Abstract
BACKGROUND:
Automated segmentation using convolutional neural networks (CNNs) have been developed using 4D flow MRI. To broaden usability for congenital heart disease (CHD), training with multi-institution data is necessary. However, the performance impact of heterogeneous multi-site and multi-vendor data on CNNs is unclear.
PURPOSE:
To investigate multi-site CNN segmentation of 4D flow MRI for pediatric blood flow measurement.
STUDY TYPE:
Retrospective.
POPULATION:
174 subjects across two sites (female:46%; N=38 healthy controls, N=136 CHD patients). Participants from site 1 (N=100), site 2 (N=74) and both sites (N=174) were divided into subgroups to conduct 10-fold cross validation (10% for testing, 90% for training).
FIELD STRENGTH/SEQUENCE:
3T/1.5T; retrospectively gated gradient recalled echo-based 4D flow MRI
ASSESSMENT:
Accuracy of the 3D CNN segmentations trained on data from single site (single-site CNNs) and data across both sites (multi-site CNN) were evaluated by geometrical similarity (Dice score, human segmentation as ground truth) and net flow quantification at the ascending aorta (Qs), main pulmonary artery (Qp), and their balance (Qp/Qs), between human observers, single-site and multi-site CNNs.
STATISTICAL TESTS:
Kruskal-Wallis test, Wilcoxon rank-sum test, and Bland-Altman analysis. A P-value <0.05 was considered statistically significant.
RESULTS:
No difference existed between single-site and multi-site CNNs for geometrical similarity in the aorta by Dice score (site 1: 0.916 vs. 0.915, P=0.55; site 2: 0.906 vs. 0.904, P=0.69) and for the pulmonary arteries (site 1: 0.894 vs. 0.895, P=0.64; site 2: 0.870 vs. 0.869, P=0.96). Qs site-1 medians were 51.0–51.3ml/cycle (P=0.81) and site-2 medians were 66.7–69.4ml/cycle (P=0.84). Qp site-1 medians were 46.8–48.0ml/cycle (P=0.97) and site-2 medians were 76.0–77.4ml/cycle (P=0.98). Qp/Qs site-1 medians were 0.87–0.88 (P=0.97) and site-2 medians were 1.01–1.03 (P=0.43). Bland-Altman analysis for flow quantification found equivalent performance.
DATA CONCLUSION:
Multi-site CNN-based segmentation and blood flow measurement are feasible for pediatric 4D flow MRI and maintain performance of single-site CNNs.
Keywords: deep learning, 4D flow, congenital heart diseases, pediatrics
INTRODUCTION
Pediatric time-resolved 3D phase-contrast MRI (4D flow MRI) is a promising tool to quantify complex flow hemodynamics in the presence of congenital heart disease (CHD) (1–3). 4D flow MRI enables a comprehensive assessment of proximal cardiovascular function, with the flexibility to perform post-hoc hemodynamic analysis, even in the presence of abnormal CHD anatomy (4–7). While the approach has the potential to reduce time via simplified planning, accurate 4D flow measurements are dependent on careful vessel identification and segmentation during post-processing. Currently, most 3D segmentation approaches need human interaction to some extent and thus lengthy processing times to identify structures, to manually segment vessels of interest, and to quantify flow metrics (for the segmentation process, studies have reported at least 10 minutes for the aorta alone (8)). This is further complicated in pediatric patients with CHD, who can present with highly complex cardiac and great vessel anatomy (9). The added complexity further increases processing time and requires specialized knowledge of CHD (10).
A recent study utilizing a deep learning-based approach has enabled fully automated and fast segmentation of the aorta in adult 4D flow MRI exams with performance similar to human observers (8). However, this initial study, which employed a convolutional neural network (CNN), has been limited to a single site and single vendor platform, and was only capable of identifying a single label or vascular territory (i.e., the thoracic aorta). Given that CHDs involve multiple vascular territories and abnormal communication between systemic and pulmonary circulation, segmentation of both the aorta and pulmonary arteries (PAs) is needed to assess the existence or contribution of shunt flow (which is quantified by pulmonary to systemic flow ratio) (4).
For CNNs to achieve high performance across a variety of CHD patients, training data with a wide range of pathologies such as Tetralogy of Fallot (TOF) and hypoplastic left heart syndrome (HLHS), including rare CHDs, is required. Combining datasets across institutions is thus necessary, especially in the pediatric setting. This need for multiple sites introduces additional complexities as 4D flow data will vary across institutions due to practice preferences, MRI vendors, standard acquisition parameters, field of view (FOV), and varying image quality and contrast.
The aim of this study was to investigate the segmentation performance of a multi-label U-net CNN trained using multi-institutional data obtained on two different vendor platforms. We hypothesized that the multi-site CNN will (1) achieve human-level segmentation accuracy, (2) segment the proximal pulmonary and systemic vasculature with equivalent performance to single site data, and (3) outperform the single-site CNN for cases with complex CHD.
MATERIALS AND METHODS
Study Cohort
This Health Insurance Portability and Accountability Act (HIPAA)–compliant study was approved by the institutional review boards at Children’s Hospital Colorado (Aurora, CO; site 1) and Ann & Robert H. Lurie Children’s Hospital of Chicago (Chicago, IL; site 2). All patients were retrospectively enrolled with waiver of consent. A total of 174 subjects across a wide range of ages (range: 0.3 – 56.3, median: 15.5 years) and pathologies who underwent 4D flow MRI from 2014 to 2020 across two pediatric heart centers were retrospectively included in this study. A total of 136 patients underwent standard of care cardiothoracic MRI: 85 from site 1 and 51 from site 2. Additionally, 38 healthy controls who underwent research 4D flow scans or patients who received standard of care exams and deemed to have no evidence of cardiovascular disease by the radiologist at each institution were included as healthy controls at site 1 (N=15) and site 2 (N=23). All healthy research controls recruited for research-only exams provided written informed consent. Patient demographics and frequently seen abnormalities are summarized in Table 1 and Table S1, respectively.
Table 1.
Patient demographics and scan parameters.
| Site 1 | Site 2 | P value | |
|---|---|---|---|
| Patients | |||
| N (m/f) | 100 (51/49) | 74 (43/31) | - |
| Controls (%) | 15 (15) | 23 (31) | - |
| Patients (%) | 85 (85) | 51 (69) | - |
| TOF | 25 (25) | 13 (18) | - |
| BAV | 10 (10) | 7 ( 9) | - |
| TGA | 0 ( 0) | 7 ( 9) | - |
| Fontan | 6 ( 6) | 0 ( 0) | - |
| Age [y] | 15.2 [0.3– 51.7] | 15.9 [ 1.8– 56.3] | 0.83 |
| Weight [kg] | 52.0 [ 6– 110.0] | 57.2 [10.2–114.8] | 0.09 |
| Height [cm] | 159.0 [ 61– 202.0] | 164.0 [16.5–186.0] | 0.18 |
| BSA [m2] | 1.5 [0.3– 2.4] | 1.65 [ 0.4– 2.4] | 0.17 |
| Scan parameters | |||
| Echo time [ms] | 2.0–3.6 | 2.1–3.0 | 0.65 |
| Repetition time [ms] | 3.8–5.9 | 4.7–5.7 | <0.001 |
| Flip angle [deg] | 6–10 | 7–25 | <0.001 |
| Voxel size [mm] | 1.3×1.3×2.5 – 2.7×2.7×2.8 | 1.4×1.3×1.3 – 2.8×2.4×2.4 | <0.001 |
| Velocity encoding [cm/s] | 120–400 | 80–350 | 0.002 |
| Temporal resolution [ms] | 19.8–75.1 | 37.8–45.6 | <0.001 |
| Acquisition matrix | 64×64×35 – 224×224×104 | 160×70×26 – 160×140×96 | <0.001 |
| Field of view [mm3] | 150×150×72 – 500×500×100 | 200×150×101 – 380×295×160 | 0.26 |
| Acceleration factor | 3x SENSE (2x (phase),1.5x (slice)) | 2x GRAPPA | - |
P<0.05 is shown by bold numbers. Age, weight, height, and body surface area (BSA) are shown as median [range]. Scan parameters are shown as min-max. TOF, Tetralogy of Fallot; BAV, bicuspid aortic valve; TGA, transposition of the great arteries.
MRI
All patients underwent clinical cardiothoracic MRI including a retrospective ECG-gated and gradient recalled echo 4D flow MRI with full volumetric coverage of the heart, thoracic aorta, and first branch of the PAs (sagittal-oblique 3D volume). A respiratory navigator beam was placed on the lung-liver interface to accept data weighted to the expiration phase. Site 1 involved Philips scanners (Ingenia and Ingenia Elition X, Philips Healthcare, Best, Netherlands) and site 2 used Siemens (Aera, Siemens Medical Systems, Erlangen, Germany). Specifically, scans were performed using either 1.5T (N = 18; Philips Ingenia / 74; Siemens Aera), or 3T (N=82; Philips Ingenia and Ingenia Elition X). Individual site scan parameters are presented in Table 1. Of note, site 1 used an acceleration factor of SENSE = 2 in the phase encoding direction, and 1.5 in the slice direction while site 2 used GRAPPA = 2, leading to narrower FOV in site 2 for acceptable scan time. Data for all subjects were acquired during free-breathing with respiratory navigator gating placed on the lung-liver interface.
Data Analysis
The data preparation, training, validation, and testing steps are illustrated in Fig. 1. All 4D flow data were processed by in-house MATLAB (R2019b; Mathworks, Natick, MA, USA) scripts to remove eddy current effects, to mask noise, and to derive phase-contrast MRA (PC-MRA) (11) , computed from magnitude images and phase images using the following equation:
| (1) |
where T is total number of cardiac time frames of the dataset; are the velocity components in x, y and z directions; is magnitude data. Based on the PC-MRA, 3D segmentations of both the aorta and PAs were manually created by observers (AB, 3 years MS human anatomy; HB, 3 years MS Bioengineering) from each institution trained and overseen by practicing Radiologists specializing in CMR (LPB, 15 years; CR, 19 years. For 3D segmentation, site 1 used 3D slicer (http://www.slicer.org), while site 2 used Mimics (Materialise, Leuven Belgium). Prior to segmentation, a consensus approach was determined to maintain consistency across the sites for the segmentations, as is depicted in Fig. 2. Briefly, the aorta was segmented from the aortic valve plane to the furthest visible portion of the abdominal aorta in the FOV. The brachiocephalic trunk, right carotid artery, and left subclavian arteries were segmented from the aortic arch takeoff to approximately the distance of one ascending aortic diameter length downstream. PA segmentation included the right ventricular outflow tract, main pulmonary artery (MPA), and left and right pulmonary arteries (LPA and RPA). The right ventricular outflow tract was transected orthogonal to the outflow tract axis where the tangent of the axis was vertical in the sagittal orientation. The RPA was segmented to the edge of the FOV while the LPA was segmented as far as could be visualized without exceeding the length of the RPA. The first branches of the RPA and LPA were included when PC-MRA contrast was sufficient. In addition, due to the wide variability of CHDs, case-specific criteria treatment was required. For example, in the presence of signal void artifact caused by stents or artificial valves, the observers segmented the vessel shape by inference using the proximal and distal vessel wall adjacent to the artifact. Additionally, the PAs were not segmented for patients who underwent the Fontan procedure, due to absence of a functional MPA.
Fig. 1.
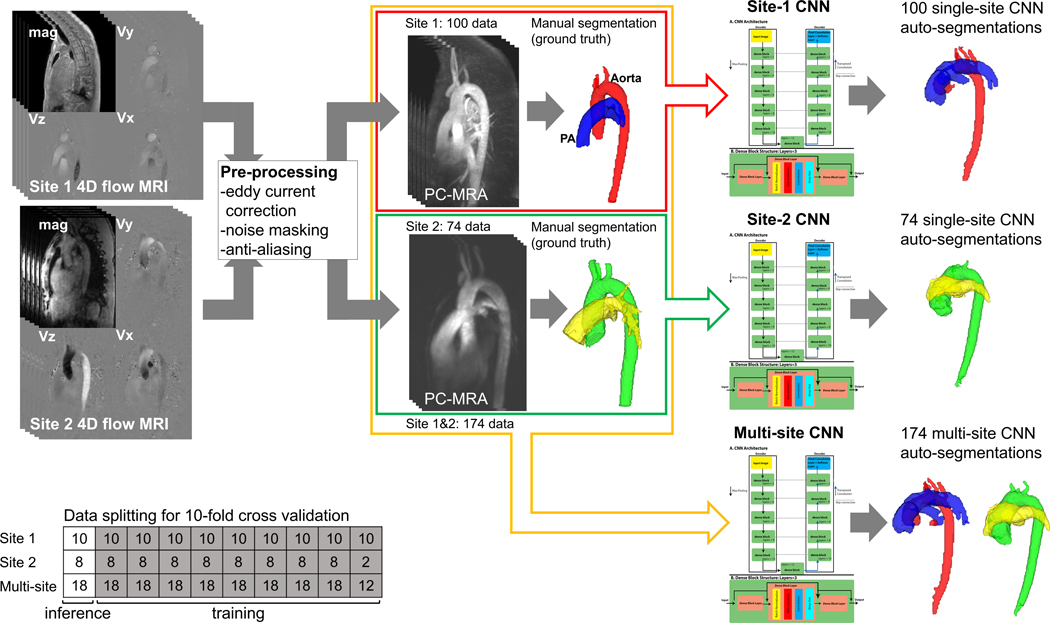
Flowchart for CNN training. 4D flow data were processed to derive a phase-contrast MRA (PC-MRA), and the ground truth segmentation was performed manually. The CNN parameters were tuned based on the PC-MRA and ground truth to automatically segment the test data. Geometrical similarity (Dice score) and net flows using manual and CNN segmentations were then compared.
Fig. 2.
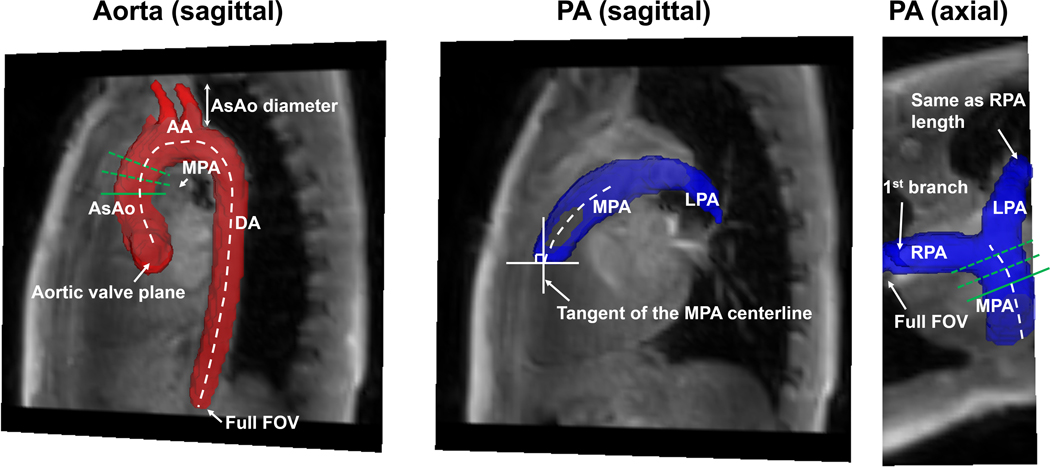
Criteria for segmentation and subsequent flow quantification. The aorta was transected at the aortic valve and the abdominal aorta (include full field of view). The branches were truncated at one ascending aortic (AsAo) diameter downstream the bifurcation. The pulmonary artery was cut perpendicular to the main pulmonary artery (MPA) centerline at the level of the pulmonary valve (when present), where the tangent of the MPA centerline was approximately vertical. The right pulmonary artery (RPA) was included for the full field of view and the left pulmonary artery (LPA) was included to approximately the same length. Only the first branches were included for the branch PAs. A plane (a solid green line) was positioned manually on each centerline, and the other two planes (dotted green lines) were automatically placed distal to the manual plane with equal spacing to quantify net flows at the ascending aorta and MPA.
Convolutional Neural Network For Multi-label Segmentation
Manual segmentations of the aorta and PA were used as the ground truth to train a modified version of a previously reported 3D DenseNet U-net CNN (8). The U-net (12) was modified to extend the capability from a segmentation of the aorta alone to a segmentation of the aorta and PAs (i.e., “multi-label” segmentation). The input and output of the CNN were matrices of the 3D PC-MRAs (input) and the labeled segmentations (output) with ‘0’ indicating background, ‘1’ indicating the aorta, and 2 indicating the PAs. To allow for uniform input to the CNN, and due to limited GPU capacity, the input images were center cropped or padded to have identical dimensions of 128×96×70. Dense blocks were included which consisted of convolution, dropout, batch normalization and the rectified linear unit (ReLU) functions. 3D convolution was conducted with a 3×3×3 kernel, and the channel size was set to 12. In each dense block, all the prior feature maps were concatenated to all subsequent layers to strengthen feature propagation (13). Max pooling and transposed convolution were used for the down-sampling and up-sampling, respectively. The final layer conducted convolution with a 1×1×1 kernel and output 3 feature maps, followed by a softmax function that output the probability of each class. The class with the largest probability per voxel was considered as the final segmentation. A combination of softmax-cross entropy and Dice loss was used as a loss function during the training. Note that here the Dice loss excludes the background (‘0’) label. The network was coded with Python 3.6.10 (Python Software Foundation, Beaverton, OR) and run with GPU version of Tensorflow 1.8.0 (Google, Mountain View, CA). An Intel Corei7–6700 processor with Nvidia Quadro P2000 GPU was used for all the training and testing.
Three separate models were trained to understand if multi-site data improved performance: (1) the CNN was trained by data from site 1 (n=100, site-1 CNN), (2) the CNN was trained by site 2 (n=74, site-2 CNN), and (3) the CNN was trained by all data (n=174, multi-site CNN). A 10-fold cross validation was conducted for each training to determine the performance of the CNN on all patients from the single-site CNNs and the multi-site CNN. The hyperparameters were set as follows: epoch number=200, batch size=1, learning rate=0.0001, and drop-out rate = 0.1.
Performance Metrics
CNN vs manual segmentation results were evaluated by geometrical similarity and accuracy of flow measurements. Dice score (14) was used to evaluate geometrical similarity between the ground truth and output from the single-site CNNs, as well as between the ground truth and output from the multi-site CNN.
To assess the performance of flow hemodynamic measurements obtained from the CNN segmentations, we quantified net flow in the ascending aorta (Qs) and MPA (Qp). For this purpose, three planes were semiautomatically positioned at the mid-ascending aorta and MPA, and flow was averaged across these three planes (Fig. 2). The averaging of multiple planes was used to mitigate effects of noise or flow artifacts in the original 4D flow data (15).
The semiautomatic plane placement process was conducted on in-house MATLAB script and Geom3d tool (16). This process involved the following: 1) centerlines of the aorta and PA were skeletonized using the manual segmentations (Fig 2), 2) a point depicting the mid-ascending aorta position (level of the RPA) and a position slightly downstream from the pulmonary valve was manually defined on each centerline, 3) two points on the centerline were then automatically placed distal to the manual location with equal spacing. The measurement planes were created at these locations orthogonal to the centerline. The spacing between planes was determined based on median diameter of the aorta using the following equation:
| (2) |
Here is reference diameter of normal children and adolescents and was set to 2cm (17). is the baseline spacing and was set to 1cm. D is the median aortic diameter as calculated from the manual 3D segmentation. To do this, the aorta or PA segmentation was converted to a stereolithography mesh. The intersection points between normal vectors from all faces and the mesh itself were then derived. The distance between each face and the intersection point was computed and its median was considered as D. Variable plane spacing was implemented to avoid using excessively large spacing for younger patients with small aortic diameters.
The exact coordinates of the specified three planes from the semiautomatic measurements made on the manual segmentations were then used for plane placement and flow measurement of CNN auto-segmentations. Note that the difference in flow measurements between manual and auto-segmentation comes only from difference of segmentation masks, since identical velocity data and measurement plane locations were used. Custom MATLAB code was used for this step (MATLAB, Natick, MA).
Due to the small sizes of the great vessels in younger pediatric patients, the skeletonization process can fail, generating unrealistic centerlines. In such cases, manual centerlines were drawn, and measurement planes were placed with EnSight (Ansys Inc., Canonsburg, PA). Based on the measured Qp and Qs, the pulmonary to systemic flow ratio (Qp/Qs) were computed.
To understand how the CNNs performed in the presence of different types of CHD, five sub-cohorts were selected based on the most prevalent anatomy: normal anatomy, TOF, bicuspid aortic valve (BAV), Fontan circulation, and transposition of the great arteries (TGA) (Table 1). TOF and BAV were chosen due to their high incidence in the 4D flow data set. Because of the abnormal physiology and vascular anatomy, TGA and Fontan subgroups were chosen as examples of particularly difficult cases for the CNN models.
Human Interobserver Error And Comparison To Single Site And Multi-site CNNs
To assess CNN vs. human performance, 40 subjects (20 from each site) were selected from the study datasets to assess inter-observer variability. The 40 patients were randomly chosen to match age and pathological background between sites. For these selected patients, a second observer from the original segmenting institution, who has at least two years of experience in vessel segmentation, conducted a repeat segmentation of the aorta and PA as observer 2 (TF from site 1; MS from site 2). Human interobserver variability was quantified using Dice scores and compared to the three CNN models. Dice scores were compared between two humans (observer 1 vs. observer 2) and between a human and the various CNN models (observer 1 vs. single-site CNNs; observer 1 vs. multi-site CNN).
Statistical Analysis
Dice scores and flow quantification results were reported as median [interquartile range]. The Dice scores between manual segmentation and single-site CNNs, and between manual segmentation and the multi-site CNN were tested by Wilcoxon rank-sum tests. Three-group comparison such as interobserver dice scores and quantified net flows using manual segmentation, single-site CNN, and multi-site CNN segmentations were evaluated by Kruskal-Wallis tests and post-hoc tests with Bonferroni correction. In addition, Bland-Altman analysis was conducted for the net flow measurements, and limits of agreement, bias, and their 95% confidence intervals (95% CI) (18) were calculated for the site-1, site-2, and multi-site CNNs as well as for sub-cohorts at each site. Furthermore, a difference in the CNN Qp and Qs measurements larger than 10ml/cycle (approximately 20% error when compared to the mean of the Qp and Qs manual measurements) was defined as a CNN “segmentation failure” and root cause was investigated. These cases were summarized, visually inspected, and classified into two failure modes: i.e., misidentification of a region as the aorta or PA, or, as an unintentional omission of a vessel region. A P-value < 0.05 was considered statistically significant (MATLAB R2019b, Natick MA).
RESULTS
Study Cohort
The combined data set of 174 study subjects included 38 patients with TOF, 17 with BAV, 8 with coarctation, 7 with TGA, and 6 with Fontan circulation (5 HLHS; 1 double outlet right ventricle with dextrocardia and heterotaxy). Note that Fontan patients were only included in the site 1 data while patients with TGA were available only for site 2.
The median processing time for segmentation was 1.01 seconds. Of the 100 subjects from site 1, the aorta was successfully segmented for all subjects while the PAs were identified in 94 subjects (6 patients were identified with Fontan circulation). Both the aorta and PAs were segmented for all patients from site 2. One patient from site 1 was excluded from the flow analysis due to insufficient FOV with incomplete coverage of the entire aorta and PA. Another patient from site 1 was excluded due to a failed 4D flow consisting of excessive noise of unknown origin, thereby resulting in unrealistic flow measurements, even with manual segmentation. Thus, Qs and Qp were quantified for 98 and 92 subjects from site 1 and 74 subjects from site 2.
Interobserver Comparison
The human interobserver Dice scores and CNN comparisons are shown in Table 2. In site-1 data, segmentations by both single-site and multi-site CNN showed significantly better agreement with observer 1 (ground truth) than observer 2 for both aorta and PA while the multi-site CNN performance exhibited similar performance compared to the single-site CNN (P=1 for both aorta and PA in post-hoc analysis). On the other hand, site-2 CNN segmentations did not show any significant differences (P=0.06 for aorta, P=0.39 for PA in multi-group comparison). The combined results (n=40) showed no statistically significant differences (P=0.49 for aorta, P=0.35 for PA in multi-group comparison), Comparing across sites, site 2 exhibited significantly higher Dice scores between observer 1 and observer 2.
Table 2.
Dice scores for interobserver analysis on the selected 40 subjects (median [IQR]).
| Observer 1 vs. | Post-hoc tests (P value) | ||||||
|---|---|---|---|---|---|---|---|
| Observer 2 | Single-site CNN | Multi-site CNN | P value | Observer 2 vs. single | Observer 2 vs. multi | Single vs. multi | |
| Site 1 (n=20) | |||||||
| Aorta | 0.875 [0.859–0.904] | 0.918 [0.907–0.928] | 0.919 [0.912–0.931] | <0.001 | <0.001 | <0.001 | 1 |
| PA | 0.868 [0.802–0.885] | 0.898 [0.856–0.910] | 0.897 [0.886–0.919] | 0.002 | 0.01 | 0.003 | 1 |
| Site 2 (n=20) | |||||||
| Aorta | 0.931 [0.909–0.953] | 0.915 [0.877–0.945] | 0.905 [0.868–0.937] | 0.06 | - | - | - |
| PA | 0.887 [0.857–0.919] | 0.884 [0.859–0.906] | 0.869 [0.801–0.913] | 0.39 | - | - | - |
| All (n=40) | |||||||
| Aorta | 0.908 [0.869–0.935] | 0.924 [0.896–0.938] | 0.922 [0.902–0.937] | 0.49 | - | - | - |
| PA | 0.879 [0.842–0.908] | 0.893 [0.868–0.912] | 0.898 [0.857–0.914] | 0.35 | - | - | - |
P<0.05 is shown by bold numbers. PA, pulmonary arteries; CNN, convolutional neural network.
Segmentation Performance Of The Single-site CNNs And Multi-site CNN
Dice scores comparing manual segmentation to single-site CNNs or multi-site CNN for all patients and patient subgroups are shown in Table 3. No statistical differences were found in overall Dice scores for the aorta (site 1: 0.916 vs. 0.915, P=0.55; site 2: 0.906 vs. 0.904, P=0.69) and the pulmonary arteries (site 1: 0.894 vs. 0.895, P=0.64; site 2: 0.870 vs. 0.869, P=0.96) (Table 3). Likewise, no clear difference or tendency was seen in Dice scores from subgroup analysis.
Table 3.
Dice scores of single-site and multi-site CNN for all patients and disease sub-cohorts (median [IQR]).
| Single-site vs. manual | Multi-site vs. manual | P value | |
|---|---|---|---|
| Site 1 (n=98 for aorta, n=92 for PA) | |||
| All | |||
| Aorta | 0.916 [0.883–0.930] | 0.915 [0.885–0.934] | 0.55 |
| PA | 0.894 [0.857–0.912] | 0.895 [0.856–0.915] | 0.64 |
| Control (n=15) | |||
| Aorta | 0.924 [0.912–0.929] | 0.922 [0.910–0.937] | 0.52 |
| PA | 0.891 [0.866–0.907] | 0.885 [0.865–0.912] | 0.76 |
| TOF (n=25) | |||
| Aorta | 0.897 [0.820–0.914] | 0.897 [0.864–0.914] | 0.86 |
| PA | 0.864 [0.823–0.902] | 0.874 [0.816–0.896] | 0.85 |
| BAV (n=10) | |||
| Aorta | 0.927 [0.910–0.933] | 0.929 [0.924–0.940] | 0.68 |
| PA | 0.902 [0.896–0.915] | 0.901 [0.892–0.910] | 0.82 |
| Fontan (n=6) | |||
| Aorta | 0.735 [0.676–0.790] | 0.797 [0.762–0.824] | 0.33 |
| Site 2 (n=74) | |||
| All | |||
| Aorta | 0.906 [0.877–0.939] | 0.904 [0.873–0.936] | 0.69 |
| PA | 0.870 [0.815–0.905] | 0.869 [0.826–0.909] | 0.96 |
| Control (n=23) | |||
| Aorta | 0.942 [0.911–0.950] | 0.932 [0.906–0.949] | 0.51 |
| PA | 0.905 [0.890–0.926] | 0.911 [0.882–0.930] | 0.91 |
| TOF (n=13) | |||
| Aorta | 0.872 [0.791–0.892] | 0.866 [0.832–0.881] | 0.86 |
| PA | 0.804 [0.776–0.842] | 0.833 [0.792–0.867] | 0.76 |
| BAV (n=7) | |||
| Aorta | 0.863 [0.828–0.898] | 0.890 [0.854–0.925] | 0.43 |
| PA | 0.823 [0.719–0.882] | 0.870 [0.793–0.871] | 1.00 |
| TGA (n=7) | |||
| Aorta | 0.887 [0.870–0.911] | 0.876 [0.858–0.899] | 0.46 |
| PA | 0.796 [0.723–0.828] | 0.832 [0.765–0.847] | 0.54 |
TOF, Tetralogy of Fallot; BAV, bicuspid aortic valve; TGA, transposition of the great arteries; PA, pulmonary artery.
Performance of Flow Quantification
Table 4 shows the hemodynamic measurements using 3D segmentations from human observers, the single-site CNN, and multi-site CNN. Centerline detection by skeletonization was successful in all but 7 patients (manual centerline placement occurred in 6 patients from site 1, and 1 patient from site 2). Five patients had an abnormal pulmonary artery due to TOF or an obstructed right ventricular outflow tract while the other two had BAV and/or HLHS. No significant differences between manual segmentation and the CNN models were found for entire population (Qs site-1 medians 51.0–51.3ml/cycle (P=0.81) and site-2 medians 66.7–69.4ml/cycle (P=0.84); Qp site-1 medians 46.8–48.0ml/cycle (P=0.97) and site-2 medians 76.0–77.4ml/cycle (P=0.98); Qp/Qs site-1 medians 0.87–0.88 (P=0.97) and site-2 medians 1.01–1.03 (P=0.43)) (Table 4). Similarly, no significant differences were found for all sub-cohorts. The results of Bland-Altman analysis are shown in Table 5 (bias and limits of agreement) and Table 6 (95% CI of bias). Biases and/or limits of agreement showed the multi-site CNN exhibited a similar performance to the single-site CNN for all the sub-cohorts except for Qp for the BAV patients at site 1, where the bias was found to be significantly larger than zero by the 95% CI (Table 6, [0.49 – 2.15]). In the entire cohort at each site, significant biases were found for Qs in both single-site and multi-site CNNs at site 1 (95% CI: [0.9–3.8] for single-site CNN, [0.5–3.3] for multi-site CNN) and site 2 (95% CI: [0.3–4.6] for single-site CNN, [0.5–3.4] for multi-site CNN) (Table 6).
Table 4.
Net flow for all patients and disease sub-cohorts (median [IQR]).
| Manual | Single-site | Multi-site | P value | ||
|---|---|---|---|---|---|
| Site 1 (n=98 for aorta, n=92 for PA) | |||||
| All | |||||
| Qs | [ml/cycle] | 51.3 [32.6– 73.3] | 51.1 [26.9– 71.4] | 51.0 [27.5– 71.8] | 0.81 |
| Qp | [ml/cycle] | 46.8 [31.9– 65.9] | 48.0 [31.4– 65.2] | 47.2 [31.7– 65.2] | 0.97 |
| Qp/Qs | [ - ] | 0.87 [0.77– 0.98] | 0.88 [0.78– 1.00] | 0.88 [0.77– 0.98] | 0.97 |
| Control (n=15) | |||||
| Qs | [ml/cycle] | 63.9 [45.2– 81.3] | 62.3 [46.4– 79.6] | 65.4 [46.1– 80.5] | 1.00 |
| Qp | [ml/cycle] | 55.7 [43.2– 73.4] | 52.2 [43.8– 72.8] | 54.2 [43.9– 72.5] | 0.99 |
| Qp/Qs | [ - ] | 0.90 [0.86– 0.97] | 0.89 [0.85– 1.00] | 0.90 [0.85– 0.99] | 0.92 |
| TOF (n=25) | |||||
| Qs | [ml/cycle] | 38.0 [24.6– 59.8] | 31.6 [22.2– 55.3] | 31.2 [21.8– 54.5] | 0.81 |
| Qp | [ml/cycle] | 32.4 [16.4– 52.1] | 32.3 [16.8– 54.1] | 32.1 [17.3– 54.3] | 0.99 |
| Qp/Qs | [ - ] | 0.82 [0.60– 0.90] | 0.83 [0.68– 1.09] | 0.82 [0.69– 0.90] | 0.76 |
| BAV (n=10) | |||||
| Qs | [ml/cycle] | 70.8 [46.4– 87.7] | 65.2 [47.4– 87.7] | 65.4 [47.4– 82.0] | 0.89 |
| Qp | [ml/cycle] | 60.4 [51.3– 78.6] | 58.1 [49.3– 78.2] | 59.1 [50.3– 78.4] | 0.88 |
| Qp/Qs | [ - ] | 0.91 [0.83– 1.08] | 0.92 [0.82– 1.04] | 0.96 [0.82– 1.10] | 0.92 |
| Fontan (n=6) | |||||
| Qs | [ml/cycle] | 17.6 [11.3– 27.5] | 10.1 [ 5.6– 11.9] | 12.4 [ 6.5– 18.0] | 0.34 |
| Site 2 (n=74) | |||||
| All | |||||
| Qs | [ml/cycle] | 69.4 [55.8– 90.9] | 66.7 [54.9– 90.6] | 69.0 [55.1– 89.2] | 0.84 |
| Qp | [ml/cycle] | 76.0 [54.6– 97.9] | 77.4 [55.3–100.9] | 76.2 [55.4– 97.5] | 0.98 |
| Qp/Qs | [ - ] | 1.01 [0.94– 1.10] | 1.03 [0.97– 1.23] | 1.03 [0.96– 1.15] | 0.43 |
| Control (n=23) | |||||
| Qs | [ml/cycle] | 66.2 [57.1– 81.3] | 65.3 [57.2– 81.0] | 66.2 [55.4– 80.4] | 0.97 |
| Qp | [ml/cycle] | 75.1 [59.0– 79.8] | 75.5 [59.5– 80.1] | 75.8 [59.6– 79.5] | 0.99 |
| Qp/Qs | [ - ] | 1.00 [0.96– 1.05] | 1.01 [0.98– 1.04] | 1.02 [0.98– 1.07] | 0.69 |
| TOF (n=13) | |||||
| Qs | [ml/cycle] | 69.0 [59.3– 86.9] | 70.4 [55.6– 89.5] | 70.8 [57.7– 86.3] | 0.99 |
| Qp | [ml/cycle] | 76.4 [68.0– 96.3] | 75.8 [68.6– 90.7] | 76.1 [67.7– 93.8] | 0.97 |
| Qp/Qs | [ - ] | 1.09 [1.02– 1.40] | 1.13 [0.98– 1.56] | 1.12 [1.00– 1.34] | 0.99 |
| BAV (n=7) | |||||
| Qs | [ml/cycle] | 65.0 [50.5–104.7] | 54.8 [40.3– 80.1] | 65.1 [46.7– 98.2] | 0.74 |
| Qp | [ml/cycle] | 71.5 [42.8– 97.4] | 74.9 [43.2–101.2] | 71.3 [43.4–100.5] | 0.90 |
| Qp/Qs | [ - ] | 0.97 [0.88– 0.99] | 1.14 [0.99– 1.33] | 1.04 [0.96– 1.10] | 0.14 |
| TGA (n=7) | |||||
| Qs | [ml/cycle] | 55.7 [38.3– 63.1] | 56.3 [37.5– 63.7] | 56.5 [38.7– 61.6] | 0.96 |
| Qp | [ml/cycle] | 47.4 [43.2– 73.3] | 48.3 [43.8– 86.1] | 49.3 [46.2– 85.2] | 0.91 |
| Qp/Qs | [ - ] | 1.03 [0.96– 1.20] | 1.17 [0.93– 1.26] | 1.15 [0.91– 1.25] | 0.87 |
TOF, Tetralogy of Fallot; BAV, bicuspid aortic valve; TGA, transposition of the great arteries; Qs, net flow at the ascending aorta; Qp, net flow at the main pulmonary artery; Qp/Qs, pulmonary-systemic flow ratio.
Table 5.
Bland-Altman analysis for flow quantification (mean bias ± limits of agreement).
| Site 1 | All | Control (n=15) | TOF (n=25) | BAV (n=10) | Fontan (n=6) | |
|---|---|---|---|---|---|---|
| Manual vs. single-site CNN | ||||||
| Qs | [ml/cycle] | 2.4±14.2 | 1.12±7.51 | 2.67±14.63 | 1.50±5.72 | 14.41±36.61 |
| Qp | [ml/cycle] | 0.6±10.6 | 0.60±5.10 | −1.25±18.29 | 1.32±2.28 | - |
| Qp/Qs | [ - ] | −0.2± 3.1 | −0.05±0.49 | −0.64± 5.84 | −0.00±0.11 | - |
| Manual vs. multi-site CNN | ||||||
| Qs | [ml/cycle] | 1.9±14.1 | 0.16±2.66 | 2.71±17.25 | 2.28±7.33 | 4.92±23.95 |
| Qp | [ml/cycle] | 0.3± 6.1 | 0.57±3.87 | −0.89± 9.44 | 0.18±3.99 | - |
| Qp/Qs | [ - ] | −0.1± 0.7 | 0.00±0.11 | −0.15± 0.99 | −0.03±0.10 | - |
|
| ||||||
| Site 2 | All | Control (n=23) | TOF (n=13) | BAV (n=7) | TGA (n=7) | |
|
| ||||||
| Manual vs. single-site CNN | ||||||
| Qs | [ml/cycle] | 2.4±18.2 | −0.04±2.08 | 0.13±21.32 | 12.90± 37.16 | 0.61± 3.85 |
| Qp | [ml/cycle] | −0.9±10.1 | −0.40±3.62 | 2.38±16.63 | −1.91± 4.24 | −3.87±13.02 |
| Qp/Qs | [ - ] | −0.1± 0.9 | −0.01±0.04 | 0.12± 1.34 | −0.28± 0.82 | −0.07± 0.24 |
| Manual vs. multi-site CNN | ||||||
| Qs | [ml/cycle] | 2.0±12.0 | 0.44±3.62 | 0.22± 9.11 | 4.20± 9.44 | 0.76± 4.62 |
| Qp | [ml/cycle] | −0.9± 9.4 | −0.53±3.01 | 1.50± 7.08 | −1.89± 5.44 | −6.45±18.43 |
| Qp/Qs | [ - ] | −0.1± 0.5 | −0.01±0.05 | −0.01± 0.22 | −0.09± 0.22 | −0.08± 0.33 |
TOF, Tetralogy of Fallot; BAV, bicuspid aortic valve; TGA, transposition of the great arteries Qs, net flow at the ascending aorta; Qp, net flow at the main pulmonary artery; Qp/Qs, pulmonary-systemic flow ratio; CNN, convolutional neural network.
Table 6.
Bland-Altman analysis for flow quantification (95% confidence intervals for bias).
| Site 1 | All | Control (n=15) | TOF (n=25) | BAV (n=10) | Fontan (n=6) | |
|---|---|---|---|---|---|---|
| Manual vs. single-site CNN | ||||||
| Qs | [ml/cycle] | 0.9 – 3.8 | −1.00 – 3.25 | −0.41 – 5.76 | −0.58 – 3.59 | −5.19 – 34.01 |
| Qp | [ml/cycle] | −0.5 – 1.7 | −0.84 – 2.04 | −5.10 – 2.60 | 0.49 – 2.15 | - |
| Qp/Qs | [ - ] | −0.5 – 0.1 | −0.18 – 0.09 | −1.87 – 0.59 | −0.04 – 0.04 | - |
| Manual vs. multi-site CNN | ||||||
| Qs | [ml/cycle] | 0.5 – 3.3 | −0.59 – 0.91 | −0.93 – 6.34 | −0.40 – 4.95 | −7.90 – 17.74 |
| Qp | [ml/cycle] | −0.3 – 1.0 | −0.51 – 1.67 | −2.88 – 1.09 | −1.27 – 1.64 | - |
| Qp/Qs | [ - ] | −0.1 – 0.0 | −0.03 – 0.03 | −0.36 – 0.05 | −0.07 – 0.01 | - |
|
| ||||||
| Site 2 | All | Control (n=23) | TOF (n=13) | BAV (n=7) | TGA (n=7) | |
|
| ||||||
| Manual vs. single-site CNN | ||||||
| Qs | [ml/cycle] | 0.3 – 4.6 | −0.50 – 0.42 | −6.44 – 6.71 | −4.63 – 30.44 | −1.21 – 2.42 |
| Qp | [ml/cycle] | −2.1 – 0.3 | −1.20 – 0.39 | −2.75 – 7.51 | −3.91 – 0.09 | −10.01 – 2.28 |
| Qp/Qs | [ - ] | −0.2 – 0.0 | −0.01 – 0.00 | −0.30 – 0.53 | −0.67 – 0.10 | −0.18 – 0.05 |
| Manual vs. multi-site CNN | ||||||
| Qs | [ml/cycle] | 0.5 – 3.4 | −0.35 – 1.24 | −2.59 – 3.03 | −0.26 – 8.65 | −1.42 – 2.94 |
| Qp | [ml/cycle] | −2.1 – 0.2 | −1.19 – 0.13 | −0.68 – 3.68 | −4.46 – 0.68 | −15.15 – 2.25 |
| Qp/Qs | [ - ] | −0.1 – 0.0 | −0.02 – 0.00 | −0.08 – 0.05 | −0.19 – 0.01 | −0.23 – 0.08 |
Biases significantly larger than zero are shown by bold numbers. TOF, Tetralogy of Fallot; BAV, bicuspid aortic valve; TGA, transposition of the great arteries Qs, net flow at the ascending aorta; Qp, net flow at the main pulmonary artery; Qp/Qs, pulmonary-systemic flow ratio; CNN, convolutional neural network.
Multi-site vs Single-site CNN Flow Quantification
Figure 3 shows Bland-Altman plots of flow measurement results for the single-site CNNs and multi-site CNN. For the single-site CNNs, 7 and 13 subjects with Qp and Qs differences >10ml/cycle were found, respectively. The segmentation ‘failures’ consisted of patients with TOF (5 cases), BAV (3 cases), Fontan (3 cases), ASD (2 cases), Marfan (2 case), TGA (1 case), double outlet right ventricle (1 cases), dilated aorta (1 case), and a healthy control. Application of the multi-site CNN reduced the number of failures to 6 for Qp and 9 for Qs (5 TOF, 2 ASD, 1 TGA, 1 Marfan, 1 Fontan, 1 BAV, and 1 dilated aorta patients), contributing to smaller biases and limits of agreement. Qp/Qs also showed a tendency toward smaller biases and limits of agreement, particularly for site 1 (see Table 5). However, even with the multi-site CNN, patients less than 10 years old (N=44) still had a relatively larger median error of 4.8% for Qs and 4.0% for Qp compared to those larger than 10 years old (1.5% for Qs and 2.1% for Qp).
Fig. 3.
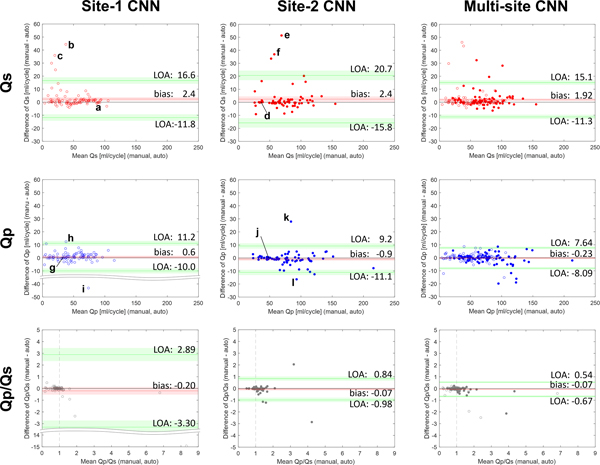
Bland-Altman plots of Qs (upper), Qp (middle), and Qp/Qs (lower) for the site-1 CNN (right), site-2 CNN (middle), and multi-site CNN (left). Site 1 data and site 2 data are shown by open and filled plots. Biases (mean) and limits of agreement (LOA) are shown by red and green lines with 95% confidence intervals (area shaded by red and green). The vertical dotted lines (Qp/Qs=1) represent a standard Qp/Qs value, which is seen in subjects without cardiovascular shunt flow. Example segmentations of the aorta and pulmonary arteries for selected successful and failed cases are marked by letters and shown in Fig. 4.
The failure modes for the single-site CNNs were: misidentification of unintended region as the aorta or PA (14 subjects), or the omission of vessel regions (10 subjects). Four subjects were classified to both mode 1 and 2 due to coexistence of the two error modes. Figure 4 shows examples of the segmentations of both successful (i.e., differences < 10ml/cycle) and failed (differences > 10ml/cycle) flow quantification in single-site CNNs. While the successful cases exhibited visually equivalent segmentations, failures by the single-site CNN can be seen in parts of the ascending aorta of patients with BAV, TOF and DORV. Here segmentation can be seen to omit or misrecognize the PA (Fig. 4b,c,f). On the other hand, the PA was misrecognized as the aorta in one of the TOF patients (Fig. 4i). In another case, a recirculation zone downstream the pulmonary valve in a patient with TOF is missing in the single site CNN segmentation due to low signal (Fig.4h). These problems were partly (c, i, h) or completely (b, f) resolved by the multi-site CNN. However, in the other cases (e, k, l), the segmentation performance worsened rather than improved.
Fig. 4.
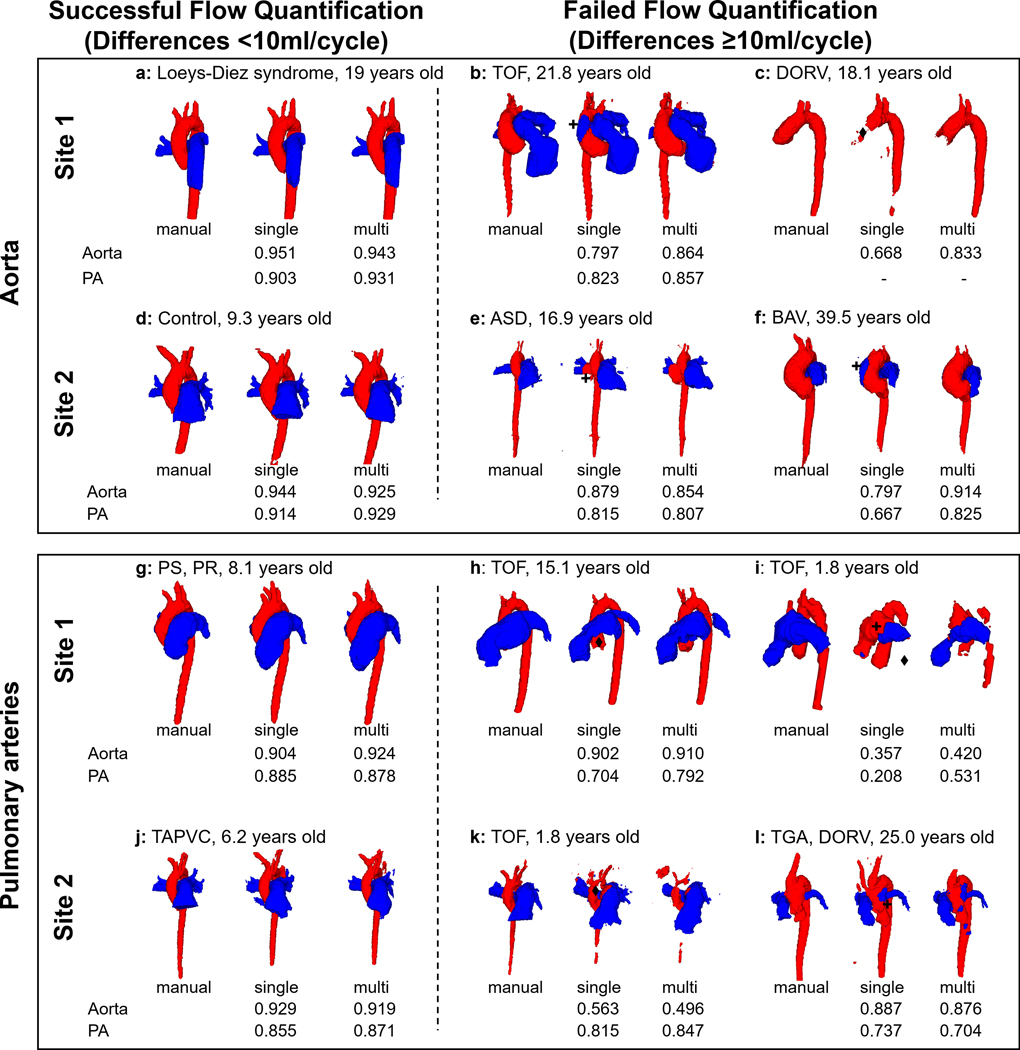
Example segmentations for selected successful, and failed flow quantifications with single-site CNN segmentations, as identified in Fig 3. Successes and failures for Qs are shown in upper rows while those for Qp are in the lower rows. The aorta and pulmonary arteries are colored red and blue, respectively, and their Dice scores are presented below the segmentations. Failed cases can be classified into two modes: mode 1: misidentification, mode 2: omission of vessel. Failed parts and its modes are marked by asterisks (+ for mode 1, ♦ for mode 2). TOF, Tetralogy of Fallot; DORV, double outlet right ventricle; TGA, transposition of the great arteries; PS, pulmonary stenosis; PR, pulmonary regurgitation; TAPVC, total anomalous pulmonary venous connection; ASD, atrial septal defect.
DISCUSSION
This study has three main findings: 1) a CNN trained on multi-site 4D flow data performed multi-label vessel segmentation as accurately as human observers, (2) the multi-site CNN segmented the proximal pulmonary and systemic vasculature with equivalent performance compared to single site data, and (3) the multi-site CNN showed equivalent performance for patients with complex CHD. The most important outcome of this effort was the ability to segment complex 4D flow datasets in the setting of CHD from data acquired on different vendor MRI systems and different sites around 1 second – with performance equivalent to human observers.
It is important to note that segmentation performance was strongly degraded when data from site 1 was input into the site 2 CNN (median Dice score for aorta and PAs are 0.83 and 0.78), or vice versa (median Dice score for aorta and PAs are 0.72 and 0.58). This was due to vendor-related differences in image signal intensities, contrast, and noise, as well as site related protocol differences (e.g., FOV) as demonstrated and suggested in previous multi-site studies (19–21). Thus, the multi-site CNN increased the potential for broader use of this approach across institutions and vendors and may have the potential to improve Dice scores and flow quantification for complex CHD cases with the addition of multisite patient data. This is especially important as it will allow to increase the number of datasets for rarer diseases and has the potential to be less reliant on expert availability for time-consuming segmentation.
We found the quality of the CNN-based approach was equivalent to human segmentation for both the aorta and PA. For flow quantification, failed cases did occur in which flow quantification for the multi-site CNN showed differences from human segmentation by more than 10ml per cycle. This represents 15 out of 338 measurements that were outside of our acceptance limits (approximately 4% of the measurements). Considering that the dataset was from two separate vendors and consisted of a large variation in cardiovascular physiology, ages, and body habitus, we hypothesize that larger training datasets could improve the performance of this pilot effort and future efforts will investigate larger datasets. It should be noted that these failed cases were all manually analyzable for clinical reporting purposes. Thus, in cases where the CNN was deemed to fail, the clinical workflow could take longer, but would not be impeded.
Although the single-site CNNs performed well for segmentation of data from their respective sites, marked segmentation errors were seen, contributing to flow measurement errors, such as for Qp/Qs of the TOF subgroup from site 1. Errors in the single-site CNNs were likely due to abnormal geometry or insufficient contrast of the target vessels in the PC-MRA images. The errors seen in the visual comparisons are possibly because of abnormal shape such as enlargement or mirrored ascending aorta or low signal due to recirculation zone downstream the pulmonary valve . Geometry and flow at these specific abnormalities are of particular interest, highlighting the potential added value of a large ‘N’ multi-site CNN.
The flexibility to use multi-site data improves the chances that sufficient data can be collected to train future CNN’s to identify the variety of CHD abnormalities found in the pediatric setting. The fact that the overall performance was not degraded by the variability in quality, planning, and FOV of the 4D flow data suggests datasets across institutions or vendors can be used for this purpose. While a trend toward improved performance was seen with our multi-site CNN model, we acknowledge that additional improvement for this relatively small patient cohort (n<200) is necessary for clinical adaptation, particularly for younger patients, which presented larger error in the net flow quantification. Nonetheless, we postulate that increasing the number of patients may further improve CNN performance.
Recent work has also achieved fully automated segmentation of the aorta in with alternative approaches. Gamechi et al. established fully automated segmentation for CT images combining an atlas-based approach (22), centerline extraction, and an optimal surface segmentation. Fantazzini et al. applied multiple 2D U-net to multi-planar CT images and combined the outputs to obtain 3D segmentation (23). Although these novel approaches also do not require human interaction, their framework was validated in adult cohorts and only the aorta was segmented without the additional added-value of the 4D flow MRI velocity data. A 4D flow MRI atlas-based approach was also proposed for fully-automated segmentation of great vessels and its performance was compared in terms of geometrical similarity and measured net flow (24). Although the aorta and PAs were successfully segmented over the cardiac cycle, it may be difficult to apply to a wide variety of CHDs as the atlas was created based on healthy subjects and from a single site. In contrast, our work confirmed the proposed multi-label segmentation algorithm can be applied to a younger patient cohort, across a wide variety of ages and CHD status.
Compared to previous work using the same CNN architecture presented here (8), the median Dice score in this study for the aorta was decreased (0.95 vs. 0.90). Likewise, larger bias and wider limits of agreement were observed in ascending aortic net flow quantification (0.1±5.3 vs. 1.9±13.2 ml/cycle). As mentioned, this could be the result of the smaller number of training datasets, multiple vendor differences in signal and contrast, as well as the greater number of pediatric and complex CHD cases in this study. The previous study used 499 training datasets on a single vendor platform, at a single site, and did not have patients with complicated geometry such as Fontan or TGA. With these considerations in mind, our study confirmed good agreement between human and automated segmentation in flow quantification for most cases on two scanner platforms acquired at two separate pediatric hospitals.
Limitations
There were only a limited number of complex CHD cases (particularly, Fontan and TGA, n=13); we anticipate that additional complex cases will further improve the CNN performance. Our results are limited to a Dense U-net CNN architecture. A previous study trained different variants of CNNs (SegNet, U-net, and pix2pix) using single- and multi-site datasets and compared their performance on 2D prostate zonal segmentation (25). The authors reported U-net showed better performance when trained by multi-site datasets. Although our CNN was based on a U-net approach, additional investigations are needed to determine the optimal network choice for multi-site training. The use of a static segmentation is also a limitation. Neglecting cyclic vessel motion may affect the Qp/Qs calculation, which is more sensitive to measurement errors than single Qp or Qs measurement. Future efforts will focus on time-resolved segmentation. Nonetheless, realistic Qp/Qs ratios were obtained in healthy controls both with manual and automated segmentations. Therefore, we considered static segmentation sufficient for the flow metrics measured in this preliminary study. The larger bias (~0.1) found in Qp/Qs quantification for the healthy controls from site 1 could result from separate factors. One possible factor for worse bias performance could be larger acceleration factor (3x) used in site 1 which may degrade SNR. Evidence of poorer performance by Dice scores is also seen between observer 1 and 2 for the site 1 data, which could also be due to lower SNR caused by higher acceleration factors. We intend to investigate this finding further and note that it is the semiautomated approach here that enabled this large cohort study, which led to unique insight to site and/or vendor specific features.
Conclusions
The use of a multi-site, multi-label CNN performed comparable to manual human measurements and single site CNN’s. A statistically significant improvement in complex CHD cases was not observed in this limited number of datasets. However, the results demonstrate the availability of a dense U-net CNN trained by 4D flow data of pediatric patients with a variety of congenital heart diseases from multiple institutions and vendors. The multi-site CNN provides the flexibility to train and analyze patient data acquired across multiple sites.
Supplementary Material
Acknowledgments
Grant support: NIH R01HL133504, R01HL115828
REFERENCES
- 1.Rizk J. 4D flow MRI applications in congenital heart disease. Eur Radiol 2021;31:1160–1174. doi: 10.1007/s00330-020-07210-z [DOI] [PubMed] [Google Scholar]
- 2.Jacobs KG, Chan FP, Cheng JY, Vasanawala SS, Maskatia SA. 4D flow vs. 2D cardiac MRI for the evaluation of pulmonary regurgitation and ventricular volume in repaired tetralogy of Fallot: a retrospective case control study. Int J Cardiovasc Imaging 2020;36:657–669. doi: 10.1007/s10554-019-01751-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isorni MA, Moisson L, Moussa NB, et al. 4D flow cardiac magnetic resonance in children and adults with congenital heart disease: Clinical experience in a high volume center. Int J Cardiol 2020;320:168–177. doi: 10.1016/j.ijcard.2020.07.021 [DOI] [PubMed] [Google Scholar]
- 4.Vasanawala SS, Hanneman K, Alley MT, Hsiao A. Congenital heart disease assessment with 4D flow MRI. J Magn Reson Imaging 2015;42:870–886. doi: 10.1002/jmri.24856 [DOI] [PubMed] [Google Scholar]
- 5.Piatti F, Pirola S, Bissell M, et al. Towards the improved quantification of in vivo abnormal wall shear stresses in BAV-affected patients from 4D-flow imaging: Benchmarking and application to real data. J Biomech 2017;50:93–101. doi: 10.1016/j.jbiomech.2016.11.044 [DOI] [PubMed] [Google Scholar]
- 6.Lorenz R, Bock J, Barker AJ, et al. 4D flow magnetic resonance imaging in bicuspid aortic valve disease demonstrates altered distribution of aortic blood flow helicity. Magn Reson Med 2014;71:1542–1553. doi: 10.1002/mrm.24802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirtler D, Garcia J, Barker AJ, Geiger J. Assessment of intracardiac flow and vorticity in the right heart of patients after repair of tetralogy of Fallot by flow-sensitive 4D MRI. Eur Radiol 2016;26:3598–3607. doi: 10.1007/s00330-015-4186-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berhane H, Scott M, Elbaz M, et al. Fully automated 3D aortic segmentation of 4D flow MRI for hemodynamic analysis using deep learning. Magn Reson Med 2020;84:2204–2218. doi: 10.1002/mrm.28257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiene G, Frescura C. Anatomical and pathophysiological classification of congenital heart disease. Cardiovasc Pathol 2010;19:259–274. doi: 10.1016/j.carpath.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 10.Imazio M, Andriani M, Lobetti Bodoni L, Gaita F. Learning Cardiac Magnetic Resonance: A Case-Based Guide. Cham: Springer; 2019: p 155–169. doi: 10.1007/978-3-030-11608-8 [DOI] [Google Scholar]
- 11.Bock J, Kreher B, Hennig J, Markl M. Optimized pre-processing of time-resolved 2D and 3D phase contrast MRI data. Proceedings of 15th Annual Meeting of International Society for Magnetic Resonance in Medicine 2007;3138. [Google Scholar]
- 12.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, volume 9351. Cham: Springer; 2015 p 234–241. doi: 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 13.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017:4700–4708. doi: 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 14.Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15. doi: 10.1186/s12880-015-0068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aristova M, Vali A, Ansari SA, et al. Standardized Evaluation of Cerebral Arteriovenous Malformations Using Flow Distribution Network Graphs and Dual-venc 4D Flow MRI. J Magn Reson Imaging 2019;50:1718–1730. doi: 10.1002/jmri.26784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legland D. (2021) geom3d (https://www.mathworks.com/matlabcentral/fileexchange/24484-geom3d), MATLAB Central File Exchange. Retrieved Oct. 14, 2015.
- 17.Kaiser T, Kellenberger CJ, Albisetti M, Bergstrasser E, Valsangiacomo Buechel ER. Normal values for aortic diameters in children and adolescents--assessment in vivo by contrast-enhanced CMR-angiography. J Cardiovasc Magn Reson 2008;10:1–8. doi: 10.1186/1532-429X-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb) 2015;25:141–151. doi: 10.11613/BM.2015.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Dou Q, Yu L, Heng PA. MS-net: multi-site network for improving prostate segmentation with heterogeneous MRI data. IEEE Trans Med Imaging 2020;39:2713–2724. doi: 10.1109/TMI.2020.2974574 [DOI] [PubMed] [Google Scholar]
- 20.Styner MA, Charles HC, Park J, Gerig G. Multisite validation of image analysis methods: assessing intra- and intersite variability. Proceedings of SPIE 4684, Medical Imaging 2002: Image Processing, San Diego, CA, 2002. doi: 10.1117/12.467167 [DOI] [Google Scholar]
- 21.AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med Phys 2018;45:1150–1158. doi: 10.1002/mp.12752 [DOI] [PubMed] [Google Scholar]
- 22.Sedghi Gamechi Z, Bons LR, Giordano M, et al. Automated 3D segmentation and diameter measurement of the thoracic aorta on non-contrast enhanced CT. Eur Radiol 2019;29:4613–4623. doi: 10.1007/s00330-018-5931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantazzini A, Esposito M, Finotello A, et al. 3D Automatic Segmentation of Aortic Computed Tomography Angiography Combining Multi-View 2D Convolutional Neural Networks. Cardiovasc Eng Technol 2020;11:576–586. doi: 10.1007/s13239-020-00481-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustamante M, Petersson S, Eriksson J, et al. Atlas-based analysis of 4D flow CMR: automated vessel segmentation and flow quantification. J Cardiovasc Magn Reson 2015;17:87. doi: 10.1186/s12968-015-0190-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rundo L, Han C, Zhang JH, R., et al. CNN-based prostate zonal segmentation on T2-weighted MR image: a cross-dataset study. In: Esposito A, Faundez-Zanuy M, Morabito F, Pasero E, editors. Neural Approaches to Dynamics of Signal Exchanges. Smart Innovation, Systems and Technologies, volume 151. Singapore: Springer; 2020. p 269–280. doi: 10.1007/978-981-13-8950-4_25 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


