Abstract
The lantibiotic Pep5 is produced by Staphylococcus epidermidis 5. Within its biosynthetic gene cluster, the immunity gene pepI, providing producer self-protection, is localized upstream of the structural gene pepA. Pep5 production and the immunity phenotype have been found to be tightly coupled (M. Reis, M. Eschbach-Bludau, M. I. Iglesias-Wind, T. Kupke, and H.-G. Sahl, Appl. Environ. Microbiol. 60:2876–2883, 1994). To study this phenomenon, we analyzed pepA and pepI transcription and translation and constructed a number of strains containing various fragments of the gene cluster and expressing different levels of immunity. Complementation of a pepA-expressing strain with pepI in trans did not result in phenotypic immunity or production of PepI. On the other hand, neither pepA nor its product was found to be involved in immunity, since suppression of the translation of the pepA mRNA by mutation of the ATG start codon did not reduce the level of immunity. Moreover, homologous and heterologous expression of pepI from a xylose-inducible promoter resulted in significant Pep5 insensitivity. Most important for expression of the immunity phenotype was the stability of pepI transcripts, which in the wild-type strain, is achieved by an inverted repeat with a free energy of −56.9 kJ/mol, localized downstream of pepA. We performed site-directed mutagenesis to study the functional role of PepI and constructed F13D PepI, I17R PepI, and PepI 1-65; all mutants showed reduced levels of immunity. Western blot analysis indicated that F13D PepI and PepI 1-65 were not produced correctly or were partially degraded, while I17R PepI apparently was less efficient in providing self-protection than the wild-type PepI.
Lantibiotics are antibiotic peptides that contain the thioether amino acids lanthionine and/or methyllanthionine (37). The tricyclic peptide Pep5 (38) is produced by Staphylococcus epidermidis 5 and is classified along with nisin (19), subtilin (18), and epidermin (1) as a type A lantibiotic. This group of peptides comprises screw-shaped, positively charged, amphipathic molecules which exert their primary bactericidal action by the formation of pores in the cytoplasmic membrane of sensitive bacteria (36). Like all other known lantibiotics Pep5 is ribosomally synthesized. The biosynthetic gene cluster is located on the 20-kb plasmid pED503 (14, 22) and consists of the structural gene pepA as well as the genes encoding proteins required for posttranslational modification (pepB and pepC), proteolytic processing (pepP), transport (pepT), and immunity (pepI) (28). The Pep5 immunity gene pepI codes for a 69-amino acid peptide, which is characterized by a hydrophobic N-terminal segment and a strongly hydrophilic C-terminal part. PepI displays a high degree of similarity (74.2%) to EciI, the epicidin 280 immunity peptide (20), and confers cross-immunity to epicidin 280, suggesting a similar molecular self-protection mechanism for both lantibiotics. The nisin and subtilin immunity proteins NisI and SpaI consist of 245 and 165 amino acids, respectively, and have typical lipoprotein consensus sequences (23, 26). However, NisI and SpaI share no sequence homology and do not provide cross-immunity.
An important contribution to producer protection is apparently provided by dedicated ATP-binding cassette transporter systems. Such transporters have been identified in the nisin (43), subtilin (23), epidermin (29), and lacticin 481 (34) gene clusters and typically consist of two or three separate proteins, LanE, LanF, and/or LanG. NisFEG, SpaFG, EpiFEG, and LctFEG are thought to be located in the membrane and to act by exporting the lantibiotic out of the cytoplasmic membrane, thus keeping the concentration below a critical level (29). In contrast to these transporters, there are currently no theories that address how the lipoproteins NisI and SpaI or the small immunity peptides PepI and EciI could function at the molecular level to reduce producer strain sensitivity to the respective lantibiotics. However, PepI and EciI do not seem to be the only members of this unique class of immunity peptides. Recently, in the gene clusters of the structurally unrelated lantibiotic lactocin S (44) and the nonlantibiotic divergicin A (48) two genes were discovered which code for peptides of similar size, charge distribution, and significant sequence similarity (20). Particularly the presence of a related gene in a nonlantibiotic gene cluster suggests that such an immunity mechanism could be of general importance for bacteriocins of gram-positive bacteria.
Here, we report the molecular characterization of the Pep5 immunity system. One striking feature of the Pep5 system is the apparent coupling of the immunity phenotype with the production of Pep5, which led us to assume that pepA or its product, i.e., the unmodified prepeptide or as reported for nisin (25, 26) the mature lantibiotic, could be involved in the expression of this phenotype (32). We now demonstrate that pepI is sufficient for expression of Pep5 immunity. Coupling to Pep5 production is achieved at the transcriptional level through the stabilization of pepI-containing transcripts by means of an inverted repeat, which in the wild type, is located downstream of pepA. The presence of the terminator element rather than its position in the transcript was found to be important for mRNA stabilization, allowing the construction of hyperimmune strains and eventually hyperproducer strains for biotechnological purposes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1. All staphylococcal strains were maintained on blood agar or tryptone soy agar supplemented with the appropriate antibiotic. Staphylococcus carnosus TM300 (17) was used for heterologous expression, Escherichia coli BMH 71-18 (carrying mutS) was used as the host for recombinant DNA. The vector pALTER-1 (Altered Sites in vitro mutagenesis system; Promega, Madison, Wis.) was used for site-directed mutagenesis. The E. coli-Staphylococcus shuttle vector pCU1 and the staphylococcal expression vector pCX15 were kindly provided by R. Rosenstein and F. Götz, Tübingen, Germany.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| S. epidermidis 5 | Imm+ Pep+, wild-type producer of Pep5, harbors pED503 (20 kb) | 38 |
| S. epidermidis 25 | Imm− Pep− of S. epidermidis 5, pED503 removed | 14 |
| S. carnosus TM 300 | Sensitive to Pep5, cloning host | 41 |
| E. coli BMH 71-18 (carrying mutS) | thi supE+ Δ(lac-proAB) [mutS::Tn10] [F′ proAB+ lacIqlacZΔM15], cloning host | 50 |
| E. coli JM 109 | supE+ Δ(lac-proAB) hsdR17 dam [F′ traD36 proAB+ lacIqlacZΔM15], cloning host | 49 |
| pCU1 | Ampr Cmr, 4.95-kb shuttle vector | 3 |
| pT181mcs | Tetr, 4.76-kb staphylococcal vector | 3 |
| pCX15 | Cmr, 5.8-kb staphylococcal expression vector | 46 |
| pALTER-1 | Amps Tetr, 5.68-kb phagemid | Promega |
| pMR2 | Imm+ Pep− Ampr Cmr, contains 1.39-kb KpnI fragment of pED503 in pCU1 | 33 |
| pMR7 | Imm− Pep− Ampr Cmr, contains 540-bp PCR-pepA cloned in pCU1 | 33 |
| pMR9 | Imm− Pep− Ampr Cmr, contains 438-bp PCR-pepI cloned in pCU1 | 33 |
| pGB8 | Imm+ Pep− Ampr Cmr, contains a 6.8-kb SphI-BamHI fragment from pGB3 in pCU1 | 5 |
| pTMR9 | Imm− Pep− Tetr, contains 438-bp PCR-pepI cloned in pT181mcs | This study |
| pAG1/1 | Ampr Cmr, analog to pMR2, contains a mutated pepI gene in which Ile17 has been exchanged for Arg | This study |
| pAG2/1 | Ampr Cmr, analog to pMR2, contains a mutated pepI gene in which Lys65 has been exchanged for a stop codon | This study |
| pAG4/1 | Ampr Cmr, analog to pMR2, contains a mutated pepI gene in which Phe13 and Ile17 have been exchanged for Asp and Arg, respectively | This study |
| pAG5/1 | Ampr Cmr, analog to pMR2, contains a mutated pepA gene in which Met1 has been exchanged for Gly | This study |
| pUP2 | Cmr, contains a 284-bp PCR-pepI cloned in pCX15 | This study |
| pUP4 | Ampr Cmr, contains a 530-bp PCR-pepI and a 340-bp PCR-pepA cloned in pCU1 | This study |
| pUP6 | Ampr Cmr, contains a 530-bp PCR-pepI and a 250-bp PCR terminator region downstream of pepA cloned in pCU1 | This study |
| pUP7 | Ampr Cmr, contains a 460-bp PCR-pepI and a 340-bp PCR-pepA cloned in pCU1 | This study |
DNA cloning and sequencing.
Staphylococcal plasmid DNA was isolated by the method of Feliciello and Chinali (15). Lysis of staphylococci was achieved by adding 200 μg of lysostaphin per ml to solution I (50 mM glucose, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and incubating the cells for 30 to 60 min at 37°C. Plasmid DNA of E. coli was purified with QIAprepspin columns (Qiagen, Hilden, Germany). E. coli and S. epidermidis 25 were transformed by electroporation (2), and S. carnosus protoplasts were transformed by the method of Götz and Schumacher (17). Double-stranded plasmid DNA was sequenced on an A.L.F. DNA sequencer (Pharmacia, Uppsala, Sweden) by the dideoxynucleotide chain termination method (40) with the AutoRead sequencing kit (Pharmacia). Restriction enzymes and T4 DNA ligase were supplied by Boehringer (Mannheim, Germany), and synthetic oligonucleotides were obtained from Eurogentec (Seraing, Belgium).
PCR amplification.
For PCR amplification of pepI, pepA, and the terminator structure, we used pMR2 as a DNA template, Pwo DNA polymerase (Boehringer) or Goldstar Red DNA polymerase (Eurogentec), and the following primer pairs: pepI-SalI 5′ [5′(CAATAATAAATGTCGACTTAGGCCATTTAATTTTTG)3′] and pepI-XbaI 3′ [5′(CATTTTCTAGAATTAATTATTTAAACATACAAAG)3′], pepI-XbaI 5′ [5′(ATTTTCTAGAAGGTATTAAAAAAATTTTAC)3′] and pepI-SalI 3′ [5′(TATAATGTCGACACAATATAGAAAAAAAC)3′], pepA-EcoRI 5′ [5′(GTTTAAAGAATTCATTATAAAAAATGTATTG)3′] and pepA-XbaI 3′ [5′(GATTACTCTAGATTTTTTCTCCTGCATAC)3′], and Term-XbaI 5′ [5′(CAGTGTCTAGAAAAGGAAAAAACGGATG)3′] and Term-EcoRI 3′ [5′(TATTTGAATTCCATGCCCAGTGTAATCAC)3′] for amplifying the terminator structure (bold letters indicate deviations from the original sequence; the generated restriction sites are underlined). The correct introduction of the mutations was verified by sequencing the entire PCR product.
Site-directed mutagenesis.
Site-directed mutagenesis of PepI and PepA was performed with a commercial phagemid system, as previously described for Pep5 mutant peptides (5). The following mutagenic oligonucleotides were employed (mismatches are underlined): 5′(CTTTATTTTTTGCTTTAAGAATTTTTATTGTAACTTAT)3′ changes Ile17 of PepI into Arg, 5′(AAGAATAAATAGCAACTAAAAAGATAAACTTTAG)3′ mutates Lys65 into a stop codon, and 5′(GTAATTTTAACTTCTTTAGATTTTGCTTTAAGAATTTT)3′ was designed to change Phe13 into Asp. The oligonucleotide 5′(GAGGAGGTGGTTATATGGGAAAAATAACAAAAATT)3′ exchanges the Met start codon of PepA for a Gly codon.
RNA isolation and Northern hybridization.
Total RNA was prepared by using the RNeasy minikit (Qiagen) with the following modifications for isolation of RNA from staphylococci. The wild-type strain S. epidermidis 5 and mutants were grown in tryptic soy broth (Oxoid, Wesel, Germany) to an absorbance at 600 (A600) nm of 1. Cultures (10 ml) were harvested by centrifugation (5,000 × g for 5 min at 4°C). The cells were resuspended in 600 μl of Tris-EDTA (pH 8.0) and, after addition of 0.4 mg of lysostaphin per ml and 32 U of RNAguard (Pharmacia), were incubated for 15 min at 37°C. After addition of 2.1 ml of lysis buffer RLT (Qiagen), the sample was mixed vigorously and centrifuged (2 min at 8,000 × g) to remove unlysed cells. The supernatant was mixed with 1.5 ml of ethanol (100%) and applied onto a spin column in several centrifugation steps. The following steps were performed according to the manufacturer’s instructions.
Total RNA was denatured with glyoxal-dimethylsulfoxide (27) and separated on a 1.2% agarose gel with 10 mM sodium phosphate, pH 7, as a running buffer. Vacuum blotting was performed with a Vacu Gene XL (Pharmacia) within 2.5 h and a suction of 65 cm of H2O.
Expression of pepI from the xylose-inducible pCX15 vector.
For cloning in pCX15, pepI was amplified by PCR with pMR2 as a template and the following primers: pepI-BglII 5′ [5′(CTATAAGATCTTCTAAATATATTTAAAAAGGG)3′] and pepI-EcoRI 3′ [3′(CATTTTTTAGAATTCATTATTTAAACATACTAAAG)3′] (nucleotides in bold face are mutations introduced for the generation of restriction sites, which are underlined). After sequencing, the 0.284-kb BglII-EcoRI pepI fragment was cloned in pCX15 that had been restricted with BamHI and EcoRI.
Staphylococcal recombinant strains were grown in medium without glucose (10 g of casein hydrolysate, 5 g of yeast extract, 5 g of NaCl, and 1 g of K2HPO4 per liter; pH 7.3) to an A600 of 0.5 and induced with 0.5% xylose. Cells were harvested by centrifugation and disrupted by intervals of boiling and ultrasonic treatment. Intact cells were removed by centrifugation at 13,500 × g for 20 min. The supernatant was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotting with anti-maltose-binding protein (MBP)-PepI antiserum (32).
Preparation of staphylococcal cell fractions.
The preparation of membrane and soluble cytoplasmic fractions of S. epidermidis 5 and mutants was done by disruption of cells with glass beads and differential centrifugation, as previously described (32).
MIC determinations.
The determination of MICs was performed in a microtiter plate assay with half-concentrated tryptone soy broth, as described by Bierbaum et al. (5).
RESULTS
Transcriptional analysis of the Pep5 biosynthetic gene cluster.
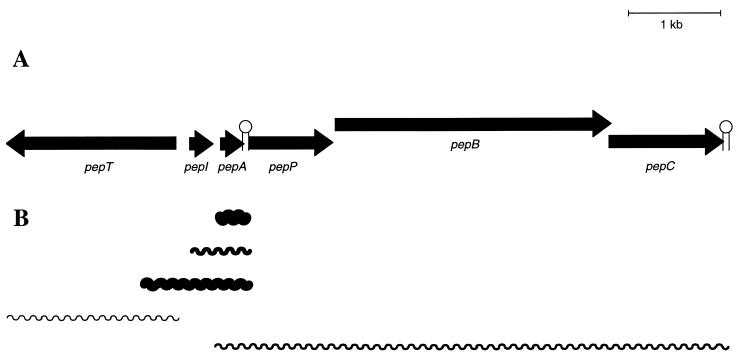
The production of Pep5 is encoded by the 20-kb plasmid pED503 of the producer strain S. epidermidis 5 (14). The Pep5 biosynthetic gene cluster covers approximately 7.9 kb and comprises pepTIAPBC. A putative terminator (−56.9 kJ/mol) that may allow partial readthrough is located in the short noncoding segment between pepA and pepP (32). We identified five different mRNA transcripts within the biosynthetic gene cluster (Fig. 1) by Northern blot analysis. A single pepA mRNA of 0.3 kb was detected in large amounts. A pepT transcript of 0.9 kb and a transcript of 5.3 kb, covering pepA and the downstream genes for proteolytical processing and modification (pepP, pepB, and pepC), were identified in low concentrations. Two further transcripts hybridized with the pepI probe. One transcript (0.6 kb) contained the immunity gene pepI and the structural gene pepA, and the other transcript (1.0 kb) additionally covered a region upstream of pepI; a small transcript of approximately 0.3 kb covering only pepI was not detectable.
FIG. 1.
Organization of the Pep5 gene cluster (A) and transcription products (B). The arrows represent the direction of transcription relative to the structural gene pepA. The relative amounts of transcripts as judged from Northern blotting are indicated by the thicknesses of the lines.
Is PepA involved in the expression of immunity?
Previous studies had shown that S. epidermidis 25(pMR9), containing only pepI of the Pep5 gene cluster, was sensitive to Pep5 to the same extent as its parent strain, S. epidermidis 25, which had had the Pep5 production plasmid pED503 removed (32). This was taken to indicate a role for pepA or its product in the expression of the immunity phenotype. In order to investigate whether both genes have to be transcribed in cis, pepI was cut out of pMR9 and cloned into the staphylococcal vector pT181mcs in S. carnosus TM300, generating pTMR9. We then transferred pTMR9 into S. epidermidis 25(pMR7), which harbors pepA in pCU1 and accumulates unmodified Pep5 prepeptide but is fully sensitive to Pep5. The resulting clone showed the same degree of sensitivity to Pep5 as the plasmidless strain S. epidermidis 25 (MIC, 0.6 μg of Pep5 per ml), demonstrating that immunity cannot be established by the complementation of pepA with pepI in trans.
S. epidermidis 25(pMR2) contained both pepI and pepA in cis and was as immune as the wild-type S. epidermidis 5 (32) (Table 2). Similar to S. epidermidis 25(pMR7), this strain produces the inactive Pep5 prepeptide (Fig. 2). In order to determine whether the Pep5 prepeptide is necessary for the expression of immunity either in a regulating or direct function, the translation of pepA was suppressed by changing the Met start codon into GGG (Gly). Site-directed mutagenesis was performed on the 1.39-kb KpnI fragment isolated from pMR2 containing pepI and pepA, and subsequently this fragment was inserted into pCU1 at the KpnI site. The resulting clone, S. epidermidis 25(pAG5/1), did not produce the Pep5 prepeptide (Fig. 2) but showed the same degree of insensitivity to Pep5 as S. epidermidis 25(pMR2), demonstrating that the unmodified Pep5 prepeptide is not taking part in the expression of immunity.
TABLE 2.
MICs of Pep5 for different staphylococci and mutant strains generated in this study
| Bacterial strain | MIC (μg of Pep5 per ml) |
|---|---|
| S. epidermidis 5 | 18.0 |
| S. epidermidis 25 | 0.6 |
| S. epidermidis 25(pMR2) | 18.0 |
| S. epidermidis 25(pMR9) | 0.6 |
| S. epidermidis 25(pMR7, pTMR9) | 0.6 |
| S. epidermidis 25(pUP2) | 7.0 |
| S. carnosus | 0.02 |
| S. carnosus(pMR2) | 0.07 |
| S. carnosus(pUP2) | 0.6 |
| S. epidermidis 25(pAG1/1) | 2.3 |
| S. epidermidis 25(pAG2/1) | 3.5 |
| S. epidermidis 25(pAG4/1) | 4.5 |
| S. epidermidis 25(pAG5/1) | 18.0 |
| S. epidermidis 25(pGB8) | 75.0 |
| S. epidermidis 25(pUP4) | 18.0 |
| S. epidermidis 25(pUP6) | 7.0 |
| S. epidermidis 25(pUP7) | 75.0 |
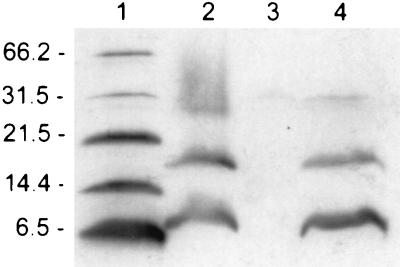
FIG. 2.
Identification of Pep5 prepeptide by Western blot analysis. Total cell extracts of the respective strains were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-Pep5 leader peptide antiserum. SDS-stable Pep5 prepeptide dimers and trimers (lanes 2 and 4) have been described previously (39). Lanes: 1, molecular size marker (numbers to the left of the gel indicate sizes in kilodaltons); 2, membrane fraction of S. epidermidis 25(pMR7) used as the standard; 3, S. epidermidis 25(pAG5/1); 4, S. epidermidis 25(pMR7, pTMR9).
We amplified pepI by PCR, introducing a BglII site and an EcoRI site, and subsequently cloned the 284-bp fragment into pCX15 at the BamHI and EcoRI restriction sites. This placed the expression of pepI under the transcriptional control of the xylA promoter and the repressor XylR. The recombinant plasmid, pUP2, was then transferred into S. carnosus and S. epidermidis 25. After induction with 0.5% xylose, PepI was detected by immunoblotting with anti-MBP-PepI antiserum in both strains (Fig. 3). PepI was visible 2 h after induction, and the amount of PepI increased with further incubation. In the presence of xylose, the sensitivity of both cloning hosts decreased from 0.02 to 0.6 μg of Pep5 per ml for S. carnosus and from 0.6 to 7 μg of Pep5 per ml for S. epidermidis 25. Thus, the producer self-protection against Pep5 appeared to solely depend on the functional expression of PepI.
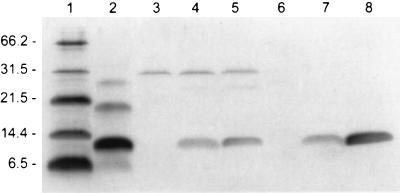
FIG. 3.
Detection of PepI in cell extracts of induced S. carnosus and S. epidermidis cells by Western blotting. Lanes: 1, molecular size marker (numbers to the left of the gel indicate sizes in kilodaltons); 2, synthetic PepI (synthetic PepI was found to form SDS-stable multimers); 3, noninduced S. carnosus(pUP2); 4 and 5, induced S. carnosus(pUP2) (2 and 19 h after induction with 0.5% xylose, respectively); 6, noninduced S. epidermidis 25(pUP2); 7 and 8, induced S. epidermidis 25(pUP2) (2 and 19 h after induction with 0.5% xylose, respectively).
Role of the inverted repeat downstream of pepA.
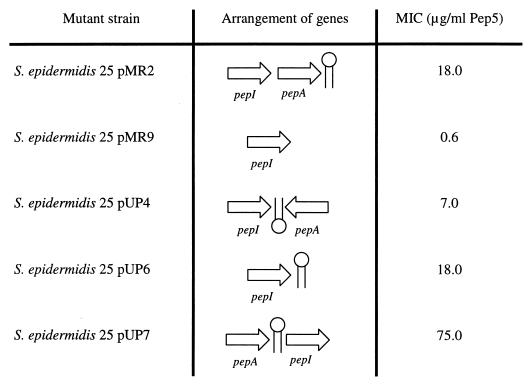
To identify the element that is essential for the self-protection mechanism, in addition to the immunity gene pepI, we constructed various gene arrangements (Fig. 4). First, the polarity of the structural gene was reversed, in order to test whether pepI has to be transcribed together with pepA as one transcript for the production of a functional mRNA. Both genes were amplified by PCR, introducing SalI-XbaI and EcoRI-XbaI restriction sites, respectively, and subsequently both PCR products were subcloned into pCU1 digested with SalI-EcoRI. The resulting recombinant plasmid pUP4 contained pepI and, on the complementary strand in a head-to-tail arrangement, pepA (Fig. 4). This variant strain was only slightly less insensitive to Pep5 than the wild-type strain.
FIG. 4.
pepI-containing S. epidermidis 25 variants and their sensitivities to Pep5. The arrangement of pepI and pepA and the locations of the pepA terminator element in different S. epidermidis 25 strains are correlated with their respective MICs.
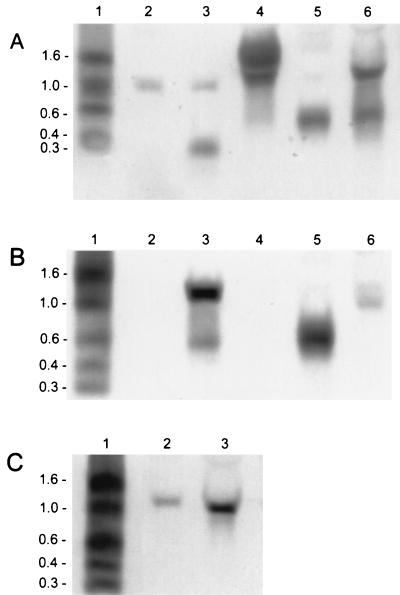
A possible regulating element, which is present in S. epidermidis 25(pUP4) and in S. epidermidis 25(pMR2) but is missing in the sensitive strain S. epidermidis 25(pMR9), is the inverted repeat downstream of pepA (Fig. 5) which allows partial readthrough. Since such palindromic structures can stabilize mRNAs by protecting them from degradation by ribonucleases, we amplified the region downstream of pepA, including the terminator, by PCR. The resulting XbaI-EcoRI PCR fragment was cloned downstream of the SalI-XbaI pepI PCR product into SalI-EcoRI restricted pCU1. The resulting plasmid, pUP6, was able to promote significant protection against Pep5 (Table 2) to S. epidermidis 25. The pepI mRNA seemed to be stabilized by the terminator, facilitating the translation of the transcript into the immunity peptide. We performed Northern blot analysis (Fig. 6) to confirm the mRNA-stabilizing function of the terminator element. As already mentioned two pepI-containing transcripts of 0.6 and 1.0 kb, respectively, were detected in the wild-type producer S. epidermidis 5 (Fig. 6B). In the immune mutant S. epidermidis 25(pMR2) only the 0.6-kb pepI mRNA was produced, because the respective DNA region for transcription of the second pepI mRNA is not present in this clone. However, the 0.6-kb transcript was produced in larger amounts than the wild type, because the cloning vector pCU1 has a higher copy number than the wild-type plasmid pED503, thus compensating for the missing second pepI transcript. The same result was obtained with the mutant harboring pAG5/1, which differs from pMR2 only by a mutation of the pepA start codon. In the Pep5-sensitive strain S. epidermidis 25(pMR9) no stable pepI transcript, which should have a size of about 0.3 kb, could be detected. In contrast, a stable 0.3-kb pepI mRNA was detected in S. epidermidis 25(pUP6), in which the terminator was placed downstream of the immunity gene, demonstrating the importance of the terminator for the stability of the pepI mRNA.
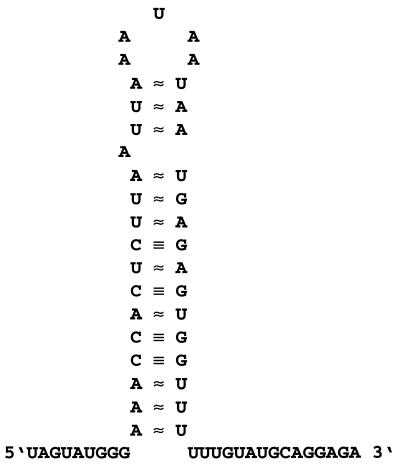
FIG. 5.
Terminator element downstream of pepA. The terminator downstream of pepA has a calculated free energy of −56.9 kJ/mol.
FIG. 6.
Northern blot analysis of pepI transcription. (A) Lanes: 1, size standard (numbers to the left of the gels indicate sizes in kilobases); 2, S. epidermidis 25(pUP4); 3, S. epidermidis 25(pUP6); 4, S. epidermidis 25(pUP7); 5, S. epidermidis 25(pAG5/1); 6, S. epidermidis 25(pGB8). A pepI-specific probe was used for detection. In S. epidermidis 25(pUP7) the identical pattern was obtained with a pepA-specific probe; additionally, a single pepA transcript of 0.3 kb was detectable (data not shown). (B) Lanes: 1, size standard; 2, S. epidermidis 25; 3, S. epidermidis 5; 4, S. epidermidis 25(pCU1); 5, S. epidermidis 25(pMR2); 6, S. epidermidis 25(pMR9). A pepI-specific probe was used for detection. (C) Northern blot analysis of the 1.1 kb pepI transcript, with a probe that hybridizes with a region upstream of pepI. Lanes: 1, size standard; 2, S. epidermidis 5; 3, S. epidermidis 25(pGB8).
We then changed the juxtaposition of pepI and pepA, placing the terminator upstream of pepI. PCR products of both genes were ligated to linearized pCU1 in reversed order relative to the wild type (pUP7). MIC determination (Table 2) revealed that the resulting variant, S. epidermidis 25(pUP7), was hyperimmune to Pep5 when compared to the wild-type producer strain. Also, in this clone the pepAI mRNA signal was detected in significantly increased amounts (Fig. 6A), demonstrating the high stability of the transcript. This result points to a direct correlation of the amount of pepI mRNA, the concentration of PepI, and the level of immunity; this interpretation is also supported by Northern blot analysis of S. epidermidis 25(pGB8), which is as hyperimmune as S. epidermidis 25(pUP7) (Table 2). This clone harbors pepIAPBC and the 5′ segment of pepT in pCU1. As in the wild-type strain S. epidermidis 5, two pepI transcripts of 0.6 and 1.1 kb, respectively, could be detected in S. epidermidis 25(pGB8); however, these transcripts were produced in larger amounts because the copy number of pCU1 was higher than that of the wild-type plasmid pED503 (Fig. 6A and C).
Site-directed mutagenesis of PepI.
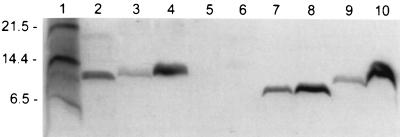
PepI is characterized by a striking charge distribution. Whereas the N-terminal segment contains a 20-amino acid stretch of apolar residues, the C-terminal region is very hydrophilic, with a net positive charge. These features are shared by the recently discovered epidicin 280 immunity peptide, EciI, which confers cross-immunity to Pep5 (20). Interestingly, the degree of similarity between PepI and EciI is even higher in the N-terminal region (80%), which was proposed to mediate interactions with the cytoplasmic membrane (32). The exchange of Ile17, which is conserved in both peptides, for Arg (pAG1/1) caused a significant reduction in the level of immunity (Table 2). Nevertheless, the mutant strain was less sensitive to Pep5 than the cloning host, S. epidermidis 25. Western blot analysis of membrane and soluble cell fractions (Fig. 7) did not indicate that the decreased level of immunity was due to reduced PepI production or to a reduced association of PepI with the membrane. With both native PepI and I17R PepI, about one-third of the peptide was detected in the soluble cytoplasmic fraction and two-thirds was detected in the membrane fraction. An additional mutation, introduced in the N-terminal region by exchanging the conserved Phe13 for Asp (pAG4/1), resulted in partially improved immunity compared to that caused by the single I17R mutation. By Western blot analysis the shortened F13D-I17R PepI was detected, indicating a proteolytic truncation of the peptide. We also introduced a stop codon at position 65 of PepI, thereby deleting the four terminal, charged amino acids, three of which are not present in EciI. Again, the strain displayed a significantly decreased level of immunity. The amount of PepI 1-65 was reduced to a level below the detection limit in Western blots, although S. epidermidis 25(pAG2/1) was less sensitive than S. epidermidis 25.
FIG. 7.
Western blot analysis of PepI. Immunoblot analyses of wild-type PepI (lanes 3 and 4), PepI 1-65 (lanes 5 and 6), F13D-I17R PepI (lanes 7 and 8), and I17R PepI (lanes 9 and 10) in soluble and membrane cytoplasmic fractions of cell extracts, respectively, are shown. Standard peptides (sizes are indicated to the left of the gel in kilodaltons) and synthetic PepI are shown in lanes 1 and 2, respectively.
DISCUSSION
An efficient self-protection system is vital for a bacteriocin-producing bacterial strain. Channel-forming colicins (e.g., colicins A, E1, and B) produced by E. coli are known to be specifically antagonized by stoichiometric complex formation with immunity proteins residing in the cytoplasmic membrane (16, 42, 45). The structural genes are located upstream of the respective immunity genes, which are transcribed, however, in the opposite direction (9, 31). In the lantibiotic gene clusters, specific immunity peptides and/or ATP-binding cassette transporter systems are encoded (23, 26, 29, 32, 34, 43). Regulation of immunity to nisin, subtilin, and epidermin is mediated through the two-component regulators NisR-SpaR and NisK-SpaK and the transcriptional activator EpiQ, respectively (13, 23, 29). An intact nisin structural gene was found to be necessary for the expression of full immunity (26), which was later shown to be due to the fact that mature nisin activates transcription of the biosynthetic gene cluster via NisR and NisK (11, 25). Also, disruption of the spaA structural gene, which is the last gene in the spaBTCA operon, resulted in subtilin-sensitive cells (23). Coordinate expression of the structural and immunity genes had been established for Pep5 as well (32) and is shown here to be achieved through a specific stabilization of the mRNA transcripts. In the presence of an inverted repeat, which in wild-type S. epidermidis 5 is localized downstream of the structural gene pepA, pepI transcripts are stable and can be translated into the immunity peptide.
mRNA stability plays an important role in regulation of bacterial gene expression. While the majority of ribonucleases are involved in the maturation of rRNA and tRNA, a few ribonucleases, acting either as exonucleases or endonucleases, have been shown to be implicated in mRNA decay. Processive degradation by the 3′ exonucleases RNaseII and polynucleotide phosphorylase is impaired by RNA secondary structures (4, 30). Similarly, stable pepI transcripts could not be detected when the terminator was missing, while placing the terminator downstream of the immunity gene (pUP6 [Fig. 6]) led to the production of a stable pepI mRNA. Another example was observed by Klug and Cohen (24), who found two hairpins at the 3′ terminus of the puf operon of Rhodobacter capsulatus that function in both transcription termination and stabilization of upstream puf mRNA sequences. The transcriptional terminator of the crystal protein gene (cry) from Bacillus thuringiensis versus Kurstaki HD-1 was shown to function as a positive retroregulator in controlling gene expression in both Bacillus subtilis and E. coli (47). The replacement of the penP (penicillinase gene) terminator with the cry terminator resulted in enhanced mRNA stability. The stabilizing function of the retroregulator was observed to be independent of its orientation of insertion with respect to the target gene. This is also true for the pepA terminator, since it was able to stabilize the pepI mRNA in spite of a reversed orientation, as in the case of the mutant pUP4. However, the pepA inverted repeat lost its function as a transcription termination element. The terminator function is strongly dependent on a stretch of at least four U at the 3′ end (35), which after the change of orientation of the pepA repeat, is placed at the 5′ end.
Cloning of the pepA terminator upstream of the immunity gene (pUP7 [Fig. 6]) resulted in the appearance of a stable pepAI mRNA and a small (0.3-kb) pepA transcript (data not shown). Several structural elements, such as 5′ untranslated regions (7, 12) or polypurine sequences (21), also function as mRNA stabilizers. These probably act by blocking access of 5′ binding endonucleases like RNase E to the transcript, presumably by stalling ribosomes at the 5′ end. In pUP7 an equivalent structural element, which could explain the high stability of the transcript comprising pepA and pepI in this order, is not present at the 5′ end of pepA. The pepA terminator was placed upstream of pepI, but since it was also localized upstream of the pepI promoter sequences, it could not be included in a potential pepI transcript; consequently a small transcript containing only pepI was not detectable. The presence of a pepAI mRNA in this construct also suggests that the progressive degradation in the 5′-to-3′ direction does not play a crucial role in the decay of the pepI transcripts. These transcripts rather seem to be degraded to a high extent by the endonucleolytic attack of ribonucleases, which do not necessarily bind to the 5′ end of a transcript but are blocked by intercistronic secondary structures. Additionally, 3′-to-5′ exonucleases may be involved in degradation. Intercistronic stem-loop structures in the puf operon were also shown to be involved in stabilizing mRNA (10); however, these loops influenced only upstream sequences and not, as in the case of pepI, downstream sequences.
The identification of the stem-loop structure as a stabilizing element and the apparent correlation between RNA stability, quantity of PepI produced, and resulting levels of immunity have several interesting implications for biotechnology and may provide clues to how PepI and related immunity peptides work. The construction of hyperimmune strains is a prerequisite for increasing the production rate of bacteriocins. In this respect, placing bacteriocin structural gene expression under the control of efficient promoters and increasing mRNA half-life, by placing the hairpin in appropriate positions, are valuable tools for increasing the amount of transcript for these genes. Moreover, such stem loops, which are found in many gene clusters of unmodified and modified bacteriocins, could be generally useful for enhancing protein yields, e.g., in heterologous expression hosts such as the food-grade S. carnosus bacteria used here.
The obvious correlation between the level of immunity and the amount of PepI may indicate a direct interaction between Pep5 and PepI and stoichiometric antagonization similar to the mechanism found for the channel-forming colicins (16, 42, 45). In this case, the colicins bind specifically to an immunity protein which is anchored within the cytoplasmic membrane by means of four membrane-spanning helices. The channel-forming domain interacts with an external loop of the immunity protein which prevents this domain from folding into a functional pore. It is tempting to assume by analogy that PepI, as well as the related peptides encoded by eciI, orf57, and dviA, could stoichiometrically antagonize Pep5 or the respective peptide bacteriocins. The N-terminal stretch of hydrophobic amino acids could provide the membrane anchor, while the C-terminal segment of PepI could mediate the interaction with Pep5. However, our mutagenesis results do not necessarily favor such a model. Although the introduction of an Arg residue in the hydrophobic segment strongly reduced the level of immunity, the distribution of the mutant peptide between soluble fractions and membrane-associated fractions was not changed; even the peptide containing a second charge in this segment was still mostly associated with the membrane.
Generally, the interpretation of the mutagenesis experiments was hampered by the lack of a molecular model of the activity of PepI and related peptides, which in turn seems difficult to establish because of our limited knowledge of the mode of action of the bacteriocins. While it is widely accepted that the peptide antibiotics destabilize the cytoplasmic membrane, it remains largely unclear which molecular interactions take place at or within a bacterial membrane. Recently, we were able to demonstrate that, in vivo, nisin and epidermin use the undecaprenol-bound cell wall precursors, the so-called lipid I and lipid II, as docking molecules for subsequent pore formation (8). The high-affinity binding of the lantibiotics to lipids I and II seems to facilitate energetically the formation of pores and may explain the high sensitivity of some bacterial species, e.g., Micrococcus luteus, which has MICs in the nanomolar range. Pep5 does not make use of the cell wall precursors (8) and is not very active against Micrococcus. However, Staphylococcus simulans and S. carnosus are sensitive in nanomolar concentrations to this lantibiotic. As such low MICs are generally not observed with amphipathic peptides and may only be achievable through high-affinity binding to particular components of bacterial membranes, we are currently trying to identify a docking molecule for Pep5. Such a concept for the molecular activity of at least some lantibiotics could not only help to explain the enormous variation in sensitivities of gram-positive bacteria to different bacteriocins but may also provide a new role for an immunity peptide, in that it may function by preventing the molecular target from binding the lantibiotic.
We currently do not have a satisfactory explanation for the observation that in the double mutant the immunity was partially restored in spite of an apparent truncation of the twofold mutated peptide; also, the instability of PepI after shortening of the C terminus remains to be explained. PepI was localized outside the cells (32), and it is conceivable that the mutations interfere with the export of the peptides, resulting in prolonged exposure to intracellular proteases, as observed with some altered Pep5 prepeptides (6). However, the processes that lead to the translocation of PepI across the membrane are also not understood. Finally, it could also be of functional importance that the PepI-type immunity peptides seem to be associated with lantibiotics which are processed inside the cell (28, 44). It is obvious that many questions about these peptides remain unanswered and that, in general, producer self-protection of gram-positive bacteria at the molecular level is the least understood topic in bacteriocin research. Certainly, these phenomena are of interest for fundamental research and applied disciplines and deserve more attention in the future.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Sa 292/8-1) and the BONFOR-Programm of the Medical Faculty, University of Bonn.
We thank A. Surovoy and G. Jung for making synthetic PepI available, G. Klemm for help with the figures, and M. Reis-Pauken for valuable discussions.
REFERENCES
- 1.Allgaier H, Jung G, Werner R G, Schneider U, Zähner H. Epidermin: sequencing of a heterodet tetracyclic 21-peptide amide antibiotic. Eur J Biochem. 1986;160:9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- 2.Augustin J, Götz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;66:203–208. doi: 10.1016/0378-1097(90)90283-v. [DOI] [PubMed] [Google Scholar]
- 3.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, Engelke G, Entian K-D, Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 4.Belasco J G, Higgins C F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988;72:15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- 5.Bierbaum G, Reis M, Szekat C, Sahl H-G. Construction of an expression system for engineering of the lantibiotic Pep5. Appl Environ Microbiol. 1994;60:4332–4338. doi: 10.1128/aem.60.12.4332-4338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, Sahl H-G. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl Environ Microbiol. 1997;62:385–392. doi: 10.1128/aem.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet P, Belasco J G. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 8.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl H-G. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan P T, Ohmori H, Tomizawa J, Lebowitz J. Nucleotide sequence and gene organisation of ColE1 DNA. J Biol Chem. 1985;260:8925–8935. [PubMed] [Google Scholar]
- 10.Chen C-Y A, Beatty J T, Cohen S N, Belasco J G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988;52:609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- 11.Dodd H M, Horn N, Chan W C, Giffard C J, Bycroft B W, Roberts G C K, Gasson M J. Molecular analysis of the regulation of nisin immunity. Microbiology. 1996;142:2385–2392. doi: 10.1099/00221287-142-9-2385. [DOI] [PubMed] [Google Scholar]
- 12.Emory S A, Bouvet P, Belasco J G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 13.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K-D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ersfeld-Dreßen H, Sahl H-G, Brandis H. Plasmid involvement in production of and immunity to the staphylococcin-like peptide Pep5. J Gen Microbiol. 1984;130:3029–3035. doi: 10.1099/00221287-130-11-3029. [DOI] [PubMed] [Google Scholar]
- 15.Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli. Anal Biochem. 1993;212:394–401. doi: 10.1006/abio.1993.1346. [DOI] [PubMed] [Google Scholar]
- 16.Geli V, Baty D, Pattus F, Lazdunski C. Topology and function of the integral membrane protein conferring immunity to colicin A. Mol Microbiol. 1989;3:679–687. doi: 10.1111/j.1365-2958.1989.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 17.Götz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 18.Gross E, Kiltz H, Nebelin E. Subtilin. VI. Die Struktur des Subtilins. Hoppe-Seyler’s Z Physiol Chem. 1973;354:810–812. [PubMed] [Google Scholar]
- 19.Gross E, Morell J L. The structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 20.Heidrich C, Pag U, Josten M, Metzger J, Jack R W, Bierbaum G, Jung G, Sahl H-G. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hue K K, Cohen S D, Bechhofer D H. A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J Bacteriol. 1995;177:3465–3471. doi: 10.1128/jb.177.12.3465-3471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaletta C, Entian K-D, Kellner R, Jung G, Reis M, Sahl H-G. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch Microbiol. 1989;152:16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- 23.Klein C, Entian K-D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klug G, Cohen S N. Combined actions of multiple hairpin loop structures and sites of rate-limiting endonucleolytic cleavage determine differential degradation rates of individual segments within polycistronic puf operon mRNA. J Bacteriol. 1990;172:5140–5146. doi: 10.1128/jb.172.9.5140-5146.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 26.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for producer immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 27.McMaster G K, Carmichael G G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer C, Bierbaum G, Heidrich C, Reis M, Süling J, Iglesias-Wind M I, Kempter C, Molitor E, Sahl H-G. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- 29.Peschel A, Götz F. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J Bacteriol. 1996;178:531–536. doi: 10.1128/jb.178.2.531-536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992;6:277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley A P. Nucleotide sequencing of the structural gene for colicin N reveals homology between the catalytic, C-terminal domains of colicins A and N. Mol Microbiol. 1987;1:317–325. doi: 10.1111/j.1365-2958.1987.tb01938.x. [DOI] [PubMed] [Google Scholar]
- 32.Reis M, Eschbach-Bludau M, Iglesias-Wind M I, Kupke T, Sahl H-G. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl Environ Microbiol. 1994;60:2876–2883. doi: 10.1128/aem.60.8.2876-2883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reis M, Sahl H-G. Genetic analysis of the producer self-protection mechanism (“immunity”) against Pep5. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom; 1991. pp. 320–332. [Google Scholar]
- 34.Rince A, Dufour A, Uguen P, Le Pennec J-P, Haras D. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl Environ Microbiol. 1997;63:4252–4260. doi: 10.1128/aem.63.11.4252-4260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 36.Sahl H-G. Pore formation in bacterial membranes by cationic lantibiotics. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom; 1991. pp. 347–358. [Google Scholar]
- 37.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 38.Sahl H-G, Brandis H. Production, purification and chemical properties of an antistaphylococcal agent produced by Staphylococcus epidermidis. J Gen Microbiol. 1981;127:377–384. doi: 10.1099/00221287-127-2-377. [DOI] [PubMed] [Google Scholar]
- 39.Sahl H-G, Reis M, Eschbach M, Szekat C, Beck-Sickinger A G, Metzger J, Stevanovic S, Jung G. Isolation of Pep5 prepeptides in different stages of modification. In: Jung G, Sahl H-G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: Escom; 1991. pp. 332–346. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleifer K H, Fischer U. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int J Syst Bacteriol. 1982;32:153–156. [Google Scholar]
- 42.Schramm E, Ölschläger T, Tröger W, Braun V. Sequence, expression and localization of the immunity protein for colicin B. Mol Gen Genet. 1988;21:176–182. doi: 10.1007/BF00338410. [DOI] [PubMed] [Google Scholar]
- 43.Siegers K, Entian K-D. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol. 1995;61:1082–1089. doi: 10.1128/aem.61.3.1082-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skaugen M, Abildgaard C I M, Nes I F. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet. 1997;253:674–686. doi: 10.1007/s004380050371. [DOI] [PubMed] [Google Scholar]
- 45.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wieland K-P, Wieland B, Götz F. A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene. 1995;158:91–96. doi: 10.1016/0378-1119(95)00137-u. [DOI] [PubMed] [Google Scholar]
- 47.Wong H C, Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci USA. 1986;83:3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 50.Zell R, Fritz H. DNA mismatch repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987;6:1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]