Abstract
Ninety-two strains of lactic acid bacteria (LAB) were isolated from a Malaysian food ingredient, chili bo, stored for up to 25 days at 28°C with no benzoic acid (product A) or with 7,000 mg of benzoic acid kg−1 (product B). The strains were divided into eight groups by traditional phenotypic tests. A total of 43 strains were selected for comparison of their sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) whole-cell protein patterns with a SDS-PAGE database of LAB. Isolates from product A were identified as Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus farciminis, Pediococcus acidilactici, Enterococcus faecalis, and Weissella confusa. Five strains belonging to clusters which could not be allocated to existing species by SDS-PAGE were further identified by 16S rRNA sequence comparison. One strain was distantly related to the Lactobacillus casei/Pediococcus group. Two strains were related to Weissella at the genus or species level. Two other strains did not belong to any previously described 16S rRNA group of LAB and occupied an intermediate position between the L. casei/Pediococcus group and the Weissella group and species of Carnobacterium. The latter two strains belong to the cluster of LAB that predominated in product B. The incidence of new species and subspecies of LAB in chili bo indicate the high probability of isolation of new LAB from certain Southeast Asian foods. None of the isolates exhibited bacteriocin activity against L. plantarum ATCC 14917 and LMG 17682.
Lactic acid bacteria (LAB) are utilized in the production and preservation of various fermented foods in Southeast Asia. Examples of such foods include soybean tempeh (20), tape ketan (5), fermented rice cake (puto) (16), and various foods produced in Thailand (35). In addition, LAB may occur as indigenous contaminants in a wide range of nonfermented foods in the region. One example is the Malaysian food ingredient chili bo (19). Compared with Western foods, the identity and distribution of LAB in Southeast Asian foods have not, with some exceptions (16, 21, 30, 33–35), been extensively examined.
Identification of LAB mostly depends on traditional phenotypic analyses, although molecular biology-based methods have become available (14, 26, 37). Hence, until now modern identification techniques have not been used to a large degree for the identification to the species level of LAB from Southeast Asian foods. Two recent reports, however, included pulsed-field gel electrophoresis (16) and randomly amplified polymorphic DNA analyses (30) for characterization at the subspecies level of various species of LAB in fermented rice cakes and Indonesian soy mash, respectively. For identification at the species level, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins has the advantage of being fairly simple and rapid to perform. However, for identification purposes, this technique requires an extensive database covering all known target species. Because such a database was developed for LAB (25, 27, 37), SDS-PAGE can be used for the demonstration of new LAB taxa, although DNA-DNA hybridization and/or rRNA sequence analysis is required for species confirmation in quantitative genotypic terms (37).
Interest in microorganisms as a component of biological diversity has been renewed in recent years (2). The interest in microorganisms occurring in foods is primarily due to the biotechnological potential of new bacterial species and strains. In the present study we selected chili bo as a potential source of new species or types of LAB because chili bo, as a low-pH product with an added carbohydrate source, will support the predominance of LAB. Chili bo is not a fermented product, but samples were stored for extended periods beyond the time required for spoilage in order to select for LAB. Chili bo is made on a cottage industry scale, offering ample opportunity for the introduction of a wide variety of microbes, including LAB, from the surrounding environment.
The aim of this study was to isolate and characterize LAB with potential biotechnological uses from the traditional Malaysian food chili bo.
MATERIALS AND METHODS
Processing and storage conditions of chili bo.
Chili bo was prepared by a commercial producer located in Petaling Jaya, Selangor, D.E., Malaysia. The manufacturing process has been described elsewhere (19), and the variable parameters were the addition of 7,000 mg of benzoic acid kg−1 and a fine-grinding step, where the temperature of the product rose to above 45 to 50°C for an extended period of time (product B), or a process without added benzoic acid or any fine grinding (product A). Two storage trials were done for each type of product. The packaged samples were stored at ambient temperatures (28 ± 2°C) for up to 25 days.
Isolation of strains.
A total of 92 single colonies of LAB were selected randomly from pinpoint colonies in plate count agar (PCA) (Oxoid) or from all colonies on de Man, Ragosa, and Sharpe-sorbic acid (MRS-S) agar (3), restreaked on either PCA or MRS-S agar, and examined for purity. Isolates were stored at 4°C in Cooked Meat Medium broth (Oxoid) or in a Protect bead storage system (STC, Heywood, Lancashire, United Kingdom) at −20°C. Isolation was done on days 0, 1, 3 (trial I; PCA only), and 25 for product A and days 0 and 25 for product B.
Phenotypical characterization of isolates.
LAB strains used as reference strains for the characterization and identification of the chili bo LAB isolates included Enterococcus faecalis ATCC 19433, Enterococcus faecium ATCC 19434, Lactobacillus acidophilus ATCC 4356, Lactobacillus zeae ATCC 393, Lactobacillus fermentum ATCC 9338, Lactobacillus plantarum ATCC 14917, Lactococcus lactis ATCC 11454, and Leuconostoc mesenteroides subsp. mesenteroides ATCC 23386.
Before being tested, the strains were subcultured twice overnight in MRS broth at 30°C. The following phenotypical tests were conducted: Gram reaction; oxidase-reaction; acid from glucose under anaerobic conditions (32); reduction of 1% (wt/vol) nitrate in MRS broth; phase-contrast microscopy of cell shape and motility; production of NH3 from arginine (18); production of gas from glucose or gluconic acid (tested with Durham tubes in APT broth [11] with trisodium citrate omitted); production of polymers from sucrose (32); reduction of tetrazolium (42); pH of <4.15 in La broth (31); growth on acetate agar (29); growth on MRS agar with 6.5 or 8% NaCl; growth on MRS agar at 4, 42, or 45°C; growth on MRS agar with 1,000 or 7,000 mg of sodium benzoate kg−1; and sensitivity to 30 μg of vancomycin g−1 in MRS agar. Sensitivity during growth on soft MRS agar (0.75%) plates to nisin produced by L. lactis ATCC 11454 was examined by the spot-on-lawn method (1). Production of antimicrobial compounds was tested by the use of the deferred test (1) against selected indicators among the ATCC reference strains and chili bo isolates.
The strains selected for SDS-PAGE analysis and a few additional strains were tested for production of acids from carbohydrates and related compounds by use of API (Montalieu, Vercieu, France) 50 CH strips and API CHL medium. The tests were done according to the instructions of the manufacturer, and the results were read after incubation of the strains at 37°C for 2 and 3 days. Identification was done by the computerized database program provided by the manufacturer.
SDS-PAGE of proteins and identification of isolates.
Preparation of cell extracts and polyacrylamide gel electrophoresis were done as described previously (27). Identification of the isolates was performed by comparison of their protein patterns with a database of normalized protein fingerprints derived from reference strains for almost all known species of LAB (25, 28). Pattern storage and comparison were performed on an MS-DOS-compatible PC with the software package GelCompar (version 4.0 [39]).
Sequencing of 16S rRNA genes.
Bacteria were cultured overnight in MRS broth, and 1 ml of culture was harvested by centrifugation. The cells were washed in 50:50 TE buffer (50 mM Tris, 50 mM EDTA, pH 8) by centrifugation and resuspended in 0.5 ml of 50:50 TE. Lysis was initiated by the addition of 50 μl of 10-mg/ml lysozyme. After incubation at 37°C for 30 min, 50 μl of 10-mg/ml proteinase K in 50:50 TE and 20 μl of 10% SDS (pH 7.2) were added and gently mixed, and the mixture was further incubated for 2 h at 56°C. Cell debris and protein were precipitated with 297 μl of 3 M potassium acetate. After centrifugation for 10 min at 16,000 × g the supernatant was added to 0.54 volume of isopropanol and gently mixed, and the mixture was spun down at 16,000 × g for 10 min. The pellet was washed twice in ice-cold ethanol by centrifugation, vacuum dried, and resuspended in ultrapure water (Millipore quality).
PCR amplification was performed as described by Vogel et al. (40). Oligonucleotides for both PCR amplification and sequencing were synthesized according to sequences and 16S rRNA positions given by Dewhirst et al. (10) and Paster and Dewhirst (23).
The PCR-amplified fragments were purified on Microspin columns (Pharmacia Biotech) and cycle sequenced (Thermo Sequenase fluorescence-labelled primer cycle sequencing kit; Amersham, Little Chalfont, England) on an A.L.F. sequencer (Pharmacia Biotech) with fluorescein-labelled primers.
Searches for 16S rRNA sequences were performed by FastA and BLAST in the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.). Sequences were aligned manually to the Escherichia coli 16S rRNA sequence and to the consensus sequence given by Lane (17). Maximum-likelihood analysis was performed by the fastDNAml program (22), including bootstrap analysis (12) run on an HP9000/819 computer.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this report have been deposited with GenBank under accession no. AF086706, AF086707, AF049743, AF049745, and AF949742 for the strains LMG 17702, LMG 17707, LMG 17676, LMG 17710, and LMG 17714, respectively.
RESULTS
Phenotypical identification of isolates.
A total of 69 strains of LAB were obtained from product A and 23 from product B, resulting in a total of 92 strains. Table 1 presents the distribution of isolates by type of product, trial, and storage time. All 92 isolates were shown to be LAB by their positive Gram reactions, absence of catalase and oxidase activity, fermentative catabolism of glucose, and lack of reduction of nitrate. They all grew under aerobic conditions, none produced spores in MRS broth, and they were all nonmotile, with the exception of one presumptive Streptococcus isolate. The isolates could be differentiated into eight major groups by a few key traditional phenotypic tests listed in Table 1. A total of 43 isolates were selected from these groups (except group 8) for identification by use of API 50 CH and SDS-PAGE of whole-cell proteins.
TABLE 1.
Sources of isolation and phenotypic characteristics used to divide the major groups of chili bo LAB
| Source or characteristic | Values
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 (22)a | 2 (9) | 3 (16) | 4 (23) | 5 (9) | 6 (7) | 7 (4) | 8 (2) | |
| Source of isolation | ||||||||
| Product | B | A | A | A | A | A | Ab | A |
| Day | 25 | 1, 3, 25 | 3, 25 | 0, 1, 25 | 0, 1 | 1, 3 | 0 | 0 |
| Trial | I, II | I, II | I | II | II | I | I, II | I |
| Phenotypic characteristics | ||||||||
| Rods | 22 | 9 | 16 | 23 | 8 | 0 | 0 | 0 |
| NH3 from arginine | 0 | 4 | 0 | 23 | 0 | 7 | 2 | 0 |
| Gas from glucose | 0 | 0 | 0 | 23 | 9 | 0 | 0 | 0 |
| Gas from gluconic acid | 0c | 0 | 16 | 21 | 9 | 0 | 0 | 0 |
| pH < 4.15 in La broth | 22 | 9 | 16 | 0 | 0 | 7 | 2 | 0 |
| Growth with vancomycind | 22 | 9 | 16 | 23 | 9 | 7 | 0 | 2 |
| Sensitivity to nisin | 1 | 9 | 14 | 23 | 9 | 6 | 3 | 2 |
Group (number of isolates examined).
One isolate from product B.
No growth.
At a concentration of 30 μg g−1.
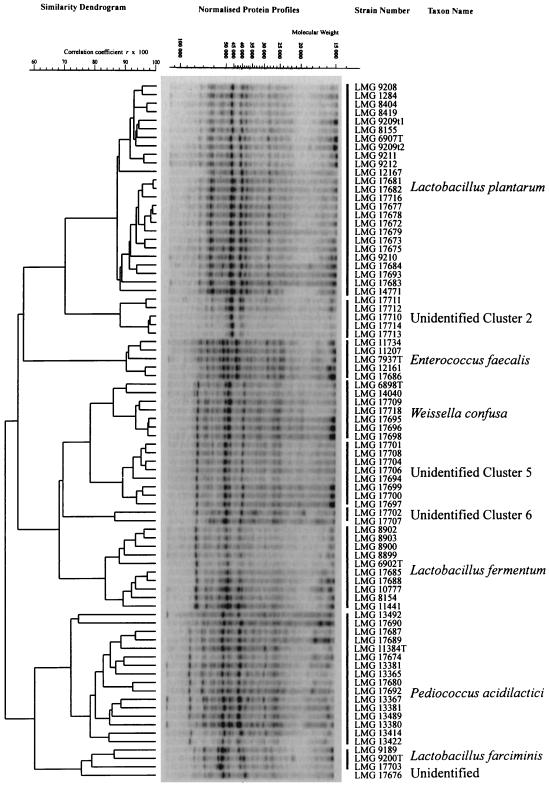
Group 1 consisted of 22 strains of an unidentified LAB isolated from product B. In comparison with other isolates, they grew slowly in APT broth at 30°C (data not shown) as well as in sterilized chili bo (19). No gas was produced from glucose. They did not grow at 45°C or with 6.5% NaCl. They were aciduric microorganisms, because they grew on acetate agar. The group 1 isolates were in general resistant to nisin (Table 1). When examined by SDS-PAGE, five of the group 1 isolates formed a separate cluster (Fig. 1, cluster 2) with a correlation level above 88% (r × 100). The cluster could not be identified at the species level; the highest correlation values obtained were with Lactobacillus brevis reference strains (r × 100 < 85%) (results not shown), but there was not a clear overall visual similarity between the protein patterns.
FIG. 1.
Dendrogram for SDS-PAGE profiles, including selected strains of LAB isolated from chili bo and reference strains. The left-hand side of the figure shows the mean correlation coefficient expressed as a percentage (r × 100) and represented as a dendrogram calculated by the unweighted average pair grouping method for 43 strains from chili bo. The points 15 to 320 of the 400 point traces, indicated by a gray bar on the molecular weight axis, were used for the calculation of similarities between the individual pairs of traces. The central part of the figure represents a gel with protein patterns of all of the strains compared. On the right-hand side of the figure are LMG numbers (Culture Collection of the Laboratory of Microbiology Gent, Department of Physiology, Biochemistry and Microbiology, Faculty of Sciences, University of Gent, Gent, Belgium) and cluster delineation by species. T, type strain.
Group 2 was a heterogeneous group of Lactobacillus spp. obtained from product A which did not produce gas from glucose. Two isolates were identified as L. plantarum by SDS-PAGE analysis although they did not produce gas from gluconic acid (Table 1). An additional isolate was identified as Lactobacillus farciminis, and one isolate (LMG 17676) remained unidentified (Table 2 and Fig. 1). LMG 17676 clustered among L. farciminis reference strains (Table 2 and Fig. 1) but differed within the molecular weight range of 40,000 to 50,000 from those of the few available reference strains of this species. Clearly, more reference strains of L. farciminis are necessary to estimate the heterogeneity of this species based on electrophoresis of their proteins.
TABLE 2.
Chili bo LAB isolates included in SDS-PAGE and API 50 CH analyses
| Group (API 50 CH characteristics) | Species | Strain |
|---|---|---|
| 1 | Unidentified, group 1 | LMG 17710 |
| Unidentified, group 1 | LMG 17711 | |
| Unidentified, group 1 | LMG 17712 | |
| Unidentified, group 1 | LMG 17713 | |
| Unidentified, group 1 | LMG 17714 | |
| 2a | L. plantarum | LMG 17672 |
| L. plantarum | LMG 17678 | |
| Unidentified, group 2 | LMG 17676 | |
| L. farciminis | LMG 17703 | |
| 3b | L. plantarum | LMG 17673 |
| L. plantarum | LMG 17675 | |
| L. plantarum | LMG 17677 | |
| L. plantarum | LMG 17679 | |
| L. plantarum | LMG 17681 | |
| L. plantarum | LMG 17682 | |
| L. plantarum | LMG 17683 | |
| L. plantarum | LMG 17684 | |
| L. plantarum | LMG 17693 | |
| L. plantarum | LMG 17716 | |
| 4c | L. fermentum | LMG 17685 |
| L. fermentum | LMG 17688 | |
| W. confusa | LMG 17695 | |
| W. confusa | LMG 17696 | |
| W. confusa | LMG 17698 | |
| W. confusa | LMG 17709 | |
| W. confusa | LMG 17718 | |
| Unidentified, group 4 | LMG 17694 | |
| Unidentified, group 4 | LMG 17697 | |
| Unidentified, group 4 | LMG 17699 | |
| Unidentified, group 4 | LMG 17700 | |
| Unidentified, group 4 | LMG 17701 | |
| Unidentified, group 4 | LMG 17704 | |
| Unidentified, group 4 | LMG 17706 | |
| Unidentified, group 4 | LMG 17708 | |
| 5d | Unidentified, group 5 | LMG 17702 |
| Unidentified, group 5 | LMG 17707 | |
| 6 | P. acidilactici | LMG 17674 |
| P. acidilactici | LMG 17680 | |
| P. acidilactici | LMG 17687 | |
| P. acidilactici | LMG 17689 | |
| P. acidilactici | LMG 17690 | |
| P. acidilactici | LMG 17692 | |
| 7 | E. faecalis | LMG 17686 |
Characteristics of L. plantarum isolates are as in group 3 except negative for rhamnose in API 50 CH.
Characteristics are the same as those of L. plantarum isolates in group 2 except positive for rhamnose in API 50 CH.
API 50 CH characteristics: L. fermentum and W. confusa are negative for l-arabinose but unidentified isolates are positive; L. fermentum is positive for raffinose, and other isolates are negative.
Both isolates are raffinose positive in API 50 CH.
Sixteen strains of Lactobacillus (group 3) all showed similar phenotypes (Table 1). They all produced gas from gluconic acid but not from glucose. A selection of 10 strains were all identified as L. plantarum by use of API 50 CH as well as the SDS-PAGE databases. Including the two L. plantarum isolates from group 2, SDS-PAGE grouped all of these strains at a correlation level above 87% (r × 100) (Fig. 1).
A large group of LAB (group 4) including 23 isolates was distinguished by their production of gas from glucose and NH3 from arginine and their inability to acidify La broth below pH 4.15. A total of 15 isolates were examined by SDS-PAGE, resulting in their identification as L. fermentum (two strains), Weissella confusa (five strains), and eight unidentified isolates (Table 2). The unidentified isolates exhibited a correlation coefficient r of >0.89 (Fig. 1, unidentified cluster 5) and showed protein patterns which are similar to the patterns obtained from W. confusa (Fig. 1), resulting in a correlation of almost 79% (r × 100). This group most likely represents a subspecies of W. confusa. However, all strains could be phenotypically distinguished from W. confusa by the production of acid from l-arabinose in the API 50 CH kit (Table 2).
Group 5 comprised nine strains of heterofermentative LAB able to produce gas from glucose but not NH3 from arginine. Seven of these showed similar carbohydrate fermentation patterns, did not produce polymers from sucrose (data not shown), produced gas from gluconate, and, with one exception, grew in the presence of 8% NaCl. They were tentatively identified as Weissella paramesenteroides when compared with information given by Garvie (13), Barreau and Wagener (6), and Collins et al. (8). However, this identification could not be confirmed by SDS-PAGE for two of the isolates investigated (Fig. 1, unidentified cluster 6). The strains resembled each other (r × 100 > 86%) but were not closely related to any other LAB species, as the correlation level with the nearest neighbor was <70% (r × 100).
Two isolates in group 5 differed from the other strains by having a distinct rod shape. They produced polymers from sucrose but their carbohydrate fermentation patterns did not resemble those of any Leuconostoc spp., Weissella spp., or Lactobacillus viridescens possessing this phenotypic trait (data not shown). They were not examined by SDS-PAGE and remain unidentified.
Group 6 consisted of Pediococcus strains that were differentiated from other LAB by their cell morphology and from Aerococcus by their resistance to vancomycin, production of NH3 from arginine, and ability to grow at 45°C. Six of the isolates were further identified as Pediococcus acidilactici by SDS-PAGE (Table 2), but the species was found to be considerably heterogeneous (r × 100 < 80%) (Fig. 1).
Isolates belonging to group 7 were identified as Enterococcus spp. due to their cell morphologies and sensitivity to vancomycin. One isolate was identified both by API 50 CH and SDS-PAGE as E. faecalis. One additional isolate was presumptively identified as Enterococcus avium by the fermentation pattern of carbohydrates and by the absence of NH3 production from arginine. The two remaining isolates were not further identified.
Two strains of presumptive streptococci (group 8) could not be assigned to any known species by use of the traditional phenotypic analyses. These strains were not further examined by SDS-PAGE of whole-cell proteins.
Bacteriocin production.
All of the strains presented in Table 2, except for one strain of the group 1 LAB and one strain of L. plantarum, were examined by the deferred tests for antimicrobial activity towards L. plantarum ATCC 14917. None exhibited antimicrobial activity towards this strain. Thirty-nine of these strains were also tested against the chili bo isolate L. plantarum LMG 17682 with the same negative result.
Furthermore, 19 strains of the group 1 LAB exhibited no antagonistic activity against five additional strains: L. casei ATCC 393, L. fermentum ATCC 9338, L. lactis ATCC 11454, E. faecium ATCC 19434, and L. mesenteroides subsp. mesenteroides ATCC 23386.
16S rRNA gene sequence analysis.
Two isolates from the unidentified group 1 (LMG 17710 and LMG 17714), one unidentified isolate from group 2 (LMG 17676), and two unidentified isolates from group 5 (LMG 17702 and LMG 17707) were selected for analysis. Sequences were obtained for the region 24 to 1492 (E. coli positions) of the 16S rRNA gene. The sequences were 1,477, 1,479, and 1,488 bases in LMG 17710, LMG 17714, and LMG 17676, respectively, and 1,497 bases in LMG 17702 and LMG 17707. The highest similarity was found between the group 5 isolates LMG 17702 and LMG 17707 (99.9%). A high similarity was also found between the group 1 isolates LMG 17710 and LMG 17714 (99.3%). LMG 17710 was related to LMG 17676 with 91.1% similarity and to LMG 17702 and LMG 17707 with 89 or 88.9% similarity. LMG 17714 was related to LMG 17676 with 91.4% similarity and to LMG 17702 and LMG 17707 with 90 or 89.9% similarity. LMG 17676 was related to LMG 17702 and LMG 17707 with 87 or 86.9% similarity.
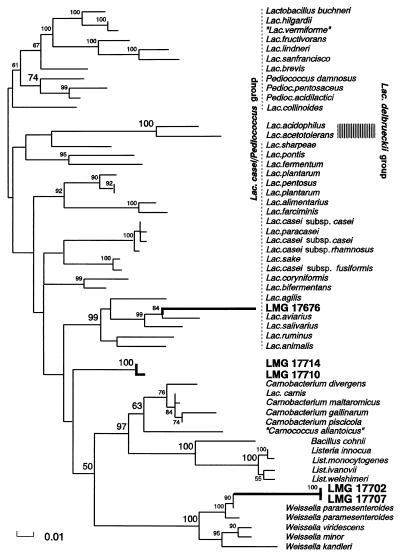
The sequence of each of the five strains was used to search the GenBank and EMBL databases. The phylogenetic analysis was performed with the 16S rRNA gene sequences of the 50 taxa most closely related to the five presumed LAB. The region 29 to 1481 (E. coli positions) was aligned, leaving 1,457 positions for phylogenetic analysis. The phylogenetic tree based on maximum likelihood is shown in Fig. 2.
FIG. 2.
Phylogenetic relationship of LMG 17702, LMG 17707, LMG 17710, LMG 17714, and LMG 17676 based on maximum-likelihood analysis of 16S rRNA sequences. The numbers indicate significance values higher than 50% for support of specific nodes based on bootstrap analysis. The thick lines represent sequences obtained in the present study.
The group 2 strain LMG 17676 was distantly related to a group consisting of Lactobacillus agilis, Lactobacillus aviarius, Lactobacillus salivarius, Lactobacillus ruminis, and Lactobacillus animalis, with similarities of 89.3 to 91.2%.
The group 5 strains LMG 17702 and LMG 17707 were highly similar to each other, with only one difference in the 16S rRNA sequence at E. coli position 1265. These strains were most closely related to Weissella, with 93 to 95% similarity to W. paramesenteroides and 91% similarity to W. viridescens.
The group 1 strains LMG 17710 and LMG 17714 did not fall within previously recognized groups of LAB and occupied an intermediate position between the L. casei/Pediococcus group (with 88.8 to 91.3% similarity to the subgroup of L. agilis, L. aviarius, L. salivarius, L. ruminus, L. animalis, and LMG 17676) and the Weissella group (86.6 to 90.6% similarity) and the Carnobacterium group (88.1 to 90.2% similarity). The two strains represent a new group of LAB, based on 16S rRNA sequence comparison.
DISCUSSION
The results reported here constitute part of a study focusing on identification of LAB derived from a source of food products hitherto not well examined: Malaysian fermented and nonfermented foods produced on a cottage industry scale. The high potential for isolating new types or species of LAB was demonstrated in this study by SDS-PAGE screening of 43 isolates from two variants of the product chili bo. It was observed from SDS-PAGE that 16 strains (37%) belonged to four taxa not covered by the very extensive database for known species of LAB (25, 27). The significance of this finding can be illustrated by the fact that screening of 157 strains of LAB from a variety of traditional Greek dairy products (36), 30 strains from Portuguese wines and must (24), and 200 strains from traditional fermented products (38) and from sourdough (41) always resulted in only a few strains (<2%) which could not be identified by the database used for the study. Additional 16S sequences obtained from strains within three of the four taxa that were not identifiable by SDS-PAGE revealed that they represented new genera or species based on the lack of match with sequences stored in the GenBank and EMBL databases. Of these, LMG 17676 was distantly related to species of the L. casei/Pediococcus group, representing both obligately homofermentative species and facultative heterofermentative and obligately heterofermentative species (7, 37). The two strains LMG 17702 and LMG 17707 were related to various species within the genus Weissella. The two strains LMG 17710 and 17714 of group 1 represent a new group of LAB distantly related to the groups L. casei/Pediococcus, Weissella, and Carnobacterium.
Two types of chili bo were examined in this study: with and without 7,000 mg of benzoic acid kg−1. Although this additive is only allowed in amounts up to 1,000 mg of benzoic acid kg−1 according to Malaysian food legislation (4), local manufacturers frequently violate this regulation by adding benzoic acid up to 7,000 mg kg−1 (41a). Product B, with 7,000 mg of benzoic acid kg−1, would therefore allow a growth of LAB representative for a commercial product, whereas product A, without added benzoic acid, would allow for growth of a wider range of LAB initially present in the product. Both products spoiled after 1 to 2 days of storage at 28°C (19). Thus, samples from day 3 and day 25 represent spoiled products which were included in the study to facilitate selection of LAB.
The two types of chili bo products differed dramatically in the composition of the LAB flora. Product A contained high levels of LAB, ranging from log 6 CFU g−1 initially to log 8 CFU g−1 after 2 to 25 days of storage (19) and contained LAB of several species. Within each species considerable heterogeneity was observed when analyzed by SDS-PAGE, indicating that the majority, if not all, of the isolates within each species represented different clonal lines. The species encountered in product A differed in two trials (Table 1) although the products were from the same manufacturer. This might be due to variations in raw material or in the production process.
Product B initially contained low levels of LAB at log 2 CFU g−1, which slowly increased to log 7 to log 8 CFU g−1 by day 25 (19). All of the strains isolated at day 25 belonged to a single species (group 1), whereas one strain of E. faecalis was isolated at day 0. Although one species of LAB predominated in product B after 25 days, the SDS-PAGE data revealed more than one clonal line to be present. The lack of species diversity of LAB in product B can be explained by the addition of 7,000 mg of benzoic acid kg−1 and the fine-grinding procedure, which exposes the microbial flora to temperatures above 45 to 50°C for an extended period. The reason for the predominance of the group 1 LAB in product B is not known, but it might be related to resistance to benzoic acid as well as to the mild heat treatment procedure used for product B. The fact that this predominant group 1 LAB was not encountered in product A was probably due to the slow growth of this species compared with the other LAB isolated (reference 19 and data not shown).
The predominant species and types of LAB in both products were able to grow on acetate agar (data not shown), indicating that they are aciduric microorganisms. Nonaciduric LAB might not grow well on the selective MRS-S agar (15), which was used as one of the isolation media in this study. The low pH values (<4.5) in chili bo probably selected for aciduric LAB.
The results obtained by Leisner et al. (19) show that chili bo provides a substrate for the growth of several species of LAB in the absence of inhibitory parameters (product A). It is interesting that only L. farciminis out of a total of 43 isolates examined was able to catabolize starch, the single carbohydrate source added to chili bo (data not shown). Fermentable carbohydrates probably are present in chili bo as a result of amylases generated by other microorganisms, e.g., Bacillus subtilis, or as a result of the breakdown of polysaccharides other than starch, e.g., the cellulose, hemicelluloses, and pectic substances present in the chili.
The potential biotechnological applications of the chili bo LAB were not examined to a wide extent. None of the isolates produced antagonistic activity towards L. plantarum ATCC 14917 and LMG 17682. The possibility exists that such an activity could be observed against other species of LAB although no such result was obtained by screening group 1 LAB for activity against five additional LAB species.
The rapid spoilage of the chili bo products was not caused by the LAB (19). The incidence of heterofermentative LAB might be a problem on some occasions due to production of gas and/or slime. Their spoilage potential might easily be controlled by various methods, including low pH, the presence of 1,000 mg of benzoic acid kg−1, storage at refrigeration temperature, or the addition of nisin (19).
This study demonstrates the high potential for isolating new species and types of LAB from Malaysian foods. We are currently expanding this research to other Malaysian foods made on a cottage industry scale, such as tapai, durian paste, tempeh, and various fermented seafoods. Further investigations into the taxonomic classification of the new taxa demonstrated in this study are in progress.
ACKNOWLEDGMENTS
This study was partly funded by the Malaysian Government through the IRPA mechanism (Project no. 51094). B.P. is indebted to the Federal Office for Scientific, Technical and Cultural Affairs for research and personnel grants as a node of the Belgian Coordinated Collections of Microorganisms (BCCM/LMG).
Gitte Frederiksen and Stina Holm are thanked for technical assistance with the 16S rRNA analysis.
REFERENCES
- 1.Ahn C, Stiles M E. Antibacterial activity of lactic acid bacteria isolated from vacuum-packaged meats. J Appl Bacteriol. 1990;60:302–310. doi: 10.1111/j.1365-2672.1990.tb01520.x. [DOI] [PubMed] [Google Scholar]
- 2.Alsopp D, Colwell R R, Hawksworth D L. Proceedings of the IUBS/IUMS workshop held at Egham, U.K., 10–13 August 1993 in support of the IUBS/UNESCO/SCOPE “DIVERSITAS” programme. Cambridge, United Kingdom: CAB International, University Press; 1995. [Google Scholar]
- 3.Anonymous. de Man, Rogosa and Sharpe agar with sorbic acid (MRS-S agar) Int J Food Microbiol. 1987;5:230–232. [Google Scholar]
- 4.Anonymous. Details on food regulations amendments from 1987 to June, 1990. Kuala Lumpur, Malaysia: MDC. Sdn. Bhd.; 1990. Food act 1983 and food regulations 1985. [Google Scholar]
- 5.Ardhana M M, Fleet G H. The microbial ecology of tape-ketan fermentation. Int J Food Microbiol. 1989;9:157–165. doi: 10.1016/0168-1605(89)90086-x. [DOI] [PubMed] [Google Scholar]
- 6.Barreau B, Wagener W. Characterization of Leuconostoc lactis strains from human sources. J Clin Microbiol. 1990;28:1728–1733. doi: 10.1128/jcm.28.8.1728-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins M D, Rodrigues U, Ash C, Aguirre M, Farrow J A E, Martinez-Murcia A, Phillips B A, Williams A M, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett. 1991;77:5–12. [Google Scholar]
- 8.Collins M D, Samelis J, Metaxopoulos J, Wallbanks S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J Appl Bacteriol. 1993;75:595–603. doi: 10.1111/j.1365-2672.1993.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 9.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 10.Dewhirst F E, Paster B J, Bright P L. Chromobacterium, Eikenella, Kingella, Neisseria, Simonsiella, and Vitreoscilla species comprise a major branch of the beta group Proteobacteria by 16S ribosomal ribonucleic acid sequence comparison: transfer of Eikenella and Simonsiella to the family Neisseriaceae (emend.) Int J Syst Bacteriol. 1988;38:258–266. [Google Scholar]
- 11.Difco Laboratories, Inc. Difco manual. 10th ed. Detroit, Mich: Difco Laboratories Inc.; 1984. p. 1071. [Google Scholar]
- 12.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 13.Garvie E. Genus Leuconostoc. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: Williams & Wilkins; 1986. pp. 1071–1075. [Google Scholar]
- 14.Hertel C, Ludwig W, Pot B, Kersters K, Schleifer K-H. Differentiation of lactobacilli in fermented milk products by using oligonucleotide probes and electrophoretic protein profiles. Syst Appl Microbiol. 1993;16:463–467. [Google Scholar]
- 15.Holzapfel W H. Culture media for non-sporulating Gram-positive food spoilage bacteria. Int J Food Microbiol. 1992;17:113–133. doi: 10.1016/0168-1605(92)90110-o. [DOI] [PubMed] [Google Scholar]
- 16.Kelly W J, Asmundson R V, Harrison G L, Huang C M. Differentiation of dextran-producing Leuconostoc strains from fermented rice cake (puto) using pulsed-field gel electrophoresis. Int J Food Microbiol. 1995;26:345–352. doi: 10.1016/0168-1605(94)00146-w. [DOI] [PubMed] [Google Scholar]
- 17.Lane D. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–147. [Google Scholar]
- 18.Leisner J J, Millan J C, Huss H H, Larsen L M. Production of histamine and tyramine by lactic acid bacteria isolated from vacuum-packed sugar-salted fish. J Appl Bacteriol. 1994;76:417–423. doi: 10.1111/j.1365-2672.1994.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 19.Leisner J J, Rusul G, Wee B W, Boo H C, Muhammad K. Microbiology of chili bo, a popular Malaysian food ingredient. J Food Prot. 1997;60:1235–1240. doi: 10.4315/0362-028X-60.10.1235. [DOI] [PubMed] [Google Scholar]
- 20.Nout M J R, Beernink G, Bonants-van Laarhoven T M G. Growth of Bacillus cereus in soybean tempeh. Int J Food Microbiol. 1987;4:293–301. [Google Scholar]
- 21.Okada S, Daengsubha W, Uchimura T, Ohara N, Kozaki M. Flora of lactic acid bacteria in miang produced in northern Thailand. J Gen Appl Microbiol. 1986;32:57–65. [Google Scholar]
- 22.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 23.Paster B J, Dewhirst F E. Phylogeny of campylobacters, wollinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 24.Patarata L, Pimentel M S, Pot B, Kersters K, Faia A M. Identification of lactic acid bacteria isolated from Portuguese wines and musts by SDS-PAGE. J Appl Bacteriol. 1994;76:288–293. [Google Scholar]
- 25.Pot B, Janssens D. The potential of a culture collection for identification and maintenance of lactic acid bacteria. In: Foo E L, Griffin H G, Mollby R, Heden C G, editors. The lactic acid bacteria. Proceedings of the first Lactic Acid Bacteria Computer Conference. Norfolk, Va: Horizon Scientific Press; 1993. pp. 81–87. [Google Scholar]
- 26.Pot B, Hertel C, Ludwig W, Descheemaeker P, Kersters K, Schleifer K-H. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J Gen Microbiol. 1993;139:513–517. doi: 10.1099/00221287-139-3-513. [DOI] [PubMed] [Google Scholar]
- 27.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organisms protein fingerprints. In: Goodfellow M, O’Donnel A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, England: John Wiley & Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 28.Pot B, Ludwig W, Kersters K, Schleifer K-H. Taxonomy of lactic acid bacteria. In: de Vuyst L, Vandamme E J, editors. Bacteriocins of lactic acid bacteria. New York, N.Y: Blackie Academic & Professional; 1994. pp. 13–89. [Google Scholar]
- 29.Rogosa M, Mitchell J A, Wiseman R F. A selective medium for the isolation of oral and fecal lactobacilli. J Bacteriol. 1951;62:132–133. doi: 10.1128/jb.62.1.132-133.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Röling W F M, van Verseveld H W. Characterization of Tetragenococcus halophila populations in Indonesian soy mash (Kecap) fermentations. Appl Environ Microbiol. 1996;62:1203–1207. doi: 10.1128/aem.62.4.1203-1207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw B G, Harding C D. A numerical taxonomic study of lactic acid bacteria from vacuum-packed beef, pork, lamb and bacon. J Appl Bacteriol. 1984;56:25–40. doi: 10.1111/j.1365-2672.1984.tb04693.x. [DOI] [PubMed] [Google Scholar]
- 32.Shaw B G, Harding C D. Atypical lactobacilli from vacuum packaged meats: comparison by DNA hybridization, cell composition and biochemical tests with description of Lactobacillus carnis sp. nov. Syst Appl Microbiol. 1985;6:291–297. [Google Scholar]
- 33.Tanasupawat S, Daengsubha W. Pediococcus species and related bacteria found in fermented foods and related materials in Thailand. J Gen Appl Microbiol. 1983;29:487–506. [Google Scholar]
- 34.Tanasupawat S, Ezaki T, Suzuki K-I, Okada S, Komagata K, Kozaki M. Characterization and identification of Lactobacillus pentosus and Lactobacillus plantarum strains from fermented foods in Thailand. J Gen Appl Microbiol. 1992;38:121–134. [Google Scholar]
- 35.Tanasupawat S, Komagata K. Lactic acid bacteria in fermented foods in Thailand. World J Microbiol Biotechnol. 1995;11:253–256. doi: 10.1007/BF00367094. [DOI] [PubMed] [Google Scholar]
- 36.Tsakalidou E, Manolopoulou E, Kabaraki E, Zoidou E, Pot B, Kersters K, Kalantzopoulos G. The combined use of whole-cell protein extracts for the identification (SDS-PAGE) and enzyme activity screening of acid bacteria isolated from traditional Greek dairy products. Syst Appl Microbiol. 1994;17:444–458. [Google Scholar]
- 37.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Berg D J C, Smits A, Pot B, Ledeboer A M, Kersters K, Verbakel J M A, Verrips C T. Isolation, screening and identification of lactic acid bacteria from traditional food fermentation processes and culture collections. Food Biotechnol. 1993;7:189–205. [Google Scholar]
- 39.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 40.Vogel B F, Jørgensen K, Christensen H, Olsen J E, Gram L. Differentiation of Shewanella putrefaciens and Shewanella alga on the basis of whole-cell protein profiles, ribotyping, phenotypic characterization, and 16S rRNA gene sequence analysis. Appl Environ Microbiol. 1997;63:2189–2199. doi: 10.1128/aem.63.6.2189-2199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel R, Böcker G, Stolz P, Ehrmann M, Fanta D, Ludwig W, Pot B, Kersters K, Schleifer K-H, Hammes W P. Identification of lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int J Syst Bacteriol. 1994;44:223–229. doi: 10.1099/00207713-44-2-223. [DOI] [PubMed] [Google Scholar]
- 41a.Wee, B. W. Unpublished results.
- 42.Wilkinson B J, Jones D. A numerical taxonomic survey of Listeria and related bacteria. J Gen Microbiol. 1977;98:399–421. doi: 10.1099/00221287-98-2-399. [DOI] [PubMed] [Google Scholar]