Abstract
Schizophrenia is a serious mental disorder with considerable somatic and psychiatric morbidity. It is unclear whether comorbid health conditions predominantly arise due to shared genetic risk or consequent to having schizophrenia. To explore the contribution of genetic risk for schizophrenia, we analysed the effect of schizophrenia polygenic risk scores (PRS) on a broad range of health problems in 406 929 individuals with no schizophrenia diagnosis from the UK Biobank. Diagnoses were derived from linked health data including primary care, hospital inpatient records, and registers with information on cancer and deaths. Schizophrenia PRS were generated and tested for associations with general health conditions, 16 ICD10 main chapters, and 603 diseases using linear and logistic regressions. Higher schizophrenia PRS was significantly associated with poorer overall health ratings, more hospital inpatient diagnoses, and more unique illnesses. It was also significantly positively associated with 4 ICD10 chapters: mental disorders; respiratory disease; digestive disease; and pregnancy, childbirth and the puerperium, but negatively associated with musculoskeletal disorders. Thirty-one specific phenotypes were significantly associated with schizophrenia PRS, and the 19 novel findings include several musculoskeletal diseases, respiratory diseases, digestive diseases, varicose veins, pituitary hyperfunction, and other peripheral nerve disorders. These findings extend knowledge of the pleiotropic effect of genetic risk for schizophrenia and offer insight into how some conditions often comorbid with schizophrenia arise. Additional studies incorporating the genetic basis of hormone regulation and involvement of immune mechanisms in the pathophysiology of schizophrenia may further elucidate the biological mechanisms underlying schizophrenia and its comorbid conditions.
Introduction
Individuals with schizophrenia are at increased risk for numerous additional health problems, including both psychiatric disorders and somatic diseases. Anxiety, depression, obsessive-compulsive disorder (OCD), post-traumatic stress disorder (PTSD), eating disorders, and substance abuse disorders are all observed more often in individuals with schizophrenia compared with the general population 1–3. Moreover, numerous non-psychiatric disorders are also elevated in individuals with schizophrenia including: stroke, diabetes mellitus, influenza or pneumonia, chronic obstructive pulmonary disease (COPD), autoimmune disorders, metabolic disturbances, and dementia 4–7. However, the etiology of most of these comorbidities remains elusive. Although there has been growing evidence recently suggesting SNP-based genetic correlations between schizophrenia and multiple traits using linkage disequilibrium score regression (LDSC) 8, 9, questions remain regarding the cause of commonly comorbid conditions due to the difficulties in quantifying a dose-response relationship from LDSC. Furthermore, it remains unclear to what extent the comorbidities stem from underlying shared genetic risk or arise consequent to having schizophrenia (e.g., from poor dietary habits or antipsychotic medication effects).
Strong evidence supports an important role of genetics in the etiology of schizophrenia with heritability estimated at 64%−81% 10, 11. Numerous common genetic variants, copy number variants (CNVs), rare genetic variants, and their interactions are all found to contribute to the risk for schizophrenia 12–16. Although the contribution of single genetic variants or CNVs to schizophrenia liability is small (e.g., CNVs explain 0.85% of the variance) 13, the accumulation of common variants i.e., polygenic risk scores (PRS) are robustly associated with schizophrenia and explains a significant portion of variance (increased from 3.2% to 22.1%) 12, 17–19. Thus, PRS provide a tool to quantify and index the genetic risk of schizophrenia in investigations of comorbidity.
The pleotropic effect of schizophrenia PRS on psychiatric disorders and other medical syndromes was observed in a patient population from the US electronic health system 20. Even with limited follow-up time and a selected patient sample which included schizophrenia patients, schizophrenia PRS was found to be positively associated with several psychiatric phenotypes including anxiety, mood, and substance use disorders, which previously demonstrated significant positive genetic correlation with schizophrenia 21–24. Obesity was found to be inversely related to schizophrenia PRS, also supporting a prior negative genetic effect 20. Moreover, most of the phenotypes remained significant after adjusting for schizophrenia status.
Restricting analyses to individuals without schizophrenia may provide insight into whether the relationships between genetic risk for schizophrenia and other health outcomes are due to the direct effect of shared genetic risk or indirect effects from medications and lifestyle differences that follow from having schizophrenia. This approach is central to providing a better understanding of the biological mechanisms giving rise to comorbid conditions.
The present study uses large datasets for both a wide range of phenotypes and the schizophrenia PRS generation to investigate the influence of schizophrenia PRS (as an index of genetic risk for schizophrenia) on various health outcomes in individuals without a schizophrenia diagnosis. Specifically, we investigated the associations between schizophrenia PRS and general health, broad domains of medical conditions indexed by ICD10 main chapters, and a wide range of psychiatric and somatic illnesses in over 400 000 individuals with no schizophrenia diagnosis in UK Biobank.
Materials and Methods
Data Source and Study Population
The UK Biobank is a large-scale biomedical research resource consisting of over 500 000 participants aged 39–73 years recruited from England, Wales, and Scotland between 2006 and 2010 25. The linkage to the health-related records including primary care, hospital in-patient records, cancer register data, and death records makes it possible to investigate a broad range of health outcomes.
Of 488 264 individuals with available genotype data and linked health data, we restricted analyses to individuals with consistent sex information and White-British ancestry determined on the basis of self-report and genetic data 25. Individuals (N=2087) with any record of a schizophrenia diagnosis or other type of psychosis or psychotic illness from primary care records, hospital in-patient records, and/or death records demonstrated a 0.53 (95% CI: 0.48–0.58, p-value: 2. 65×10−93) significantly higher schizophrenia PRS than those without these conditions. They were excluded from further analyses, yielding a final sample of 406 929 individuals.
The use of these data has been approved by the research ethics committee of UK Biobank. All participants included in this study have provided informed consent when enrolled in UK Biobank. This research was conducted under application number 54928.
Schizophrenia-PRS
The PRS were computed using summary statistics from the PGC schizophrenia GWAS as the discovery dataset 12 using PLINK1.9. Detailed information for genotyping and quality control of UK Biobank data was described previously 26. SNP quality control, including filtering for minor allele frequency (MAF>0.10) and imputation quality (INFO>0.9), resulted in a subset of 5,471,645 SNPs. These were clumped into clusters of approximate linkage disequilibrium (R2<0.1, within 1000kb distance). The classic thresholding + clumping approach was used to generate PRS with 10 different p-value thresholds to select genetic variants, p < 5e-08, p < 1e-06, p < 1e-04, p < 0.001, p < 0.01, p < 0.05, p < 0.1, p < 0.2, p < 0.5, and p < 1 27.
To consolidate predictive power across PRS thresholds and minimize the multiple testing burden, we performed principal component analysis (PCA) on the set of ten PRS 28 in R, version 4.0.3. The first principal component, PRS-PC1, was then standardized and used for further association testing.
Assessment of outcomes
The linked health data cover primary care data (Read code), hospital inpatient data (ICD-9 and ICD-10 codes), cancer register records (ICD-9 and ICD-10 codes), death register records (ICD-10 codes), and self-reported medical condition codes disclosed at baseline or follow-up 29–31. A wide range of health outcomes across the linked health data included self-reported phenotypes have been mapped into a 3-digit code of ICD-10 as described in the UK Biobank document 31. Together with the records from the cancer register, around 1200 unique 3-character ICD10 codes covering ICD10 chapters I to XVII were identified to indicate the first occurrence of health outcomes for each individual. The mapped ICD10 codes were then converted into phenotype codes (phecodes) which were considered to be better aligned with the terminology used in clinical practice 32, 33. A total of 768 phecodes were matched in the current study.
Outcomes of interest for this study from broad to narrow definitions were obtained from the linked health data including general health, ICD10 main chapters corresponding to phenotypic groupings, and specific somatic and psychiatric illnesses (phecodes). General health measures included self-report overall health rating at baseline (scored 1–4, where a higher number indicates poorer self-rated overall health), total number of all hospital inpatient diagnoses received, total number of all primary care diagnoses received, and total number of all unique phecodes identified throughout the coverage period of UK Biobank registers 29, 30. In order to enhance the generalizability of the results, we removed a small number of people with extremely high numbers of diagnoses. The upper limit of total number of all hospital in-patient diagnoses received was therefore set to 100. The upper limit for primary care diagnoses was set to 1500, and the upper limit for unique phecodes was set to 60 (Figure S1). In the next stage of health outcomes, participants who ever received at least one relevant diagnosis in a certain ICD-10 main chapter were defined as cases for that chapter. Thus, 17 binary variables for each of the ICD-10 main chapters were created following the same procedures. When narrowing down to the specific somatic and psychiatric illnesses, participants with at least one relevant ICD-10 code mapped to a phecode were defined as cases, whereas individuals without the phecode of interest were defined as controls. After excluding phecodes with fewer than 200 cases 34, a total of 603 phecodes were retained for further analyses (Supplementary Excel Sheet 1).
Statistical Analyses
Data management and analyses were performed using R, version 4.0.3, and PRS were computed using PLINK1.9. In the first stage of the analysis, linear regressions (the ‘drgee’ package in R) were used for overall health rating at baseline, total number of all hospital inpatient diagnoses received, total number of all primary care diagnoses received, and total number of all unique illnesses identified from the linked health data adjusting for birth year, sex, attained age, region of the UK (England, Scotland, or Wales), and the first 10 genetic principal components. Regression coefficients with 95% confidence intervals (CIs) and corresponding p-values were reported. In these linear regressions we used robust (“sandwich”) standard errors, which avoid assuming normality and homoscedasticity of the outcomes 35.
In the second stage of the analysis, logistic regressions (the ‘drgee’ package in R) were used for 17 ICD10 main chapters (except for chapter XVI, due to low prevalence of cases) while adjusting for birth year, sex, attained age, region, and the first 10 genetic principal components. Odds ratios (ORs) with 95% CIs were reported as risk estimates for ICD10 main chapters per unit increase of PRS-PC1.
In the third stage of the analysis, we conducted a PheWAS for PRS-PC1, using all phecodes with at least 200 cases. Logistic regressions were carried out through the ‘PheWAS’ package in R of each phecode against the PRS-PC1 adjusting for sex, birth year, attained age, region, and 10 genetic principal components provided by UK Biobank.
For all models, the regression analyses were restricted to participants with complete information on all variables in the model resulting in slight differences in the sample size of each analysis (Table S1). Correction for multiple tests was implemented using Bonferroni corrections (p<0.003 for ICD main chapters, p<8.29×10−5 for phecodes).
Despite removing individuals with schizophrenia or other psychotic illnesses from analyses, a number of people remained who had been prescribed antipsychotic medications (N=9 687). In order to investigate the extent to which individuals using antipsychotic medications impact the results, we performed further sensitivity analyses in a sub-population with General Practitioner (GP) registration records (N=157 018), and with available GP prescription information (N=69 294) using the same regression models as above and additionally adjusting for antipsychotic use in the models. Since 17.9% of the 3-character codes were not able to be matched to phecodes, we also performed a sensitivity analysis using original ICD-9 and ICD-10 codes from hospital inpatient records as an internal validation.
Furthermore, due to the genetic correlation between psychiatric disorders36, 37, we performed a sensitivity analysis additionally adjusting for mental and behavioral disorder (chapter V) status for all the other ICD10 main chapters to check whether the associations between schizophrenia PRS and those chapters are mediated by mental disorders.
To examine the degree of correlation between the effect estimates from the previous study 20 and ours, we ran a Pearson’s correlation test of ORs between our main PheWAS results and the sensitivity analysis (Phecode 295 cases excluded) results from the previous paper matched by phecodes. Pearson’s correlation coefficient with 95% CI and p value were reported.
Code availability
All code used for data preparation and analysis are available upon request.
Results
The demographic and clinical characteristics of the study population are presented in Table 1. Of the 406 929 UKB individuals, 54.1% were females. The average attained age was 56.9 years. All individuals with available primary care records and hospital inpatient information had collectively received over 101.4 million primary care diagnoses and 6.8 million hospital inpatient diagnoses. The prevalence of cases for each of the ICD10 main chapters is presented in Figure S2.
Table 1.
Descriptive characteristics of the study population
| N =406 929 | |
|---|---|
| Sex | |
| Female (%) | 220 153 (54.1) |
| Male (%) | 186 776 (45.9) |
| Birth year (mean±SD ) | 1951±8.0 |
| Attained age 1 (mean±SD ) | 56.9±8.0 |
| Region of the UK | |
| England (%) | 296 248 (72.8) |
| Scotland (%) | 25 768 (6.3) |
| Wales (%) | 13 868 (3.4) |
| Missing (%) | 71 045 (17.5) |
| Self-report overall health rating | |
| 1 = Excellent (%) | 67 672 (16.6) |
| 2 = Good (%) | 238 009 (58.5) |
| 3 = Fair (%) | 82 998 (20.4) |
| 4 = Poor (%) | 16 826 (4.1) |
| Missing (%) | 1 424 (0.4) |
| Total inpatient diagnoses | 6 881 462 |
| Total primary care diagnoses | 101 441 015 |
Attained age is defined as the participants’ age at their enrollment in the UK Biobank.
Associations of SCZ PRS-PC1 with general health conditions
The estimated differences with 95% CIs and corresponding p-values for overall health rating, cumulative measurements including total inpatient diagnoses, total primary care diagnoses, and total unique phecodes per standard deviation increase of schizophrenia PRS-PC1 are presented in Table 2.
Table 2.
Linear regression of general health conditions and schizophrenia PRS-PC1
| Regression coefficient 5 (95% CI) | p value | |
|---|---|---|
| Overall health rating 1 | 0.02 (0.013, 0.018) | 2.02×10−32 |
| Total diagnoses | ||
| Hospital inpatient 2 | 0.12 (0.06, 0.19) | 5.24×10−5 |
| Primary care 3 | 0.80 (−0.89, 2.49) | 0.35 |
| Unique phecodes 4 | 0.09 (0.05, 0.13) | 3.03×10−5 |
Number of individuals with complete information for each analysis: N1=334 586; N2=324 511; N3=151 643; N4=324 493
Effect sizes are in standard deviations (SDs) of PRS-PC1.
Higher schizophrenia PRS-PC1 was associated with poorer self-reported overall health rating (coef= 0.02, P =2.02 × 10−32). One standard deviation of schizophrenia PRS-PC1 was significantly associated with receiving 0.12 more inpatient diagnoses (P= 5.24 × 10−5) and 0.09 more unique diagnoses (P=3.03 × 10−5) on average. A similar association was also detected for higher schizophrenia PRS-PC1 and receiving more total primary care diagnoses (coef= 0.80, P=0.35), albeit not statistically significant, which might be due to the smaller sample size (Table S1).
Primary associations of SCZ PRS-PC1 with ICD10 main chapters
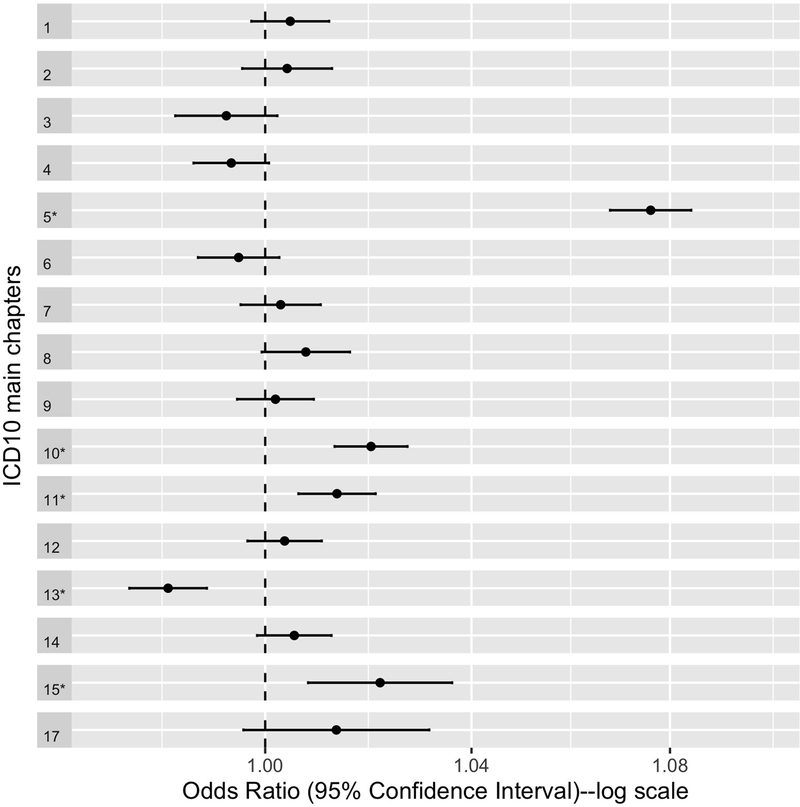
We tested the associations between schizophrenia PRS-PC1 and 16 ICD10 main chapters for 335 874 individuals with complete information on mapped ICD10 codes from primary care data, hospital inpatient data, cancer register records, and death register records, and all covariates. Chapter XVI was not tested for association due to the extremely low prevalence of diagnoses in the study population (Figure S2). Figure 1 presents the ORs with 95% confidence intervals for ICD10 main chapters corresponding to 1 standard deviation increase in schizophrenia PRS-PC1.
Figure 1. Odds ratios for ICD10 main chapters corresponding to 1 standard deviation increase in schizophrenia PRS-PC1.
The vertical line (odds ratio=1) reflects no change in risk. Error bars indicate 95% confidence intervals. * indicates significant associations after Bonferroni correction (p value <0.003). Chapter names corresponding to chapter numbers: 1= Certain infectious and parasitic diseases; 2 = Neoplasms; 3 = Blood and blood-forming organs and certain disorders involving the immune mechanism; 4 = Endocrine, nutritional and metabolic diseases; 5 = Mental and behavioral disorders; 6 = Nervous system disorders; 7 = Eye and adnexa disorders; 8 = Ear and mastoid process disorders; 9 = Circulatory system disorders; 10 = Respiratory system disorders; 11 = Digestive system disorders; 12 = Skin and subcutaneous tissue disorders; 13 = The musculoskeletal system and connective tissue disorders; 14 = The genitourinary system disorders; 15 = Pregnancy, childbirth and the puerperium; 17 = Congenital malformations, deformations and chromosomal abnormalities.
Five ICD10 main chapters remained statistically significant after Bonferroni correction (P<0.003) (Table S2). The strongest and most significant association with schizophrenia PRS-PC1 was Chapter V, mental and behavioral disorders (OR = 1.08, SE = 0.004, P < 2 × 10−16), followed by Chapter X, diseases of the respiratory system (OR = 1.02, SE = 0.004, P = 1.24 × 10−8), Chapter XV, pregnancy, childbirth and the puerperium (OR = 1.02, SE = 0.007, P = 1.76 × 10−3), and Chapter XI, diseases of the digestive system (OR = 1.01, SE = 0.004, P = 2.68 × 10−4). We observed a significant negative association between schizophrenia PRS-PC1 and Chapter XIII, diseases of the musculoskeletal system and connective tissue (OR = 0.98, SE = 0.004, P < 9.08 × 10−7).
PheWAS of SCZ PRS-PC1
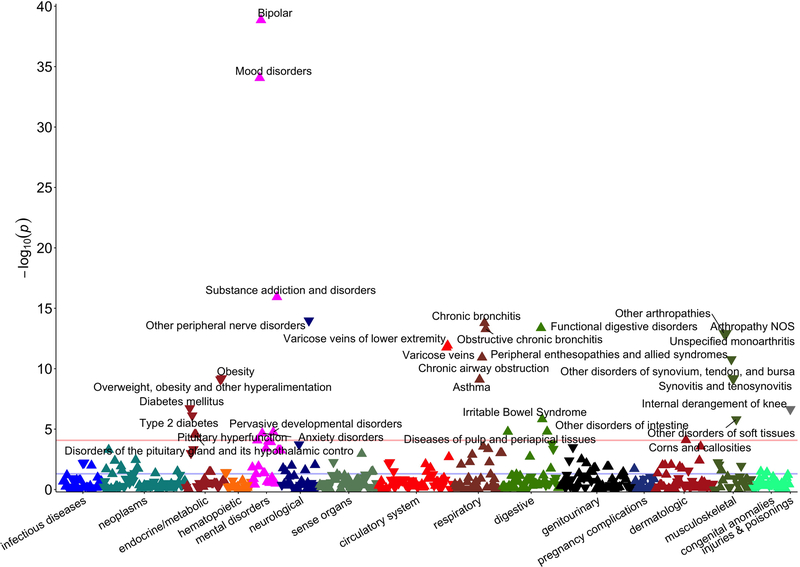
Figure 2 shows the association between 603 phenotypes and schizophrenia PRS-PC1.
Figure 2. Schizophrenia PRS-PC1 phenome-wide association study Manhattan plot.
This depicts association results for schizophrenia PRS-PC1 in UK Biobank (603 phenotypes, 325 992 individuals). The horizontal axis indicates broad disease category and the vertical axis indicates the significance (–log10 p) of the association derived by logistic regression. The horizontal red line within the graph indicates phenome-wide significance (p=8.29×10−5) using Bonferroni correction, and all phenotypes that pass this threshold are labeled. The vertex of each triangle represents the direction of the association.
Thirty-one phenotypes including 7 musculoskeletal, 6 endocrine/metabolic, 5 mental disorders, 4 respiratory, 4 digestive, 2 circulatory system, 1 neurological, 1 dermatologic, and 1 injuries & poisonings phenotypes were significantly associated with schizophrenia PRS-PC1 after Bonferroni correction for the 603 phenotypes tested (P < 8.29 × 10−5). Bipolar disorders yielded the most significant association with the schizophrenia PRS-PC1 (Ncases = 1323, OR = 1.45, SE = 0.03, P = 1.39 × 10−39), followed by mood disorders (Ncases = 2623, OR = 1.28, SE = 0.02, P = 8.56 × 10−35), substance addiction and disorders (Ncases = 8851, OR = 1.10, SE = 0.01, P = 1.17 × 10−16), other peripheral nerve disorders (Ncases = 22 987, OR = 0.95, SE = 0.01, P = 1.05 × 10−14), and chronic bronchitis (Ncases = 17 046, OR = 1.06, SE = 0.01, P = 1.64 × 10−14). Corresponding to the primary associations with ICD10 main chapters, all four phenotypes within the functional digestive disorders group (Ncases = 46 351, OR = 1.04, SE = 0.006, P = 1.16 × 10−13) were significantly positively associated with schizophrenia PRS-PC1. Negative associations were observed for all seven significant phenotypes from the musculoskeletal group under the other arthropathies heading (Ncases = 34 687, OR = 0.96, SE = 0.006, P = 1.16 × 10−13). Five additional phenotypes including obesity, overweight, diabetes mellitus, internal derangement of knee, and type 2 diabetes were significantly inversely associated with schizophrenia PRS-PC1. Although no statistically significant evidence was found in the primary association with the circulatory system chapter, varicose veins (Ncases = 20111, OR = 1.05, SE = 0.007, P = 1.74 × 10−12) were found to be significantly positively associated with schizophrenia PRS-PC1 after Bonferroni correction. The complete results table with effect sizes for all significant phenotypes can be found in the Supplementary Excel Sheet 1.
Sensitivity analysis
As an internal validation, we performed a sensitivity analysis using original ICD-9 and ICD-10 codes from hospital inpatient records to assist mapping the 17.9% previously unmatched 3-character codes. In 332 855 individuals with available information, 43 phenotypes remained significant after Bonferroni correction for 1214 phenotypes tested (P <4.12 × 10−5) (Supplementary Excel Sheet 2). Nearly all associations remained significant with minor variability for most phenotypes. Nonetheless, mental disorders, respiratory, musculoskeletal, and digestive were still the groups with the most significant associated phenotypes (Figure S3A).
We also explored whether some of the observed associations might be mediated through antipsychotic use via sensitivity analyses additionally adjusting for antipsychotic use in the models. In the sensitivity analysis of a sub-population of 157 018 individuals with GP registration records, 22 phenotypes remained significant after Bonferroni correction for 532 phenotypes tested (P< 9.40 × 10−5) additionally adjusting for antipsychotic use (Supplementary Excel Sheet 3). And in a smaller sub-population of 69 294 individuals with GP prescription records, 4 phenotypes remained significant after Bonferroni correction for 427 phenotypes tested (P < 1.17 × 10−4) (Supplementary Excel Sheet 4). Bipolar, mood disorders, other disorders of synovium, tendon, and bursa, and synovitis and tenosynovitis remained significant in every sensitivity analysis (Figure S3B & S3C). Associations with diabetes, anxiety, irritable bowel syndrome, and other phenotypes which were statistically significant in the main analysis were less robust and significant after adjusting for antipsychotic medication use. However, they consistently remained highly ranked within the results (Supplementary Excel Sheet 3 & 4).
After adjusting for mental disorder status, four ICD10 main chapters remained statistically significant after Bonferroni correction (P<0.003) (Figure S4). Chapter XIII, diseases of the musculoskeletal system and connective tissue, was still strongly negatively associated with schizophrenia PRS, while chapter III (Blood and blood-forming organs and certain disorders involving the immune mechanism, IV (Endocrine, nutritional and metabolic diseases), VI (Nervous system disorders) were also found to be significantly negatively associated with schizophrenia PRS independent of mental disorder status. Other chapters which were associated with schizophrenia PRS in the main analyses including chapter X (Respiratory system disorders), XI (Digestive system disorders), and XV (Pregnancy, childbirth and the puerperium diseases) were no longer significant after adjusting for mental disorder status after Bonferroni correction, even though the effect sizes were only slightly diminished.
Compared with the previous study 20, there was a significant positive correlation between the effect estimates (r= 0.25). On average, the effect estimates from our main analysis were stronger than previous findings (Figure S5).
Discussion
We investigated associations between schizophrenia PRS and medical conditions from broad to narrow definitions in over 400,000 individuals with no schizophrenia diagnosis in UK Biobank. Our results demonstrate that individuals with higher schizophrenia PRS were more likely to have poorer self-reported overall health ratings, receive more hospital inpatient diagnoses, and suffer more unique diseases. In addition to mental and behavioral disorders, schizophrenia PRS was also found to be significantly associated with four other ICD10 chapters: respiratory system disorders; digestive system disorders; musculoskeletal system and connective tissue disorders; and pregnancy, childbirth and the puerperium. Moreover, we identified 31 diseases and disorders (out of 603) significantly associated with schizophrenia PRS from the PheWAS analysis with ORs ranging from 1.02–1.41 for positive associations and 0.94–0.97 for negative associations, revealing greater pleiotropic effects of schizophrenia PRS than previously known.
The strongest associations we found were within the mental and behavioral disorder chapter including bipolar disorder, mood disorder, substance addiction and disorders, and anxiety disorders, which is in line with the previous reports of positive genetic correlations and shared genetic risk loci with schizophrenia 21–23. These findings indicate that genetic factors that are not specific to schizophrenia contribute to the occurrence of the other disorders, supporting the concept of a general psychopathology factor 38. Prior work attributed associations with several other phenotypes within this chapter, including neurological disorders, personality disorders, suicidal behavior, and memory loss, to being consequences of a schizophrenia diagnosis rather than to shared risk 20. However, in our analysis, restricting to hospital inpatient diagnoses (which incorporates additional phenotypes and probably more severe cases), all significant associations except for memory loss were replicated in individuals with no schizophrenia diagnosis (Supplementary Excel Sheet 2, Figure S3A), implying that shared genetics may play a role in the development of neurological disorders, personality disorders, and suicidal behavior, but not memory loss.
Novel associations were detected between schizophrenia PRS and respiratory diseases including chronic bronchitis, chronic airway obstruction, and asthma. Prior studies had reported elevated risk of chronic bronchitis and COPD in individuals with schizophrenia 39, 40. Moreover, asthma was previously found to be associated with later risk of developing schizophrenia 41. It is known that the rate of smoking is markedly higher in individuals with schizophrenia, which might mediate some associations between schizophrenia and respiratory diseases 39, 42. Notably, the previously reported genetic correlation between smoking behaviors 9 and our observed positive association between tobacco use and schizophrenia PRS in individuals with no schizophrenia diagnosis (Figure S3A) indicates that the association between schizophrenia PRS and respiratory diseases might be mediated by smoking. However, the analyses in the subpopulation with available tobacco smoking information from our study population (24%) suggested that the direct effect of schizophrenia PRS on respiratory diseases exists independent of tobacco smoking (Table S3). Furthermore, asthma and other respiratory diseases are linked to the immune system and inflammation 43. The earliest and strongest GWAS associations for schizophrenia have been in genomic loci with important immune functions 12, 17. Shared immune mechanisms might partly explain the association between schizophrenia PRS and asthma.
The associations between schizophrenia PRS and digestive disorders found in our study are also novel. Specifically, functional digestive disorder, irritable bowel syndrome (IBS), disease of pulp and periapical tissues, and other disorders of the intestine were significantly positively associated with schizophrenia PRS. Substantial gastrointestinal comorbidities in schizophrenia and increased psychiatric comorbidities in individuals with gastrointestinal disorders, including IBS, have been reported previously 44–47. Consistent with previous findings, our results provide evidence for shared genetic factors between schizophrenia and digestive disorders.
Endocrine and metabolic diseases were not found to be significantly associated with schizophrenia PRS at the chapter level. However, significant associations with opposite directions were detected for specific diseases. The positive phenotypic association despite the intriguing negative genetic correlation between schizophrenia and obesity/diabetes has been described in several studies previously 20, 48–51. Our results corroborate the negative genetic associations between schizophrenia PRS and obesity, overweight, and diabetes with individual-level phenotypic data, suggesting the increased rates of obesity and diabetes among individuals with schizophrenia arise consequent to having this disorder (e.g., medication effects). Further exploration of this paradoxical relationship may assist case definition and treatment improvement for both traits.
Additionally, novel positive associations were observed for pituitary hyperfunction and disorders of the pituitary gland and its hypothalamic control with schizophrenia PRS. Larger pituitary volume, which is thought to reflect a greater activation of the hypothalamic–pituitary–adrenal (HPA) axis 52, 53, has been previously reported in individuals with psychosis 54. Our findings provide evidence for a genetic link between schizophrenia and HPA axis activation. Future studies focusing on the genetic basis of hormone regulation in the pathophysiology of schizophrenia may provide new insights into mechanisms which could be leveraged for treatment development for schizophrenia.
Remarkable negative associations between schizophrenia PRS and musculoskeletal disease were observed in our study. The inverse association between rheumatoid arthritis (RA) and non-inflammatory disease of the joints (e.g., osteoarthritis) and schizophrenia has been described in several epidemiological studies 55–57. One potential explanation for the negative phenotypic association is that individuals with schizophrenia may expose their joints to less damage and subsequent osteoarthritis since they are more likely to be sedentary 58. However, our results favor an etiological link between schizophrenia and musculoskeletal diseases on a genetic level. RA is an immune-mediated inflammatory joint disease for which the MHC region is the most important genetic locus 59. As mentioned, previous schizophrenia GWAS also observed MHC associations enriched in tissue with important immune functions 12. Our results expand the previous findings indicating a possible involvement of immune mechanisms in the pathophysiology of schizophrenia.
The main strengths of this study are the large sample size with long medical follow-up and high-quality phenotype definitions, which enabled screening of a large number of medical conditions and greater reliability for the results. Our design of testing medical conditions from a broad to narrow definitions makes it less constrained by phenotype selection, yielding greater opportunities to identify disease associations despite an incomplete understanding of mechanistic relationships. Moreover, associations based on genotypes fixed from birth renders reverse causality virtually impossible. Furthermore, restricting analyses to individuals with no schizophrenia diagnosis allows us to investigate the direct effect of schizophrenia PRS independent of the disorder, which provides new insights into the etiology of comorbid conditions as well as previously undocumented associations. In addition, compared to the methods using summary statistics from GWAS (e.g., LDSC) to assess genetic correlation, associations based on schizophrenia PRS enable measurement of dose-response relationships with the possibility of adjusting for environmental confounders. This approach offers individual indices relevant to the etiology which might assist future application of PRS testing in clinical practice.
There are several limitations to consider. First, the coverage period of diagnosis records varied from 20–38 years across different registers. However, the coverage period highly correlated with the region of UK, which was adjusted for in all analysis. We also used all possible diagnoses from different sources including primary and secondary care, cancer, and death registers to define phenotypes, which makes under-detection less likely for most of the diseases. Second, around 18% of the 3-character harmonized codes from multiple registers were not able to be matched to phecodes, but the results using original ICD-9 and ICD-10 codes from hospital inpatient records only (which results in more severe cases) were still robust (Figure S3A). Third, less than 50% of the individuals had available GP information and only 20% had recorded prescriptions. Nevertheless, the results adjusting for antipsychotic use were still largely consistent despite diminished power and concomitant changes in significance (Figure S3B & S3C, Supplementary Excel Sheet 3 & 4). Fourth, collider bias could be a potential concern due to restricting the analyses to individuals with no schizophrenia diagnosis or adjusting for mental disorder status in the sensitivity analysis 60. This could happen if a common cause (other than shared genetics) of both schizophrenia/mental disorders and the tested outcome phenotype/chapter exists. Fifth, since all UK Biobank participants were middle-aged at enrollment, we were not able to capture diseases that commonly occur in early life. Moreover, since our analyses were restricted to individuals with White-British ancestry, the generalizability to individuals of other ancestries merits further exploration. Furthermore, although most of the individuals in the study cohort have passed the risk period of schizophrenia, there is still a small possibility that some people with schizophrenia or related disorders remain undetected. However, given that participants in UK Biobank were found to be healthier and wealthier than the general UK population 61, the number of individuals with undetected schizophrenia or other types of psychotic illness is probably extremely low and unlikely to impact the results. On the other hand, individuals with severe diseases or who died soon after diagnosis were likely not captured by the UK Biobank. The interaction between schizophrenia PRS and education level or socioeconomic status on medical conditions may deserve further investigation.
By studying associations between schizophrenia PRS and multiple medical conditions in ~400 000 individuals with no schizophrenia diagnosis in the UK Biobank, we found that higher genetic loading for schizophrenia was associated with poorer overall health and higher cumulative somatic and psychiatric disease burden. In addition to the replication of associations with mental illness, obesity, diabetes, and synovitis, we also observed novel associations between schizophrenia PRS and respiratory diseases, musculoskeletal diseases, digestive diseases, pituitary hyperfunction, and varicose veins. Additional studies incorporating the genetic basis of hormone regulation and involvement of immune mechanisms in the pathophysiology of schizophrenia are needed to better understand the underlying etiology of schizophrenia, which might provide new insights facilitating treatment development. Further investigations examining the effect of potential mediating factors including smoking, alcohol consumption, and/or socioeconomic status may assist future application of schizophrenia PRS in disease prediction or clinical practice.
Supplementary Material
Acknowledgments
This work is supported by the US National Institute of Mental Health to SEB (R21MH116188). RZ receives support from the Chinese Scholarship Council (grant number CSC201700260258); CMB is supported by NIMH (R01MH120170; R01MH119084; R01MH118278; U01MH109528); Brain and Behavior Research Foundation Distinguished Investigator Grant; Swedish Research Council (Vetenskapsrådet, award: 538-2013-8864); Lundbeck Foundation (Grant no. R276-2018-4581).
We acknowledge and thank Prof. Patrick Sullivan for advice regarding the study design. Participation of the UK Biobank subjects is gratefully appreciated. We also acknowledge UK Biobank team for collecting and preparing data for analyses.
Conflict of interest
CM Bulik reports: Shire (grant recipient, Scientific Advisory Board member); Idorsia (consultant); Lundbeckfonden (grant recipient); Pearson (author, royalty recipient).
Footnotes
Supplementary information is available at MP’s website
References
- 1.Buckley PF, Miller BJ, Lehrer DS, Castle DJ. Psychiatric comorbidities and schizophrenia. Schizophr Bull 2009; 35(2): 383–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braga RJ, Reynolds GP, Siris SG. Anxiety comorbidity in schizophrenia. Psychiatry Res 2013; 210(1): 1–7. [DOI] [PubMed] [Google Scholar]
- 3.Cantor-Graae E, Nordstrom LG, McNeil TF. Substance abuse in schizophrenia: a review of the literature and a study of correlates in Sweden. Schizophr Res 2001; 48(1): 69–82. [DOI] [PubMed] [Google Scholar]
- 4.Crump C, Winkleby MA, Sundquist K, Sundquist J. Comorbidities and Mortality in Persons With Schizophrenia: A Swedish National Cohort Study. American Journal of Psychiatry 2013; 170(3): 324–333. [DOI] [PubMed] [Google Scholar]
- 5.Benros ME, Eaton WW, Mortensen PB. The epidemiologic evidence linking autoimmune diseases and psychosis. Biol Psychiatry 2014; 75(4): 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert TJ, Velakoulis D, Pantelis C. Medical comorbidity in schizophrenia. Med J Aust 2003; 178 Suppl: S67–70. [DOI] [PubMed] [Google Scholar]
- 7.Ku H, Lee EK, Lee KU, Lee MY, Kwon JW. Higher prevalence of dementia in patients with schizophrenia: A nationwide population-based study. Asia Pac Psychiatry 2016; 8(2): 145–153. [DOI] [PubMed] [Google Scholar]
- 8.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47(11): 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartz SM, Horton AC, Hancock DB, Baker TB, Caporaso NE, Chen LS et al. Genetic correlation between smoking behaviors and schizophrenia. Schizophr Res 2018; 194: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373(9659): 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60(12): 1187–1192. [DOI] [PubMed] [Google Scholar]
- 12.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 2017; 49(1): 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergen SE, Ploner A, Howrigan D, O’Donovan MC, Smoller JW, Sullivan PF, et al. Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. American Journal of Psychiatry 2019; 176(1): 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gratten J Rare variants are common in schizophrenia. Nat Neurosci 2016; 19(11): 1426–1428. [DOI] [PubMed] [Google Scholar]
- 16.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 2016; 19(4): 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460(7256): 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 2014; 55(10): 1068–1087. [DOI] [PubMed] [Google Scholar]
- 19.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Walters JT, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv 2020: 2020.2009.2012.20192922. [Google Scholar]
- 20.Zheutlin AB, Dennis J, Karlsson Linnér R, Moscati A, Restrepo N, Straub P et al. Penetrance and Pleiotropy of Polygenic Risk Scores for Schizophrenia in 106,160 Patients Across Four Health Care Systems. Am J Psychiatry 2019; 176(10): 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360(6395). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018; 359(6376): 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381(9875): 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mistry S, Harrison JR, Smith DJ, Escott-Price V, Zammit S. The use of polygenic risk scores to identify phenotypes associated with genetic risk of schizophrenia: Systematic review. Schizophr Res 2018; 197: 2–8. [DOI] [PubMed] [Google Scholar]
- 25.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562(7726): 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv 2017: 166298. [Google Scholar]
- 27.Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 2020; 15(9): 2759–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coombes BJ, Ploner A, Bergen SE, Biernacka JM. A principal component approach to improve association testing with polygenic risk scores. Genet Epidemiol 2020; 44(7): 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.UK Biobank primary care linked data version 1.0. https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/primary_care_data.pdf, 2019, Accessed Date Accessed 2019 Accessed.
- 30.UK Biobank hospital inpatient data version 3.0. https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/HospitalEpisodeStatistics.pdf, 2020, Accessed Date Accessed 2020 Accessed.
- 31.UK Biobank first occurrence of health outcomes defined by 3-character ICD10 code. https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/first_occurrences_outcomes.pdf, 2019, Accessed Date Accessed 2019 Accessed.
- 32.Wei WQ, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 2017; 12(7): e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Inform 2019; 7(4): e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA et al. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinformatics 2018; 19(1): 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saul BC, Hudgens MG. The Calculus of M-Estimation in R with geex. J Stat Softw 2020; 92(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45(9): 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019; 179(7): 1469–1482.e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S et al. The p Factor: One General Psychopathology Factor in the Structure of Psychiatric Disorders? Clin Psychol Sci 2014; 2(2): 119–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zareifopoulos N, Bellou A, Spiropoulou A, Spiropoulos K. Prevalence of Comorbid Chronic Obstructive Pulmonary Disease in Individuals Suffering from Schizophrenia and Bipolar Disorder: A Systematic Review. Copd 2018; 15(6): 612–620. [DOI] [PubMed] [Google Scholar]
- 40.Partti K, Vasankari T, Kanervisto M, Perälä J, Saarni SI, Jousilahti P et al. Lung function and respiratory diseases in people with psychosis: population-based study. Br J Psychiatry 2015; 207(1): 37–45. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen MS, Benros ME, Agerbo E, Børglum AD, Mortensen PB. Schizophrenia in patients with atopic disorders with particular emphasis on asthma: a Danish population-based study. Schizophr Res 2012; 138(1): 58–62. [DOI] [PubMed] [Google Scholar]
- 42.Lohr JB, Flynn K. Smoking and schizophrenia. Schizophr Res 1992; 8(2): 93–102. [DOI] [PubMed] [Google Scholar]
- 43.Holtzman MJ. Asthma as a chronic disease of the innate and adaptive immune systems responding to viruses and allergens. J Clin Invest 2012; 122(8): 2741–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fadgyas-Stanculete M, Buga AM, Popa-Wagner A, Dumitrascu DL. The relationship between irritable bowel syndrome and psychiatric disorders: from molecular changes to clinical manifestations. J Mol Psychiatry 2014; 2(1): 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, Masand PS, Kaplan D, Bhandary A, Hendricks S. The relationship between schizophrenia and irritable bowel syndrome (IBS). Schizophr Res 1997; 23(3): 265–268. [DOI] [PubMed] [Google Scholar]
- 46.Vu J, Kushnir V, Cassell B, Gyawali CP, Sayuk GS. The impact of psychiatric and extraintestinal comorbidity on quality of life and bowel symptom burden in functional GI disorders. Neurogastroenterol Motil 2014; 26(9): 1323–1332. [DOI] [PubMed] [Google Scholar]
- 47.Filipovic BR, Filipovic BF. Psychiatric comorbidity in the treatment of patients with inflammatory bowel disease. World J Gastroenterol 2014; 20(13): 3552–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Annamalai A, Kosir U, Tek C. Prevalence of obesity and diabetes in patients with schizophrenia. World J Diabetes 2017; 8(8): 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.So HC, Chau KL, Ao FK, Mo CH, Sham PC. Exploring shared genetic bases and causal relationships of schizophrenia and bipolar disorder with 28 cardiovascular and metabolic traits. Psychol Med 2019; 49(8): 1286–1298. [DOI] [PubMed] [Google Scholar]
- 50.Mamakou V, Thanopoulou A, Gonidakis F, Tentolouris N, Kontaxakis V. Schizophrenia and type 2 diabetes mellitus. Psychiatriki 2018; 29(1): 64–73. [DOI] [PubMed] [Google Scholar]
- 51.Leppert B, Millard LAC, Riglin L, Davey Smith G, Thapar A, Tilling K et al. A cross-disorder PRS-pheWAS of 5 major psychiatric disorders in UK Biobank. PLoS Genet 2020; 16(5): e1008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Axelson DA, Doraiswamy PM, Boyko OB, Rodrigo Escalona P, McDonald WM, Ritchie JC et al. In vivo assessment of pituitary volume with magnetic resonance imaging and systematic stereology: relationship to dexamethasone suppression test results in patients. Psychiatry Res 1992; 44(1): 63–70. [DOI] [PubMed] [Google Scholar]
- 53.Pariante CM, Vassilopoulou K, Velakoulis D, Phillips L, Soulsby B, Wood SJ et al. Pituitary volume in psychosis. Br J Psychiatry 2004; 185: 5–10. [DOI] [PubMed] [Google Scholar]
- 54.Nordholm D, Krogh J, Mondelli V, Dazzan P, Pariante C, Nordentoft M. Pituitary gland volume in patients with schizophrenia, subjects at ultra high-risk of developing psychosis and healthy controls: a systematic review and meta-analysis. Psychoneuroendocrinology 2013; 38(11): 2394–2404. [DOI] [PubMed] [Google Scholar]
- 55.Oken RJ, Schulzer M. At issue: schizophrenia and rheumatoid arthritis: the negative association revisited. Schizophr Bull 1999; 25(4): 625–638. [DOI] [PubMed] [Google Scholar]
- 56.Mors O, Mortensen PB, Ewald H. A population-based register study of the association between schizophrenia and rheumatoid arthritis. Schizophr Res 1999; 40(1): 67–74. [DOI] [PubMed] [Google Scholar]
- 57.Sellgren C, Frisell T, Lichtenstein P, Landèn M, Askling J. The association between schizophrenia and rheumatoid arthritis: a nationwide population-based Swedish study on intraindividual and familial risks. Schizophr Bull 2014; 40(6): 1552–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vancampfort D, Firth J, Schuch FB, Rosenbaum S, Mugisha J, Hallgren M et al. Sedentary behavior and physical activity levels in people with schizophrenia, bipolar disorder and major depressive disorder: a global systematic review and meta-analysis. World Psychiatry 2017; 16(3): 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet 2016; 388(10055): 2023–2038. [DOI] [PubMed] [Google Scholar]
- 60.Cole SR, Platt RW, Schisterman EF, Chu H, Westreich D, Richardson D et al. Illustrating bias due to conditioning on a collider. Int J Epidemiol 2010; 39(2): 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am J Epidemiol 2017; 186(9): 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.