Abstract
Prostate cancer (PCa) is a prevalent malignancy that occurs primarily in old males. Prostate tumors in different patients manifest significant inter-patient heterogeneity with respect to histo-morphological presentations and molecular architecture. An individual patient tumor also harbors genetically distinct clones in which PCa cells display intra-tumor heterogeneity in molecular features and phenotypic marker expression. This inherent PCa cell heterogeneity, e.g., in the expression of androgen receptor (AR), constitutes a barrier to the long-term therapeutic efficacy of AR-targeting therapies. Furthermore, tumor progression as well as therapeutic treatments induce PCa cell plasticity such that AR-positive PCa cells may turn into AR-negative cells and prostate tumors may switch lineage identity from adenocarcinomas to neuroendocrine-like tumors. This induced PCa cell plasticity similarly confers resistance to AR-targeting and other therapies. In this review, I first discuss PCa from the perspective of an abnormal organ development and deregulated cellular differentiation, and discuss the luminal progenitor cells as the likely cells of origin for PCa. I then focus on intrinsic PCa cell heterogeneity in treatment-naïve tumors with the presence of prostate cancer stem cells (PCSCs). I further elaborate on PCa cell plasticity induced by genetic alterations and therapeutic interventions, and present potential strategies to therapeutically tackle PCa cell heterogeneity and plasticity. My discussions will make it clear that, to achieve enduring clinical efficacy, both intrinsic PCa cell heterogeneity and induced PCa cell plasticity need to be targeted with novel combinatorial approaches.
Keywords: Cancer cell heterogeneity, Prostate cancer, Androgen receptor, Cancer stem cells, Cancer cell plasticity
1. Introduction
Human prostate cancer (PCa) is a prevalent malignancy with >1.4 million new cases and >375,000 deaths worldwide in 2020. In the U.S, more than 250,000 new PCa cases are diagnosed each year with ~30,000 PCa-associated deaths. Why is the adult prostate so susceptible to cancer development? The normal human prostate (NHP) is a pseudostratified two-layer epithelial glandular organ that consists of basal and luminal epithelial cells together with rare neuroendocrine (NE) cells [1] (Fig. 1A–B). Luminal cells, characterized by the expression of cytokeratin (CK) 8 (CK8), CK18, androgen receptor (AR), prostate-specific antigen (PSA), are ‘functional’ cell types that secrete prostatic fluid into the lumen, which constitutes a major part of the ejaculate. Basal cells express CK5, CK14 and the p53 family member, p63, and interface the luminal compartment with the underlying stroma through basal lamina (Fig. 1A–B).
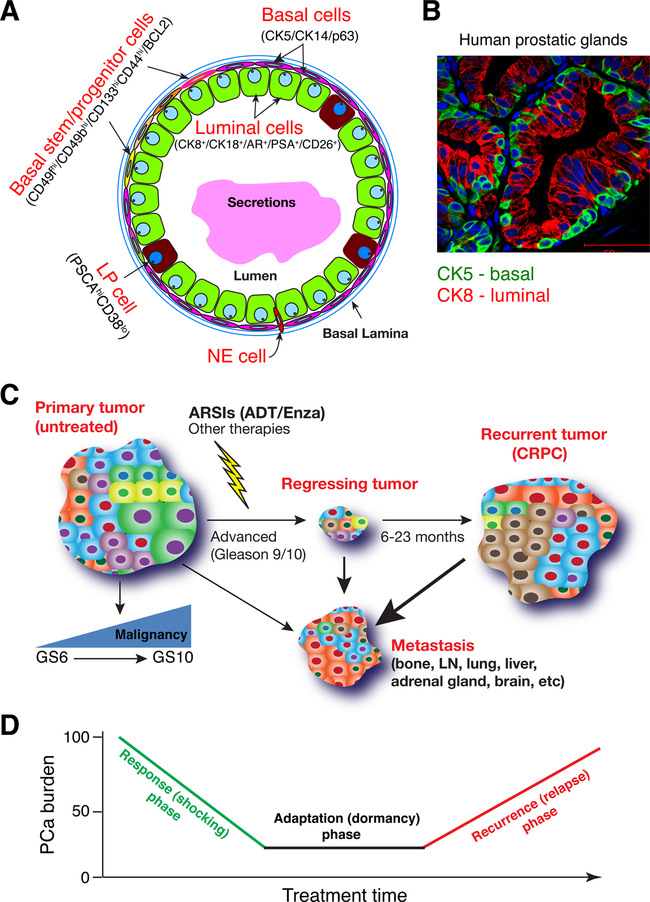
Fig. 1. Normal human prostatic glands and PCa.
A. Schematic of human prostatic glands. Both basal and luminal cell compartments harbor differentiated as well as less mature stem/progenitor cells. LP, luminal progenitor; NE, neuroendocrine.
B. Double immunofluorescence staining of CK5 and CK8 in benign human prostatic glands.
C. Human PCa at different clinical stages of development. Shown are 4 representative types of PCa, i.e., primary, regressing tumors (during therapy), recurrent tumors (i.e., CRPC in the prostate) and metastasis in different organs.
D. PCa response to clinical treatments goes through 3 typical phases, i.e., response, adaptation and recurrence. ‘PCa burden’ is measured by serum PSA levels, primary tumor sizes, and imaging analyses. see Text for details.
Primary PCa is pathologically graded in the clinic by a combined Gleason score (GS) system – the higher the GS the more malignant (less differentiated) the tumor is (Fig. 1C). Most prostate tumors are diagnosed as adenocarcinomas, meaning that tumors contain glandular structures that resemble the normal prostatic glands (see Fig. 2; below). Although the majority of low-grade tumors (GS6–7) can be ‘cured’ by the surgical procedure called radical prostatectomy, many patients with high-grade tumors (GS9–10) have extra-prostatic involvement and lymph node (LN) metastasis and are treated with drugs called AR signaling inhibitors or ARSIs (Fig. 1C). There are two broad classes of ARSIs: those that block the production of AR ligands (mainly testosterone, T) in the testis (e.g., Lupron) or adrenal gland (e.g., abiraterone acetate) and those that block AR nuclear translocation and AR functions through direct interactions with AR (e.g., enzalutamide, Enza). In most clinics, PCa patients with advanced tumors and ineligible for surgery are first treated with androgen synthesis inhibitors, a protocol called androgen-deprivation therapy (ADT), which generally is effective for ~6–23 months (Fig. 1C). The ADT-resistant PCa are then treated, as the second-line therapy, with Enza or Enza together with chemotherapeutic drugs such as docetaxel. Tumors that have failed ADT or ADT/Enza are often ‘generically’ called castration-resistant PCa or CRPC (Fig. 1C) although sometimes tumors that have failed the first-line ADT are called primary CRPC whereas tumors that have become resistant to both ADT and second-line antiandrogens such as Enza are called secondary CRPC. Advanced primary PCa and, in particular, CRPC patients often have metastases in many organs including bone, LN, lung, liver, adrenal gland and brain (Fig. 1C). PCa patients who have only a few metastases in a limited number of organs are frequently said to have oligometastatic disease. Regardless, metastatic CRPC (mCRPC) patients have a poor prognosis and very limited treatment options, which generally include chemotherapeutics such as cisplatin and docetaxel.
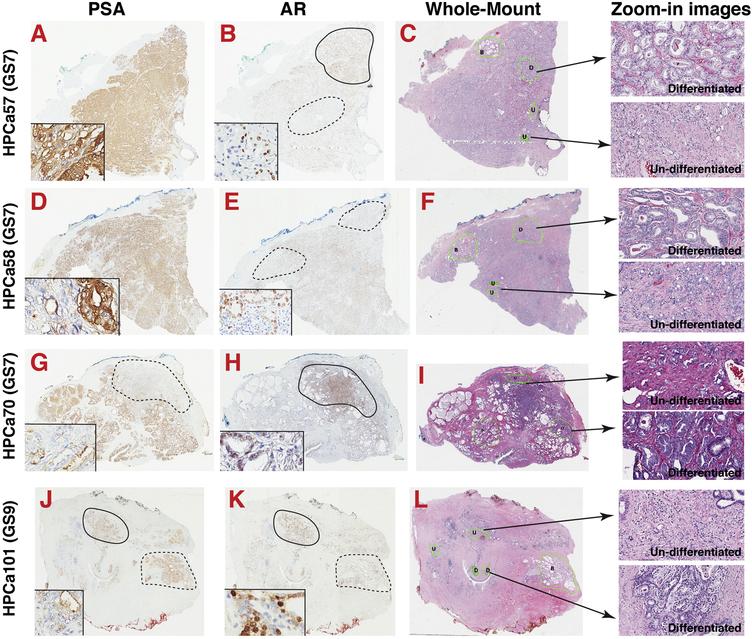
Fig. 2. Histo-structural, cellular and molecular heterogeneity in human PCa.
Shown are the Aperio ScanScope images of whole-mount sections (4x) of HPCa57 (A–C), HPCa58 (E–F), HPCa70 (G–I) and HPCa101 (J–L) stained for HE (C, F, I, L), AR (B, E, H, K), and PSA (A, D, G, J). HPCa 57, 58 and 70 are Gleason score 7 (GS7) tumors while HPCa101 is a Gleason 9 tumor. Serially transplantable PDX have been established from all 4 HPCa samples. Shown on the right are representative enlarged (zoom-in) HE images (200x) of differentiated (D) and poorly or undifferentiated (U) areas. For AR/PSA IHC images, a higher magnification (400x) inset was shown for each HPCa sample to illustrate AR−/lo and PSA−/lo PCa cells, and to illustrate the frequently discordant AR and PSA expression. For instance, in HPCa57, ARhi (demarcated by a solid circle) and AR−/lo (dashed line) areas could be discerned. In HPCa58, two AR−/lo areas are indicated by dashed circles. In HPCa70 and HPCa101, areas marked by dashed lines indicate discordant expression of AR and PSA. Note that AR IHC was performed using an antibody against the N-terminus, which recognizes both full-length AR and all c-terminally truncated splice variants.
ARSIs represent among the most remarkable and most effective targeted anti-cancer therapeutics [2], with the majority of PCa patients responding impressively with clear clinical evidence of tumor regression (Fig. 1C–D). The response of PCa patients to ARSIs follows the 3 ‘pro-totypical’ phases, i.e., the response (or shocking), adaptation and recurrence (or relapse) phases (Fig. 1D). The dramatic initial response to ARSIs can be explained by the fact that the majority of prostate tumors are adenocarcinomas with most tumor cells being AR+ and dependent on AR signaling for their survival. It is unclear what types of PCa cells survive ADT/Enza and enter the adaptation phase (Fig. 1D); however, as this phase is characterized by dormancy, it’s possible that a population of dormant or slow-cycling cells [3] such as PCa stem cells (PCSCs; see below) preferentially survive ADT/Enza and mediate subsequent tumor relapse (Fig. 1D). Interestingly, recent studies suggest that chemotherapy-treated tumors during ‘drug holiday’ enter a dormant stage resembling the developmental program called diapause, during which subsets of cancer cells downregulate mTOR signaling but upregulate autophagy to survive [4–7].
2. Cellular heterogeneity of the normal prostate and luminal progenitor cells as potential cells of origin of human prostate cancer
The adult prostate, a relatively dormant organ during homeostasis, nevertheless, consists of heterogeneous subpopulations of cells in both basal and luminal cell compartments (Fig. 1A) that may undergo dynamic changes during aging and inflammatory responses. Thus, the basal cell layer consists of differentiated CK5+/CK14+/p63+ basal cells as well as the basal stem/progenitor cells (i.e., cells with certain stem cell properties such as bi-potential or multi-lineage differentiation) expressing high levels of CD44, CD49b (integrin α2), CD49f (integrin α6) CD133, and BCL-2 (Fig. 1A). Likewise, the luminal cell layer contains not only differentiated CK8+/CK18+/AR+/PSA+/CD26+ luminal cells but also the luminal progenitor (LP) cells (Fig. 1A) that are frequently double-positive for both CK5 and CK8 (i.e., CK5+CK8+), express high levels of CK19 (CK19+/hi), and retain significant proliferative capacity [8–23]. The LPs are also endowed with an enhanced ability to form organoids ex vivo [18,21,23] and express high levels of prostate stem cell antigen (PSCA) and low levels of CD38 (i.e., CD38lo) [22–25]) (Fig. 1A). Recent single-cell RNA-seq (scRNA-seq) studies in human and mouse prostate have also revealed heterogeneous cell populations in the epithelial and stromal compartments and, particularly, have uncovered plastic LP cell populations that might be involved in mediating castration resistance [26–37].
Studying the NHP epithelial cell heterogeneity and hierarchy has an important bearing on understanding the potential cell(s) of origin of human PCa.
Like other glandular organs such as the breast and pancreas, development of the prostate follows the basic guiding principles of branching morphogenesis [38]. ‘Lineage tracing’ studies in the adult human prostate using mitochondrial DNA mutations (identified by sequencing) combined with cytochrome c oxidase/succinate dehydrogenase enzyme histochemistry (to identify areas of respiratory chain deficiency) have suggested that both basal and luminal epithelial (as well as the NE) cells are derived from a common stem/progenitor cell [39,40]. Further mapping studies reveal that primitive prostate stem cells are localized in the basal cell layer proximal to the urethra, which then direct stem cell differentiation along the prostate ductal tree with diminishing numbers of progenitors towards the distal acini [41]. scRNA-seq studies in the NHP have also implicated less mature prostate epithelial cells close to the urethra [26]. Recent whole-genome sequencing of targeted microdissections of normal/benign prostate from fetuses to aged adults suggests that the fetal organ development starts with 24–30 ductal subunits with each duct deriving from 5 to 10 embryonic progenitor cells [42]. During puberty, hormones and growth factors act on local stem cells in ductal units to promote branching morphogenesis and increase branching complexity [42]. Interestingly, somatic mutations generally accumulate slowly in the adult prostate at ~16/year/cell [42], consistent with the human adult prostate being a rather dormant organ with slow cellular turnover.
Exactly which cells, basal or luminal, differentiated or stem/progenitor cells, become tumorigenically transformed to develop into PCa remains an important outstanding question.
RNA expression profiling [17,43], coupled with prospective tumor transformation studies [e.g., 18], suggests that some cells in both basal and luminal cell compartments can function as the cells of origin for human PCa, although basal-originated PCa appears to be associated with the minor but more aggressive subtypes of PCa (such as neuroendocrine PCa, NEPC) whereas the luminal-originated PCa tend to be adenocarcinomas [17,43], the major type of PCa. Evidence is accumulating that LPs in the luminal cell compartment (Fig. 1A) may represent the preferred cellular targets of tumorigenic transformation [18,20–25]. The NHP LPs represent <2% of the CD26+CD49lo (or TROP2+CD49lo) luminal cell population, can readily establish organoids containing both luminal and basal cells (i.e., bipotentiality; [18,21,23]), and have been phenotyped as PSCAhiCD38lo [15]. Increasing numbers of LP cells have been observed to accumulate in the aging prostate [24]. Significantly, the LPs freshly purified from the CD26+ luminal cell population and propagated in 3D organoid conditions can be transformed to PCa by lentiviruses expressing c-MYC together with constitutively active AKT [21]. We have similarly shown that organoid-forming LPs in TROP2+CD49flo isolates from the NHP can be transformed by AR, AKT and ERG to form typical AR+PSA+ adenocarcinomas [18]. Notably, we have developed a novel 2D culture system that could extend the lifespan of the LPs beyond 100 population doublings (PDs) [18]. The 2D-propagated LPs possess cardinal features of stem/progenitor cell properties (e.g., forming more and larger holoclones, spheres and organoids; regenerating prostatic glands in vivo containing both CK5+P63+ basal cells and CK8+AR+PSA+ luminal cells), and are phenotyped as CK5+CK18+CK19hi with increased mRNA (but not protein) levels of AR and PSA and increased AR activity (compared to basal cells). These LP cells express a unique stem/progenitor cell-enriched gene expression profile linked to PCa aggressiveness, castration resistance, metastatic propensity and poor patient survival, and, significantly, can be oncogenically transformed by AR/AKT/ERG overexpression as well as by c-MYC together with PTEN-KO [18].
How do the LP cells (or other prostate epithelial cells) become trans- formed into tumor cells?
As PCa manifests a distinct association with age and LPs accumulate and expand in the aging prostate [24], it is reasonable to speculate that PCa etiology may be linked to immune senescence and impaired immune surveillance in the aging organ [44, 45]. Mounting evidence starts to link chronic inflammation to development of PCa precursor lesions and to prostate tumorigenesis [46,47]. There are two putative precursor lesions to PCa [48,49]: prostatic intraepithelial neoplasia (PIN) and prostate inflammatory atrophy (PIA). PIN is characterized by a hyperplasia phenotype such that proliferative luminal cells, likely LPs, would often fill up the lumen. Genomics studies using micro-dissected tissues to track clonal mutations [50–52] suggest that high-grade PIN (HGPIN) represents a genomically distant precursor to invasive adenocarcinomas and that the clinically common GS7 PCa are derived from branched evolution of clonal lesions with Gleason 3 and 4 patterns. On the other hand, PIA displays an atrophic phenotype with a single layer of smaller luminal cells lining the prostatic ducts and acini, which are frequently associated with prominent inflammatory infiltration [48,49]. Although the PIA glands are atrophic, luminal cells, which have the cardinal features of LPs being CK5+CK8+ (i.e., intermediate cells), are nevertheless highly proliferative in association with, among others, reduced levels of p27Kip1 [48,49, 53]. There is evidence that at least some HGPIN and early adenocarcinomas may arise from the PIA, likely driven by inflammatory cytokines and inflammation-derived oxidants, electrophiles, and genotoxins [46–49,53]. A recent study from the same group provided striking evidence to directly link bacterial prostatitis to PIA and TMPRSS2-ERG fusion, the most prevalent (up to 50 %) genomic alterations in PCa [54]. The authors identified ERG+ PIA transitioning to early invasive cancer and also identified pathogen-derived genotoxin colibactin as a potential source of DNA damage [54]. This remarkable new work [54] suggests that infection-induced ERG fusions represent an early alteration during prostate tumorigenesis and that PIA indeed may serve as a direct precursor to PCa.
One hypothetical scenario is that inflammatory insults induce apoptotic death of differentiated luminal cells pushing LP cells to the cell cycle and leading to expansion of the LP cell population and transitioning from PIA to PIN. Uncontrolled proliferation of LPs in HGPIN, driven by the chronic inflammation and activated oncogenes such as c-MYC and AKT (due to PTEN loss or mutations), would lead to telomere attrition and subsequent genetic, genomic and chromosomal instability, which, together with additional genetic and epigenetic alterations, would facilitate transition of HGPIN to adenocarcinomas and promote PCa progression. In support of this hypothesis, ~25% of the HGPIN are DNA aneuploid [55] and telomere shortening, with or without concomitant telomerase activation, represents an early and the most frequently observed and consistent somatic genomic alteration in HGPIN and early-stage adenocarcinoma [56–68]. In fact, as much as ~96 % of precursor lesions to multiple epithelial cancers (including those in prostate, bladder, breast, pancreas, esophagus, large intestine, oral cavity and uterine cervix) manifest shortened telomeres and abnormalities in telomere length [59]. Notably, telomere erosion occurs exclusively in luminal (but not basal) cells in HGPIN [56], different HGPIN lesions often exhibit different levels of telomere shortening [57], telomere loss in HGPIN is correlated with abnormalities on chromosome 8 and also with the levels of oxidative stress as measured by malondialdehyde [62], HGPIN and PCa often display prominent telomere fusions and anaphase bridges [63], and prostatic glands with shortened telomeres (e.g., due to reduced TRF2 expression) developed age-and p53-dependent hyperplasia whereas prostate tumors with TRF2 loss were mostly high-grade AR-negative adenocarcinomas [67].
Recent whole-genome sequencing studies [69–72] have identified many somatic clonal mutations in apparently normal tissues, especially fast-renewing tissues such as the skin, esophagus and colon, some of which accumulate with age and become driver clones of cancer. It is not clear whether apparently normal prostate or PCa precursor lesions also harbor such driver mutations although cells in PCa precursor lesions that have sustained TMPRSS2-ERG fusion appear to be clonal. It also remains unknown whether prostate tumorigenesis involves the phenomenon of (stem) cell competition and clonal fitness [73–77].
3. PCa cell heterogeneity
3.1. Genomic, transcriptomic, histological and cellular heterogeneity of PCa
Intra-tumor heterogeneity (ITH) makes significant contributions to therapy resistance [78,79]. PCa manifests ITH at every interrogated level [80,81]. At the genomic level, multiple genomic sequencing studies [82–96] in various type of PCa, including localized indolent, localized non-indolent, early-onset, metastatic castration-sensitive (mCSPC), and metastatic castration-resistant tumors (mCRPC) have made it clear that primary prostate tumors are multi-focal containing genomically distinct clones and subclones whereas PCa metastases are clonal, meaning that each metastasis in different organs was derived from the outgrowth of a single ‘metastatic stem cell’. At the epigenome level, the epigenetic landscape of PCa can be correlated with germline genomic features [97]. Whole-genome bisulfite sequencing in 100 mCRPC revealed that ~20 % of the metastases exhibited a novel epigenomic subtype associated with hypermethylation and somatic mutations in TET2, DNMT3B and IDH1 [98]. Notably, differential methylation during CRPC progression preferentially occurs at somatic mutational hotspots and putative regulatory regions [98]. Furthermore, DNA methylation-mediated 3D genome architecture acts, synergistically, with some risk SNPs (single nucleotide polymorphism) to confer predispositions to PCa [99]. At the transcriptional level, prostate tumors are found to harbor abnormal RNA transcripts such as circular RNAs (circRNAs) [100] as well as a myriad of unspliced mRNAs [101], and, to display tremendous heterogeneity in the global transcriptomes even in tumors pathologically classified as the same Gleason Grade [102]. Of clinical significance, molecular subtyping of PCa based on transcriptional profiling has identified a luminal subtype of adenocarcinomas that have an aggressive progression trajectory and are associated with poor patient outcomes [103–108]. It is likely that this aggressive subtype of luminal PCa is driven by LPs [20].
Accompanying the genomic, epigenomic and transcriptomic heterogeneity, PCa also exhibit wide variations in histological presentations. As illustrated in Fig. 2, the 3 human PCa (HPCa57, 58, 70) that are all diagnosed pathologically as GS7 look nothing alike histologically. Finally, prostate tumors are characterized with pervasive cellular heterogeneity, manifested as variegated morphologies, nuclear-to-cytoplasm ratio, metabolic activity, and antigenic marker expression (see discussions below). Recent scRNA-seq studies [109–115] have also revealed heterogeneous cell subpopulations in LNCaP cultures [109], carcinoma-associated fibroblasts (CAFs) [110] and cancer-associated macrophages [115] in patient tumors, and in primary prostate tumors [111–113] as well as mCRPC [114]. In fact, even circulating tumor cells (CTCs) in PCa patients can be epithelial, mesenchymal or a hybrid phenotype and similarly exhibit heterogeneity in their transcriptomes at the single-cell levels [116].
3.2. PCa as an abnormally differentiated organ and PCa cell heterogeneity in AR and PSA
Since 2002, our lab has been studying PCa from the perspective of deregulated organ differentiation by systematically and meticulously dissecting phenotypic and functional properties of heterogeneous PCa cell populations and studying molecular regulators in HPCa samples, PDX, xenografts and cell cultures [2,3,16–18,20,117–169] (Figs. 2–13). Nearly 20 years of focused studies in our lab on PCa cell heterogeneity and plasticity, and their impact on therapy response, have uncovered several fundamental principles relevant to our understanding of tumor progression and development of therapy resistance in patients (Fig. 14; see discussions below). PCa development and progression are accompanied by a decrease and loss of normal differentiation program at the organ and cellular levels. Hence, in low-grade (GS6–7) tumors, both differentiated (characterized by the presence of glandular structures) and undifferentiated (lack of glandular structures) tumor regions can be observed (Fig. 2C, F, I; zoom-in images). The ‘differentiated’ areas in these tumors are populated by significantly increased numbers of glands, as if these prostatic glands had undergone fission (Fig. 2C, F, I; zoom-in images). In contrast, most high-grade (GS9–10) tumors present an undifferentiated or a poorly differentiated phenotype that lacks significant glandular structures (Fig. 2L) [131,144,149]. The concept of prostate tumors as an organ of increasingly deregulated differentiation is also supported by the gradual loss of normal prostatic basal cells expressing CK5, CK14 and p63.
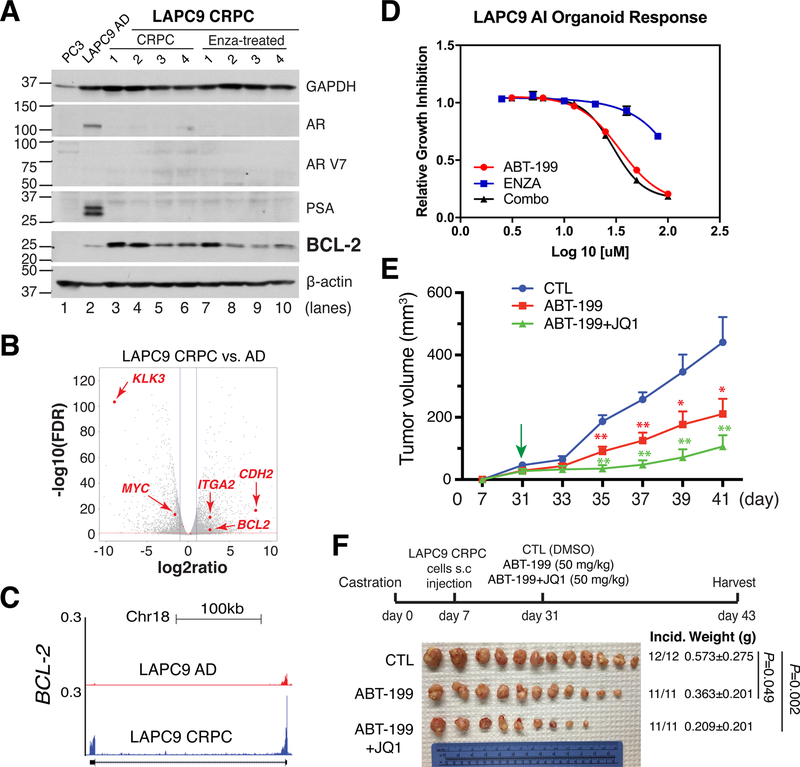
Fig. 13. BCL-2 represents a therapeutic target in AR−/lo subtype of CRPC.
A. BCL-2 was upregulated in LAPC9-CRPC and remained at increased levels in LAPC9-CRPC further treated with Enza.
B-C. BCL-2 mRNA was significantly upregulated in LAPC9-CRPC compared to LAPC-AD. Also upregulated in LAPC9-CRPC were integrin α2 (ITA2) and N-cadherin (CDH2) (B).
D. BCL-2 inhibitor, ABT-199, dose-dependently inhibited the growth of LAPC9-CRPC (i.e., LAPC9-AI) organoids whereas Enza demonstrated minimal effects and combination of ABT-199 and Enza (Combo) exhibited similar inhibitory effects to ABT-199 alone.
E–F. BCL-2 antagonist, ABT-199, alone inhibited LAPC9-CRPC in vivo, which was further inhibited by the combination of ABT-199 and JQ1.
All panels in this Figure were adapted with permission from reference [163].
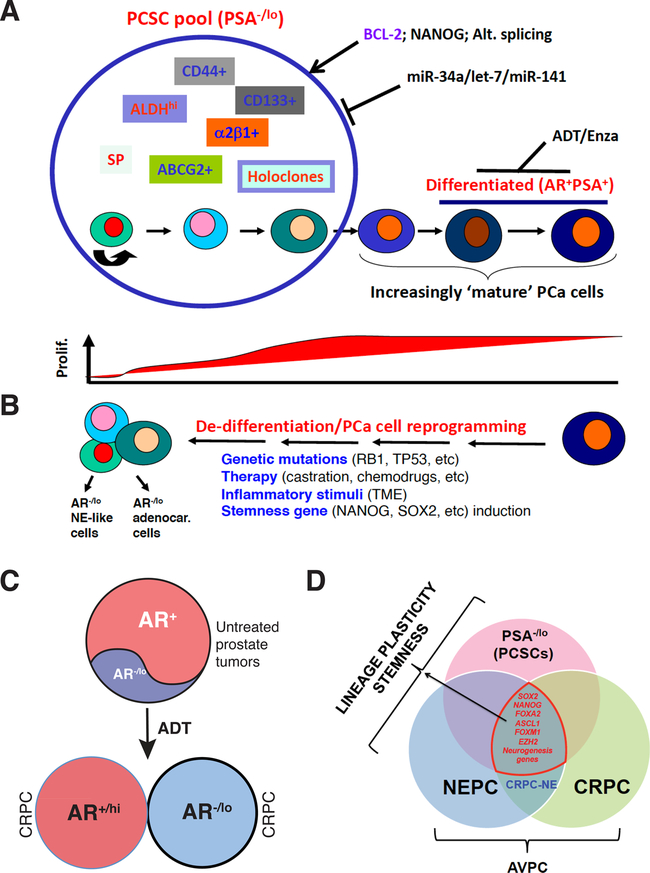
Fig. 14. Potential strategies to tackle PCa cell heterogeneity and plasticity.
A. A hypothetical model of intrinsic PCa cell heterogeneity. Primary (treatment-naïve) PCa consist of both bulk differentiated (AR+PSA+) cells as well as a subset of phenotypically undifferentiated (PSA−/lo) cells. The latter cells represent the PCSC pool in which other subsets of tumorigenic cells can be further identified and purified. While AR+PSA+ PCa cells are highly proliferative, most cells in the PSA−/lo PCSC pool remain dormant (below). ARSIs including ADT and Enza primarily and preferentially target the differentiated AR+PSA+ PCa cells while largely ignoring the PSA−/lo cells. We have identified BCL-2, NANOG and deregulated alternative splicing program as positive regulators of PCSCs and several tumor-suppressive miRNAs including miR-34a, let-7 and miR-141 as negative regulators of PCSCs. In principle, these tumor-suppressive miRNAs or the inhibitors of positive regulators can be used in conjunction with ADT/Enza to tackle PCa cell heterogeneity and plasticity.
B. PCa cell plasticity (or de-differentiation) can be induced by genetic mutations, therapeutic treatments, inflammatory cytokines in the TME and by stemness gene overexpression. The reprogrammed PCa cells can be AR−/lo adenocarcinoma or AR−/lo NE-like cells (left).
C. Persistent ADT leads to two subtypes of CRPC, AR+/hi and AR−/lo.
D. Many of the stemness and neurogenesis genes are commonly enriched in PSA−/lo PCSCs, NEPC, CRPC and CRPC-NE, which are often called AVPC.
Panel A was adapted with permission from reference [144].
At the cellular level, significant heterogeneity in the expression of AR and PSA and prominent discordance between AR and PSA expression have been observed (Figs. 2; 3) [131,144,147,163]. For example, in GS7 and GS9 tumors, both AR+ and AR−/lo regions and PCa cells can be readily observed in whole-mount (WM) sections (Fig. 2B, E, H, K). Similarly, both PSA+ and PSA−/lo PCa regions and PCa cells can be observed in GS7 tumors (Fig. 2A, D, G), and, in GS9 tumors, most PCa cells appear to be PSA−/lo(Fig. 2J). Heterogeneity in AR and PSA expression in human PCa has also been observed at the mRNA levels (Fig. 3). In fact, heterogeneous expression in AR and PSA has been reported for decades in the literature [2,147,170–214]. AR and PSA heterogeneity, and, AR−/lo and PSA−/lo PCa cells have been observed across the entire spectrum of PCa development, progression, therapy resistance and metastasis from treatment-naïve primary tumors to CRPC to mCRPC (Figs. 2; 3B; 4) [2,131,132,144,147,164]. Importantly, discordant AR and PSA expression has been observed at both mRNA and protein levels and at the tumor as well as the cellular levels (Figs. 3B; 4A). Indeed, at the tumor level, discordance in AR and PSA mRNA expression is apparent across GS6–9 tumors (Fig. 3B) [131,144]. Similarly, discordance in AR and PSA protein expression also exists in treatment-naïve primary HPCa such that although the majority cells in these adenocarcinomas, as expected, have the AR+PSA+ (i.e., double-positive) pheno-type, AR+PSA−/lo, AR−/loPSA+ and AR−/loPSA−/lo PCa cells are also observed at different abundance (Fig. 4A–B) [144]. Of note, PCa cells of the above 4 AR/PSA phenotypes have also been observed in xenograft models and shown to manifest distinct responses to ARSIs and other therapeutics (Fig. 5) [144] (see discussions below).
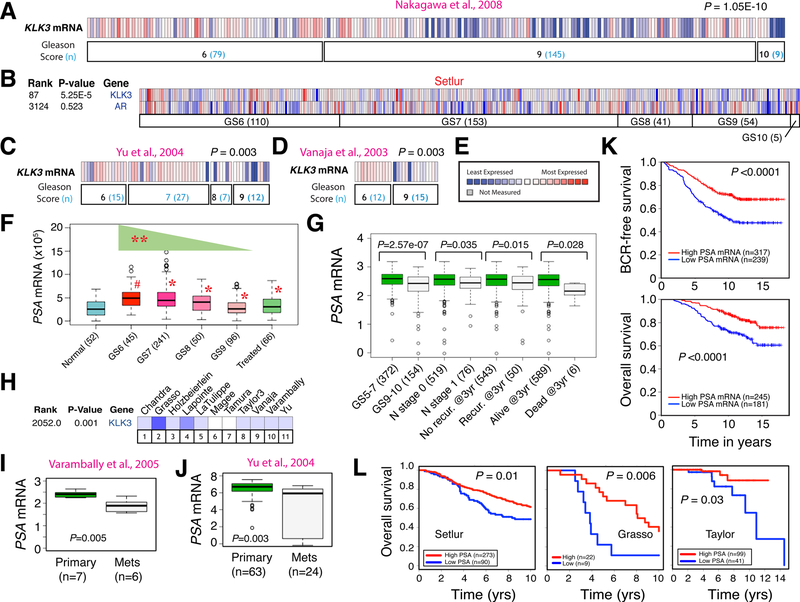
Fig. 3. PCa heterogeneity: Inverse correlation between tumor PSA (KLK3) mRNA levels and tumor grade, metastasis, and patient survival.
A-E. Tumor KLK3 mRNA levels are inversely correlated with the tumor grade. Shown are results from 4 PCa cohorts (A–D) in Oncomine. E, the heatmap legend. Data in B also illustrates discordant PSA and AR mRNA expression in patient tumors.
F. Reducing PSA (KLK3) mRNA levels with increasing Gleason grade (GS) of prostate tumors in TCGA. Patient numbers are indicated in parentheses. #, P < 0.001 compared with normal and *P < 0.00001 compared with GS6 tumors (paired Student’s t-test). The PSA mRNA levels display a statistically significant decreasing trend with increasing tumor grade (**P < 0.0001, Proportion Trend test).
G. Tumor PSA mRNA levels inversely correlate with tumor grade, metastasis, recurrence and survival in the Nakagawa data set. Shown are reduced PSA mRNA levels in high-grade (GS9–10) tumors, patients with lymph node (N) metastasis, patients that had recurrent diseases within 3 years, and in patients who were dead 3 years after diagnosis.
H-J. Tumor PSA mRNA levels were reduced in metastases in most Oncomine data sets (H) as illustrated in Varambally (I) and Yu (J) data sets. P values were determined by Student’s t-test.
K-L. High tumor PSA mRNA levels correlate with better biochemical recurrence (BCR) free and overall survival of PCa patients in the Nakagawa data set (K) and with better patient survival in 3 other data sets (L). P values were determined by log-rank test.
Panels A, C–E, G, I–K were adapted with permission from reference [131]. Panels B, H and L were adapted with permission from reference [144].
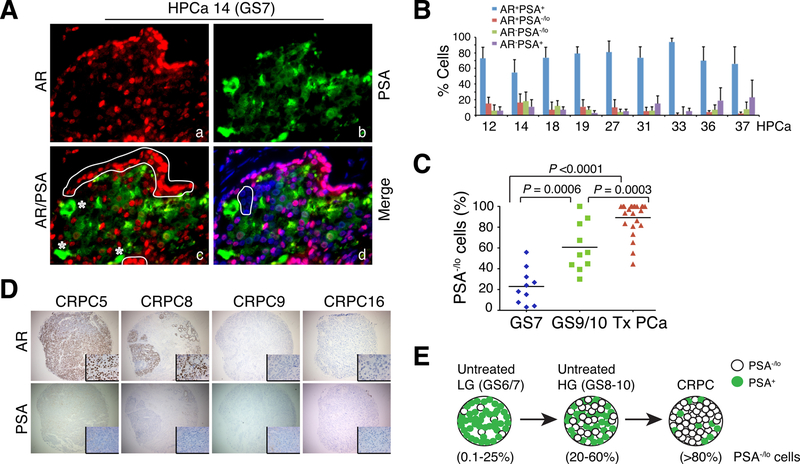
Fig. 4. PCa cell heterogeneity: Pre-existence of PSA−/lo PCa cells in treatment-naive HPCa and enrichment of PSA−/lo cells in advanced and treatment-failed PCa.
A-B. Representative immunofluorescence images (x400) illustrating discordant PSA and AR protein expression and the presence of PSA−/lo PCa cells in untreated HPCa 14 (GS7; A) and quantification of 4 subpopulations of PCa cells in the 9 HPCa samples (B). In A, AR+PSA+ PCa cells are marked by red nuclei and green cytoplasm, AR+PSA−/lo cells by red staining (panel c, white circled areas), AR-PSA+ cells by green staining (panel c, asterisks), and AR-PSA−/lo /lo cells by being negative (or low) for both red and green staining (panel d, white circle).
C. The abundance of PSA−/lo tumor cells is increased in high-grade (GS9/10) and in treatment-failed (Tx) PCa compared to untreated low/intermediate grade (GS7) tumors (n = 20–24 for each type of tumor samples).
D. IHC analysis of AR and PSA in the TMA samples. Shown are 4 CRPC samples illustrating homogeneous loss of PSA in all 4 samples and heterogeneous expression of AR (insets: 400x).
E. Schematic illustration of increasing PSA−/lo PCa cells from low-grade (LG) tumors to untreated high-grade (HG) tumors and to CRPC. Panels A, B and D were adapted with permission from reference [144].
Panel C was adapted with permission from reference [131].
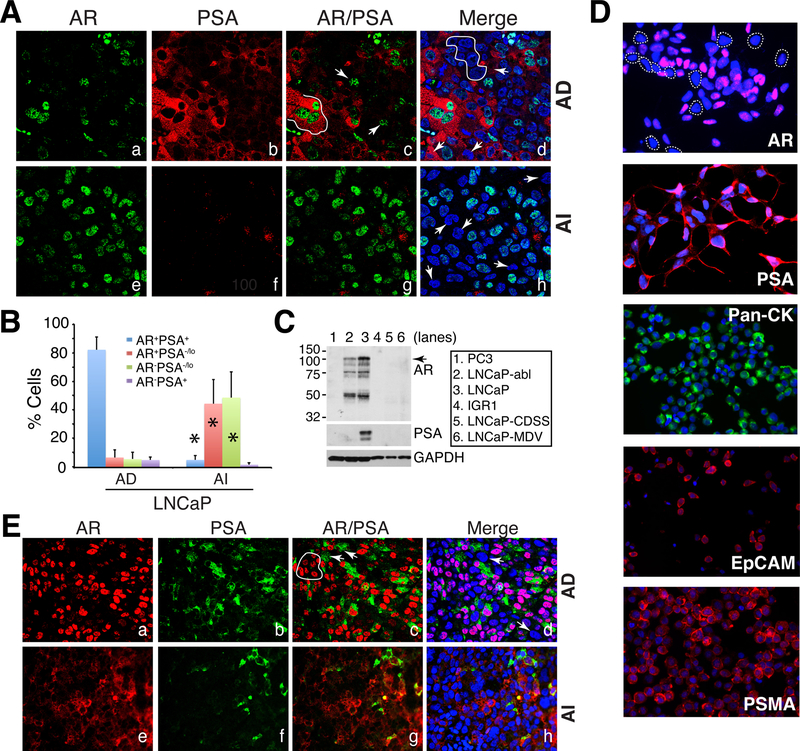
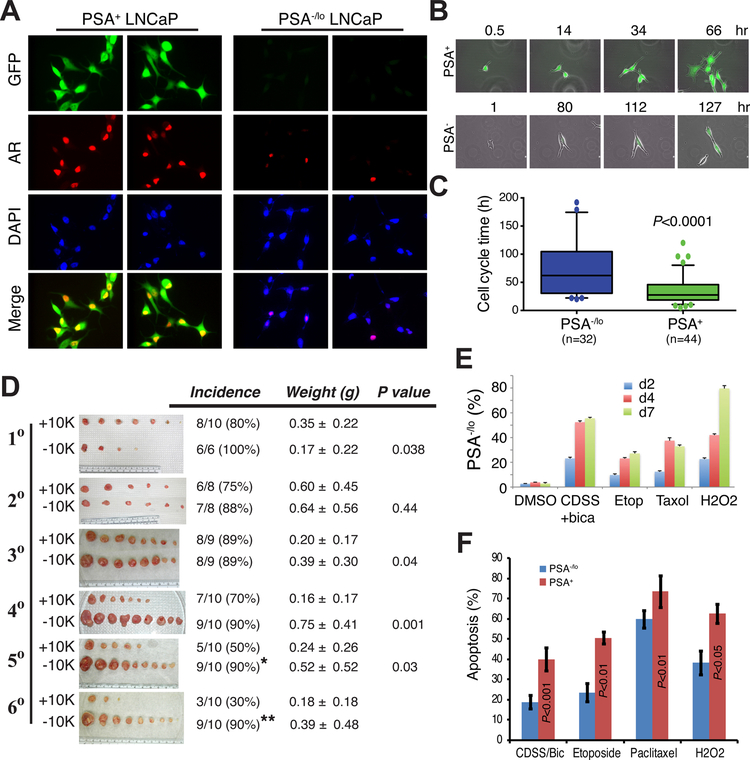
Fig. 5. Studying PCa cell heterogeneity in PCa cell and xenograft models: PSA−/lo PCa cells accumulate in experimental CRPC models.
A–B. Accumulation of PSA−/lo cells in LNCaP-AI tumors. (A) Double immunofluorescence (IF) staining of AR and PSA in AD (androgen-dependent) vs. AI (androgen-independent) LNCaP xenograft tumors. In panel c, the white line demarcates 3 AR+PSA+ cells and the arrows point to 2 AR+PSA−/lo cells. In panel d, the white circle demarcates several AR-PSA−/lo cells and the arrows point to 3 AR-PSA+ cells. In panel h, the arrows illustrate several AR-PSA−/lo cells. Shown are representative confocal images (original magnification; x400). (B) Quantification of the 4 subtypes of PCa cells in AD and AI LNCaP xenograft tumors. A total of 809 and 907 cells were counted from several AD and AI tumors, respectively. *P < 0.001 in AI compared in AD tumors.
C. WB analysis of AR and PSA heterogeneity in cultured LNCaP cell sublines. PC3 and IGR1 cells, which are negative for both proteins, were used as controls. Note that the wild-type LNCaP cells (lane 3) were AR+PSA+ whereas LNCaP-abl cells AR+PSA− (lane 2). LNCaP-CDSS and LNCaP-MDV cells were both AR−PSA− (lanes 5–6). The arrow indicates the ~114 kD full-length AR and lower bands represent AR splice variants (top panel).
D. Heterogeneity in expression of phenotypic markers in cultured LNCaP cells. Shown are representative IF images of AR, PSA, pan-cytokeratin, EpCAM and PSMA.
E. Double IF staining of AR and PSA in AD vs. AI LAPC4 xenograft tumors. In panel c, the white circle indicates several AR+PSA−/lo cells and arrows indicate AR-PSA+ cells. In panel d, arrows point to AR-PSA−/lo cells. Note significantly increased PSA−/lo LAPC4 cells in the AI tumor (f). Shown are representative confocal images (original magnification; x400).
All panels (except most of D) were adapted with permission from reference [144].
3.3. Tumor cell PSA is a PCa differentiation marker and tumor PSA levels correlate with better patients' survival
Do phenotypically distinct PCa cells display different biological and functional properties? We started addressing this question by focusing on PSA+ and PSA−/lo PCa cells. Serum PSA levels have been routinely utilized in the clinic to help diagnose PCa and, upon ADT/Enza treatment, rising (rebound) serum PSA levels represent a biomarker for tumor relapse. However, PSA, normally regulated by AR, a master pro-differentiation transcription factor (TF), represents probably one of the most lineage-specific differentiation markers in the human epithelia, as it is only expressed in fully differentiated human prostatic luminal cells. PSA is not expressed in rodents. As a prostate-specific differentiation marker, tumor cell PSA (KLK3) mRNA levels gradually decrease with increasing tumor malignancy, i.e., Gleason grade (Fig. 3A–E) [131,144]. This inverse correlation between the PSA mRNA levels and tumor grade can be best appreciated by analyzing the TCGA PRAD data set (Fig. 3F). As expected, early-stage (GS6) tumors showed significantly increased PSA mRNA levels than the normal prostate tissues (Fig. 3F), likely due to increased AR activity early during prostate tumorigenesis initiated and driven by the transformed LPs [20]. However, as tumors progress to higher Gleason grades, the tumor PSA mRNA levels gradually declined (Fig. 3F). This tumor grade-associated decrease in PSA mRNA levels may, again, be related to AR signaling since during PCa progression, AR signaling intensity becomes attenuated [215,216] due to increasing numbers of genetic and epigenetic mutations and alterations such as MYC and AKT overexpression and loss of tumor suppressors. Importantly, reduced tumor (cell) PSA mRNA levels were also observed in PCa metastases and recurrence (Fig. 3G–J), and prostate tumor PSA mRNA levels positively correlate with patients’ overall and BCR (Biochemical Recurrence) free survival (Fig. 3K–L) [131,144].
Consistent with the changes in KLK3 mRNA, intra-tumor PSA protein levels also decrease accompanying PCa progression (i.e., increasing Gleason grade) and become largely lost in CRPC (e.g., Fig. 4D) [131,144, 147]. Similar to PSA, another prostatic luminal differentiation marker, CD26 (also called DPP4; Fig. 1A), is also downregulated and lost during PCa progression and its loss causally promotes CRPC development [217]. It should be noted that our discussions herein linking tumor PSA (mRNA and protein) levels to overall tumor (cell) differentiation and to better PCa patient survival should NOT be confused with increased serum PSA levels as a PCa diagnostic and prognostic biomarker. PSA, as a secreted protein, is normally restricted to prostatic ducts and constitutes part of the seminal fluid, and should not be detected in the blood at significant levels. Increasing blood PSA levels only signal that the organ and vascular integrity of the prostate has been compromised allowing the access of PSA to the circulation.
3.4. The undifferentiated (PSA−/lo) compartment harbors selfrenewing and therapy-resistant PCSCs
Associated with the heterogeneity in PSA expression are (differentiated) PSA+ and (undifferentiated) PSA−/lo PCa cells that are readily observed in treatment-naïve human prostate tumors (e.g., Fig. 2A,D,G,J; Fig. 4A–B) as well as in PCa cell cultures (Fig. 5D) and xenografts (Fig. 5A,B,E) [131,144,149]. The PSA−/lo PCa cells are significantly increased in high-grade (GS9/10) tumors compared to low-grade (GS6/7) tumors (e.g., compare Fig. 2J and A,D,G) and become the predominant cell population in treatment-failed PCa and CRPC (Fig. 4C–E). Similarly, all castrated xenografts such as LNCaP-AI (Fig. 5A–B) and LAPC4-AI (Fig. 5E), as well as LNCaP cells subjected to chronic castration in vitro (Fig. 5C; also see Fig. 11, below), are enriched in PSA−/lo cells [131,144,149,152].
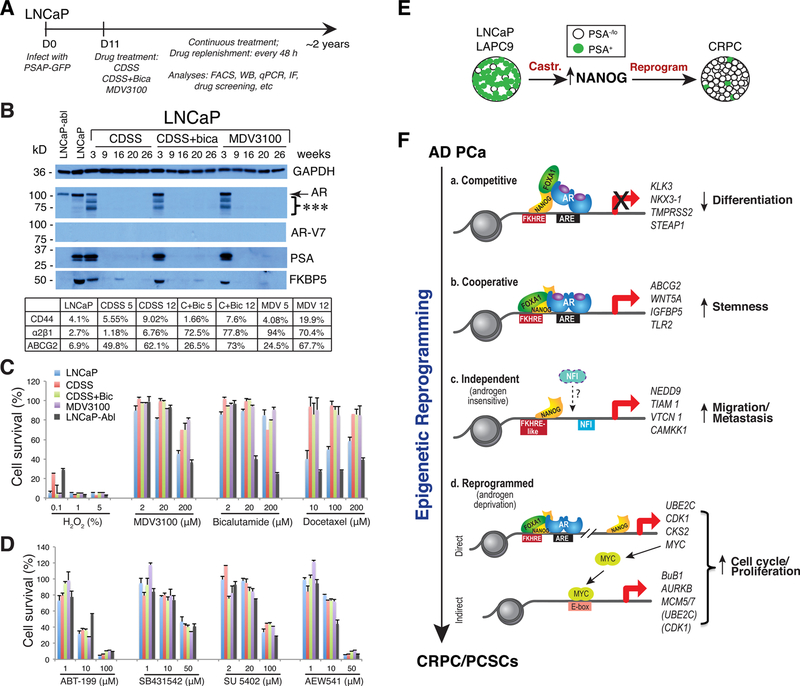
Fig. 11. PCa cell plasticity induced by ARSIs and stemness factor NANOG.
A–D. LNCaP cell plasticity and reprogramming by ARSIs. Shown in (A) is the experimental schema, in which bulk LNCaP cells were infected with the PSAP-GFP lentiviral reporter. In (B), LNCaP cells treated with 3 regimens of castration, i.e., CDSS, CDSS plus bicalutamide, and MDV3100 (Enza) for up to 26 weeks were used in Western blot analysis of AR, AR-V7 and two AR downstream targets, PSA and FKBP5. Regular LNCaP cells and LNCaP-abl cells were used as controls. Shown below is the summary of flow analysis of 3 PCSC markers in regular LNCaP and in 3 LNCaP-CRPC cells treated with CDSS, CDSS/bicalutamide (C + Bic) and MDV3100 (MDV) for 5 or 12 weeks. LNCaP-CRPC cells, in contrast to LNCaP and LNCaP-abl cells, were highly refractory to Enza, Bicalutamide and docetaxel (C) but were sensitive to BCL-2 inhibitor (ABT-199) and inhibitors of ALK (SB431542), VEGFR2/FGFR/PDGFR (SU 5402) and IGFR1 (AEW541).
E–F. Inducible NANOG expression time-dependently reprograms LNCaP-AD cells to the CRPC state. Shown in E is a schematic to illustrate that in LNCaP and LAPC9, PSA−/lo PCa cells normally comprise the minor population but castration upregulates NANOG that subsequently contributes to the generation of CRPC in which PSA−/lo cells now constitute the majority. Shown in F is a model depicting modes of operation of NANOG during reprogramming of androgen-dependent PCa cells to the CRPC state (see Text).
Panels A–D were adapted with permission from reference [152] and panels E–F from reference [150].
Do PSA+ and PSA−/lo PCa cells have distinct biological behavior, functional properties and therapeutic responses? To directly address this question, we developed a series of lentiviral-based lineage reporters in which a compound PSA promoter (PSAP) and enhancer was used to drive GFP (or other fluorescent reporter) expression. The PSAP-GFP lentiviral reporter was found to faithfully report and separate PCa cells that express abundant endogenous PSA (GFP+) vs. those that express little/no PSA (GFP−/lo) (Fig. 6) [131,144,152]. As expected, most PSA+ PCa cells express high levels of nuclear AR whereas most PSA−/lo cells are AR−/lo (Fig. 6A). Comprehensive biological studies and functional interrogations reveal that the PSA−/lo PCa cell population harbors authentic PCSCs that can long-term self-renew and propagate tumors in vivo, and are inherently therapy-resistant [131,144,149,152]. First of all, the PSA−/lo PCa cells are enriched in stem cell gene expression profiles, much like the normal (AR−/loPSA-) basal/stem cells [17,43]. Secondly, PSA−/lo PCa cells also possess some of the epigenetic makeups of normal stem cells such as poised bivalent chromatin characterized by simultaneous association of genes with the H3K4me3 and H3K27me3 histone marks [144]. Thirdly, 5–18 % of the cells in PSA−/lo PCa cell population can undergo Notch-regulated asymmetric cell division (ACD; Fig. 6B) [131,144,149], a cardinal feature of epithelial stem cells. Fourthly, similar to many normal adult stem cells, PSA−/lo PCa cells are much more quiescent and have longer cell-cycle transit times than PSA+ cells (Fig. 6C). The dormant nature of PSA−/lo cells can also be appreciated from the single-cell tracing: while the single GFP+ LNCaP cell developed into a clone of 6 cells within 66 h, the single GFP- LNCaP cell only generated a 3-cell progeny in 127 h (Fig. 6B). Fifthly, the PSA−/lo PCa cells possess much greater serial tumor-propagating capacity than corresponding PSA+ cells in that the PSA−/lo PCa cells can initiate xenograft tumors that can be propagated indefinitely whereas PSA+ PCa cell-initiated xenografts can only be propagated for a limited number of generations (Fig. 6D) [131,144,149]. Sixthly, PSA−/lo PCa cells are much more resistant to apoptosis induction and become greatly enriched by castration, prooxidants and chemotherapeutic drugs (Fig. 6E–F) [131, 144,152]. Lastly, purified PSA−/lo PCa cells could readily initiate and propagate CRPC in castrated mice at low cell input [131,144,149]. Together, these observations indicate that the undifferentiated PSA−/lo PCa cell population harbors long-term self-renewing CSCs (i.e., PCSCs), which can indefinitely propagate xenograft tumors, are inherently insensitive to castration and other therapeutics, and may function as cell (s) of origin for CRPC.
Fig. 6. The PSA−/lo PCa cell population harbors cells that exhibit CSC properties.
A. Most PSA−/lo PCa cells are also AR−. Shown are IF images of LNCaP cells separated by the PSAP-GFP lentiviral reporter and stained for AR. Note that all GFP+ (PSA+) LNCaP have high nuclear AR whereas the majority of PSA−/lo cells are also AR−/lo.
B. PSA−/lo PCa cells are more dormant and can undergo ACD (bottom) whereas PSA+ PCa cells are more proliferative and only undergo symmetrical cell division. Shown are time-lapse images of LNCaP cells infected with the PSAP-GFP reporter and recorded at the indicated time points.
C. PSA−/lo LNCaP cells have much longer cell-cycle transit times than the corresponding PSA+ cells. The number of cells analyzed by time lapse videomicroscopy are indicated in parentheses.
D. The PSA−/lo PCa cells can initiate long-term xenografts that can be serially propagated. PSA+(GFP+) and PSA−/lo (GFP−/lo) LAPC9 cells were purified from reporter tumors and 10,000 cells each (+10 K or −10 K) were implanted subcutaneously in intact male NOD-SCID mice. The first-generation (1 °) tumors were harvested ~2 months later. One +10 K tumor and two −10 K tumors were utilized to purify GFP+ and GFP-/lo LAPC9 cells, respectively, which were used in secondary (2 °) tumor transplantation experiments. One +10 K and one −10 K 2 ° tumors were utilized to purify GFP+ and GFP- LAPC9 cells, respectively, for 3 ° transplantation. The 4 °, 5 ° and 6 ° transplantation experiments were carried out in a similar fashion. For each generation, tumor images (shown not to the same scale), tumor incidence (%; *P = 0.045 and **P = 0.006, when compared to the corresponding +10 K groups, χ2 test), time (d) of harvest, tumor weights (mean ± S.D), and P values (Student’s t-test) were indicated. For the 6 ° generation, statistics was not applicable due to the small number of tumors developed in the PSA+ group.
E. PSA−/lo LNCaP cells are resistant to androgen deprivation (i.e., CDSS plus bicalutamide) as well as chemotherapeutics and hydrogen peroxide. Shown are % PSA−/lo cells in PSAP-GFP infected LNCaP cells treated with the conditions indicated for 2, 4, and 7 days (d). Differences between all individual treatments and DMSO are statistically significant (P < 0.01).
F. PSA−/lo PCa cells are more resistant to apoptosis as assessed by the Vybrant apoptosis assays. Bulk cultured LNCaP cells infected with the PSAP-GFP lentiviral reporter were plated at 120,000 cells/well in 6-well plates. Cells were treated for 4 days with either DMSO, 2% CDSS plus 20 μM Bicalutamide (CDSS/Bic), 20 nM Paclitaxel, 1 μM etoposide, or 1 μM H2O2. The % apoptosis represents the mean ± S.D (n = 3) and P values determined by Student’s t-test. No difference in apoptosis was observed in the two populations in response to vehicle DMSO (not shown).
Panels A–C and F were adapted with permission from reference [144]. Panels D and E were adapted with permission from reference [131].
3.5. Heterogeneity in PCSC phenotypes and challenges of studying PCSCs in treatment-naïve primary prostate tumors
Our studies on PCa cell subpopulations and their molecular regulation [117–169] in multiple xenograft and PDX models as well as >200 patient tumors and tumor-derived cells and organoids (e.g., Figs. 2, 4, 7, 8) suggest that human PCSCs that meet at least some of the CSC definitions [3,120,129,140,145,160] are phenotypically heterogeneous. For example, ABCG2+ [171], CD44+/hi [118], CD44+α2β1+ [119], holoclone-forming [121], and NANOG-expressing [122,125] PCa cells all possess some of the classically defined CSC properties and identify overlapping and non-identical subpopulations of tumor-initiating and tumor-propagating cells depending on specific cell types and genetic backgrounds of the model and patient tumors [144]. Interestingly, these phenotypically diverse subsets of PCSCs all seem to be encompassed in the PSA−/lo PCSC pool [144] (see Fig. 14A, below). Importantly, despite the phenotypic diversity, the functionally defined PCSCs share one common property, i.e., they are intrinsically resistant to various therapeutics including castration, antiandrogens, radiation, chemotherapy drugs, and prooxidants. I should point out that the cell(s) of origin for cancer and CSCs are two different concepts that should not be mixed up and used inter-changeably [145].
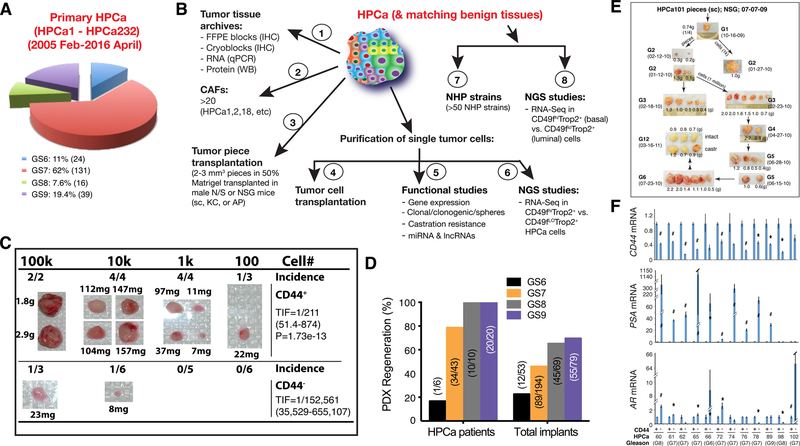
Fig. 7. Studying PCa cell heterogeneity in treatment-naïve patient primary prostate tumors.
A. Pie chart presentation of the Gleason grade distribution of 210 HPCa samples out of a total of 232 (please change the number to 232) HPCa our lab used and studied from Feb. of 2005 to April of 2016 at the M.D Anderson Cancer Center. HPCa, human prostate cancer (specimens or samples).
B. Experimental uses for the HPCa (1–6) and matching benign samples (7–8) in our lab. CAFs, carcinoma-associated fibroblasts; FFPE, formalin-fixed and paraffin embedded; NGS, next-generation sequencing (studies); NHP, normal human prostate (epithelial) cells; N/S, NOD/SCID mice; NSG, NOD/SCID-γ mice.
C. An example of tumor regeneration from purified CD44+ HPCa cells. CD44+ and CD44− HPCa52 cells were MACS-purified from patient tumor (GS8) and co-injected, at the indicated cell numbers, with 10k Hs5 cells in 50 % Matrigel s.c into irradiated NOD/SCID-γ mice. The 10k and 100k tumors were harvested at ~4 months whereas 100 and 1k tumors were harvested at 7 months after implantation. Shown on the right are TIF (tumor-initiating cell frequency). The difference in CD44+ HPCa 52 cell TIF and the corresponding CD44- cell TIF is highly statistically significant (P = 1.73e-13).
D. Success rate (%) of PDX (patient-derived xenograft) regeneration plotted based on total # of HPCa patient specimens (left bars; e.g., for GS7 tumors, 34 of the 43 patient HPCa pieces implanted in immunodeficient mice regenerated PDX [79 %]) or total # of implants (right bars; e.g., 89 of the 194 GS7 HPCa pieces implanted in all 3 sites (i.e., sc, KC, and AP) regenerated PDX [thus 46 %]).
E. Serially transplantable HPCa101 (GS9[4 + 5]) PDX derived from tumor piece implantation. Shown are representative tumor images of HPCa101 PDX at multiple generations (G). castr, castrated; sc, subcutaneously.
F. qPCR analysis of CD44, AR, and PSA mRNAs in CD44+ and CD44− HPCa cells freshly purified from untreated primary prostate tumors. The results are expressed as the mRNA levels in CD44+ HPCa cells relative to those in the matched CD44− HPCa cells. *P < 0.05; #P < 0.01 (Student’s t-test).
Panels F was adapted with permission from reference [144].
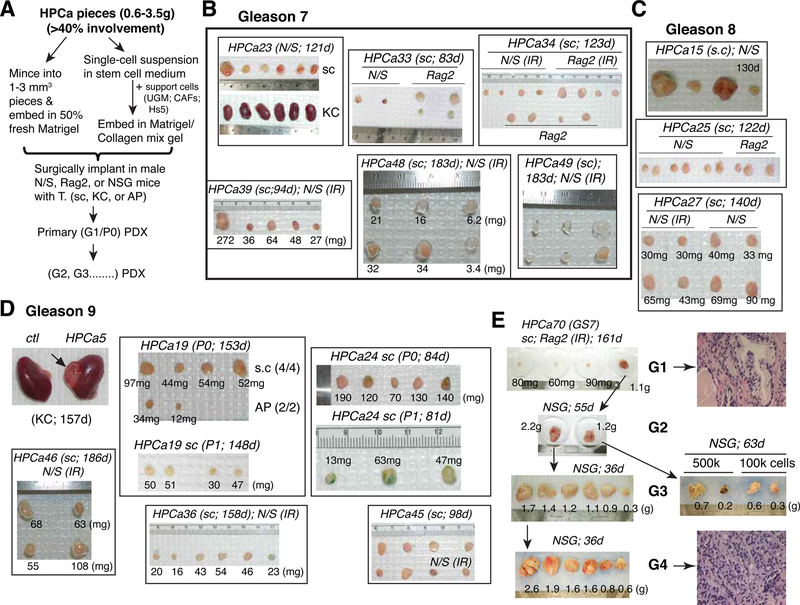
Fig. 8. Studying PCa cell heterogeneity in treatment-naïve patient primary prostate tumors.
Shown are examples of PDXs established in our lab from treatment-naïve patient prostate tumors using tumor pieces xenotransplanted in immune deficient mice.
A. Basic experimental flow. AP, anterior prostate; CAFs, carcinoma-associated fibroblasts; Hs5, immortalized human BM mesenchymal stem cells; KC, kidney capsule; sc, subcutaneously; UGM, urogenital mesenchyme.
B-D. Examples of PDXs established from Gleason 7 (B; 6 HPCa examples shown), Gleason 8 (C; 3 HPCa examples shown), and Gleason 9 (D; 6 HPCa samples shown) HPCa samples. IR, irradiated; P, passage.
E. Four generations (G) of HPCa70 PDX tumors are illustrated. A representative HE staining for G1 and G4 tumor was shown on the right.
Along with our efforts in dissecting the functional properties of PCa cell subpopulations and PCSCs, numerous groups worldwide have reported on PCSCs [218–324]. A Pubmed search with ‘Prostate Cancer Stem Cells’ as the Keyword would turn up >3000 references. These reported PCSCs manifest a wide diversity of phenotypes including, among others, ABCG2+ [218], CD133+ [219,220,236], CD133+α2β1+ [219], CD44+ [219,232,299,311], ALDH+/hi [224], MHC−/lo (and PSA−/lo) [226], LP-like [227,238] or basal-like [231], Trop2+/hi [230], TOP2A-[243], hTERThi [250], Bmi1+Sox2+ [260], CD24+/hi [261], CXCR2+ [272], Zeb1+ [280], holoclone-forming cells [291], and CD117(KIT)+ [310]. Many of these PCSC phenotypes overlap with the PCSCs reported by our lab. Of clinical relevance, expression of some PCSC phenotypic markers has been linked to PCa aggressiveness and poor patient prognosis and survival [220,240,294].
Studying PCSCs or tumor-initiating PCa cells in treatment-naïve patient primary tumors are generally challenging, based on our working experience on 232 primary HPCa at the M.D Anderson Cancer Center (from Feb. of 2005 to April of 2016; Figs. 7, 8) and 48 primary HPCa at the Roswell Park Comprehensive Cancer Center (May of 2016 – present). Unlike CSCs freshly purified out from some other aggressive human cancers (e.g., colorectal and lung cancers and GBM) that can readily initiate serially transplantable xenografts, putative PCSCs acutely purified from patient primary tumors, especially the low-grade tumors, frequently do not manifest significant tumor-regenerating capabilities, even in the presence of ‘helper’ cells such as CAFs and mesenchymal stem cells or MSCs [123,132,144]. The difficulty in experimentally reconstituting the tumorigenicity in immunodeficient mice of freshly purified HPCa cells and/or PCSCs may be related to the well-known indolent nature of most low-grade tumors (i.e., GS6–7 tumors, representing >70 % of the HPCa; Fig. 7A) and the complex tumor microenvironment (TME) of PCa [155]. In support of these points, primary prostate tumor pieces, when xeno-transplanted into various types of immune-deficient mice (regular or irradiated (IR) NOD/SCID, NSG, Rag2, etc) at different sites including subcutaneum (s.c), anterior prostate (AP) or subrenal capsule in Matrigel or together with rUGM (rat urogenital sinus mesenchyme), frequently initiated PDX that could often be serially passaged (Figs. 7D–E; 8). As expected, high-grade tumors (≥GS8) generally exhibit higher tumor take rates (Fig. 7D) although we have succeeded in generating multiple PDX lines from primary prostate tumors of GS7-GS9 (Fig. 8), and many PDX lines, e.g., HPCa101 from a GS9 (4 + 5) patient tumor (Fig. 7E) and HPCa70 from a GS7 patient tumor (Fig. 8E), have been passaged for many generations (G) in immune-deficient mice. Occasionally, we could even reconstitute the high tumorigenicity in PCSCs freshly isolated from primary tumors. For instance, the CD44+/hi PCa cells acutely purified from the GS8 HPCa52 patient tumor demonstrated significantly higher tumor-initiating frequency (TIF) than the corresponding CD44−HPCa52 cells (Fig. 7C). Significantly, we have employed HPCa cells freshly purified from many high-volume primary tumors (i.e., with ≥80 % tumor involvement) in our dissection of PCa cell heterogeneity and studies on PCSCs [17,18, 118–120,122,123,125,126,130–132,139,144,149,151,152,154,160, 163–165]. As illustrated in Fig. 7F, most of the CD44+ HPCa cell populations freshly purified from a dozen of GS7-GS9 HPCa were found to express much lower levels AR and PSA mRNA than the corresponding CD44− HPCa cells [144]. These observations in patient primary tumors are consistent with our data showing that the CD44+/hi PCa cell population is enriched in PCSCs [118,119,126,130,131,144] (Fig. 7C) and with the concept that PCSCs are less differentiated expressing lower levels of AR and PSA [131,144,149,152].
3.6. Therapy resistance and metastatic propensity in PCSCs and molecular regulators of PCSCs
Studying the genetic, transcriptomic and cellular heterogeneity in PCa [2,80–116,325] and in-depth elucidation of PCSCs [117–169, 218–324] have direct relevance to understanding the mechanisms of therapy resistance [326,327]. Recent studies in PCa patients undergoing neoadjuvant ADT trials have directly linked the clonal heterogeneity, i. e., ITH, to distinct clinical responses [328–330]. For example, primary tumor clones with alterations in multiple oncogenic drivers are preferentially selected for by neoadjuvant-intensive ADT [328]. In a case of PCa with two primary tumor foci, a large GS9 tumor and an adjacent GS7 nodule, DNA sequencing showed that the GS9 tumor had suffered single-copy loss of both PTEN and TP53 [329]. Strikingly, neoadjuvant intensive ADT completely eliminated the GS7 nodule whereas subclones from the GS9 tumor that had sustained mutations to the remaining copies of PTEN and TP53 emerged to mediate ADT resistance [329]. These studies [328–330] highlight the importance of nascent PCa cell genetic heterogeneity in dictating tumor response to clinical treatments.
PCSCs, in spite of phenotypic diversity, are commonly endowed with high clonogenic and tumor-initiating/tumor-propagating activities. Importantly, PCSCs have been reported to be intrinsically therapy-resistant, including resistance to castration, chemotherapy, radiotherapy and, likely, immunotherapy [2,3,125,131,138,218,226,227, 251–253,265,278,299,300,310,313,315]. For example, the PSA−/lo PCa cells are resistant to castration, antiandrogens, prooxidants and chemodrugs [131,144] and ABCG2+ PCSCs are resistant to ARSIs due to increased efflux of androgens [218]. Similarly, the PSA−/lo/MHC−-/lo PCSCs are resistant to docetaxel as a result of enhanced Notch and Hedgehog signaling [226], and the CD44+ [299], LP-like [227,252] and hypoxia-induced [265] PCSCs are all inherently castration- and chemo-resistant. Finally, sphere-derived PCSCs are resistant toγ?d T cell-mediated cytotoxicity [300] and PCSCs, like many other CSCs, are generally ‘immuno-deficient’ lacking the expression of many immunogenic molecules [160].
PCSCs are also invasive and possess high metastatic potential [3,118, 126,128,155,159,228,309,313]. For example, our early studies indicate that the CD44+ PCSCs are highly metastatic [118] and their metastatic capacity can be suppressed by miR-34a [126] and miR-141 [154]. Of interest, PCSCs metastasized to the bone are rather dormant [3], and this dormancy is maintained, at least partly, by BMP7 [225]. The high invasive and metastatic capabilities in PCSCs may be intrinsic to PCSCs [3,155,159] and/or are conferred by TME-derived inflammatory and cytokine molecules that induce epithelial-mesenchymal transition or EMT [3,273–275]. For example, TGF-β in the TME has been shown to promote PCa stemness [287,311] whereas signaling from IL-30/IL-27p28 (an IL-6 family member) shapes PCSC behavior and regulates their metastasis [254]. Similarly, CXCL12γ promotes the development of metastatic CRPC [255], CCL5 generated by tumor-associated macrophages (TAM) promotes PCSCs and their metastasis via β-catenin/STAT3 signaling [286], and MSC-derived IL-28 drives selection of apoptosis-resistant bone-homing PCSCs [313]. It’s noteworthy that PCSCs, slow-cycling (dormant) PCa cells, and PCa cells undergoing EMT share many biological properties (e.g., dormancy) and frequently identify overlapping but non-identical cell populations [reviewed in 3].
Studies on PCSCs have identified many signaling pathways that positively regulate the PCSC properties, which include PTEN/PI3K/AKT [222,228], NF-kB [224], Notch [137,158,226], Hedgehog [226], Wnt/β-catenin [230,250,257,259,286,314], JAK/STAT [235], β4 integrin [239], hypoxia through HIF1α [246], IL-30/IL-27p28 [254, 266], BRCA1/EZH2 [271], and FGF/FGFR [308]. Also reported are positive molecular regulators of PCSCs including, among others, MYC [234],STAT3[235,237,241,245,286,296],ERG[236,297],BMI-1[139, 249], Skp2 [251], YAP1 [253], CXCL12γ [255], LIN28B [248,295], ASPM [259], NANOG [122,125,135,137,142,143,150,156,262,263], SOX2 [270,276], OCT4 [270], CXCR2 [272], SOX9 [277], HOTAIR [296], BMP5 [298], and MUC-1 [279,306,318]. Finally, our studies have uncovered several tumor-suppressive microRNAs (miRNAs) as the negative regulators of PCSCs [127], which include miR-34a [126,167], let-7 [130,248], miR-128 [139], miR-199–3p [151], and miR-141 [154].
3.7. PCa cell heterogeneity in AR expression: Relationship to PCSCs and direct impact on tumor response to ARSIs
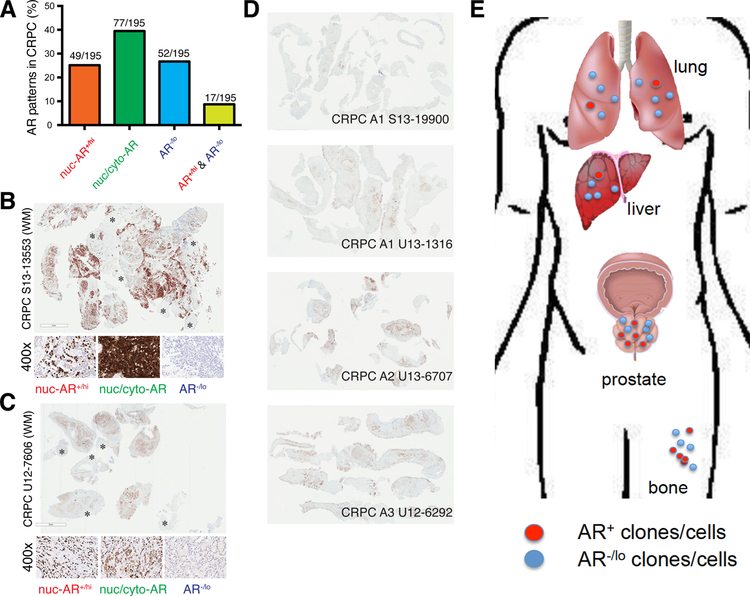
AR signaling has been the clinical therapeutic target, and different forms of ARSIs have been used to treat PCa patients for nearly 80 years. In principle, in order for ARSIs to exhibit significant and long-lasting therapeutic efficacy, most, if not all, PCa cells in patient tumors should express AR (i.e., AR+) and rely on AR activity for their survival and wellbeing. In reality, however, AR expression and AR activity are highly heterogeneous in PCa. FIRST, shortly after the AR gene was cloned and antibodies against AR were developed, numerous studies have reported the heterogeneity in AR expression and existence of AR−/lo PCa cells across the entire spectrum of PCa, from treatment-naïve primary tumors to tumors treated with neoadjuvant ADT to mCRPC [2, 147,175–190,192–200,202–209]. SECOND, transcriptomic profiling also defined a de novo low AR-activity subclass in treatment-naïve PCa [204]. THIRD, studies from PCa metastases, especially in specimens obtained from warm autopsy patients, have revealed many subtypes of mCRPC with respect to AR expression including the AR−NE− metastases called double-negative PCa or DNPC [201,203,207,325,331]. FOURTH, in support, our recent study [163] showed that ~27 % of 195 CRPC TMA cores were classified as AR−/lo (Fig. 9A). AR IHC analysis in 6 WM sections also revealed the AR−/lo PCa cells as the prominent (Fig. 9B–C) or even predominant (Fig. 9D) cell population in castration-resistant tumors in the prostate [163]. FIFTH, recent dual-modality PET/CT imaging studies using 18F-DHT (to image AR expression) and 18F-deoox-yglucose (to image overall tumor glycolytic activity) similarly revealed both AR+ and AR−/lo metastatic foci in PCa patients with mCRPC [213, 214] (Fig. 9E). FINALLY, transcriptional profiling in treatment-failed tumors identified an AR activity-low stemness program that was linked to Enza resistance [288]. This last study [288] implies that ARSIs, while blocking AR activity (as expected), also upregulate the stemness in patient PCa cells, consistent with many other studies (see below).
Fig. 9. AR heterogeneity in CRPC and PCa metastases.
A. Quantitative summary of the 4 AR expression patterns in CRPC based on the AR IHC analysis in 195 TMA cores.
B-C. Whole-mount (WM) sections showing AR heterogeneity in both expression levels and subcellular distribution patterns in two cases of patient CRPC. AR was stained using an antibody recognizing an N-terminal epitope that will capture full-length AR and all C-terminally truncated AR splice variants. AR−/lo cells/areas are indicated by asterisks (*). Shown below the WM sections are enlarged images of 3 main patterns of AR expression, i.e., AR−/lo, nuclear AR (nuc-AR+/hi) and mixed nuclear/cytoplasmic AR (nuc/cyto-AR).
D. AR IHC in another 4 cases of patient CRPC. AR staining in WM sections was conducted as in B–C.
E. Metastases in PCa patients also exhibit AR+ and AR−/lo clonal patterns, based on recent imaging analysis [212,213].
Panels A–D were adapted with permission from reference [163].
What are these AR−/lo PCa cells and how are they molecularly regulated?
There is substantial evidence that the AR−/lo cells in untreated PCa models and patient tumors represent ‘undifferentiated’ cells and possess many PCSC properties, and therapies including chemotherapy and radiotherapy frequently enrich the AR−/lo PCSCs [reviewed in 147]. For example, we have reported that the CD44+ [118] and PSA−/lo [131,144, 149] PCSCs in untreated PCa models and patient tumors are mostly AR−/lo (e.g., Fig. 7F). Similarly, many PCSC populations reported by others in untreated PCa models or treatment-naïve primary tumors are also AR−/lo, including ABCG2+ [218], CD133+ [219,220], CD133+α2β1+ [219], CD44+ [219,224,232], (PSA−/lo)MHC−/lo [226], LP-like [227, 238], basal-like [231], hTERThi [250], CD24+/hi [261], and CXCR2+ [272] cells. In fact, repression of AR expression/activity has been shown to reprogram bulk PCa cells to generate AR−/lo PCSCs [e.g., 244,245,264, 289,292] (also see Fig. 11, below). For example, loss of AR expression was shown to promote PCSCs via STAT3 signaling [245], and repression of AR signaling by cytokines secreted from the infiltrated BM MSCs led to significant expansion of AR−/lo PCSCs [244]. Notably, preferential expression of the oncogene MDM2 in AR−/lo PCa cells led to AR degradation, which helped maintain their PCSC properties [264]. Finally, androgen deprivation has been shown to reprogram bulk PCa cells, both functionally and transcriptionally, into PCSCs [289,292], consistent with our studies [152,163] (also see below) and with recent transcriptional profiling of treated patient tumors that identified a low-AR activity stemness program to be associated with Enza resistance [288].
How would the PCa cell heterogeneity, particularly, the AR−/lo PCa cells, impact tumor response to ARSIs and other clinical treatments?
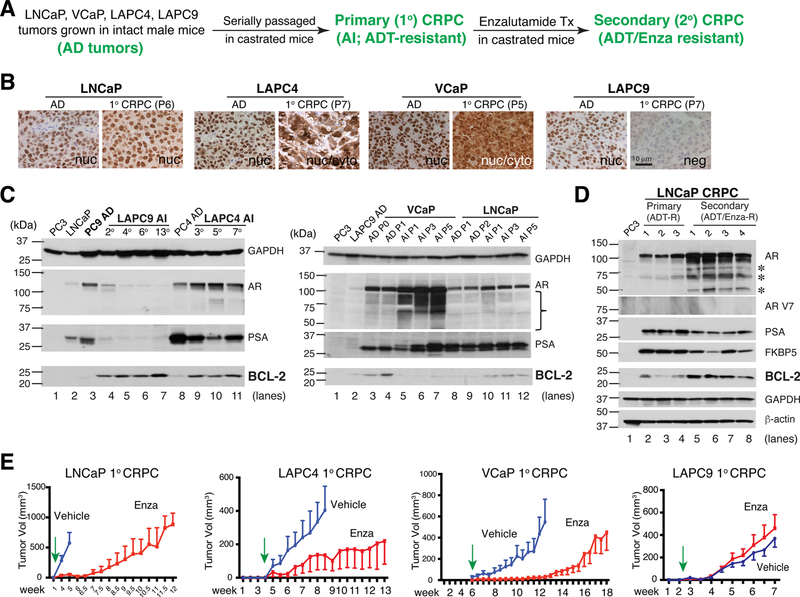
To directly answer this question, we have established several paired AD/AI xenograft models that mimic the primary (1 °; ADT resistant) and secondary (2 °; ADT/Enza-resistant) CRPC in treated PCa patients (see Introduction; Fig. 10A) [163]. Interestingly, the 4 primary CRPC models displayed unique changes in AR (and PSA) during their evolution to becoming fully castration-resistant (Fig. 10B–D). Specifically, LNCaP 1 ° CRPC showed upregulated nuclear AR, LAPC9 1 ° CRPC showed loss of AR, and LAPC4 and VCaP 1° CRPC displayed mostly cytoplasmic AR (Fig. 10B). Therapeutic studies in mice bearing these 1° CRPC revealed that the 3 AR+ CRPC models (LNCaP, LAPC4 and VCaP) responded to Enza treatment although Enza-resistant tumors started to emerge after ~6 (for LNCaP and LAPC4) and ~13 (for VCaP) weeks of Enza treatment (Fig. 10E) [163]. In contrast, the AR−/lo LAPC9 1° CRPC showed de novo resistance to Enza (Fig. 10E) [163]. The insensitivity of AR−/lo PCa/CRPC to Enza was also demonstrated using isogenic AR+ and AR-KO LNCaP clones [163]. These results suggest that persistent castration (ADT) would eventually lead to two subtypes of CRPC, the AR+/hi LNCaP-like CRPC with most cells expressing high nuclear AR and the AR−/lo LAPC9-like CRPC with most cells losing AR expression [163] (Fig. 14C). Notably, our xenograft-based CRPC modeling studies have pinpointed BCL-2 as a vitally important therapeutic target for both AR+/hi and AR−/lo CRPC [163] (Fig. 10C–D; also see Figs. 12, 13, below).
Fig. 10. AR−/lo CRPC is insensitive to Enza and BCL-2 is upregulated by castration.
A. Schema in generating paired xenograft AD tumors, primary CRPC (ADT-resistant) and secondary CRPC (both ADT/Enza-resistant) in LNCaP, VCaP, LAPC4 and LAPC9 models.
B. The above 4 xenografts develop unique AR expression patterns as they evolve towards CRPC. P, passage (of tumors in mice).
C. BCL-2 was elevated in 3 of the 4 CRPC xenograft (except VCaP) models.
D. BCL-2 was further increased in the LNCaP secondary CRPC model.
E. Distinct responses of the 4 xenograft CRPC models to Enza (see Text). Green arrows indicate the time when treatment of tumor-bearing mice was started.
All panels in the Figure were adapted with permission from reference [163].
Fig. 12. BCL-2 represents a therapeutic target in AR+/hi subtype of CRPC.
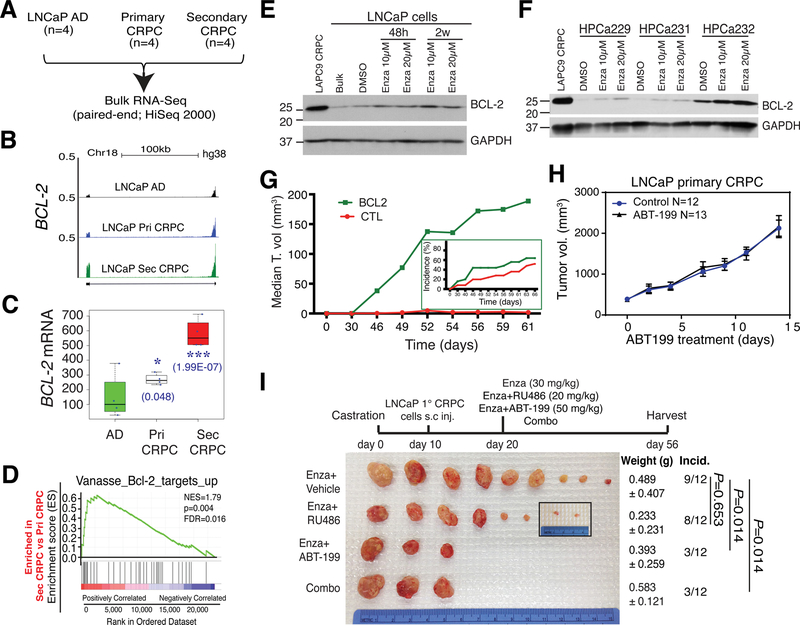
A–D. In whole-genome RNA-seq experiments (A), BCL-2 mRNA was increased in LNCaP primary (Pri; ADT-resistant) CRPC but dramatically upregulated in LNCaP secondary (Sec; both ADT/Enza-resistant) CRPC (B–C). Consequently, a BCL-2 target signature was enriched in LNCaP secondary CRPC compared to the LNCaP primary CRPC (D).
E–F. BCL-2 protein was upregulated by Enza in cultured LNCaP cells (E) as well as in primary tumor cells freshly purified from 3 HPCa patients.
G. BCL-2 overexpression is sufficient to promote LNCaP tumor growth (i.e., median tumor (T) volume) and also increase tumor incidence (inset). Please introduce a space between this sentence and the next sentence (in front of H).
H-I. BCL-2 inhibitor, ABT-199, did not appreciably affect the LNCaP primary CRPC growth (H) but, when used in combination with Enza, dramatically curbed development of castration/Enza-resistant secondary CRPC (I).
All panels in this Figure were adapted with permission from reference [163].
How are AR−/lo PCa cells/clones maintained and what unique signaling mechanisms drive their non-responsiveness to ARSIs and other therapeutics?
The inherent PCSC properties in these cells may partly answer these questions although we have just begun to understand these cells at the detailed molecular and cellular levels. Our early studies found that the PSA−/lo(AR−/lo) PCSCs actually require the corresponding PSA+(AR+) PCa cells for their survival and for maintenance of their properties such as the relative dormancy [131,144]. Others have reported that both AR+ and AR−/lo PCa cells cooperate to establish and maintain the metastatic ecosystem (niche) in the bone marrow [200]. These observations are consistent with the co-existence of both AR−/lo and AR+ PCa cells in treatment-naïve tumors as well as in castration-resistant primary (prostate) tumors and metastases. These observations also suggest that the two subpopulations of PCa cells may constantly interact with each other to form a tumor-sustaining cellular network, which will be disrupted by, and also dynamically evolves in response to, therapeutic interventions. Recent studies begin to unravel signaling pathways and molecules that may play uniquely important roles in supporting the AR−/lo PCa cells, clones and metastases, including, e.g., β-catenin [197], FGF/FGFR [201], the polycomb repressor complex-1 [203], T-type calcium channels [206], and stem cell regulators [163].
4. PCa cell plasticity and PCa lineage plasticity
PCa cell heterogeneity in expression of AR, the therapeutic target, and the presence of AR−/lo PCa cells and PCSCs in primary tumors are not the only problems in PCa treatment. Like most other cancer cells, PCa cells are also phenotypically and functionally plastic due to their fluid epigenome and genetic alterations. The PCa cell plasticity can be best appreciated from our studies in LNCaP cells [e.g., 150,152] (Fig. 11), one of the most frequently used PCa cell line model that has manifested heterogeneity in every phenotypic marker (including AR) we have analyzed (e.g., Fig. 5D), that has been used to generate dozens of sublines including LNCaP-abl (Figs. 5C; 11C–D), and that has been shown, by scRNA-seq analysis, to be very heterogeneous containing a subpopulation with attenuated androgen response [109]. Chronic exposure of LNCaP cells to 3 regimens of castration, i.e., CDSS, CDSS plus bicalutamide and Enza (also called MDV3100), in 2–3 months (Fig. 11A) turned the bulk AR+ (and PSA+) LNCaP cultures into AR-PSA-cells [152] (Fig. 11B). Importantly, the reprogrammed AR−PSA− LNCaP cells acquired many PCSC phenotypic markers including CD44, integrin α2β1 and ABCG2 [152] (Fig. 11B, table below). These AR−PSA−LNCaP cells were resistant to not only antiandrogens (Enza and bicalutamide) but also docetaxel (Fig. 11C) but, interestingly, displayed sensitivities to the BCL-2 inhibitor (ABT-199) and several kinase inhibitors [152] (Fig. 11D). This study [152] vividly illustrates PCa cell plasticity caused by a clinically relevant treatment regimen, i.e., castration (Fig. 14B). Numerous studies by others have also shown that repression of AR expression/activity can reprogram bulk PCa cells to generate AR−/lo PCSCs [e.g., 244,245,264,289,292].
PCa cell plasticity can also be induced by stemness factor reactivation or overexpression. Our studies have implicated NANOG in PCSCs (and other CSCs) and in regulating cancer aggressiveness and castration resistance [122,125,135,137,142,143,150,156]. Interestingly, castration of LNCaP and LAPC9 models, in which most cells are differentiated (AR+PSA+), led to upregulation of NANOG that subsequently contributed to CRPC development [125] (Fig. 11E). Strikingly, induction of NANOG expression in bulk LNCaP cells through a transgene, in a time-dependent manner (~3–4 weeks), gradually reprogrammed these cells into androgen-insensitive CRPC cells [150] (Fig. 11F). Time-resolved RNA-seq coupled with NANOG ChIP-seq, together with other molecular studies reveal that NANOG-induced reprogramming of LNCaP cells to the castration-resistant state involved 4 sequential steps: inhibition of the AR-mediated differentiation program due to competitive NANOG binding to many canonical pro-differentiation AR/FOXA1 sites (step 1), increased expression of other stemness genes including ABCG2 and WNT5A as a result of NANOG cooperation with AR/FOXA1 (step 2), AR/FOXA1-independent NANOG binding to the genome leading to increased transcription of genes associated with migration/invasion/metastasis (step 3), and, finally, transactivation of genes associated with cell-cycle progression and proliferation [150] (Fig. 11F). This reprogramming process, resembling the reprogramming of fibroblasts to induced pluripotent cells, leads, eventually, to the generation of highly proliferative, aggressive and castration-resistant LNCaP cells [150]. This study [150] provides a prime example of PCa cell plasticity induced by a single stemness factor (Fig. 14B).
PCa cell plasticity can also be induced by genetic perturbations and diffusible inflammatory cytokines in the TME (Fig. 14B). For example, oncogenic Kras can cooperate with AR signaling to induce PCSCs and tumor-propagating cells [229], and mutant Kras may induce CD24 upregulation leading to the expansion of PCSCs and increased PCa stemness [261]. C-MYC in combination with activated AKT1 can induce de novo plasticity in human prostate luminal epithelial cells [302]. Infiltrating BM MSCs have been reported to secrete cytokines to repress AR signaling and expand PCSCs [244]. Other TME factors such as TGF-β1 (please change to symbol of beta) [287,311], elevated ROS levels [237] and hypoxia [265] may also induce PCa cell plasticity.
The above examples of PCa cell plasticity generally refer to the ‘de-differentiation’ of AR+ PCa cells to AR−/lo cells of the same lineage, i.e., adenocarcinoma [Fig. 14B]. Frequently, however, treated and reprogrammed PCa cells may change the lineage identity to become NE-like cells in a process often dubbed as PCa lineage plasticity or transdifferentiation [129,279,280,283,295,307,314,327,332–334]. As much as 20–25 % of the treatment-failed patient CRPC may contain NE-like cells and sometimes the entire CRPC clone or tumor may be populated by NE-like cells [327,334]. Such ‘transdifferentiated’ tumors are called CRPC-NE or NEPC [327]. Obviously, development of resistance to ARSIs does not necessarily always lead to CRPC-NE [335], and most metastases in mCRPC patients have the adenocarcinoma phenotype whether AR+ or AR−/lo. Indeed, in our xenograft modeling studies, both AR+/hi LNCaP-type of CRPC and AR−/lo LAPC9-type of CRPC, although exhibiting increase NE score (a cohort of neurogenesis and NE differentiation related genes), have the typical adenocarcinoma cell phenotypes without increased NE cells [163]. One of the best examples of both PCa cell plasticity and PCa lineage plasticity is provided in a genetic mouse model of PCa [332]. When only Pten was deleted in the mouse prostate, mice developed indolent prostate adenocarcinomas with homogeneous AR+ PCa cells. However, when Pten and Rb1 were co-deleted, the double knockout (DKO) prostate developed more aggressive adenocarcinomas with significantly increased AR heterogeneity and increased AR−/lo PCa cells [332]. The DKO tumors were initially sensitive to castration, which then gradually became castration-resistant. Strikingly, the castration-resistant DKO tumors turned completely to AR−/lo phenotype and some tumors even became NE-like [332]. When p53 was also deleted together with Pten and Rb1, the triple knockout (TKO) prostate tumors were typical AR− de novo NEPC [332]. This study [332] elegantly illustrates both PCa cell plasticity and PCa lineage plasticity elicited by genetic perturbations together with therapeutic interventions (castration). In a similar fashion, the NHP epithelial cells can be reprogrammed by different genetic drivers to form AR+ adenocarcinomas [18,21,302] or AR- NE-like cancers [258].
In addition to Rb1 and p53, whose mutations have been convincingly linked to PCa cell/lineage plasticity and to therapy resistance [326,327, 332–336], other molecules and signaling pathways have also been implicated in PCa lineage switch to NE, which include MUC-1 [279, 306], ASPM [259], LIN28B [248,295], c-MYC/AKT1 [302], N-MYC [314], SOX2 [270,276,307], ONECUT2 [337], etc. Interestingly, many of these lineage infidelity-inducing genes (N-MYC, SOX2, LIN28B, ASPM, ASCL1, etc) belong to the functional class of neural development/neurogenesis genes that are also preferentially expressed in AR− PSA− normal prostate basal/stem cells [17,43], PSA−/lo PCSCs [131,144], and in de novo NEPC, CRPC-NE and CRPC-adeno [163,327] (Fig. 14D). De novo NEPC, large-cell neuroendocrine PCa, and CRPC (including both CRPC-NE and CRPC-adeno), as a group, are sometimes called aggressive variant PCa or AVPC (Fig. 14D). We propose that the lineage-regulating and neurogenesis genes may present a common cohort of genes required to establish the ‘ground state’ of the aggressiveness, stemness and therapy resistance associated with the PCSCs and AVPC cells (Fig. 14D).
5. Therapeutic targeting of PCa cell heterogeneity and plasticity: focus on PCSCs and PCa stemness
Xenograft-based CRPC modeling and retrospective studies in patient CRPC specimens suggest many phenotypically divergent subtypes of endpoint CRPC [163,327,331] including the AR+/hi and AR−/lo CRPC. As the AR+ PCa cells in treatment-naïve tumors are exquisitely sensitive to ARSIs and AR−/lo PCa cells possess PCSC properties and are inherently resistant to ARSIs, the focus of the therapeutic development should be on identifying key regulators in PCSCs that can become drug targets. In principle, inhibitors of positive PCSC regulators such as Wnt/β-catenin and pluripotency molecules such as NANOG could be developed and used together with ARSIs to target both PCSCs and differentiated (AR+) PCa cells (Fig. 14A). In support, studies have shown that inhibition of pluripotency regulators such as NANOG [262,263] and SOX2/OCT4 [270] or critical signaling nodes including STAT3 [241], GSK-3β [242] and Wnt/β-catenin (please change to symbol of beta) [257] could lead to depletion of PCSCs and help overcome PCa resistance to ASRIs. As many of the TME-derived cytokines could reprogram PCSCs (discussed above), neutralizing antibodies could potentially be deployed together with ARSIs to treat PCa. PCSCs might also be targeted from different angles, e. g., by exploiting their unique metabolism [161,267] associated with the mitochondrial fission [268]. Some PCSC-specific surface markers, such as CD133 [338], could be developed into targeted imaging/therapeutic ligands for lethal CRPC. Finally, tumor-suppressive miRNAs such as miR-34a [126,167] and miR-141 [154], which are normally devoid in PCSCs, could potentially be developed into replacement therapeutics to be used in the clinic in conjunction with ARSIs (Fig. 14A).
Our recent studies [163] have nominated BCL-2 as a critical therapeutic target in both AR+/hi LNCaP-type (Fig. 12) and AR−/lo LAPC9-type (Fig. 13) of CRPCs. Indeed, in the LNCaP AD/AI models, BCL-2 was slightly induced in primary CRPC but significantly upregulated in ADT/Enza-resistant secondary CRPC (Figs. 10C–D; 12B–D). In vitro, LNCaP and freshly purified HPCa cells upregulated BCL-2 in response to Enza treatment (Fig. 12–E–F) [163]. Overexpression of BCL-2 in LNCaP cells was sufficient to increase tumor incidence and promote tumor growth [163] (Fig. 12G). Consequently, the BCL-2 inhibitor, ABT-199, although not appreciably inhibiting the primary LNCaP CRPC (Fig. 12H), significantly blocked the development of LNCaP secondary CRPC when used in combination with Enza (Fig. 12I) [163]. ABT-199 also sensitized in vitro reprogrammed (AR−) LNCaP-CRPC cells (Fig. 11D) [152]. Similarly, BCL-2 was significantly upregulated in AR−/lo LAPC9 primary CRPC (Fig. 13A–C), which did not respond to Enza (Fig. 10E) [163]. ABT-199 alone inhibited the growth of both LAPC9-AI organoids (Fig. 13D) and tumors (Fig. 13E–F), and its tumor-inhibitory effect was further enhanced by the BET inhibitor JQ1 (Fig. 13E–F) [163]. These observations promoted us to initiate a phase Ib/II clinical trial of treating Enza-naïve CRPC patients with a combination of Enza and BCL-2 inhibitor venetoclax (NCT03751436; clinical PI, Dr. Gurkamal Chatta). A recent study has also implicated BCL-2 as a therapeutic target in small-cell NEPC [339].
Of interest, one PCa treatment regimen, called SPT (supraphysiological testosterone) or SPA (supraphysiological androgens), exploits cyclic treatment of PCa patients with ADT followed by giving patients high doses of testosterone [340–344]. The clinical benefit of the SPT protocol was thought to be derived, mostly, from DNA damage caused by high doses of T. It will be interesting to elucidate the detailed cellular mechanisms of the SPT regimen under the framework of our current understanding of PCa cell heterogeneity and plasticity and PCa development as deregulated prostate differentiation, and in the context of potentially very different biological effects of T on AR+ vs. AR−/lo PCa cells.
Finally, can we potentially target PCSCs and AVPC with immunological approaches? PCSCs have been evinced to be ‘immune-privileged’ lacking the expression of MHC [226] and other immune recognition molecules [160] while overexpressing immune-evading molecules such as Dickkopf-1 [211]. Therefore, PCSCs may not be very susceptible to immune-mediated cell killing [160]. For example, PCSCs have been shown to be refractory to γδ T cell-mediated cytotoxicity [300]. On the other hand, a recent phase I/II clinical trial hints that vaccination in PCa patients against RhoC could elicit long-lasting immune response [305]. Also, radioresistant PCSCs [138] could be targeted by B7-H3 CAR-T cells [315]. Furthermore, inhibition of N-cadherin, preferentially expressed in PSA−/lo PCSCs [131] and in AR−/lo CRPC [163], appears to ‘normalize’ the PCa TME that protects tumor-infiltrated lymphocytes (TILs) from immune checkpoints and Tregs [316]. A recent study also suggests that epigenetic reprogramming through EZH2 inhibition may be able to enhance PCa response to anti-PD-1 due to the activation of STING-interferon signaling axis [345].
6. Perspectives & outstanding questions
It has long been known that the differentiation level of human tumors is inversely correlated with their malignancy. An embryonic stem cell gene expression signature characterizes undifferentiated human cancers [346], human adult stem cell signature marks aggressive subtypes of epithelial cancers [256], and induction of core stem cell and pluripotency master regulators in cancers defines poor clinical outcomes and therapy resistance [269]. My discussions proposed the LP cells as potential cells of origin for human PCa. LP-like PCSCs are intrinsically castration-resistant and survive castration [227], and LPs mediate the progression of basal cell-originated tumors to aggressive adenocarcinomas [238]. Understanding PCa etiology, progression, treatment response and development of therapy resistance from the perspective of abnormal organ and cellular differentiation has a great significance in helping identify both naturally therapy-resistant and induced treatment-refractory PCa cell populations, and subsequently develop novel therapeutics and therapeutic combinations to target these cells. With this goal in mind, I identify many outstanding questions in the field and list several below:
What exactly is the phenotype of LP cells in the human prostate and how do their phenotypes and functions change during organ development, maturation and aging and in response to inflammatory challenges?
How do LPs (or other prostate epithelial cells) become tumorigenically transformed and what epigenetic mechanisms are involved in prostate tumorigenesis instigated by inflammation?
How can we culture and propagate differentiated AR+PSA+ human prostate epithelial and PCa cells?
What PCa cells survive ADT/Enza, enter the adaptation phase and mediate tumor relapse (Fig. 1D)? Are they pre-existent AR−/lo PCa cells or de-differentiated and reprogrammed AR+PCa cells? Are they dormant slow-cycling cells and PCSCs [3]? Do prostate tumors in the adaptation phase enter a diapause-like state to survive?
There are notable challenges in reconstituting the tumorigenicity and stemness of PCSCs from primary prostate tumors. In contrast to CSCs freshly isolated from aggressive human tumors such as lung cancer, colorectal cancer and glioblastoma multiforme that can readily regenerate tumors in immune-deficient mice, putative PCSCs freshly purified from treatment-naïve PCa are generally deficient in initiating tumor development in mice. Why?
Modeling of CRPC development in xenografts has revealed distinct changes in AR expression and subcellular distribution patterns [163] (Fig. 10B). How do the AR+ and AR−/lo PCa cell populations in treatment-naïve tumors reciprocally interact with and regulate each other? How are AR+/hi and AR−/lo PCa cells generated during CRPC development? How does cytoplasmic AR function?
What exactly are the cellular and molecular mechanisms underlying the therapeutic efficacy of intermediate ADT using cyclic ADT and SPT regimen in addition to the proposed DNA damage response?
What strategies can we adopt to turn the PCa TME, generally considered as immunologically ‘cold’ with limited infiltration of CD8+ cytotoxic T cells, into a ‘hot’ proinflammatory TME? What PCSC-specific tumor antigens or neoantigens can be uncovered to help design novel immunological approaches to target PCSCs and AVPC? Can the deregulated alternative splicing program and prominently increased intron retention in aggressive PCa [165] be exploited to identify tumor neoantigens and develop novel anti-PCa immunotherapies?
Acknowledgements
Work in the author’s lab was supported, in part, by grants from the U. S National Institutes of Health (NIH),National Cancer Institute (NCI) R01CA237027, R01CA240290, R21CA237939, and R21CA218635, and Department of Defense (W81XWH-16-1-0575), and by the Roswell Park Comprehensive Cancer Center (RPCCC) and NCI center grantP30CA016056. We acknowledge the support of RPCCC Shared Resources (SR) including BSGSR, ETM, FICSR, GSR, TISR, LASR, and PNSR. I thank all Tang lab members (past and present) and numerous collaborators at the MD Anderson Cancer Center, RPCCC, in the U.S and across the world for their contributions to our lab projects and for helpful discussions. I apologize to the colleagues whose work could not be cited due to space constraint.
Abbreviations:
- ACD
asymmetric cell division
- AD
androgen-dependent
- ADT
androgen deprivation therapy
- AI
androgen-independent
- AP
anterior prostate
- AR
androgen receptor
- AR-KO
AR knockout
- ARSIs
AR signaling inhibitors
- AR-Vs
AR splice variants
- AVPC
aggressive variant prostate cancer
- BCR
biochemical recurrence
- BM
bone marrow
- BMP
bone morphogenic protein
- circRNA
circular RNA
- CAF
carcinoma associated fibroblasts
- CDSS
charcoal and dextran-stripped serum
- CK
cytokeratin
- CRPC
castration-resistant prostate cancer
- CSCs
cancer stem cells
- CTCs
circulating tumor cells
- DKO
double knockout (i.e., Pten−/1−Rb1−/−)
- DNPC
double negative prostate cancer
- DHT
5α-dihydrotestosterone
- DNPC
double-negative PCa
- EMT
epithelial mesenchymal transition
- Enza
enzalutamide
- GR
glucocorticoid receptor
- GS
Gleason Score
- HGPIN
high grade prostate intraepithelial neoplasia
- HPCa
human PCa (specimens)
- IF
immunofluorescence
- IHC
immunohistochemistry
- IL
interleukin
- IR
irradiated
- ITH
intra-tumor heterogeneity
- KO
knockout
- LN
lymph node
- LP
luminal progenitor cells
- mCSPC
metastatic castration-sensitive PCa
- mCRPC
metastatic castration resistant prostate cancer
- MDV3100
e nzalutamide
- miRNA
microRNA
- MSCs
mesenchymal stem cells
- NE
neuroendocrine
- NEPC
neuroendocrine prostate cancer
- NHP
normal human prostate
- PCa
prostate cancer
- PCSCs
prostate cancer stem cells
- PDX
patient (tumor) Derived Xenografts
- PIA
prostate inflammatory atrophy
- PIN
prostate intraepithelial neoplasia
- PSAP-GFP
PSA Promoter-driven GFP expressing (lentiviral reporters)
- PSCA
prostate stem cell antigen
- PSA
prostate specific antigen
- ROS
reactive oxygen species
- s.c
subcutaneously
- SCCs
slow-cycling cells
- scRNA-seq
single-cell RNA sequencing
- SNP
single nucleotide polymorphism
- SPA
supraphysiological androgens
- SPT
supraphysiological testosterone
- T
testosterone
- TAM
tumor-associated macrophages
- TILs
tumor-infiltrating lymphocytes
- TF
transcription factor
- TMA
tissue microarray
- TME
tumor microenvironment
- Tregs
regulatory T cells
- WM
Whole-mount (sections)
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
References
- [1].Shen MM, Abate-Shen C, Molecular genetics of prostate cancer: new prospects for old challenges, Genes Dev. 24 (18) (2010) 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jamroze A, Chatta G, Tang DG, Androgen receptor (AR) heterogeneity in prostate cancer and therapy resistance, Cancer Lett. 518 (October 10) (2021) 1–9, 10.1016/j.canlet.2021.06.006. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Basu S, Dong Y, Kumar R, Jeter C, Tang DG, Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis, Semin. Cancer Biol. (May 9) (2021), 10.1016/j.semcancer.2021.04.021.S1044-1579X(21)00122-X. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fenelon JC, Banerjee A, Murphy BD, Embryonic diapause: development on hold, Int. J. Dev. Biol. 58 (2014) 163–174. [DOI] [PubMed] [Google Scholar]
- [5].Bulut-Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, Ramalho-Santos M, Inhibition of mTOR induces a paused pluripotent state, Nature 540 (2016) 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgartner D, Carnevalli LS, Atzberger A, Haas S, et al. , Myc depletion induces a pluripotent dormant state mimicking diapause, Cell 164 (2016) 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rehman SK, Haynes J, Collignon E, Brown KR, Wang Y, Nixon AML, Bruce JP, Wintersinger JA, Singh Mer A, Lo EBL, et al. , Colorectal cancer cells enter a diapauselike DTP state to survive chemotherapy, Cell 184 (2021) 226–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tang SH, Bhatia B, Maldonado C, Yang P, Newman RA, Liu J, Chandra D, Traag J, Klein RD, Fischer SM, Chopra D, Shen J, Zhau H, Chung LWK, Tang DG, Evidence that arachidonate 15-lipoxygenase 2 is a negative cell-cycle regulator in normal prostate epithelial cells, J. Biol. Chem. 277 (2002) 16189–16201. [DOI] [PubMed] [Google Scholar]
- [9].Bhatia B, Maldonado C, Tang S-H, Chandra D, Klein RD, Chopra D, Shappell S, Yang P, Newman RA, Tang DG, Subcellular localization and tumor-suppressive functions of 15-lipoxygenase 2 (15-LOX2) and its splice variants, J. Biol. Chem. 278 (2003) 25091–25100. [DOI] [PubMed] [Google Scholar]
- [10].Tang SH, Bhatia B, Zhou J-J, Maldonado CJ, Chandra D, Kim E, Fischer S, Butler AF, Friedman SL, Tang DG, Evidence that Sp1 positively and Sp3 negatively regulate and androgen does not directly regulate the functional tumor suppressor 15-lipoxygenase 2 (15-LOX2) gene expression in normal prostate epithelial cells, Oncogene 23 (2004) 6942–6953. [DOI] [PubMed] [Google Scholar]
- [11].Bhatia B, Tang S, Yang P, Doll A, Aumüeller G, Newman RA, Tang DG, Cell-autonomous induction of functional tumor suppressor 15-lipoxygenase 2 (15-LOX2) contributes to replicative senescence of human prostate progenitor cells, Oncogene 24 (2005) 3583–3595. [DOI] [PubMed] [Google Scholar]
- [12].Bhatia B, Multani AS, Patrawala L, Chen X, Calhoun-Davis T, Zhou J, Schroeder L, Schneider-Broussard R, Shen J, Pathak S, Chang S, Tang DG, Evidence that senescent human prostate epithelial cells enhance tumorigenicity: cell fusion as a potential mechanism and inhibition by p16INK4a and hTERT, Int. J. Cancer 122 (2008) 1483–1495. [DOI] [PubMed] [Google Scholar]
- [13].Bhatia B, Jiang M, Suraneni M, Patrawala L, Badeaux M, Schneider-Broussard R, Multani AS, Jeter CR, Calhoun-Davis T, Hu L, Hu J, Tsavachidis S, Zhang W, Chang S, Hayward S, Tang DG, Critical and distinct roles of p16 and telomerase in regulating the proliferative lifespan of normal human prostate epithelial progenitor cells, J. Biol. Chem. 283 (2008) 27957–27972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jiang M, Strand DW, Fernandez S, He Y, Yi Y, Birbach A, Qiu Q, Schmid J, Tang DG, Hayward SW, Functional remodeling of benign human prostatic tissues in vivo by spontaneously immortalized progenitor and intermediate cells, Stem Cells 28 (2) (2010) 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Suraneni MV, Schneider-Broussard R, Moore JR, Calhoun-Davis T, Maldonado CJ, Li H, Newman RA, Kusewitt D, Hu J, Yang P, Tang DG, Transgenic expression of 15-lipoxygenase 2 (15-LOX2) in mouse prostate leads to hyperplasia and cell senescence, Oncogene 29 (2010) 4261–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Laffin B, Tang DG, An old player on a new playground: Bmi-1 as a regulator of prostate stem cells, Cell Stem Cell 7 (6) (2010) 639–640. [DOI] [PubMed] [Google Scholar]
- [17].Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, Chen X, Liu X, Chao HP, Whitney P, Calhoun-Davis T, Takata Y, Shen J, Iyer VR, Tang DG, Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer, Nat. Commun 7 (2016) 10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang D, Lin K, Lu Y, Rycaj K, Zhong Y, Chao HP, Calhoun-Davis T, Shen J, Tang DG, Developing a novel two-dimensional culture system to enrich human prostate luminal progenitors that can function as a cell of origin for prostate cancer, Stem Cells Transl. Med. 6 (2017) 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang D, Jeter C, Gong S, Tracz A, Lu Y, Shen J, Tang DG, Histone 2B-GFP label-retaining prostate luminal cells possess progenitor cell properties and are intrinsically resistant to castration, Stem Cell Rep. 10 (2018) 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang D, Zhao S, Li X, Kirk JS, Tang DG, Prostate luminal progenitor cells in development and cancer, Trends Cancer 4 (11) (2018) 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Park JW, Lee JK, Phillips JW, Huang P, Cheng D, Huang J, Witte ON, Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay, Proc. Natl. Acad. Sci. U. S. A. 113 (16) (2016) 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, Shkhyan RO, Nowroozizadeh B, Rettig MB, Sawyers CL, Elashoff D, Horvath S, Huang J, Witte ON, Goldstein AS, Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome, Cell Rep. 17 (10) (2016) 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RGJ, Cuppen E, Chen Y, Sawyers CL, Clevers HC, Identification of multipotent luminal progenitor cells in human prostate organoid cultures, Cell 159 (2014) 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Crowell PD, Fox JJ, Hashimoto T, Diaz JA, Navarro HI, Henry GH, Feldmar BA, Lowe MG, Garcia AJ, Wu YE, Sajed DP, Strand DW, Goldstein AS, Expansion of luminal progenitor cells in the aging mouse and human prostate, Cell Rep 28 (6) (2019) 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Freeland J, Crowell PD, Giafaglione JM, Boutros PC, Goldstein AS, Aging of the progenitor cells that initiate prostate cancer, Cancer Lett. 515 (2021) 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW, A cellular anatomy of the normal adult human prostate and prostatic urethra, Cell Rep. 25 (12) (2018) 3530–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kwon OJ, Zhang Y, Li Y, Wei X, Zhang L, Chen R, Creighton CJ, Xin L, Functional heterogeneity of mouse prostate stromal cells revealed by single-cell RNA-Seq, iScience 13 (2019) 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, Zhu HH, Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate, Nat. Commun 11 (1) (2020) 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, Carver B, Gopalan A, Abida W, Laudone V, Biton M, Chaudhary O, Xu T, Masilionis I, Manova K, Mazutis L, Pe’er D, Regev A, Sawyers CL, Regenerative potential of prostate luminal cells revealed by single-cell analysis, Science 368 (6490) (2020) 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MT, Malladi VS, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Roehrborn CG, Baker LA, Vezina CM, Strand DW, Urethral luminal epithelia are castration-insensitive cells of the proximal prostate, Prostate 80 (11) (2020) 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guo W, Li L, He J, Liu Z, Han M, Li F, Xia X, Zhang X, Zhu Y, Wei Y, Li Y, Aji R, Dai H, Wei H, Li C, Chen Y, Chen L, Gao D, Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips, Nat. Genet. 52 (9) (2020) 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, Ahmad I, Leung HY, Oliveira P, Blyth K, Baena E, Lacaud G, RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development, Elife 9 (October 7) (2020) e60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li H, Chaitankar V, Zhu J, Chin K, Liu W, Pirooznia M, Rodgers GP, Olfactomedin 4 mediation of prostate stem/progenitor-like cell proliferation and differentiation via MYC, Sci. Rep 10 (1) (2020) 21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R, Shen MM, A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors, Elife 9 (2020) e59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee DH, Olson AW, Wang J, Kim WK, Mi J, Zeng H, Le V, Aldahl J, Hiroto A, Wu X, Sun Z, Androgen action in cell fate and communication during prostate development at single-cell resolution, Development 148 (1) (2021) dev196048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Brady NJ, Bagadion AM, Singh R, Conteduca V, Van Emmenis L, Arceci E, Pakula H, Carelli R, Khani F, Bakht M, Sigouros M, Bareja R, Sboner A, Elemento O, Tagawa S, Nanus DM, Loda M, Beltran H, Robinson B, Rickman DS, Temporal evolution of cellular heterogeneity during the progression to advanced AR-negative prostate cancer, Nat. Commun. 12 (1) (2021) 3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Joseph DB, Henry GH, Malewska A, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Malladi VS, Roehrborn CG, Vezina CM, Strand DW, Single cell analysis of mouse and human prostate reveals novel fibroblasts with specialized distribution and microenvironment interactions, J. Pathol. (June 26) (2021), 10.1002/path.5751. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hannezo E, Scheele CLGJ, Moad M, Drogo N, Heer R, Sampogna RV, van Rheenen J, Simons BD, A unifying theory of branching morphogenesis, Cell 171 (1) (2017) 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Blackwood JK, Williamson SC, Greaves LC, Wilson L, Rigas AC, Sandher R, Pickard RS, Robson CN, Turnbull DM, Taylor RW, Heer R, In situ lineage tracking of human prostatic epithelial stem cell fate reveals a common clonal origin for basal and luminal cells, J. Pathol. 225 (October (2)) (2011) 181–188. [DOI] [PubMed] [Google Scholar]
- [40].Gaisa NT, Graham TA, McDonald SA, Poulsom R, Heidenreich A, Jakse G, Knuechel R, Wright NA, Clonal architecture of human prostatic epithelium in benign and malignant conditions, J. Pathol. 225 (2) (2011) 172–180. [DOI] [PubMed] [Google Scholar]
- [41].Moad M, Hannezo E, Buczacki SJ, Wilson L, El-Sherif A, Sims D, Pickard R, Wright NA, Williamson SC, Turnbull DM, Taylor RW, Greaves L, Robson CN, Simons BD, Heer R, Multipotent basal stem cells, maintained in localized proximal niches, support directed long-ranging epithelial flows in human prostates, Cell Rep. 20 (7) (2017) 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Grossmann S, Hooks Y, Wilson L, Moore L, O’Neill L, Martincorena I, Voet T, Stratton MR, Heer R, Campbell PJ, Development, maturation, and maintenance of human prostate inferred from somatic mutations, Cell Stem Cell 28 (7) (2021) 1262–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, Mathis C, Cheng D, Stuart JM, Witte ON, A basal stem cell signature identifies aggressive prostate cancer phenotypes, Proc. Natl. Acad. Sci. U. S. A. 112 (47) (2015) E6544–E6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Montecino-Rodriguez E, Berent-Maoz B, Dorshkind K, Causes, consequences, and reversal of immune system aging, J. Clin. Invest. 123 (3) (2013) 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ovadya Y, Landsberger T, Leins H, Vadai E, Gal H, Biran A, Yosef R, Sagiv A, Agrawal A, Shapira A, Windheim J, Tsoory M, Schirmbeck R, Amit I, Geiger H, Krizhanovsky V, Impaired immune surveillance accelerates accumulation of senescent cells and aging, Nat. Commun 9 (1) (2018) 5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].De Marzo AM, Platz EA, Sutcliffe S, Xu J, Grönberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG, Inflammation in prostate carcinogenesis, Nat. Rev. Cancer 7 (4) (2007) 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sfanos KS, Yegnasubramanian S, Nelson WG, De Marzo AM, The inflammatory microenvironment and microbiome in prostate cancer development, Nat. Rev. Urol 15 (1) (2018) 11–24. [DOI] [PubMed] [Google Scholar]
- [48].De Marzo AM, Marchi VL, Epstein JI, Nelson WG, Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis, Am. J. Pathol. 155 (6) (1999) 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].De Marzo AM, Meeker AK, Zha S, Luo J, Nakayama M, Platz EA, Isaacs WB, Nelson WG, Human prostate cancer precursors and pathobiology, Urology 62 (5 Suppl 1) (2003) 55–62. [DOI] [PubMed] [Google Scholar]
- [50].Sowalsky AG, Ye H, Bubley GJ, Balk SP, Clonal progression of prostate cancers from Gleason grade 3 to grade 4, Cancer Res. 73 (3) (2013) 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gerrin SJ, Sowalsky AG, Balk SP, Ye H, Mutation profiling indicates high grade prostatic intraepithelial neoplasia as distant precursors of adjacent invasive prostatic adenocarcinoma, Prostate 76 (13) (2016) 1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sowalsky AG, Kissick HT, Gerrin SJ, Schaefer RJ, Xia Z, Russo JW, Arredouani MS, Bubley GJ, Sanda MG, Li W, Ye H, Balk SP, Gleason score 7 prostate cancers emerge through branched evolution of clonal gleason pattern 3 and 4, Clin. Cancer Res. 23 (14) (2017) 3823–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].van Leenders GJ, Gage WR, Hicks JL, van Balken B, Aalders TW, Schalken JA, De Marzo AM, Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy, Am. J. Pathol. 162 (5) (2003) 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shrestha E, Coulter JB, Guzman W, Ozbek B, Hess MM, Mummert L, Ernst SE, Maynard JP, Meeker AK, Heaphy CM, Haffner MC, De Marzo AM, Sfanos KS, Oncogenic gene fusions in nonneoplastic precursors as evidence that bacterial infection can initiate prostate cancer, Proc. Natl. Acad. Sci. U. S. A. 118 (32) (2021) e2018976118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Koeneman KS, Pan CX, Jin JK, Pyle JM 3rd, Flanigan RC, Shankey TV, Diaz MO, Telomerase activity, telomere length, and DNA ploidy, J. Urol. 160 (4) (1998) 1533–1539. [PubMed] [Google Scholar]
- [56].Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, De Marzo AM, Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis, Cancer Res. 62 (22) (2002) 6405–6409. [PubMed] [Google Scholar]
- [57].Vukovic B, Park PC, Al-Maghrabi J, Beheshti B, Sweet J, Evans A, Trachtenberg J, Squire J, Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma, Oncogene 22 (13) (2003) 1978–1987. [DOI] [PubMed] [Google Scholar]
- [58].Bettendorf O, Heine B, Kneif S, Eltze E, Semjonow A, Herbst H, Stein H, Böcker W, Poremba C, Expression-patterns of the RNA component (hTR)and the catalytic subunit (hTERT) of human telomerase in nonneoplastic prostate tissue, prostaticintraepithelial neoplasia, and prostate cancer, Prostate 55 (2) (2003) 99–104. [DOI] [PubMed] [Google Scholar]
- [59].Meeker AK, Hicks JL, Iacobuzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, Ronnett BM, De Marzo AM, Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis, Clin. Cancer Res. 10 (10) (2004) 3317–3326. [DOI] [PubMed] [Google Scholar]
- [60].Meeker AK, Telomeres and telomerase in prostatic intraepithelial neoplasia and prostate cancer biology, Urol. Oncol. 24 (2) (2006) 122–130. [DOI] [PubMed] [Google Scholar]
- [61].Joshua AM, Vukovic B, Braude I, Hussein S, Zielenska M, Srigley J, Evans A, Squire JA, Telomere attrition in isolated high-grade prostatic intraepithelial neoplasia and surrounding stroma is predictive of prostate cancer, Neoplasia 9 (1) (2007) 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Joshua AM, Shen E, Yoshimoto M, Marrano P, Zielenska M, Evans AJ, Van der Kwast T, Squire JA, Topographical analysis of telomere length and correlation with genomic instability in whole mount prostatectomies, Prostate 71 (7) (2011) 778–790. [DOI] [PubMed] [Google Scholar]
- [63].Tu L, Huda N, Grimes BR, Slee RB, Bates AM, Cheng L, Gilley D, Widespread telomere instability in prostatic lesions, Mol. Carcinog. 55 (5) (2016) 842–852. [DOI] [PubMed] [Google Scholar]
- [64].Hubbard GK, Mutton LN, Khalili M, McMullin RP, Hicks JL, Bianchi-Frias D, Horn LA, Kulac I, Moubarek MS, Nelson PS, Yegnasubramanian S, De Marzo AM, Bieberich CJ, Combined MYC activation and pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer, Cancer Res. 76 (2) (2016) 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Graham MK, Meeker A, Telomeres and telomerase in prostate cancer development and therapy, Nat. Rev. Urol. 14 (10) (2017) 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Baena-Del Valle JA, Zheng Q, Esopi DM, Rubenstein M, Hubbard GK, Moncaliano MC, Hruszkewycz A, Vaghasia A, Yegnasubramanian S, Wheelan SJ, Meeker AK, Heaphy CM, Graham MK, De Marzo AM, MYC drives overexpression of telomerase RNA (hTR/TERC) in prostate cancer, J. Pathol. 244 (1) (2018) 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wu J, Crowe DL, Telomere DNA damage signaling regulates prostate cancer tumorigenesis, Mol. Cancer Res. 18 (9) (2020) 1326–1339. [DOI] [PubMed] [Google Scholar]
- [68].Heaphy CM, Yoon GS, Peskoe SB, Joshu CE, Lee TK, Giovannucci E, Mucci LA, Kenfield SA, Stampfer MJ, Hicks JL, De Marzo AM, Platz EA, Meeker AK, Prostate cancer cell telomere length variability and stromal cell telomere length as prognostic markers for metastasis and death, Cancer Discov. 3 (10) (2013) 1130–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ, Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin, Science 348 (6237) (2015) 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, Fitzgerald RC, Handford PA, Campbell PJ, Saeb-Parsy K, Jones PH, Somatic mutant clones colonize the human esophagus with age, Science 362 (6417) (2018) 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yizhak K, Aguet F, Kim J, Hess JM, Kubler K, Grimsby J, Frazer R, Zhang H, Haradhvala NJ, Rosebrock D, Livitz D, Li X, Arich-Landkof E, Shoresh N, Stewart C, Segre AV, Branton PA, Polak P, Ardlie KG, Getz G, RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues, Science 364 (6444) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, Georgakopoulos N, Torrente F, Noorani A, Goddard M, Robinson P, Coorens THH, O’Neill L, Alder C, Wang J, Fitzgerald RC, Zilbauer M, Coleman N, Saeb-Parsy K, Martincorena I, Campbell PJ, Stratton MR, The landscape of somatic mutation in normal colorectal epithelial cells, Nature 574 (7779) (2019) 532–537. [DOI] [PubMed] [Google Scholar]
- [73].Liu N, Matsumura H, Kato T, Ichinose S, Takada A, Namiki T, Asakawa K, Morinaga H, Mohri Y, De Arcangelis A, Geroges-Labouesse E, Nanba D, Nishimura EK, Stem cell competition orchestrates skin homeostasis and ageing, Nature 568 (7752) (2017) 344–350. [DOI] [PubMed] [Google Scholar]
- [74].Kon S, Ishibashi K, Katoh H, Kitamoto S, Shirai T, Tanaka S, Kajita M, Ishikawa S, Yamauchi H, Yako Y, Kamasaki T, Matsumoto T, Watanabe H, Egami R, Sasaki A, Nishikawa A, Kameda I, Maruyama T, Narumi R, Morita T, Sasaki Y, Enoki R, Honma S, Imamura H, Oshima M, Soga T, Miyazaki JI, Duchen MR, Nam JM, Onodera Y, Yoshioka S, Kikuta J, Ishii M, Imajo M, Nishida E, Fujioka Y, Ohba Y, Sato T, Fujita Y, Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes, Nat. Cell Biol. 19 (5) (2017) 530–541. [DOI] [PubMed] [Google Scholar]
- [75].Flanagan DJ, Pentinmikko N, Luopajärvi K, Willis NJ, Gilroy K, Raven AP, Mcgarry L, Englund JI, Webb AT, Scharaw S, Nasreddin N, Hodder MC, Ridgway RA, Minnee E, Sphyris N, Gilchrist E, Najumudeen AK, Romagnolo B, Perret C, Williams AC, Clevers H, Nummela P, Lähde M, Alitalo K, Hietakangas V, Hedley A, Clark W, Nixon C, Kirschner K, Jones EY, Ristimäki A, Leedham SJ, Fish PV, Vincent JP, Katajisto P, Sansom OJ, NOTUM from Apc-mutant cells biases clonal competition to initiate cancer, Nature 594 (7863) (2021) 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Salehi S, Kabeer F, Ceglia N, Andronescu M, Williams MJ, Campbell KR, Masud T, Wang B, Biele J, Brimhall J, Gee D, Lee H, Ting J, Zhang AW, Tran H, O’Flanagan C, Dorri F, Rusk N, de Algara TR, Lee SR, Cheng BYC, Eirew P, Kono T, Pham J, Grewal D, Lai D, Moore R, Mungall AJ, Marra MA, IMAXT Consortium A, McPherson A, Bouchard-Côté S, Aparicio S, Shah P, Clonal fitness inferred from time-series modelling of single-cell cancer genomes, Nature 595 (2021) 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Colom B, Herms A, Hall MWJ, Dentro SC, King C, Sood RK, Alcolea MP, Piedrafita G, Fernandez-Antoran D, Ong SH, Fowler JC, Mahbubani KT, Saeb-Parsy K, Gersting M, Hall BA, Jones PH, Mutant clones in normal epithelium outcompete and eliminate ebrging tumors, Nature 598 (2021) 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Marusyk A, Janiszewska M, Polyak K, Intratumor heterogeneity: the rosetta stone of therapy resistance, Cancer Cell 37 (2020) 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, Yu K, Rubanova Y, Macintyre G, Demeulemeester J, Vázquez-García I, Kleinheinz K, Livitz DG, Malikic S, Donmez N, Sengupta S, Anur P, Jolly C, Cmero M, Rosebrock D, Schumacher SE, Fan Y, Fittall M, Drews RM, Yao X, Watkins TBK, Lee J, Schlesner M, Zhu H, Adams DJ, McGranahan N, Swanton C, Getz G, Boutros PC, Imielinski M, Beroukhim R, Sahinalp SC, Ji Y, Peifer M, Martincorena I, Markowetz F, Mustonen V, Yuan K, Gerstung M, Spellman PT, Wang W, Morris QD, Wedge DC, Van Loo P, PCAWG Evolution and Heterogeneity Working Group and the PCAWG Consortium, Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes, Cell 184 (8) (2021) 2239–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Erickson A, Hayes A, Rajakumar T, Verrill C, Bryant RJ, Hamdy FC, Wedge DC, Woodcock DJ, Mills IG, Lamb AD, A systematic review of prostate cancer heterogeneity: understanding the clonal ancestry of multifocal disease, Eur Urol Oncol. 4 (3) (2021) 358–369. [DOI] [PubMed] [Google Scholar]
- [81].Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, De Marzo AM, Nelson PS, Yegnasubramanian S, Genomic and phenotypic heterogeneity in prostate cancer, Nat. Rev. Urol. 18 (2) (2021) 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, Massie CE, Kay J, Luxton HJ, Edwards S, Kote-Jarai Z, Dennis N, Merson S, Leongamornlert D, Zamora J, Corbishley C, Thomas S, Nik-Zainal S, O’Meara S, Matthews L, Clark J, Hurst R, Mithen R, Bristow RG, Boutros PC, Fraser M, Cooke S, Raine K, Jones D, Menzies A, Stebbings L, Hinton J, Teague J, McLaren S, Mudie L, Hardy C, Anderson E, Joseph O, Goody V, Robinson B, Maddison M, Gamble S, Greenman C, Berney D, Hazell S, Livni N, Group IP, Fisher C, Ogden C, Kumar P, Thompson A, Woodhouse C, Nicol D, Mayer E, Dudderidge T, Shah NC, Gnanapragasam V, Voet T, Campbell P, Futreal A, Easton D, Warren AY, Foster CS, Stratton MR, Whitaker HC, McDermott U, Brewer DS, Neal DE, Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue, Nat. Genet. 47 (4) (2015) 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, Zia A, Fox NS, Livingstone J, Shiah YJ, Wang J, Beck TA, Have CL, Chong T, Sam M, Johns J, Timms L, Buchner N, Wong A, Watson JD, Simmons TT, P’ng C, Zafarana G, Nguyen F, Luo X, Chu KC, Prokopec SD, Sykes J, Dal Pra A, Berlin A, Brown A, Chan-Seng-Yue MA, Yousif F, Denroche RE, Chong LC, Chen GM, Jung E, Fung C, Starmans MH, Chen H, Govind SK, Hawley J, D’Costa A, Pintilie M, Waggott D, Hach F, Lambin P, Muthuswamy LB, Cooper C, Eeles R, Neal D, Tetu B, Sahinalp C, Stein LD, Fleshner N, Shah SP, Collins CC, Hudson TJ, McPherson JD, van der Kwast T, Bristow RG, Spatial genomic heterogeneity within localized, multifocal prostate cancer, Nat. Genet. 47 (7) (2015) 736–745. [DOI] [PubMed] [Google Scholar]
- [84].Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC, ICGC Prostate Group DE, Neal CS, Cooper RA, Eeles T, Visakorpi P, Campbell J, McDermott U, Wedge DC, Bova GS, The evolutionary history of lethal metastatic prostate cancer, Nature 520 (7547) (2015) 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cancer Genome Atlas Research Network, The molecular taxonomy of primary prostate cancer, Cell 163 (4) (2015) 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hopkins JF, Sabelnykova VY, Weischenfeldt J, Simon R, Aguiar JA, Alkallas R, Heisler LE, Zhang J, Watson JD, Chua MLK, Fraser M, Favero F, Lawerenz C, Plass C, Sauter G, McPherson JD, van der Kwast T, Korbel J, Schlomm T, Bristow RG, Boutros PC, Mitochondrial mutations drive prostate cancer aggression, Nat. Commun. 8 (1) (2017) 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, Fox NS, Xie M, Prokopec SD, Berlin A, Lalonde E, Ahmed M, Trudel D, Luo X, Beck TA, Meng A, Zhang J, D’Costa A, Denroche RE, Kong H, Espiritu SM, Chua ML, Wong A, Chong T, Sam M, Johns J, Timms L, Buchner NB, Orain M, Picard V, Hovington H, Murison A, Kron K, Harding NJ, P’ng C, K. Houlahan E, Chu KC, Lo B, Nguyen F, Li CH, Sun RX, de Borja R, Cooper CI, Hopkins JF, Govind SK, Fung C, Waggott D, Green J, Haider S, Chan-Seng-Yue MA, Jung E, Wang Z, Bergeron A, Dal Pra A, Lacombe L, Collins CC, Sahinalp C, Lupien M, Fleshner NE, He HH, Fradet Y, Tetu B, van der Kwast T, McPherson JD, Bristow RG, Boutros PC, Genomic hallmarks of localized, non-indolent prostate cancer, Nature 541 (7637) (2017) 359–364. [DOI] [PubMed] [Google Scholar]
- [88].Taylor RA, Fraser M, Livingstone J, Espiritu SM, Thorne H, Huang V, Lo W, Shiah YJ, Yamaguchi TN, Sliwinski A, Horsburgh S, Meng A, Heisler LE, Yu N, Yousif F, Papargiris M, Lawrence MG, Timms L, Murphy DG, Frydenberg M, Hopkins JF, Bolton D, Clouston D, McPherson JD, van der Kwast T, Boutros PC, Risbridger GP, Bristow RG, Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories, Nat. Commun. 8 (2017) 13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, Fox NS, Chua MLK, Yamaguchi TN, Heisler LE, Livingstone J, Wintersinger J, Yousif F, Lalonde E, Rouette A, Salcedo A, Houlahan KE, Li CH, Huang V, Fraser M, van der Kwast T, Morris QD, Bristow RG, Boutros PC, The evolutionary landscape of localized prostate cancers drives clinical aggression, Cell 173 (4) (2018) 1003–1013, e1015. [DOI] [PubMed] [Google Scholar]
- [90].Wedge DC, Gundem G, Mitchell T, Woodcock DJ, Martincorena I, Ghori M, Zamora J, Butler A, Whitaker H, Kote-Jarai Z, Alexandrov LB, Van Loo P, Massie CE, Dentro S, Warren AY, Verrill C, Berney DM, Dennis N, Merson S, Hawkins S, Howat W, Lu YJ, Lambert A, Kay J, Kremeyer B, Karaszi K, Luxton H, Camacho N, Marsden L, Edwards S, Matthews L, Bo V, Leongamornlert D, McLaren S, Ng A, Yu Y, Zhang H, Dadaev T, Thomas S, Easton DF, Ahmed M, Bancroft E, Fisher C, Livni N, Nicol D, Tavare S, Gill P, Greenman C, Khoo V, Van As N, Kumar P, Ogden C, Cahill D, Thompson A, Mayer E, Rowe E, Dudderidge T, Gnanapragasam V, Shah NC, Raine K, Jones D, Menzies A, Stebbings L, Teague J, Hazell S, Corbishley C, Group CS, de Bono J, Attard G, Isaacs W, Visakorpi T, Fraser M, Boutros PC, Bristow RG, Workman P, Sander C, Consortium T, Hamdy FC, Futreal A, McDermott U, Al-Lazikani B, Lynch AG, Bova GS, Foster CS, Brewer DS, Neal DE, Cooper CS, Eeles RA, Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets, Nat. Genet. 50 (5) (2018) 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, Heckmann D, Sidiropoulos N, Waszak SM, Hubschmann D, Urbanucci A, Girma EG, Kuryshev V, Klimczak LJ, Saini N, Stutz AM, Weichenhan D, Bottcher LM, Toth R, Hendriksen JD, Koop C, Lutsik P, Matzk S, Warnatz HJ, Amstislavskiy V, Feuerstein C, Raeder B, Bogatyrova O, Schmitz EM, Hube-Magg C, Kluth M, Huland H, Graefen M, Lawerenz C, Henry GH, Yamaguchi TN, Malewska A, Meiners J, Schilling D, Reisinger E, Eils R, Schlesner M, Strand DW, Bristow RG, Boutros PC, von Kalle C, Gordenin D, Sultmann H, Brors B, Sauter G, Plass C, Yaspo ML, Korbel JO, Schlomm T, Weischenfeldt J, Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories, Cancer Cell 34 (6) (2018) 996–1011, e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J, Lesurf R, Shiah YJ, Vujcic T, Huang X, Espiritu SMG, Heisler LE, Yousif F, Huang V, Yamaguchi TN, Yao CQ, Sabelnykova VY, Fraser M, Chua MLK, van der Kwast T, Liu SK, Boutros PC, Bristow RG, Molecular landmarks of tumor hypoxia across cancer types, Nat. Genet. 51 (2) (2019) 308–318. [DOI] [PubMed] [Google Scholar]
- [93].Sinha A, Huang V, Livingstone J, Wang J, Fox NS, Kurganovs N, Ignatchenko V, Fritsch K, Donmez N, Heisler LE, Shiah YJ, Yao CQ, Alfaro JA, Volik S, Lapuk A, Fraser M, Kron K, Murison A, Lupien M, Sahinalp C, Collins CC, Tetu B, Masoomian M, Berman DM, van der Kwast T, Bristow RG, Kislinger T, Boutros PC, The proteogenomic landscape of curable prostate cancer, Cancer Cell 35 (3) (2019) 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mazrooei P, Kron KJ, Zhu Y, Zhou S, Grillo G, Mehdi T, Ahmed M, Severson TM, Guilhamon P, Armstrong NS, Huang V, Yamaguchi TN, Fraser M, van der Kwast T, Boutros PC, He HH, Bergman AM, Bristow RG, Zwart W, Lupien M, Cistrome partitioning reveals convergence of somatic mutations and risk variants on master transcription regulators in primary prostate tumors, Cancer Cell 36 (6) (2019) 674–689. [DOI] [PubMed] [Google Scholar]
- [95].van Dessel LF, van Riet J, Smits M, Zhu Y, Hamberg P, van der Heijden MS, Bergman AM, van Oort IM, de Wit R, Voest EE, Steeghs N, Yamaguchi TN, Livingstone J, Boutros PC, Martens JWM, Sleijfer S, Cuppen E, Zwart W, van de Werken HJG, Mehra N, Lolkema MP, The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact, Nat. Commun. 10 (1) (2019) 5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Liu LY, Bhandari V, Salcedo A, Espiritu SMG, Morris QD, Kislinger T, Boutros PC, Quantifying the influence of mutation detection on tumour subclonal reconstruction, Nat. Commun. 11 (1) (2020) 6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Houlahan KE, Shiah YJ, Gusev A, Yuan J, Ahmed M, Shetty A, Ramanand SG, Yao CQ, Bell C, O’Connor E, Huang V, Fraser M, Heisler LE, Livingstone J, Yamaguchi TN, Rouette A, Foucal A, Espiritu SMG, Sinha A, Sam M, Timms L, Johns J, Wong A, Murison A, Orain M, Picard V, Hovington H, Bergeron A, Lacombe L, Lupien M, Fradet Y, Têtu B, McPherson JD, Pasaniuc B, Kislinger T, Chua MLK, Pomerantz MM, van der Kwast T, Freedman ML, Mani RS, He HH, Bristow RG, Boutros PC, Genomewide germline correlates of the epigenetic landscape of prostate cancer, Nat. Med. 25 (10) (2019) 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, Aggarwal R, Playdle D, Liao A, Alumkal JJ, Das R, Chou J, Hua JT, Barnard TJ, Bailey AM, Chow ED, Perry MD, Dang HX, Yang R, Moussavi-Baygi R, Zhang L, Alshalalfa M, Laura Chang S, Houlahan KE, Shiah YJ, Beer TM, Thomas G, Chi KN, Gleave M, Zoubeidi A, Reiter RE, Rettig MB, Witte O, Yvonne Kim M, Fong L, Spratt DE, Morgan TM, Bose R, Huang FW, Li H, Chesner L, Shenoy T, Goodarzi H, Asangani IA, Sandhu S, Lang JM, Mahajan NP, Lara PN, Evans CP, Febbo P, Batzoglou S, Knudsen KE, He HH, Huang J, Zwart W, Costello JF, Luo J, Tomlins SA, Wyatt AW, Dehm SM, Ashworth A, Gilbert LA, Boutros PC, Farh K, Chinnaiyan AM, Maher CA, Small EJ, Quigley DA, Feng FY, The DNA methylation landscape of advanced prostate cancer, Nat. Genet. 52 (8) (2020) 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ahmed M, Soares F, Xia JH, Yang Y, Li J, Guo H, Su P, Tian Y, Lee HJ, Wang M, Akhtar N, Houlahan KE, Bosch A, Zhou S, Mazrooei P, Hua JT, Chen S, Petricca J, Zeng Y, Davies A, Fraser M, Quigley DA, Feng FY, Boutros PC, Lupien M, Zoubeidi A, Wang L, Walsh MJ, Wang T, Ren S, Wei GH, He HH, CRISPRi screens reveal a DNA methylation-mediated 3D genome dependent causal mechanism in prostate cancer, Nat. Commun. 12 (1) (2021) 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, Wang M, Yousif F, Zhang Y, Donmez N, Ahmed M, Volik S, Lapuk A, Chua MLK, Heisler LE, Foucal A, Fox NS, Fraser M, Bhandari V, Shiah YJ, Guan J, Li J, Orain M, Picard V, Hovington H, Bergeron A, Lacombe L, Fradet Y, Têtu B, Liu S, Feng F, Wu X, Shao YW, Komor MA, Sahinalp C, Collins C, Hoogstrate Y, de Jong M, Fijneman RJA, Fei T, Jenster G, van der Kwast T, Bristow RG, Boutros PC, He HH, Widespread and functional RNA circularization in localized prostate cancer, Cell 176 (4) (2019) 831–843, e22. [DOI] [PubMed] [Google Scholar]
- [101].Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W, Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer, Mol. Cancer Res. 13 (1) (2015) 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kishan AU, Romero T, Alshalalfa M, Liu Y, Tran PT, Nickols NG, Ye H, Sajed D, Rettig MB, Reiter RE, Garraway IP, Spratt DE, Freedland SJ, Zhao X, Li Z, Deek M, Livingstone J, Mahal BA, Nguyen PL, Feng FY, Schaeffer EM, Den RB, Lotan TL, Karnes RJ, Klein EA, Ross AE, Grogan T, Davicioni E, Elashoff D, Boutros PC, Weidhaas JB, Transcriptomic heterogeneity of gleason grade group 5 prostate cancer, Eur. Urol. 78 (3) (2020) 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, Davicioni E, Karnes RJ, Klein EA, Ross AE, Den RB, Schaeffer EM, Garraway IP, Kim J, Freeman MR, Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome, Cancer Res. 76 (17) (2016) 4948–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, Speers C, Cooperberg MR, Kim W, Ryan CJ, Den RB, Freedland SJ, Posadas E, Sandler H, Klein EA, Black P, Seiler R, Tomlins SA, Chinnaiyan AM, Jenkins RB, Davicioni E, Ross AE, Schaeffer EM, Nguyen PL, Carroll PR, Karnes RJ, Spratt DE, Feng FY, Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy, JAMA Oncol. 3 (12) (2017) 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhao SG, Chen WS, Das R, Chang SL, Tomlins SA, Chou J, Quigley DA, Dang HX, Barnard TJ, Mahal BA, Gibb EA, Liu Y, Davicioni E, Duska LR, Posadas EM, Jolly S, Spratt DE, Nguyen PL, Maher CA, Small EJ, Feng FY, Clinical and genomic implications of luminal and basal subtypes across carcinomas, Clin. Cancer Res. 25 (8) (2019) 2450–2457. [DOI] [PubMed] [Google Scholar]
- [106].Mazzu YZ, Armenia J, Nandakumar S, Chakraborty G, Yoshikawa Y, Jehane LE, Lee GM, Atiq M, Khan N, Schultz N, Kantoff PW, Ribonucleotide reductase small subunit M2 is a master driver of aggressive prostate cancer, Mol. Oncol. 14 (8) (2020) 1881–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yoon J, Kim M, Posadas EM, Freedland SJ, Liu Y, Davicioni E, Trock BJ, Den RB, Karnes RJ, Klein EA, Freeman MR, You S, A comparative study of PCS and PAM50 prostate cancer classification schemes, Prostate Cancer Prostatic Dis. 24 (2021) 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Han H, Lee HH, Choi K, Moon YJ, Heo JE, Ham WS, Jang WS, Rha KH, Cho NH, Giancotti FG, Choi YD, Prostate epithelial genes define therapy-relevant prostate cancer molecular subtype, Prostate Cancer Prostatic Dis. (April 26) (2021), 10.1038/s41391-021-00364-x. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Horning AM, Wang Y, Lin CK, Louie AD, Jadhav RR, Hung CN, Wang CM, Lin CL, Kirma NB, Liss MA, Kumar AP, Sun L, Liu Z, Chao WT, Wang Q, Jin VX, Chen CL, Huang TH, Single-cell RNA-seq reveals a subpopulation of prostate cancer cells with enhanced cell-cycle related transcription and attenuated androgen response, Cancer Res. 78 (4) (2018) 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Vickman RE, Broman MM, Lanman NA, Franco OE, Sudyanti PAG, Ni Y, Ji Y, Helfand BT, Petkewicz J, Paterakos MC, Crawford SE, Ratliff TL, Hayward SW, Heterogeneity of human prostate carcinoma-associated fibroblasts implicates a role for subpopulations in myeloid cell recruitment, Prostate 80 (2) (2020) 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ma X, Guo J, Liu K, Chen L, Liu D, Dong S, Xia J, Long Q, Yue Y, Zhao P, Hu F, Xiao Z, Pan X, Xiao K, Cheng Z, Ke Z, Chen ZS, Zou C, Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing, Mol. Cancer 19 (1) (2020) 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Dong B, Miao J, Wang Y, Luo W, Ji Z, Lai H, Zhang M, Cheng X, Wang J, Fang Y, Zhu HH, Chua CW, Fan L, Zhu Y, Pan J, Wang J, Xue W, Gao WQ, Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer, Commun Biol. 3 (1) (2020) 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Chen S, Zhu G, Yang Y, Wang F, Xiao YT, Zhang N, Bian X, Zhu Y, Yu Y, Liu F, Dong K, Mariscal J, Liu Y, Soares F, Loo Yau H, Zhang B, Chen W, Wang C, Chen D, Guo Q, Yi Z, Liu M, Fraser M, De Carvalho DD, Boutros PC, Di Vizio D, Jiang Z, van der Kwast T, Berlin A, Wu S, Wang J, He HH, Ren S, Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression, Nat. Cell Biol. 23 (1) (2021) 87–98. [DOI] [PubMed] [Google Scholar]
- [114].He MX, Cuoco MS, Crowdis J, Bosma-Moody A, Zhang Z, Bi K, Kanodia A, Su MJ, Ku SY, Garcia MM, Sweet AR, Rodman C, DelloStritto L, Silver R, Steinharter J, Shah P, Izar B, Walk NC, Burke KP, Bakouny Z, Tewari AK, Liu D, Camp SY, Vokes NI, Salari K, Park J, Vigneau S, Fong L, Russo JW, Yuan X, Balk SP, Beltran H, Rozenblatt-Rosen O, Regev A, Rotem A, Taplin ME, Van Allen EM, Transcriptional mediators of treatment resistance in lethal prostate cancer, Nat. Med. 27 (3) (2021) 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Siefert JC, Cioni B, Muraro MJ, Alshalalfa M, Vivié J, van der Poel HG, Schoots IG, Bekers E, Feng FY, Wessels LFA, Zwart W, Bergman AM, The prognostic potential of human prostate cancer-associated macrophage subtypes as revealed by single-cell transcriptomics, Mol. Cancer Res. 19 (10) (2021) 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA, RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance, Science 349 (6254) (2015) 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG, Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic, Cancer Res. 65 (14) (2005) 6207–6219. [DOI] [PubMed] [Google Scholar]
- [118].Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG, Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells, Oncogene 25 (12) (2006) 1696–1708. [DOI] [PubMed] [Google Scholar]
- [119].Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG, Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+a2b1+ cell population is enriched in tumor-initiating cells, Cancer Res. 67 (14) (2007) 6796–6805. [DOI] [PubMed] [Google Scholar]
- [120].Tang DG, Patrawala L, Calhoun T, Bhatia B, Choy G, Schneider-Broussard R, Jeter C, Prostate cancer stem/progenitor cells: identification, characterization, and implications, Mol. Carcinog. 46 (1) (2007) 1–14. [DOI] [PubMed] [Google Scholar]
- [121].Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG, PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells, Cancer Res. 68 (6) (2008) 1820–1825. [DOI] [PubMed] [Google Scholar]
- [122].Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ, Tang DG, Functional evidence that the self-renewal gene NANOG regulates human tumor development, Stem Cells 27 (5) (2009) 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Li H, Jiang M, Honorio S, Patrawala L, Jeter CR, Calhoun-Davis T, Hayward SW, Tang DG, Methodologies in assaying prostate cancer stem cells, Methods Mol. Biol. 568 (2009) 85–138. [DOI] [PubMed] [Google Scholar]
- [124].Yan H, Chen X, Zhang Q, Qin J, Li H, Liu C, Calhoun-Davis T, Coletta LD, Klostergaard J, Fokt I, Skora S, Priebe W, Bi Y, Tang DG, Drug-tolerant cancer cells show reduced tumor-initiating capacity: depletion of CD44 cells and evidence for epigenetic mechanisms, PLoS One 6 (9) (2011) e24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG, NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation, Oncogene 30 (36) (2011) 3833–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG, The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44, Nat. Med. 17 (2) (2011) 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Liu C, Tang DG, MicroRNA regulation of cancer stem cells, Cancer Res. 71 (18) (2011) 5950–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Li H, Tang DG, Prostate cancer stem cells and their potential roles in metastasis, J. Surg. Oncol. 103 (6) (2011) 558–562. [DOI] [PubMed] [Google Scholar]
- [129].Tang DG, Understanding cancer stem cell heterogeneity and plasticity, Cell Res. 22 (3) (2012) 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG, Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7, Cancer Res. 72 (13) (2012) 3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, Calhoun-Davis T, Li H, Palapattu GS, Pang S, Lin K, Huang J, Ivanov I, Li W, Suraneni MV, Tang DG, The PSA−/lo prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration, Cell Stem Cells Regen. Med. 10 (5) (2012) 556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Chen X, Liu B, Li Q, Honorio S, Liu X, Liu C, Multani AS, Calhoun-Davis T, Tang DG, Dissociated primary human prostate cancer cells coinjected with the immortalized Hs5 bone marrow stromal cells generate undifferentiated tumors in NOD/SCID-γ mice, PLoS One 8 (2) (2013) e56903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Chen X, Rycaj K, Liu X, Tang DG, New insights into prostate cancer stem cells, Cell Cycle 12 (4) (2013) 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Du L, Rao G, Wang H, Li B, Tian W, Cui J, He L, Laffin B, Tian X, Hao C, Liu H, Sun X, Zhu Y, Tang DG, Mehrpour M, Lu Y, Chen Q, CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer, Cancer Res. 73 (8) (2013) 2682–2694. [DOI] [PubMed] [Google Scholar]
- [135].Badeaux MA, Jeter CR, Gong S, Liu B, Suraneni MV, Rundhaug J, Fischer SM, Yang T, Kusewitt D, Tang DG, In vivo functional studies of tumor-specific retrogene NANOGP8 in transgenic animals, Cell Cycle 12 (15) (2013) 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Shi Y, Liu C, Liu X, Tang DG, Wang J, The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells, PLoS One 9 (3) (2014) e90022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Liu B, Badeaux MD, Choy G, Chandra D, Shen I, Jeter CR, Rycaj K, Lee CF, Person MD, Liu C, Chen Y, Shen J, Jung SY, Qin J, Tang DG, Nanog1 in NTERA-2 and recombinant NanogP8 from somatic cancer cells adopt multiple protein conformations and migrate at multiple M.W species, PLoS One 9 (3) (2014) e90615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Rycaj K, Tang DG, Cancer stem cells and radioresistance, Int. J. Radiat. Biol. 90 (8) (2014) 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV and Tang DG. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells, Cancer Res. 74 (15) (2014) 4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yang T, Rycaj K, Liu ZM, Tang DG, Cancer stem cells: constantly evolving and functionally heterogeneous therapeutic targets, Cancer Res. 74 (11) (2014) 2922–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi L, Ramondetta LM, Lin K, Johanning GL, Tang DG, Wang-Johanning F, Cytotoxicity of human endogenous retrovirus K-specific T cells toward autologous ovarian cancer cells, Clin. Cancer Res. 21 (2) (2015) 471–483. [DOI] [PubMed] [Google Scholar]
- [142].Jeter CR, Yang T, Wang J, Chao HP, Tang DG, NANOG in cancer stem cells and tumor development: an update and outstanding questions, Stem Cells 33 (8) (2015) 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Gong S, Li Q, Jeter CR, Fan Q, Tang DG, Liu B, Regulation of NANOG in cancer cells, Mol. Carcinog. 54 (9) (2015) 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Liu X, Chen X, Rycaj K, Chao HP, Deng Q, Jeter C, Liu C, Honorio S, Li H, Davis T, Suraneni M, Laffin B, Qin J, Li Q, Yang T, Whitney P, Shen J, Huang J, Tang DG, Systematic dissection of phenotypic, functional, and tumorigenic heterogeneity of human prostate cancer cells, Oncotarget 6 (27) (2015) 23959–23986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Rycaj K, Tang DG, Cell-of-origin of cancer versus cancer stem cells: assays and interpretations, Cancer Res. 75 (19) (2015) 4003–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Li Q, Rycaj K, Chen X, Tang DG, Cancer stem cells and cell size: a causal link? Semin. Cancer Biol. 35 (2015) 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Deng Q, Tang DG, Androgen receptor and prostate cancer stem cells: biological mechanisms and clinical implications, Endocr. Relat. Cancer 22 (6) (2015) T209–T220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Tang DG, Cancers of the breast and prostate: a stem cell perspective, Endocr. Relat. Cancer 22 (6) (2015) E9–E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Chen X, Li Q, Liu X, Liu C, Liu R, Rycaj K, Zhang D, Liu B, Jeter C, Calhoun-Davis T, Lin K, Lu Y, Chao HP, Shen J, Tang DG, Defining a population of stem-like human prostate cancer cells that can generate and propagate castration-resistant prostate cancer, Clin. Cancer Res. 22 (17) (2016) 4505–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Jeter CR, Liu B, Lu Y, Chao HP, Zhang D, Liu X, Chen X, Li Q, Rycaj K, Calhoun-Davis T, Yan L, Hu Q, Wang J, Shen J, Liu S, Tang DG, NANOG reprograms prostate cancer cells to castration resistance via dynamically repressing and engaging the AR/FOXA1 signaling axis, Cell Discov. 2 (2016) 16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Liu R, Liu C, Zhang D, Liu B, Chen X, Rycaj K, Jeter C, Calhoun-Davis T, Li Y, Yang T, Wang J, Tang DG, miR-199a-3p targets stemness-related and mitogenic signaling pathways to suppress the expansion and tumorigenic capabilities of prostate cancer stem cells, Oncotarget 7 (35) (2016) 56628–56642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Rycaj K, Cho EJ, Liu X, Chao HP, Liu B, Li Q, Devkota AK, Zhang D, Chen X, Moore J, Dalby KN, Tang DG, Longitudinal tracking of subpopulation dynamics and molecular changes during LNCaP cell castration and identification of inhibitors that could target the PSA−/lo castration-resistant cells, Oncotarget 7 (12) (2016) 14220–14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Paranjape AN, Soundararajan R, Werden SJ, Joseph R, Taube JH, Liu H, Rodriguez-Canales J, Sphyris N, Wistuba I, Miura N, Dhillon J, Mahajan N, Mahajan K, Chang JT, Ittmann M, Maity SN, Logothetis C, Tang DG, Mani SA, Inhibition of FOXC2 restores epithelial phenotype and drug sensitivity in prostate cancer cells with stem-cell properties, Oncogene 35 (46) (2016) 5963–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao HP, Rycaj K, Takata Y, Lin K, Lu Y, Zhong Y, Krolewski J, Shen J, Tang DG, MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes, Nat. Commun. 8 (2017) 14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Rycaj K, Li H, Zhou J, Chen X, Tang DG, Cellular determinants and microenvironmental regulation of prostate cancer metastasis, Semin. Cancer Biol. 44 (2017) 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Liu B, Gong S, Li Q, Chen X, Moore J, Suraneni MV, Badeaux MD, Jeter CR, Shen J, Mehmood R, Fan Q, Tang DG, Transgenic overexpression of NANOGP8 in the mouse prostate is insufficient to initiate tumorigenesis but weakly promotes tumor development in the Hi-Myc mouse model, Oncotarget 8 (32) (2017) 52746–52760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Liu X, Krawczyk E, Suprynowicz FA, Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng YL, Sripadhan P, Chen C, Lu J, Hou TW, Choudhury S, Kallakury B, Tang DG, et al. , Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens, Nat. Protoc. 12 (2017) 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Guo Y, Zhang K, Cheng C, Ji Z, Wang X, Wang M, Chu M, Tang DG, Zhu HH, Gao WQ, Numb−/low enriches a castration-resistant prostate cancer cell subpopulation associated with enhanced notch and hedgehog signaling, Clin. Cancer Res. 23 (21) (2017) 6744–6756. [DOI] [PubMed] [Google Scholar]
- [159].Rycaj K, Tang DG, Molecular determinants of prostate cancer metastasis, Oncotarget 8 (2017) 88211–88231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zhang D, Tang DG, Rycaj K, Cancer stem cells: regulation programs, immunological properties and immunotherapy, Semin. Cancer Biol. 52 (2018) 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Skvortsov S, Skvortsova II, Tang DG, Dubrovska A, Prostate cancer stem cells: current understanding, Stem Cells 36 (10) (2018) 1457–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zhang D, Tang DG, Splice" a way towards neuroendocrine prostate cancer, EBioMedicine 35 (September) (2018) 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Li Q, Deng Q, Chao H, Liu X, Lu Y, Liu B, Tang GW, Zhang D, Tracz A, Jeter C, Rycaj K, Calhoun-Davis T, Huang J, Rubin M, Shen J, Chatta G, Puznov I, Mohler J, Wang J, Zhao R, Kirk J, CHen X, Tang DG, Linking prostate cancer cell AR heterogeneity to distinct castration and enzlutamide responses, Nat. Commun. 9 (1) (2018) 3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Li Q, Liu B, Chao HP, Ji Y, Lu Y, Mehmood R, Jeter C, Chen T, Moore JR, Li W, Liu C, Rycaj K, Tracz A, Kirk J, Calhoun-Davis T, Xiong J, Deng Q, Huang J, Foster BA, Gokhale A, Chen X, Tang DG, LRIG1 is a pleiotropic androgen receptor-regulated feedback tumor suppressor in prostate cancer, Nat. Commun. 10 (1) (2019) 5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang D, Hu Q, Liu X, Ji Y, Chao HP, Liu Y, Tracz A, Kirk J, Buonamic S, Zhu P, Wanh J, Liu S, Tang DG, Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer, Nat. Commun. 11 (1) (2020) 2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Liu B, Kumar R, Chao HP, Mehmood R, Ji Y, Tracz A, Tang DG, Evidence for context-dependent functions of KDM5B in prostate development and prostate cancer, Oncotarget 11 (46) (2020) 4243–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Li W, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, Tang DG, MicroRNA-34a: potent tumor suppressor, cancer stem cell inhibitor, and potential anticancer therapeutic, Front. Cell Dev. Biol. 9 (2021), 640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Xu L, Yin Y, Li Y, Chen X, Chang Y, Zhang H, Liu J, Beasley J, McCaw P, Zhang H, Young S, Groth J, Wang Q, Locasale JW, Gao X, Tang DG, Dong X, He Y, George D, Hu H, Huang J, A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer, Proc. Natl. Acad. Sci. U. S. A. 118 (13) (2021) e2012748118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Ji Y, Kumar R, Ghokale A, Chao HP, Rycaj K, Chen X, Li Q, Tang DG, LRIG1, a regulator of stem cell quiescence and a pleiotropic feedback tumor suppressor, Semin. Cancer Biol. 18 (January) (2021). S1044–579X(20)30276–5. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Purnell DM, Heatfield BM, Trump BF, Immunocytochemical evaluation of human prostatic carcinomas for carcinoembryonic antigen, nonspecific cross-reacting antigen, beta-chorionic gonadotrophin, and prostate-specific antigen, Cancer Res. 44 (1984) 285–292. [PubMed] [Google Scholar]
- [171].Stein BS, Vangore S, Petersen RO, Immunoperoxidase localization of prostatic antigens. Comparison of primary and metastatic sites, Urology 24 (1984) 146–152. [DOI] [PubMed] [Google Scholar]
- [172].Feiner HD, Gonzalez R, Carcinoma of the prostate with atypical immunohistological features. Clinical and histologic correlates, Am J Surg Pathol. 10 (1986) 765–770. [DOI] [PubMed] [Google Scholar]
- [173].Abrahamsson PA, Lilja H, Falkmer S, Wadstrom LB,¨ Immunohistochemical distribution of the three predominant secretory proteins in the parenchyma of hyperplastic and neoplastic prostate glands, Prostate 12 (1988) 39–46. [DOI] [PubMed] [Google Scholar]
- [174].Gallee MP, Visser-de Jong E, van der Korput JA, van der Kwast TH, ten Kate FJ, Schroeder FH, Trapman J, Variation of prostate-specific antigen expression in different tumour growth patterns present in prostatectomy specimens, Urol. Res. 18 (1990) 181–187. [DOI] [PubMed] [Google Scholar]
- [175].de Winter JA, Trapman J, Brinkmann AO, Boersma WJ, Mulder E, Schroeder FH, Claassen E, van der Kwast TH, Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry, J. Pathol. 160 (1990) 329–332. [DOI] [PubMed] [Google Scholar]
- [176].Masai M, Sumiya H, Akimoto S, Yatani R, Chang CS, Liao SS, Shimazaki J, Immunohistochemical study of androgen receptor in benign hyperplastic and cancerous human prostates, Prostate 17 (1990) 293–300. [DOI] [PubMed] [Google Scholar]
- [177].van der Kwast TH, Schalken J, Ruizeveld de Winter JA, van Vroonhoven CC, Mulder E, Boersma W, Trapman J, Androgen receptors in endocrine-therapy-resistant human prostate cancer, Int. J. Cancer 48 (1991) 189–193. [DOI] [PubMed] [Google Scholar]
- [178].Oesterling JE, Prostate specific antigen: a critical assessment of the most useful tumor marker for adenocarcinoma of the prostate, J. Urol. 145 (1991) 907–923. [DOI] [PubMed] [Google Scholar]
- [179].Sadi MV, Walsh PC, Barrack ER, Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy, Cancer 67 (1991) 3057–3064. [DOI] [PubMed] [Google Scholar]
- [180].Chodak GW, Kranc DM, Puy LA, Takeda H, Johnson K, Chang C, Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate, J. Urol. 147 (3 Pt 2) (1992) 798–803. [DOI] [PubMed] [Google Scholar]
- [181].Brolin I, Lowhagen T, Skoog L, Immunocytochemical detection of the androgen receptor in fine needle aspirates from benign and malignant human prostate, Cytopathology 3 (1992) 351–357. [DOI] [PubMed] [Google Scholar]
- [182].Sadi MV, Barrack ER, Image analysis of androgen receptor immunostaining in metastatic prostate cancer. Heterogeneity as a predictor of response to hormonal therapy, Cancer 71 (1993) 2574–2580. [DOI] [PubMed] [Google Scholar]
- [183].Miyamoto K, McSherry SA, Dent OA, et al. , Immunohistochemistory of the androgen receptor in human benign and malignant prostate tissue, J. Urol. 149 (1993) 1015–1019. [DOI] [PubMed] [Google Scholar]
- [184].Ruizeveld de Winter JA, Janssen PJA, Sleddens HMEB, Verleun-Mooijman MCT, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, van der Kwast TH, Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer, Am. J. Pathol. 144 (1994) 735–746. [PMC free article] [PubMed] [Google Scholar]
- [185].Aihara M, Lebovitz RM, Wheeler TM, Kinner BM, Ohori M, Scardino PT, Prostate specific antigen and gleason grade: an immunohistochemical study of prostate cancer, J. Urol. 151 (1994) 1558–1564. [DOI] [PubMed] [Google Scholar]
- [186].TilIey WD, Lim-Tio SS, Horsfall DJ, Aspinall JO, Marshall VR, Skinner JM, Detection of discrete androgen receptor epitopes in prostate cancer by immunostaining: measurement by color video image analysis, Cancer Res. 54 (1994) 4096–4102. [PubMed] [Google Scholar]
- [187].Pertschuk LP, Macchia RJ, Feldman JG, Brady KA, Levine M, Kim DS, Eisenberg KB, Rainford E, Prins GS, Greene GL, Immunocytochemical assay for androgen receptors in prostate cancer: a prospective study of 63 cases with long-term follow-up, Ann. Surg. Oncol. 1 (1994) 495–503. [DOI] [PubMed] [Google Scholar]
- [188].Hobisch A, Culig Z, Radmayr C, Bariseli G, Klocker H, Hittmair A, Distant metastases from prostatic carcinoma express androgen receptor protein, Cancer Res. 55 (1995) 3068–3072. [PubMed] [Google Scholar]
- [189].Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL, Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone, Hum. Pathol. 34 (2003) 646–653. [DOI] [PubMed] [Google Scholar]
- [190].Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ, Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program, Cancer Res. 64 (2004) 9209–9216. [DOI] [PubMed] [Google Scholar]
- [191].Birtle AJ, Freeman A, Masters JR, Payne HA, Harland SJ, BAUS Section of Oncology Cancer Registry, Tumour markers for managing men who present with metastatic prostate cancer and serum prostate-specific antigen levels of <10 ng/ mL, BJU Int. 96 (2005) 303–307. [DOI] [PubMed] [Google Scholar]
- [192].Davis JN, Wojno KJ, Daignault S, Hofer MD, Kuefer R, Rubin MA, Day ML, Elevated E2F1 inhibits transcription of the androgen receptor in metastatic hormone-resistant prostate cancer, Cancer Res. 66 (2006) 11897–11906. [DOI] [PubMed] [Google Scholar]
- [193].Mostaghel EA, Page ST, Lin DW, Fazli L, Coleman IM, True LD, Knudsen B, Hess DL, Nelson CC, Matsumoto AM, Bremner WJ, Gleave ME, Nelson PS, Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer, Cancer Res. 67 (2007) 5033–5041. [DOI] [PubMed] [Google Scholar]
- [194].Crnalic S, Hörnberg E, Wikström P, Lerner UH, Tieva Å, Svensson O, et al. , Nuclear androgen receptor staining in bone metastases is related to a poor outcome in prostate cancer patients, Endocr. Relat. Cancer 17 (2010) 885–895. [DOI] [PubMed] [Google Scholar]
- [195].Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Chen S, Nelson PS, Liu XS, Brown M, Balk SP, Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1, Cancer Cell 20 (2011) 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Tamburrino L, Salvianti F, Marchiani S, Pinzani P, Nesi G, Serni S, Forti G, Baldi E, Androgen receptor (AR) expression in prostate cancer and progression of the tumor: lessons from cell lines, animal models and human specimens, Steroids 77 (2012) 996–1001. [DOI] [PubMed] [Google Scholar]
- [197].Wan X, Liu J, Lu JF, Tzelepi V, Yang J, Starbuck MW, Diao L, Wang J, Efstathiou E, Vazquez ES, Troncoso P, Maity SN, Navone NM, Activation of β-catenin signaling in androgen receptor-negative prostate cancer cells, Clin. Cancer Res. 18 (3) (2012) 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Decker KF, Zheng D, He Y, Bowman T, Edwards JR, Jia L, Persistent androgen receptor-mediated transcription on castration-resistant prostate cancer under androgen-deprived conditions, Nucl. Acis Res. 40 (2012) 10765–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Chu M, Chang Y, Wang N, Li W, Li P, Gao WQ, Hypermethylation-mediated transcriptional repression of TMPRSS2 in androgen receptor-negative prostate cancer cells, Exp. Biol. Med. 239 (7) (2014) 823–828. [DOI] [PubMed] [Google Scholar]
- [200].Shahriari K, Shen F, Worrede-Mahdi A, Liu Q, Gong Y, Garcia FU, Fatatis A, Cooperation among heterogenous prostate cancer cells in the bone metastatic niche, Oncogene 36 (2017) 2846–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF, Kaipainen A, Corella AN, Yang YC, Nyquist MD, Mostaghel E, Hsieh AC, Zhang X, Corey E, Brown LG, Nguyen HM, Pienta K, Ittmann M, Schweizer M, True LD, Wise D, Rennie PS, Vessella RL, Morrissey C, Nelson PS, Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling, Cancer Cell 32 (2017) 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Saeed K, Rahkama V, Eldfors S, Bychkov D, Mpindi JP, Yadav B, Paavolainen L, Aittokallio T, Heckman C, Wennerberg K, Peehl DM, Horvath P, Mirtti T, Rannikko A, Kallioniemi O, Östling P, Af Hällström TM, Comprehensive drug testing of patient-derived conditionally reprogrammed cells from castration-resistant prostate cancer, Eur. Urol. 71 (3) (2017) 319–327. [DOI] [PubMed] [Google Scholar]
- [203].Su W, Han HH, Wang Y, Zhang B, Zhou B, Cheng Y, Rumandla A, Gurrapu S, Chakraborty G, Su J, Yang G, Liang X, Wang G, Rosen N, Scher HI, Ouerfelli O, Giancotti FG, The polycomb repressor complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression, Cancer Cell 36 (2) (2019) 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Spratt DE, Alshalalfa M, Fishbane N, Weiner AB, Mehra R, Mahal BA, Lehrer J, Liu Y, Zhao SG, Speers C, Morgan TM, Dicker AP, Freedland SJ, Karnes RJ, Weinmann S, Davicioni E, Ross AE, Nguyen PL, Den RB, Feng FY, Lotan TL, Chinnaiyan AM, Schaeffer EM, Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment-naïve primary prostate cancer, Clin. Cancer Res. 25 (22) (2019) 6721–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Liu Y, Horn JL, Banda K, Goodman AZ, Lim Y, Jana S, Arora S, Germanos AA, Wen L, Hardin WR, Yang YC, Coleman IM, Tharakan RG, Cai EY, Uo T, Pillai SPS, Corey E, Morrissey C, Chen Y, Carver BS, Plymate SR, Beronja S, Nelson PS, Hsieh AC, The androgen receptor regulates a druggable translation regulon in advanced prostate cancer, Sci. Transl. Med. 11 (503) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Silvestri R, Pucci P, Venalainen E, Matheou C, Mather R, Chandler S, Aceto R, Rigas SH, Wang Y, Rietdorf K, Bootman MD, Crea F, T-type calcium channels drive the proliferation of androgen-receptor negative prostate cancer cells, Prostate 79 (13) (2019) 1580–1586. [DOI] [PubMed] [Google Scholar]
- [207].Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, Nguyen HM, Yang YC, da Costa RMG, Kaipainen A, Coleman R, Higano CS, Cheng HH, Yu EY, Mostaghel EA, Montgomery B, Schweizer MT, Hsieh AC, Lin DW, Corey E, Nelson PS, Morrissey C, Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer, J. Clin. Invest. 130 (2019) 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Sehgal PD, Bauman TM, Nicholson TM, Vellky JE, Ricke EA, Tang W, Xu W, Huang W, Ricke WA, Tissue-specific quantification and localization of androgen and estrogen receptors in prostate cancer, Hum. Pathol. 89 (2019) 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Vellky JE, Bauman TM, Ricke EA, Huang W, Ricke WA, Incidence of androgen receptor and androgen receptor variant 7 coexpression in prostate cancer, Prostate 79 (16) (2019) 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, Aggarwal R, Cetnar J, Ryan CJ, Tabatabaei S, Bailey S, Turina CB, Quigley DA, Guan X, Foye A, Youngren JF, Urrutia J, Huang J, Weinstein AS, Friedl V, Rettig M, Reiter RE, Spratt DE, Gleave M, Evans CP, Stuart JM, Chen Y, Feng FY, Small EJ, Witte ON, Xia Z, Transcriptional profiling identifies an androgen-receptor activity-low, stemness program associated with enzalutamide resistance, Proc. Natl. Acad. Sci. U. S. A. 117 (2020) 12315–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Wise DR, Schneider JA, Armenia J, Febles VA, McLaughlin B, Brennan R, Thoren KL, Abida W, Sfanos KS, De Marzo AM, Yegnasubramanian S, Fox JJ, Haas M, Heath H, Kagey MH, Newman W, Sirard CA, Fleisher M, Morris MJ, Chen Y, Larson SM, Haffner MC, Nelson PS, Schultz N, Garabedian MJ, Scher HI, Logan SK, Sawyers CL, International SU2C/PCF Prostate Cancer Dream Team, Dickkopf-1 can lead to immune evasion in metastatic castration-resistant prostate cancer, JCO Precis Oncol. 29 (September) (2020) 4. PO.20.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Pandit-Taskar N, Veach DR, Fox JJ, Scher HI, Morris MJ, Larson SM, Evaluation of castration-resistant prostate cancer with androgen receptor-axis imaging, J. Nucl. Med. 57 (2016) 73S–78S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Fox JJ, Gavane SC, Blanc-Autran E, Nehmeh S, Gönen M, Beattie B, Vargas HA, Schöder H, Humm JL, Fine SW, Lewis JS, Solomon SB, Osborne JR, Veach D, Sawyers CL, Weber WA, Scher HI, Morris MJ, Larson SM, Positron emission tomography/computed tomography-based assessments of androgen receptor expression and glycolytic activity as a prognostic biomarker for metastatic castration-resistant prostate cancer, JAMA Oncol. 4 (2) (2018) 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Vargas HA, Kramer GM, Scott AM, Weickhardt A, Meier AA, Parada N, Beattie BJ, Humm JL, Staton KD, Zanzonico PB, Lyashchenko SK, Lewis JS, Yaqub M, Sosa RE, van den Eertwegh AJ, Davis ID, Ackermann U, Pathmaraj K, Schuit RC, Windhorst AD, Chua S, Weber WA, Larson SM, Scher HI, Lammertsma AA, Hoekstra OS, Morris MJ, Reproducibility and repeatability of semiquantitative (18)F-fluorodihydrotestosterone uptake metrics in castration-resistant prostate cancer metastases: a prospective multicenter study, J. Nucl. Med. 59 (10) (2018) 1516–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM, Integrative molecular concept modeling of prostate cancer progression, Nat. Genet. 39 (1) (2007) 41–51. [DOI] [PubMed] [Google Scholar]
- [216].Stuchbery R, Macintyre G, Cmero M, Harewood LM, Peters JS, Costello AJ, Hovens CM, Corcoran NM, Reduction in expression of the benign AR transcriptome is a hallmark of localised prostate cancer progression, Oncotarget 7 (21) (2016) 31384–31392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Russo JW, Gao C, Bhasin SS, Voznesensky OS, Calagua C, Arai S, Nelson PS, Montgomery B, Mostaghel EA, Corey E, Taplin ME, Ye H, Bhasin M, Balk SP, Downregulation of dipeptidyl peptidase 4 accelerates progression to castration-resistant prostate cancer, Cancer Res. 78 (22) (2018) 6354–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Huss WJ, Gray DR, Greenberg NM, Mohler JL, Smith GJ, Breast cancer resistance protein-mediated efflux of androgen in putative benign and malignant prostate stem cells, Cancer Res. 65 (15) (2005) 6640–6650. [DOI] [PubMed] [Google Scholar]
- [219].Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ, Prospective identification of tumorigenic prostate cancer stem cells, Cancer Res. 65 (23) (2005) 10946–10951. [DOI] [PubMed] [Google Scholar]
- [220].Miki J, Furusato B, Li H, Gu Y, Takahashi H, Egawa S, Sesterhenn IA, McLeod DG, Srivastava S, Rhim JS, Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens, Cancer Res. 67 (7) (2007) 3153–3161. [DOI] [PubMed] [Google Scholar]
- [221].Gu G, Yuan J, Wills M, Kasper S, Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo, Cancer Res. 67 (10) (2007) 4807–4815. [DOI] [PubMed] [Google Scholar]
- [222].Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, Garcia-Echeverria C, Schultz PG, Reddy VA, The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations, Proc. Natl. Acad. Sci. U. S. A. 106 (1) (2009) 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G, High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer, Cancer Res. 70 (12) (2010) 5163–5173. [DOI] [PubMed] [Google Scholar]
- [224].Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI, Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling, Nat. Commun. 2 (2011) 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, Wilber A, Watabe K, Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone, J. Exp. Med. 208 (13) (2011) 2641–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, Bonal DM, Charytonowicz E, Gladoun N, de la Iglesia-Vicente J, Petrylak DP, Benson MC, Silva JM, Cordon-Cardo C, Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells, Cancer Cell 22 (3) (2012) 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Germann M, Wetterwald A, Guzman-Ramirez N, van der Pluijm G, Culig Z, Cecchini MG, Williams ED, Thalmann GN, Stem-like cells with luminal progenitor phenotype survive castration in human prostate cancer, Stem Cells 30 (6) (2012) 1076–1086. [DOI] [PubMed] [Google Scholar]
- [228].Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H, Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells, Cancer Res. 72 (7) (2012) 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Cai H, Memarzadeh S, Stoyanova T, Beharry Z, Kraft AS, Witte ON, Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer, Cancer Res. 72 (18) (2012) 4672–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Stoyanova T, Goldstein AS, Cai H, Drake JM, Huang J, Witte ON, Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via beta-catenin signaling, Genes Dev. 26 (20) (2012) 2271–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Taylor RA, Toivanen R, Frydenberg M, Pedersen J, Harewood L, Australian Prostate Cancer B, Collins AT, Maitland NJ, Risbridger GP, Human epithelial basal cells are cells of origin of prostate cancer, independent of CD133 status, Stem Cells 30 (6) (2012) 1087–1096. [DOI] [PubMed] [Google Scholar]
- [232].Saini S, Majid S, Shahryari V, Arora S, Yamamura S, Chang I, Zaman MS, Deng G, Tanaka Y and Dahiya R miRNA-708 control of CD44(+) prostate cancer-initiating cells, Cancer Res. 72 (14) (2012) 3618–3630. [DOI] [PubMed] [Google Scholar]
- [233].Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, Chen D, Seo K, Modrusan Z, Gao WQ, Settleman J, Johnson L, Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy, Cancer Res. 72 (2) (2012) 527–536. [DOI] [PubMed] [Google Scholar]
- [234].Civenni G, Malek A, Albino D, Garcia-Escudero R, Napoli S, Di Marco S, Pinton S, Sarti M, Carbone GM, Catapano CV, RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer, Cancer Res. 73 (22) (2013) 6816–6827. [DOI] [PubMed] [Google Scholar]
- [235].Kroon P, Berry PA, Stower MJ, Rodrigues G, Mann VM, Simms M, Bhasin D, Chettiar S, Li C, Li PK, Maitland NJ, Collins AT, JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells, Cancer Res. 73 (16) (2013) 5288–5298. [DOI] [PubMed] [Google Scholar]
- [236].Polson ES, Lewis JL, Celik H, Mann VM, Stower MJ, Simms MS, Rodrigues G, Collins AT, Maitland NJ, Monoallelic expression of TMPRSS2/ ERG in prostate cancer stem cells, Nat. Commun. 4 (2013) 1623. [DOI] [PubMed] [Google Scholar]
- [237].Qu Y, Oyan AM, Liu R, Hua Y, Zhang J, Hovland R, Popa M, Liu X, Brokstad KA, Simon R, Molven A, Lin B, Zhang WD, McCormack E, Kalland KH, Ke XS, Generation of prostate tumor-initiating cells is associated with elevation of reactive oxygen species and IL-6/STAT3 signaling, Cancer Res. 73 (23) (2013) 7090–7100. [DOI] [PubMed] [Google Scholar]
- [238].Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, Zhang H, Kohn DB, Huang J, Witte ON, Goldstein AS, Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells, Proc. Natl. Acad. Sci. U. S. A. 110 (50) (2013) 20111–20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Yoshioka T, Otero J, Chen Y, Kim YM, Koutcher JA, Satagopan J, Reuter V, Carver B, de Stanchina E, Enomoto K, Greenberg NM, Scardino PT, Scher HI, Sawyers CL, Giancotti FG, beta4 Integrin signaling induces expansion of prostate tumor progenitors, J. Clin. Invest. 123 (2) (2013) 682–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Fujimura T, Takahashi S, Urano T, Takayama K, Sugihara T, Obinata D, Yamada Y, Kumagai J, Kume H, Ouchi Y, Inoue S, Homma Y, Expression of androgen and estrogen signaling components and stem cell markers to predict cancer progression and cancer-specific survival in patients with metastatic prostate cancer, Clin. Cancer Res. 20 (17) (2014) 4625–4635. [DOI] [PubMed] [Google Scholar]
- [241].Han Z, Wang X, Ma L, Chen L, Xiao M, Huang L, Cao Y, Bai J, Ma D, Zhou J, Hong Z, Inhibition of STAT3 signaling targets both tumor-initiating and differentiated cell populations in prostate cancer, Oncotarget 5 (18) (2014) 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Kroon J, in’ t Veld LS, Buijs JT, Cheung H, van der Horst G, van der Pluijm G, Glycogen synthase kinase-3beta inhibition depletes the population of prostate cancer stem/progenitor-like cells and attenuates metastatic growth, Oncotarget 5 (19) (2014) 8986–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Li X, Liu Y, Chen W, Fang Y, Xu H, Zhu HH, Chu M, Li W, Zhuang G, Gao WQ, TOP2Ahigh is the phenotype of recurrence and metastasis whereas TOP2Aneg cells represent cancer stem cells in prostate cancer, Oncotarget 5 (19) (2014) 9498–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Luo J, Ok Lee S, Liang L, Huang CK, Li L, Wen S, Chang C, Infiltrating bone marrow mesenchymal stem cells increase prostate cancer stem cell population and metastatic ability via secreting cytokines to suppress androgen receptor signaling, Oncogene 33 (21) (2014) 2768–2778. [DOI] [PubMed] [Google Scholar]
- [245].Schroeder A, Herrmann A, Cherryholmes G, Kowolik C, Buettner R, Pal S, Yu H, Muller-Newen G, Jove R, Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling, Cancer Res. 74 (4) (2014) 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Marhold M, Tomasich E, El-Gazzar A, Heller G, Spittler A, Horvat R, Krainer M, Horak P, HIF1alpha regulates mTOR signaling and viability of prostate cancer stem cells, Mol. Cancer Res. 13 (3) (2015) 556–564. [DOI] [PubMed] [Google Scholar]
- [247].Rybak AP, Bristow RG, Kapoor A, Prostate cancer stem cells: deciphering the origins and pathways involved in prostate tumorigenesis and aggression, Oncotarget 6 (4) (2015) 1900–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Albino D, Civenni G, Dallavalle C, Roos M, Jahns H, Curti L, Rossi S, Pinton S, D’Ambrosio G, Sessa F, Hall J, Catapano CV, Carbone GM, Activation of the Lin28/let-7 axis by loss of ESE3/EHF promotes a tumorigenic and stem-like phenotype in prostate cancer, Cancer Res. 76 (12) (2016) 3629–3643. [DOI] [PubMed] [Google Scholar]
- [249].Bansal N, Bartucci M, Yusuff S, Davis S, Flaherty K, Huselid E, Patrizii M, Jones D, Cao L, Sydorenko N, Moon YC, Zhong H, Medina DJ, Kerrigan J, Stein MN, Kim IY, Davis TW, DiPaola RS, Bertino JR, Sabaawy HE, BMI-1 targeting interferes with patient-derived tumor-initiating cell survival and tumor growth in prostate cancer, Clin. Cancer Res. 22 (24) (2016) 6176–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Zhang K, Guo Y, Wang X, Zhao H, Ji Z, Cheng C, Li L, Fang Y, Xu D, Zhu HH, Gao WQ, Wnt/b-catenin directs self-renewal symmetric cell division of hTERThigh prostate cancer stem cells, Cancer Res. 77 (9) (2017) 2534–2547. [DOI] [PubMed] [Google Scholar]
- [251].Ruan D, He J, Li CF, Lee HJ, Liu J, Lin HK, Chan CH, Skp2 deficiency restricts the progression and stem cell features of castration-resistant prostate cancer by destabilizing Twist, Oncogene 36 (30) (2017) 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Sackmann Sala L, Boutillon F, Menara G, De Goyon-Pélard A, Leprévost M, Codzamanian J, Lister N, Pencik J, Clark A, Cagnard N, Bole-Feysot C, Moriggl R, Risbridger GP, Taylor RA, Kenner L, Guidotti JE, Goffin V, A rare castration-resistant progenitor cell population is highly enriched in Pten-null prostate tumours, J. Pathol. 243 (1) (2017) 51–64. [DOI] [PubMed] [Google Scholar]
- [253].Jiang N, Ke B, Hjort-Jensen K, Iglesias-Gato D, Wang Z, Chang P, Zhao Y, Niu X, Wu T, Peng B, Jiang M, Li X, Shang Z, Wang Q, Chang C, Flores-Morales A, Niu Y, YAP1 regulates prostate cancer stem cell-like characteristics to promote castration resistant growth, Oncotarget 8 (70) (2017) 115054–115067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Sorrentino C, Ciummo SL, Cipollone G, Caputo S, Bellone M, Di Carlo E, Interleukin-30/IL27p28 shapes prostate cancer stem-like cell behavior and is critical for tumor onset and metastasization, Cancer Res. 78 (10) (2018) 2654–2668. [DOI] [PubMed] [Google Scholar]
- [255].Jung Y, Cackowski FC, Yumoto K, Decker AM, Wang J, Kim JK, Lee E, Wang Y, Chung JS, Gursky AM, Krebsbach PH, Pienta KJ, Morgan TM, Taichman RS, CXCL12g promotes metastatic castration-resistant prostate cancer by inducing cancer stem cell and neuroendocrine phenotype, Cancer Res. 78 (8) (2018) 2026–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Smith BA, Balanis NG, Nanjundiah A, Sheu KM, Tsai BL, Zhang Q, Park JW, Thompson M, Huang J, Witte ON, Graeber TG, A human adult stem cell signature marks aggressive variants across epithelial cancers, Cell Rep. 24 (12) (2018) 3353–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Zhang Z, Cheng L, Li J, Farah E, Atallah NM, Pascuzzi PE, Gupta S, Liu X, Inhibition of the wnt/β-Catenin pathway overcomes resistance to enzalutamide in castration-resistant prostate cancer, Cancer Res. 78 (12) (2018) 3147–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, Huang J, Kurdistani SK, Graeber TG, Witte ON, Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage, Science 362 (6410) (2018) 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Pai VC, Hsu CC, Chan TS, Liao WY, Chuu CP, Chen WY, Li CR, Lin CY, Huang SP, Chen LT, Tsai KK, ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-β-catenin signaling, Oncogene 38 (8) (2019) 1340–1353. [DOI] [PubMed] [Google Scholar]
- [260].Yoo YA, Vatapalli R, Lysy B, Mok H, Desouki MM, Abdulkadir SA, The role of castration-resistant Bmi1+Sox2+ cells in driving recurrence in prostate cancer, J. Natl. Cancer Inst. 111 (3) (2019) 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Weng CC, Ding PY, Liu YH, Hawse JR, Subramaniam M, Wu CC, Lin YC, Chen CY, Hung WC, Cheng KH, Mutant Kras-induced upregulation of CD24 enhances prostate cancer stemness and bone metastasis, Oncogene 38 (12) (2019) 2005–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Zhang J, Chen M, Zhu Y, Dai X, Dang F, Ren J, Ren S, Shulga YV, Beca F, Gan W, Wu F, Lin YM, Zhou X, DeCaprio JA, Beck AH, Lu KP, Huang J, Zhao C, Sun Y, Gao X, Pandolfi PP, Wei W, SPOP promotes NANOG destruction to suppress stem cell traits and prostate cancer progression, Dev. Cell 48 (3) (2019) 329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Wang X, Jin J, Wan F, Zhao L, Chu H, Chen C, Liao G, Liu J, Yu Y, Teng H, Fang L, Jiang C, Pan W, Xie X, Li J, Lu X, Jiang X, Ge X, Ye D, Wang P, AMPK promotes SPOP-mediated NANOG degradation to regulate prostate cancer cell stemness, Dev. Cell 48 (3) (2019) 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Vummidi Giridhar P, Williams K, VonHandorf AP, Deford PL, Kasper S, Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity, Cancer Res. 79 (6) (2019) 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Reilly D, Johnson P, Buchanan PJ, Hypoxia induced canecr stem cell enrichment promotes resistance to androgen deparivation therapy in prostate cancer, Steroids 152 (2019) 108497. [DOI] [PubMed] [Google Scholar]
- [266].Sorrentino C, Yin Z, Ciummo S, Lanuti P, Lu LF, Marchisio M, Bellone M, Di Carlo E, Targeting Interleukin(IL)-30/IL-27p28 signaling in cancer stem-like cells and host environment synergistically inhibits prostate cancer growth and improves survival, J. Immunother. Cancer 7 (1) (2019) 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Lee HJ, Li CF, Ruan D, He J, Montal ED, Lorenz S, Girnun GD, Chan CH, Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion, Nat. Commun. 10 (1) (2019) 2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Civenni G, Bosotti R, Timpanaro A, Vàzquez R, Merulla J, Pandit S, Rossi S, Albino D, Allegrini S, Mitra A, Mapelli SN, Vierling L, Giurdanella M, Marchetti M, Paganoni A, Rinaldi A, Losa M, Mira-Catò E, D’Antuono R, Morone D, Rezai K, D’Ambrosio G, Ouafik L, Mackenzie S, Riveiro ME, Cvitkovic E, Carbone GM, Catapano CV, Epigenetic control of mitochondrial fission enables self-renewal of stem-like tumor cells in human prostate cancer, Cell Metab. 30 (2) (2019) 303–318. [DOI] [PubMed] [Google Scholar]
- [269].Hepburn AC, Steele RE, Veeratterapillay R, Wilson L, Kounatidou EE, Barnard A, Berry P, Cassidy JR, Moad M, El-Sherif A, Gaughan L, Mills IG, Robson CN, Heer R, The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance, Oncogene 38 (22) (2019) 4412–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Vaddi PK, Stamnes MA, Cao H, Chen S, Elimination of SOX2/OCT4-associated prostate cancer stem cells blocks tumor development and enhances therapeutic response, Cancers (Basel) 8 (9) (2019) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Gorodetska I, Lukiyanchuk V, Peitzsch C, Kozeretska I, Dubrovska A, BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype, Int. J. Cancer 145 (11) (2019) 2974–2985. [DOI] [PubMed] [Google Scholar]
- [272].Li Y, He Y, Butler W, Xu L, Chang Y, Lei K, Zhang H, Zhou Y, Gao AC, Zhang Q, Taylor DG, Cheng D, Farber-Katz S, Karam R, Landrith T, Li B, Wu S, Hsuan V, Yang Q, Hu H, Chen X, Flowers M, McCall SJ, Lee JK, Smith BA, Park JW, Goldstein AS, Witte ON, Wang Q, Rettig MB, Armstrong AJ, Cheng Q, Huang J, Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer, Sci. Transl. Med. 11 (521) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Williams ED, Gao D, Redfern A, Thompson EW, Controversies around epithelial-mesenchymal plasticity in cancer metastasis, Nat. Rev. Cancer 19 (12) (2019) 716–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Navas T, Kinders RJ, Lawrence SM, Ferry-Galow KV, Borgel S, Hollingshead MG, Srivastava AK, Alcoser SY, Makhlouf HR, Chuaqui R, Wilsker DF, Konaté MM, Miller SB, Voth AR, Chen L, Vilimas T, Subramanian J, Rubinstein L, Kummar S, Chen AP, Bottaro DP, Doroshow JH, Parchment RE, Clinical evolution of epithelial-mesenchymal transition in human carcinomas, Cancer Res. 80 (2) (2020) 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori G, Dedhar S, Derynck R, Ford HL, Fuxe J, García de Herreros A, Goodall GJ, Hadjantonakis AK, Huang RJY, Kalcheim C, Kalluri R, Kang Y, Khew-Goodall Y, Levine H, Liu J, Longmore GD, Mani SA, Massagué J, Mayor R, McClay D, Mostov KE, Newgreen DF, Nieto MA, Puisieux A, Runyan R, Savagner P, Stanger B, Stemmler MP, Takahashi Y, Takeichi M, Theveneau E, Thiery JP, Thompson EW, Weinberg RA, Williams ED, Xing J, Zhou BP, Sheng G, EMT International Association (TEMTIA), Guidelines and definitions for research on epithelial-mesenchymal transition, Nat. Rev. Mol. Cell Biol. 21 (6) (2020) 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Metz EP, Wilder PJ, Dong J, Datta K, Rizzino A, Elevating SOX2 in prostate tumor cells upregulates expression of neuroendocrine genes, but does not reduce the inhibitory effects of enzalutamide, J. Cell. Physiol. 235 (4) (2020) 3731–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Nouri M, Massah S, Caradec J, Lubik AA, Li N, Truong S, Lee AR, Fazli L, Ramnarine VR, Lovnicki JM, Moore J, Wang M, Foo J, Gleave ME, Hollier BG, Nelson C, Collins C, Dong X, Buttyan R, Transient Sox9 expression facilitates resistance to androgen-targeted therapy in prostate cancer, Clin. Cancer Res. 26 (7) (2020) 1678–1689. [DOI] [PubMed] [Google Scholar]
- [278].Li S, Goncalves KA, Lyu B, Yuan L, Hu GF, Chemosensitization of prostate cancer stem cells in mice by angiogenin and plexin-B2 inhibitors, Commun Biol. 3 (1) (2020) 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Yasumizu Y, Rajabi H, Jin C, Hata T, Pitroda S, Long MD, Hagiwara M, Li W, Hu Q, Liu S, Yamashita N, Fushimi A, Kui L, Samur M, Yamamoto M, Zhang Y, Zhang N, Hong D, Maeda T, Kosaka T, Wong KK, Oya M, Kufe D, MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer, Nat. Commun. 11 (1) (2020) 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Yin L, Hu P, Shi X, Qian W, Zhau HE, Pandol SJ, Lewis MS, Chung LWK, Wang R, Cancer cell’s neuroendocrine feature can be acquired through cell-cell fusion during cancer-neural stem cell interaction, Sci. Rep. 10 (1) (2020) 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, He Zhu H, Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate, Nat. Commun. 11 (1) (2020) 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Rossini A, Giussani M, Ripamonti F, Aiello P, Regondi V, Balsari A, Triulzi T, Tagliabue E, Combined targeting of EGFR and HER2 against prostate cancer stem cells, Cancer Biol. Ther. 21 (5) (2020) 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Carceles-Cordon M, Kelly WK, Gomella L, Knudsen KE, Rodriguez-Bravo V, Domingo-Domenech J, Cellular rewiring in lethal prostate cancer: the architect of drug resistance, Nat. Rev. Urol. 17 (5) (2020) 292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Cheng CY, Zhou Z, Stone M, Lu B, Flesken-Nikitin A, Nanus DM, Nikitin AY, Membrane metalloendopeptidase suppresses prostate carcinogenesis by attenuating effects of gastrin-releasing peptide on stem/progenitor cells, Oncogenesis 9 (3) (2020) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Brady-Nicholls R, Nagy JD, Gerke TA, Zhang T, Wang AZ, Zhang J, Gatenby RA, Enderling H, Prostate-specific antigen dynamics predict individual responses to intermittent androgen deprivation, Nat. Commun. 11 (1) (2020) 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Huang R, Wang S, Wang N, Zheng Y, Zhou J, Yang B, Wang X, Zhang J, Guo L, Wang S, Chen Z, Wang Z, Xiang S, CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling, Cell Death Dis. 11 (4) (2020) 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Jiao C, Meng T, Zhou C, Wang X, Wang P, Lu M, Tan X, Wei Q, Ge X, Jin J, TGF-β signaling regulates SPOPexpression and promotes prostate cancer cell stemness, Aging 12 (9) (2020) 7747–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, Aggarwal R, Cetnar J, Ryan CJ, Tabatabaei S, Bailey S, Turina CB, Quigley DA, Guan X, Foye A, Youngren JF, Urrutia J, Huang J, Weinstein AS, Friedl V, Rettig M, Reiter RE, Spratt DE, Gleave M, Evans CP, Stuart JM, Chen Y, Feng FY, Small EJ, Witte ON, Xia Z, Transcirptional profiling identifies an androgen-receptor activity-low, stemness program associated with enzalutamide resistance, PNAS 117 (2020) 12315–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Sánchez BG, Bort A, Vara-Ciruelos D, Díaz-Laviada I, Androgen deprivation induces reprogramming of prostate cancer cells to stem-like cells, Cells 9 (6) (2020) E1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Ci X, Hao J, Dong X, Xue H, Wu R, Choi SYC, Haegert AM, Collins CC, Liu X, Lin D, Wang Y, Conditionally reprogrammed cells from patient-derived xenograft to model neuroendocrine prostate cancer development, Cells 9 (6) (2020) E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Flynn L, Barr MP, Baird AM, Smyth P, Casey OM, Blackshields G, Greene J, Pennington SR, Hams E, Fallon PG, O’Leary J, Sheils O, Finn SP, Prostate cancer-derived holoclones: a novel and effective model for evaluating cancer stemness, Sci. Rep. 10 (1) (2020) 11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Verma S, Shankar E, Kalayci FNC, Mukunda A, Alassfar M, Singh V, Chan ER, MacLennan GT, Gupta S, Androgen deprivation induces transcriptional reprogramming in prostate cancer cells to develop stem cell-like characteristics, Int. J. Mol. Sci. 21 (24) (2020) 9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Yuan H, Han Y, Wang X, Li N, Liu Q, Yin Y, Wang H, Pan L, Li L, Song K, Qiu T, Pan Q, Chen Q, Zhang G, Zang Y, Tan M, Zhang J, Li Q, Wang X, Jiang J, Qin J, SETD2 restricts prostate cancer metastasis by integrating EZH2 and AMPK signaling pathways, Cancer Cell 38 (3) (2020) 350–365. [DOI] [PubMed] [Google Scholar]
- [294].Federer-Gsponer JR, Müller DC, Zellweger T, Eggimann M, Marston K, Ruiz C, Seifert HH, Rentsch CA, Bubendorf L, Le Magnen C, Patterns of stemness-associated markers in the development of castration-resistant prostate cancer, Prostate 80 (13) (2020) 1108–1117. [DOI] [PubMed] [Google Scholar]
- [295].Lovnicki J, Gan Y, Feng T, Li Y, Xie N, Ho CH, Lee AR, Chen X, Nappi L, Han B, Fazli L, Huang J, Gleave ME, Dong X, LIN28B promotes the development of neuroendocrine prostate cancer, J. Clin. Invest. 130 (10) (2020) 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Wang N, Jiang Y, Lv S, Wen H, Wu D, Wei Q, Dang Q, HOTAIR expands the population of prostatic cancer stem-like cells and causes Docetaxel resistance via activating STAT3 signaling, Aging 12 (13) (2020) 12771–12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Li F, Yuan Q, Di W, Xia X, Liu Z, Mao N, Li L, Li C, He J, Li Y, Guo W, Zhang X, Zhu Y, Aji R, Wang S, Tong X, Ji H, Chi P, Carver B, Wang Y, Chen Y, Gao D, ERG orchestrates chromatin interactions to drive prostate cell fate reprogramming, J. Clin. Invest. 130 (11) (2020) 5924–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Tremblay M, Viala S, Shafer ME, Graham-Paquin AL, Liu C, Bouchard M, Regulation of stem/progenitor cell maintenance by BMP5 in prostate homeostasis and cancer initiation, Elife 9 (2020), e54542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Wróbel T, Luty M, Catapano J, Karnas E, Szczygiel M, Piwowarczyk K, Ryszawy D, Drabik G, Zuba-Surma E, Siedlar M, Madeja Z, Elas M, Czyz J,? CD44+cells determine fenofibrate-induced microevolution of drug-resistance in prostate cancer cell populations, Stem Cells 38 (12) (2020) 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Miyashita M, Tomogane M, Nakamura Y, Shimizu T, Fujihara A, Ukimura O, Ashihara E, Sphere-derived prostate cancer stem cells are resistant to γδ T cell cytotoxicity, Anticancer Res. 40 (10) (2020) 5481–5487. [DOI] [PubMed] [Google Scholar]
- [301].Rivello F, Matula K, Piruska A, Smits M, Mehra N, Huck WTS, Probing single-cell metabolism reveals prognostic value of highly metabolically active circulating stromal cells in prostate cancer, Sci. Adv. 6 (40) (2020) eaaz3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Kwon OJ, Zhang L, Jia D, Zhou Z, Li Z, Haffner M, Lee JK, True L, Morrissey C, Xin L, De novo induction of lineage plasticity from human prostate luminal epithelial cells by activated AKT1 and c-Myc, Oncogene 39 (48) (2020) 7142–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, Ahmad I, Leung HY, Oliveira P, Blyth K, Baena E, Lacaud G, RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development, Elife 9 (2020), e60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Shibata M, Epsi NJ, Xuan S, Mitrofanova A, Shen MM, Bipotent progenitors do not require androgen receptor for luminal specification during prostate organogenesis, Stem Cell Rep. 15 (5) (2020) 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Schuhmacher J, Heidu S, Balchen T, Richardson JR, Schmeltz C, Sonne J, Schweiker J, Rammensee HG, Thor Straten P, Røder MA, Brasso K, Gouttefangeas C, Vaccination against RhoC induces long-lasting immune responses in patients with prostate cancer: results from a phase I/II clinical trial, J. Immunother. Cancer 8 (2) (2020) e001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Hagiwara M, Yasumizu Y, Yamashita N, Rajabi H, Fushimi A, Long MD, Li W, Bhattacharya A, Ahmad R, Oya M, Liu S, Kufe D, MUC1-C activates the BAF (mSWI/SNF) complex in prostate cancer stem cells, Cancer Res. 81 (4) (2021) 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Kwon OJ, Zhang L, Jia D, Xin L, Sox2 is necessary for androgen ablation-induced neuroendocrine differentiation from Pten null Sca-1+prostate luminal cells, Oncogene 40 (1) (2021) 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Ko J, Meyer AN, Haas M, Donoghue DJ, Characterization of FGFR signaling in prostate cancer stem cells and inhibition via TKI treatment, Oncotarget 12 (1) (2021) 22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Blagoev KB, Iordanov R, Zhou M, Fojo T, Bates SE, Drug resistant cells with very large proliferative potential grow exponentially in metastatic prostate cancer, Oncotarget 12 (1) (2021) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Harris KS, Shi L, Foster BM, Mobley ME, Elliott PL, Song CJ, Watabe K, Langefeld CD, Kerr BA, CD117/c-kit defines a prostate CSC-like subpopulation driving progression and TKI resistance, Sci. Rep. 11 (1) (2021) 1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Chen Q, Gu M, Cai ZK, Zhao H, Sun SC, Liu C, Zhan M, Chen YB, Wang Z, TGF-β1 promotes epithelial-to-mesenchymal transition and stemness of prostate cancer cells by inducing PCBP1 degradation and alternative splicing of CD44, Cell. Mol. Life Sci. 78 (3) (2021) 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Li Y, Ge C, Franceschi RT, Role of Runx2 in prostate development and stem cell function, Prostate 81 (4) (2021) 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].McGuire JJ, Frieling JS, Lo CH, Li T, Muhammad A, Lawrence HR, Lawrence NJ, Cook LM, Lynch CC, Mesenchymal stem cell-derived interleukin-28 drives the selection of apoptosis resistant bone metastatic prostate cancer, Nat. Commun. 12 (1) (2021) 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Unno K, Chalmers ZR, Pamarthy S, Vatapalli R, Rodriguez Y, Lysy B, Mok H, Sagar V, Han H, Yoo YA, Ku SY, Beltran H, Zhao Y, Abdulkadir SA, Activated ALK cooperates with N-Myc via Wnt/beta-catenin signaling to induce neuroendocrine prostate cancer, Cancer Res. 81 (8) (2021) 2157–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Zhang Y, He L, Sadagopan A, Ma T, Dotti G, Wang Y, Zheng H, Gao X, Wang D, DeLeo AB, Fan S, Sun R, Yu L, Zhang L, Wang G, Ferrone S, Wang X, Targeting radiation-resistant prostate cancer stem cells by B7-H3 CAR T cells, Mol. Cancer Ther. 20 (3) (2021) 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Sun Y, Jing J, Xu H, Xu L, Hu H, Tang C, Liu S, Wei Q, Duan R, Guo J, Yang L, N-cadherin inhibitor creates a microenvironment that protect TILs from immune checkpoints and Treg cells, J. Immunother. Cancer 9 (3) (2021) e002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Zhang W, Bado IL, Hu J, Wan YW, Wu L, Wang H, Gao Y, Jeong HH, Xu Z, Hao X, Lege BM, Al-Ouran R, Li L, Li J, Yu L, Singh S, Lo HC, Niu M, Liu J, Jiang W, Li Y, Wong STC, Cheng C, Liu Z, Zhang XH, The bone microenvironment invigorates metastatic seeds for further dissemination, Cell 184 (9) (2021) 2471–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Hagiwara M, Fushimi A, Yamashita N, Bhattacharya A, Rajabi H, Long MD, Yasumizu Y, Oya M, Liu S, Kufe D, MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells, Oncogene 40 (30) (2021) 4930–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Brady-Nicholls R, Zhang J, Zhang T, Wang AZ, Butler R, Gatenby RA, Enderling H, Predicting patient-specific response to adaptive therapy in metastatic castration-resistant prostate cancer using prostate-specific antigen dynamics, Neoplasia 23 (9) (2021) 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Hu WY, Hu DP, Xie L, Nonn L, Lu R, Abern M, Shioda T, Prins GS, Keratin profiling by single-cell RNA-Sequencing identifies human prostate stem cell lineage hierarchy and cancer stem-Like cells, Int. J. Mol. Sci. 22 (15) (2021) 8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Witte KE, Pfitzenmaier J, Storm J, Lütkemeyer M, Wimmer C, Schulten W, Czaniera N, Geisler M, Förster C, Wilkens L, Knabbe C, Mertzlufft F, Kaltschmidt B, Am Esch JS, Kaltschmidt C analysis of several pathways for efficient killing of prostate cancer stem cells: a central role of NF-kappaB RELA, Int. J. Mol. Sci. 22 (16) (2021) 8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Peña-Hernández R, Aprigliano R, Carina Frommel S, Pietrzak K, Steiger S, Roganowicz M, Lerra L, Bizzarro J, Santoro R, BAZ2A-mediated repression via H3K14ac-marked enhancers promotes prostate cancer stem cells, EMBO Rep. 17 (August) (2021), e53014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Nong S, Wei Z, Wang Z, Ma L, Guan Y, Ni J, Reduced DAPK1 expression promotes stem cell-like characteristics of prostate cancer cells by activating ZEB1 via Hippo/YAP signaling pathway, Stem Cells Dev. (August 17) (2021), 10.1089/scd.2021.0043. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- [324].Li W, Shen MM, Prostate cancer cell heterogeneity and plasticity: insights from studies of genetically-engineered mouse models, Semin. Cancer Biol. (June 18) (2021), 10.1016/j.semcancer.2021.06.016. S1044–579X(21) 00186–3. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Brady L, Kriner M, Coleman I, Morrissey C, Roudier M, True LD, Gulati R, Plymate SR, Zhou Z, Birditt B, Meredith R, Geiss G, Hoang M, Beechem J, Nelson PS, Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling, Nat. Commun. 12 (1) (2021) 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Watson PA, Arora VK, Sawyers CL, Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer, Nat. Rev. Cancer 15 (12) (2015) 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Kelly K, Yu EY, Lin D, Dicker A, Arnold J, Hecht T, Wicha M, Sears R, Rowley D, White R, Gulley JL, Lee J, Meco M. Diaz, Small EJ, Shen M, Knudsen K, Goodrich DW, Lotan T, Zoubeidi A, Sawyers CL, Rudin CM, Loda M, Thompson T, Rubin MA, Tawab-Amiri A, Dahut W, Nelson PS, The role of lineage plasticity in prostate cancer therapy resistance, Clin. Cancer Res. 25 (23) (2019) 6916–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Sowalsky AG, Ye H, Bhasin M, Van Allen EM, Loda M, Lis RT, Montaser-Kouhsari L, Calagua C, Ma F, Russo JW, Schaefer RJ, Voznesensky OS, Zhang Z, Bubley GJ, Montgomery B, Mostaghel EA, Nelson PS, Taplin ME, Balk SP, Neoadjuvant-intensive androgen deprivation therapy selects for prostate tumor foci with diverse subclonal oncogenic alterations, Cancer Res. 78 (16) (2018) 4716–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Wilkinson S, Harmon SA, Terrigino NT, Karzai F, Pinto PA, Madan RA, VanderWeele DJ, Lake R, Atway R, Bright JR, Carrabba NV, Trostel SY, Lis RT, Chun G, Gulley JL, Merino MJ, Choyke PL, Ye H, Dahut WL, Turkbey B, Sowalsky AG, A case report of multiple primary prostate tumors with differential drug sensitivity, Nat. Commun. 11 (1) (2020) 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Wilkinson S, Ye H, Karzai F, Harmon SA, Terrigino NT, VanderWeele DJ, Bright JR, Atway R, Trostel SY, Carrabba NV, Whitlock NC, Walker SM, Lis RT, Abdul Sater H, Capaldo BJ, Madan RA, Gulley JL, Chun G, Merino MJ, Pinto PA, Salles DC, Kaur HB, Lotan TL, Venzon DJ, Choyke PL, Turkbey B, Dahut WL, Sowalsky AG, Nascent prostate cancer heterogeneity drives evolution and resistance to intense hormonal therapy, Eur Urol. Mar 27 (2021). S0302–2838(21)00207–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Labrecque MP, Alumkal JJ, Coleman IM, Nelson PS, Morrissey C, The heterogeneity of prostate cancers lacking AR activity will require diverse treatment approaches, Endocr. Relat. Cancer (April 1) (2021), 10.1530/ERC-21-0002. ERC-21–0002.R1. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Swayers CL, Ellis L, Goodrich DW, Rb and p53 cooperate to suppress prostate cancer lineage plasticity, Science 355 (2017) 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Mu P, Zhang ZD, Bebelli M, Sawyers CL, et al. , SOX2 mediated lineage plasticity change confers antiandrogen resistance in advanced prostate cancer, Science 355 (2017) 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Le Magnen C, Shen MM, Abate-Shen C, Lineage plasticity in cancer progression and treatment, Annu Rev Cancer Biol. 2 (2018) 271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Brennen WN, Zhu Y, Coleman IM, Dalrymple SL, Antony L, Patel RA, Hanratty B, Chikarmane R, Meeker AK, Zheng SL, Hooper JE, Luo J, De Marzo AM, Corey E, Xu J, Yegnasubramanian S, Haffner MC, Nelson PS, Nelson WG, Isaacs WB, Isaacs JT, Resistance to androgen receptor signaling inhibition does not necessitate development of neuroendocrine prostate cancer, JCI Insight 6 (8) (2021) e146827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, Gulati R, Ang L, Chatterjee P, Lucas J, Pritchard C, Risbridger G, Isaacs J, Montgomery B, Morrissey C, Corey E, Nelson PS, Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress, Cell Rep. 31 (8) (2020) 107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang WC, Huang F, Pan X, Yáñez A, Hazelett DJ, Chu CY, Steadman K, Morrissey CM, Nelson PS, Corey E, Chung LWK, Freedland SJ, Di Vizio D, Garraway IP, Murali R, Knudsen BS, Freeman MR, ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis, Nat. Med. 24 (12) (2018) 1887–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Glumac PM, Gallant JP, Shapovalova M, Li Y, Murugan P, Gupta S, Coleman IM, Nelson PS, Dehm SM, LeBeau AM, Exploitation of CD133 for the targeted imaging of lethal prostate cancer, Clin. Cancer Res. 26 (5) (2020) 1054–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Corella AN, Cabiliza Ordonio MVA, Coleman I, Lucas JM, Kaipainen A, Nguyen HM, Sondheim D, Brown LG, True LD, Lee JK, MacPherson D, Nghiem P, Gulati R, Morrissey C, Corey E, Nelson PS, Identification of therapeutic vulnerabilities in small-cell neuroendocrine prostate cancer, Clin. Cancer Res. 26 (7) (2020) 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Mohammad OS, Nyquist MD, Schweizer MT, Balk SP, Corey E, Plymate S, Nelson PS, Mostaghel EA, Supraphysiologic testosterone therapy in the treatment of prostate cancer: models, mechanisms and questions, Cancers 9 (12) (2017) 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Chatterjee P, Schweizer MT, Lucas JM, Coleman I, Nyquist MD, Frank SB, Tharakan R, Mostaghel E, Luo J, Pritchard CC, Lam HM, Corey E, Antonarakis ES, Denmeade SR, Nelson PS, Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage, J. Clin. Invest. 129 (10) (2019) 4245–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Lam HM, Nguyen HM, Labrecque MP, Brown LG, Coleman IM, Gulati R, Lakely B, Sondheim D, Chatterjee P, Marck BT, Matsumoto AM, Mostaghel EA, Schweizer MT, Nelson PS, Corey E, Durable response of enzalutamide-resistant prostate cancer to supraphysiological testosterone is associated with a multifaceted growth suppression and impaired DNA damage response transcriptomic program in patient-derived xenografts, Eur. Urol. 77 (2) (2020) 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Nyquist MD, Corella A, Mohamad O, Coleman I, Kaipainen A, Kuppers DA, Lucas JM, Paddison PJ, Plymate SR, Nelson PS, Mostaghel EA, Molecular determinants of response to high-dose androgen therapy in prostate cancer, JCI Insight 4 (October (19)) (2019) e129715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Nyquist MD, Ang LS, Corella A, Coleman IM, Meers MP, Christiani AJ, Pierce C, Janssens DH, Meade HE, Bose A, Brady L, Howard T, De Sarkar N, Frank SB, Dumpit RF, Dalton JT, Corey E, Plymate SR, Haffner MC, Mostaghel EA, Nelson PS, Selective androgen receptor modulators activate the canonical prostate cancer androgen receptor program and repress cancer growth, J. Clin. Invest. 131 (10) (2021) e146777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, Qiu X, Cañadas I, Roehle K, Heckler M, Calagua C, Ye H, Pantelidou C, Galbo P, Panja S, Mitrofanova A, Wilkinson S, Whitlock NC, Trostel SY, Hamid AA, Kibel AS, Barbie DA, Choudhury AD, Pomerantz MM, Sweeney CJ, Long HW, Einstein DJ, Shapiro GI, Dougan SK, Sowalsky AG, He HH, Freedman ML, Balk SP, Loda M, Labbé DP, Olson BM, Ellis L, EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer, Nat. Cancer 2 (April (4)) (2021) 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA, An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors, Nat. Genet. 40 (5) (2008) 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]