 | 60.40 | −8.22 | −9.91 | 13.28 | 11.29 |
| Serine | 2.26 | −2.75 | 3.74 | −1.22 | −2.77 | 0.61 | 1.62 |
| Asparagine | 0.38 | −0.79 | −1.14 | 0.73 | −1.14 | −0.90 | −0.58 |
| Glycine | 2.75 | −6.68 | 4.13 | −5.31 | −6.72 | 1.85 | 4.48 |
| Glutamine | −17.97 | −56.61 | −99.87 | −24.96 | −55.67 | −23.06 | −18.66 |
| β-Alanine | −0.04 | −1.91 | 7.38 | 6.48 | −1.86 | 12.05 | 15.30 |
| Histidine | 2.34 | −2.63 | 17.64 | −2.64 | −2.39 | 2.67 | 0.44 |
| GABAb | 1.06 | −0.63 | 0.77 | 27.19 | −0.62 | 13.99 | 13.44 |
| Threonine | 5.39 | 25.72 | −1.96 | 29.69 | 10.08 | 13.12 | 9.21 |
| Alanine | 7.07 | −14.05 | 37.54 | −13.16 | −12.42 | 15.06 | 40.67 |
| Arginine | 0.57 | −3.72 | 2.88 | −1.10 | −4.05 | −1.76 | −1.73 |
| Proline | 2.93 | −4.66 | 1.12 | −4.83 | −4.63 | 2.55 | 4.92 |
| Tyrosine | 0.81 | −1.67 | 1.25 | 7.76 | 0.65 | 1.32 | 1.40 |
| Valine | 2.11 | −2.51 | −1.88 | −1.13 | −2.38 | 3.01 | 3.87 |
| Methionine | 1.08 | −1.53 | 1.79 | 0.27 | −1.22 | 1.36 | 2.42 |
| Isoleucine | 1.13 | −1.38 | 0.39 | 0.01 | −1.28 | 2.01 | 2.36 |
| Leucine | 2.83 | −2.62 | 2.53 | 0.27 | −2.51 | 5.69 | 8.52 |
| Phenylalanine | 1.46 | −1.85 | 2.37 | 7.90 | −1.75 | 3.33 | 4.47 |
| Tryptophane | 0.27 | −0.66 | 0.00 | 0.24 | −0.66 | 0.86 | 1.21 |
| Ornithine | 0.63 | −0.41 | 0.00 | 0.72 | −0.55 | 2.87 | 5.53 |
| Lysine | 2.90 | −3.49 | 7.72 | 1.74 | −3.75 | 2.41 | 2.42 |
| Anserine | 0.31 | −3.89 | 0.84 | −12.02 | −9.28 | −0.51 | 1.19 |
| Carnosine | −11.62 | −118.47 | −18.29 | −244.36 | −95.57 | −8.92 | −1.60 |
| Taurine | 0.52 | −24.56 | 0.25 | −14.80 | −25.26 | −9.51 | −8.41 |
Sarcoplasmic protein extracts were incubated at 37°C with whole cells, CE, and a combination of whole cells and CE.
GABA, γ-amino butyric acid.
DISCUSSION
The endoproteolytic activity responsible for the initial breakdown of protein in dry sausages has been attributed mainly to endogenous muscle cathepsins (32). The control protein patterns analyzed by SDS-PAGE did not show detectable changes according with results of previous reports (10). These protein patterns showed the ability of L. sake and L. curvatus to use the muscle sarcoplasmic proteins as substrates. The extracellular proteinase activities of both studied species seemed to have similar specificities for this substrate, although the hydrolysis caused by L. sake appeared to be also due to other intracellular enzymes. Moreover, the study of Parra et al. (22) did not reveal increases in proteolysis when activities from the CE of lactobacilli were incorporated into a model goat’s milk curds.
The peptides initially present in meat mixtures could be taken up by the cells and also hydrolyzed by the set of all cellular enzymes, since a pronounced disappearance of peaks along the chromatogram was detected only when whole cells were inoculated. This hydrolytic activity did not reflect any discrimination between hydrophobic and hydrophilic peptides, although the existence of such specificity in intact cells, as occurs in other lactic acid bacteria (16), cannot be excluded. The preferential hydrolysis of hydrophobic peptides is required, as only hydrophilic peptides are related to desirable curing flavors (2). The strongest effects on peptide changes were detected when L. curvatus CECT 904 was involved. Thus, it will be of the utmost interest to study its putative contribution to the generation of desirable peptides or the degradation of bitter ones. Then, the nature of these compounds as well as the specificity of the proteolytic system of L. curvatus must be elucidated.
The high consumption of free amino acids upon inoculation of whole cells into the meat extracts was also according to the high requirements for amino acids for the optimal growth of lactic acid bacteria (23). Carbohydrate levels present in meat were low and carbon sources were not added to the mixtures, so the activation of alternative metabolism pathways was supposed to occur. For instance, L. sake can use arginine as an energy source by the arginine deiminase pathway under glucose depletion conditions (17), and indeed, increases in arginine levels were never detected for this species. The intracellular enzymes of L. curvatus constitute a potential additive to promote the release of free amino acids such as glutamic acid, histidine, and alanine, although general increases were not detected when whole cells were present. Undoubtedly, the exoproteolytic activity of L. sake CECT 4808 is even more interesting for the generation of free amino acids, which contribute to the process either as direct flavor enhancers or as precursors of other flavor compounds (17, 30). In fact, some of the amino acids generated at higher rates, namely, glutamic acid and alanine, are known to have flavor enhancement properties and sweet taste, respectively (13, 17). The predominant release of alanine is in accordance with the specificities of the purified aminopeptidase and tripeptidase (25, 26). The final increases in the concentrations of hydrophobic and branched amino acids were lower than expected considering the broad enzyme specificity. Probably this is due to their conversion in other volatile compounds also endowed with intense aromatic characteristics (7, 17). In general, cell viability and also proteolytic events were enhanced when cell suspensions were provided with CE as an extra source of enzymes. Blom et al. (4) also found that cell growth and fermentation rates were stimulated by incorporation of a purified bacterial proteinase.
In summary, this study constitutes an initial approach to the proteolytic activities of two important species frequently present in meat fermentation. The differences in proteolytic activities observed in this study may lead to distinct flavor profiles for final products. The ability of L. curvatus to modify the peptide profile as well as the exopeptidase activity of L. sake when acting on muscle proteins should be studied in more detail in relation to their physiologies and sensory effects.
ACKNOWLEDGMENTS
This work was supported by grant ALI98-0890 from CICYT (Spain). The scholarships to S. Fadda from CONICET (Buenos Aires, Argentina) and to Y. Sanz from FPI/MEC (Madrid, Spain) are also acknowledged.
REFERENCES
- 1.Aristoy M C, Toldrá F. Deproteinization techniques for HPLC amino acid analyses in fresh pork muscle and dry cured ham. J Agric Food Chem. 1991;39:1792–1795. [Google Scholar]
- 2.Aristoy M C, Toldrá F. Isolation of flavour peptides from raw pork meat and dry cured ham. In: De Charalambous G, editor. Food flavours: generation, analysis and process influence. London, United Kingdom: Elsevier Science; 1995. pp. 1323–1343. [Google Scholar]
- 3.Bidlingmeyer B A, Cohen S A, Tarvin T L, Frost B A. A new, rapid, high sensitivity analysis of amino acids in food type samples. J Assoc Off Anal Chem. 1987;70:241–247. [PubMed] [Google Scholar]
- 4.Blom H, Hagen B F, Pedersen B O, Holck A L, Axelsson L, Naes H. Accelerated production of dry fermented sausage. Meat Sci. 1996;43:229–242. doi: 10.1016/0309-1740(96)00068-x. [DOI] [PubMed] [Google Scholar]
- 5.Brink B T, Huis In’t Veld J H J. Application of metabolic properties of lactic acid bacteria. In: Novel G, Le Querler J-F, editors. Les bacteries lactiques. Caen, France: Ardie Normandie; 1992. pp. 67–76. [Google Scholar]
- 6.De Masi T W, Wardlaw F B, Dick R L, Acton J C. Nonprotein nitrogen (NPN) and free amino acid contents of dry fermented and non fermented sausages. Meat Sci. 1990;27:1–12. doi: 10.1016/0309-1740(90)90024-Z. [DOI] [PubMed] [Google Scholar]
- 7.Demeyer D I. Meat fermentation as an integrated process. In: Smulders F J M, Toldrá F, Flores J, Prieto M, editors. New technologies for meat and meat products. Nijmegen, The Netherlands: Audet; 1992. pp. 21–36. [Google Scholar]
- 8.Díaz O, Fernández M, Carcía G D, de la Hoz L, Ordoñez J A. Proteolysis in dry fermented sausages: the effect of selected exogenous proteases. Meat Sci. 1997;46:115–128. doi: 10.1016/s0309-1740(97)00013-2. [DOI] [PubMed] [Google Scholar]
- 9.Dierick N, Vandekerckhove P, Demeyer D. Changes in non-protein nitrogen compounds during dry sausage ripening. J Food Sci. 1974;39:301–304. [Google Scholar]
- 10.Fadda S, Vignolo G, Pesce de R. Holgado A, Oliver G. Proteolytic activity in a sterile muscle sarcoplasmic extract. Microbiol Aliment Nutr. 1997;14:125–131. [Google Scholar]
- 11.Hagen B F, Berdagué J L, Holck A L, Naes H, Blom H. Bacterial proteinase reduces maturation time of dry fermented sausages. J Food Sci. 1996;61:1024–1028. [Google Scholar]
- 12.Hammes W P, Bantleon A, Min S. Lactic acid bacteria in meat fermentation. FEMS Microbiol Rev. 1990;87:165–174. [Google Scholar]
- 13.Henriksen A P, Stahnke L H. Sensory and chromatographic evaluations of water soluble fraction from dry sausages. J Agric Food Chem. 1997;45:2679–2684. [Google Scholar]
- 14.Hertel C, Schmidt G, Fischer M, Oellers K, Hammes W P. Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sake LTH677. Appl Environ Microbiol. 1998;64:1359–1365. doi: 10.1128/aem.64.4.1359-1365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson G, Berdague J L, Larsson M, Tran N, Borch E. Lipolysis, proteolysis and formation of volatile compounds during ripening of a fermented sausage with Pediococcus pentosaceus and Staphylococcus xylosus as starter cultures. Meat Sci. 1994;38:203–218. doi: 10.1016/0309-1740(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 16.Juillard V, Guillot A, Le Brass D, Gripon J L. Specificity of milk peptide utilization by Lactococcus lactis. Appl Environ Microbiol. 1998;64:1230–1236. doi: 10.1128/aem.64.4.1230-1236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato H, Rhue M R, Nishimura T. The role of free amino acids and peptides in food taste. ACS (Am Chem Soc) Symp Ser. 1989;388:158–174. [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Molina I, Toldrá F. Detection of proteolytic activity in microorganisms isolated from dry cured ham. J Food Sci. 1992;61:1308–1310. [Google Scholar]
- 20.Molly K, Demeyer D I, Johansson G, Raemaekers M, Guistelinck M, Greenen I. The importance of meat enzymes in ripening and flavour generation in dry fermented sausages. First results of a European project. Food Chem. 1997;59:539–545. [Google Scholar]
- 21.Montel M C, Seronine M P, Talon R, Hebraud M. Purification and characterization of a dipeptidase from Lactobacillus sake. Appl Environ Microbiol. 1995;61:837–839. doi: 10.1128/aem.61.2.837-839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra L, Requena T, Casal V, Gomez R. Proteolytic activity of lactobacilli in a model goat’s milk curd system. Lett Appl Microbiol. 1996;23:375–378. doi: 10.1111/j.1472-765x.1996.tb01340.x. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard G G, Coolbear T. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:179–206. doi: 10.1111/j.1574-6976.1993.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 24.Sanz Y, Hernandez M, Ferrus M A, Hernandez J. Characterization of Lactobacillus sake isolates from dry-cured sausages by restriction fragment length polymorphism analysis of the 16S rRNA gene. J Appl Microbiol. 1998;84:600–606. doi: 10.1046/j.1365-2672.1998.00387.x. [DOI] [PubMed] [Google Scholar]
- 25.Sanz Y, Mulholland F, Toldrá F. Purification and characterization of a tripeptidase from Lactobacillus sake. J Agric Food Chem. 1998;46:349–353. doi: 10.1021/jf970629u. [DOI] [PubMed] [Google Scholar]
- 26.Sanz Y, Toldrá F. Purification and characterization of an aminopeptidase from Lactobacillus sake. J Agric Food Sci. 1997;45:1552–1558. [Google Scholar]
- 27.Sanz Y, Toldrá F. Polyamines affect activity of aminopeptidases from Lactobacillus sake. J Food Sci. 1997;62:870–872. [Google Scholar]
- 28.Sanz Y, Toldrá F. Aminopeptidase activities from Lactobacillus sake in models of curing ingredients and processing conditions for dry sausages. J Food Sci. 1997;62:1211–1234. [Google Scholar]
- 29.Sanz Y, Toldrá F. Myoglobin as inhibitor of exopeptidases from Lactobacillus sake. Appl Environ Microbiol. 1998;64:2313–2314. doi: 10.1128/aem.64.6.2313-2314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toldrá F, Verplaetse A. Endogenous enzyme activity and quality for raw product processing. In: Lundstrom K, Hansson I, Wiklund E, editors. Composition of meat in relation to processing, nutritional and sensory quality. Utrecht, The Netherlands: ECCEAMST; 1995. pp. 41–55. [Google Scholar]
- 31.Twining S S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 32.Verplaetse A. Proceedings of the 40th International Congress on Meat Science and Technology. 1994. Influence of raw meat properties and processing technology on aroma quality of raw fermented meat products; pp. 45–65. [Google Scholar]